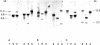Abstract
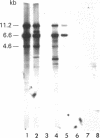
In a rat hepatoma cell line, H4-IIE-C3, a 10-fold excess of 18S and 28S rRNA genes has been found in amplified chromosome regions. Antibodies to 5-methylcytidine bound extensively to the DNA of these regions, indicating a high level of DNA methylation. Most of the amplified rRNA genes were transcriptionally inactive, as shown by their failure to stain with silver. DNAs from the tumor cells and control rat hepatocytes grown with L-[methyl-14C]methionine were digested with restriction endonuclease EcoRI; the DNA fragments were separated by agarose gel electrophoresis, denatured, transferred to nitrocellulose filters, and hybridized to 32P-labeled rRNA or cDNA. Fragments containing the 18S or 28S rRNA coding sequences occurred in three major size classes; all three were rich in 5-methylcytosine. Analysis of EcoRI fragments of DNA from the tumor and control cells after digestion with Hpa II or Msp I endonuclease indicated that the 5'-C-C-G-G-3' sequences in most of the amplified rRNA genes were methylated. Analysis of the fragments produced by digestion with Hha I endonuclease indicated a high degree of methylation within its recognition sequence in the amplified rRNA genes as well. The association of hypermethylation with restricted transcriptional activity suggests that DNA methylation may regulate the activity of the rRNA genes.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bird A. P., Southern E. M. Use of restriction enzymes to study eukaryotic DNA methylation: I. The methylation pattern in ribosomal DNA from Xenopus laevis. J Mol Biol. 1978 Jan 5;118(1):27–47. doi: 10.1016/0022-2836(78)90242-5. [DOI] [PubMed] [Google Scholar]
- Bloom S. E., Goodpasture C. An improved technique for selective silver staining of nucleolar organizer regions in human chromosomes. Hum Genet. 1976 Oct 28;34(2):199–206. doi: 10.1007/BF00278889. [DOI] [PubMed] [Google Scholar]
- Christman J. K., Weich N., Schoenbrun B., Schneiderman N., Acs G. Hypomethylation of DNA during differentiation of Friend erythroleukemia cells. J Cell Biol. 1980 Aug;86(2):366–370. doi: 10.1083/jcb.86.2.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Desrosiers R. C., Mulder C., Fleckenstein B. Methylation of Herpesvirus saimiri DNA in lymphoid tumor cell lines. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3839–3843. doi: 10.1073/pnas.76.8.3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingeras T. R., Myers P. A., Olson J. A., Hanberg F. A., Roberts R. J. A new specific endonuclease present in Xanthomonas holcicola, Xanthomonas papavericola and Brevibacterium luteum. J Mol Biol. 1978 Jan 5;118(1):113–122. doi: 10.1016/0022-2836(78)90247-4. [DOI] [PubMed] [Google Scholar]
- Glisin V., Crkvenjakov R., Byus C. Ribonucleic acid isolated by cesium chloride centrifugation. Biochemistry. 1974 Jun 4;13(12):2633–2637. doi: 10.1021/bi00709a025. [DOI] [PubMed] [Google Scholar]
- Goodpasture C., Bloom S. E. Visualization of nucleolar organizer regions im mammalian chromosomes using silver staining. Chromosoma. 1975 Nov 20;53(1):37–50. doi: 10.1007/BF00329389. [DOI] [PubMed] [Google Scholar]
- Gross-Bellard M., Oudet P., Chambon P. Isolation of high-molecular-weight DNA from mammalian cells. Eur J Biochem. 1973 Jul 2;36(1):32–38. doi: 10.1111/j.1432-1033.1973.tb02881.x. [DOI] [PubMed] [Google Scholar]
- Guntaka R. V., Rao P. Y., Mitsialis S. A., Katz R. Modification of avian sarcoma proviral DNA sequences in nonpermissive XC cells but not in permissive chicken cells. J Virol. 1980 May;34(2):569–572. doi: 10.1128/jvi.34.2.569-572.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guntaka R. V., Weiner A. J. Effect of dibutyryl cyclic AMP on intracellular levels of avian sarcoma virus specific RNA. Nature. 1978 Jul 20;274(5668):274–276. doi: 10.1038/274274a0. [DOI] [PubMed] [Google Scholar]
- Henderson A. S., Warburton D., Atwood K. C. Location of ribosomal DNA in the human chromosome complement. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3394–3398. doi: 10.1073/pnas.69.11.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofgärtner F. J., Krone W., Jain K. Correlated inhibition of ribosomal RNA synthesis and silver staining by actinomycin D. Hum Genet. 1979 Apr 5;47(3):329–333. doi: 10.1007/BF00321025. [DOI] [PubMed] [Google Scholar]
- Hofgärtner F. J., Schmid M., Krone W., Zenzes M. T., Engel W. Pattern of activity of nucleolus organizers during spermatogenesis in mammals as analyzed by silver-staining. Chromosoma. 1979 Feb 21;71(2):197–216. doi: 10.1007/BF00292823. [DOI] [PubMed] [Google Scholar]
- Hsu T. C., Spirito S. E., Pardue M. L. Distribution of 18+28S ribosomal genes in mammalian genomes. Chromosoma. 1975 Nov 20;53(1):25–36. doi: 10.1007/BF00329388. [DOI] [PubMed] [Google Scholar]
- Hsu T. W., Guntaka R. V., Taylor J. M. Specific site of action for single-strand specific nuclease on the double-stranded circular DNA intermediates of an avian RNA tumor virus. J Virol. 1978 Dec;28(3):1015–1017. doi: 10.1128/jvi.28.3.1015-1017.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P. A., Taylor S. M. Cellular differentiation, cytidine analogs and DNA methylation. Cell. 1980 May;20(1):85–93. doi: 10.1016/0092-8674(80)90237-8. [DOI] [PubMed] [Google Scholar]
- Lubit B. W., Schreck R. R., Miller O. J., Erlanger B. F. Human chromosome structure as revealed by an immunoperoxidase staining procedure. Exp Cell Res. 1974 Dec;89(2):426–429. doi: 10.1016/0014-4827(74)90815-5. [DOI] [PubMed] [Google Scholar]
- Mandel J. L., Chambon P. DNA methylation: organ specific variations in the methylation pattern within and around ovalbumin and other chicken genes. Nucleic Acids Res. 1979 Dec 20;7(8):2081–2103. doi: 10.1093/nar/7.8.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann M. B., Smith H. O. Specificity of Hpa II and Hae III DNA methylases. Nucleic Acids Res. 1977 Dec;4(12):4211–4221. doi: 10.1093/nar/4.12.4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee J. D., Ginder G. D. Specific DNA methylation sites in the vicinity of the chicken beta-globin genes. Nature. 1979 Aug 2;280(5721):419–420. doi: 10.1038/280419a0. [DOI] [PubMed] [Google Scholar]
- Miller D. A., Breg W. R., Warburton D., Dev V. G., Miller O. J. Regulation of rRNA gene expression in a human familial 14p+ marker chromosome. Hum Genet. 1978 Sep 19;43(3):289–297. doi: 10.1007/BF00278836. [DOI] [PubMed] [Google Scholar]
- Miller D. A., Dev V. G., Tantravahi R., Miller O. J. Suppression of human nucleolus organizer activity in mouse-human somatic hybrid cells. Exp Cell Res. 1976 Sep;101(2):235–243. doi: 10.1016/0014-4827(76)90373-6. [DOI] [PubMed] [Google Scholar]
- Miller O. J., Miller D. A., Dev V. G., Tantravahi R., Croce C. M. Expression of human and suppression of mouse nucleolus organizer activity in mouse-human somatic cell hybrids. Proc Natl Acad Sci U S A. 1976 Dec;73(12):4531–4535. doi: 10.1073/pnas.73.12.4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller O. J., Tantravahi R., Miller D. A., Yu L. C., Szabo P., Prensky W. Marked increase in ribosomal RNA gene multiplicity in a rat hepatoma cell line. Chromosoma. 1979 Feb 21;71(2):183–195. doi: 10.1007/BF00292822. [DOI] [PubMed] [Google Scholar]
- Peacock A. C., Dingman C. W. Resolution of multiple ribonucleic acid species by polyacrylamide gel electrophoresis. Biochemistry. 1967 Jun;6(6):1818–1827. doi: 10.1021/bi00858a033. [DOI] [PubMed] [Google Scholar]
- Perry R. P., Kelley D. E., Schibler U., Huebner K., Croce C. M. Selective suppression of the transcription of ribosomal genes in mouse-human hybrid cells. J Cell Physiol. 1979 Mar;98(3):553–559. doi: 10.1002/jcp.1040980313. [DOI] [PubMed] [Google Scholar]
- Pollock J. M., Jr, Swihart M., Taylor J. H. Methylation of DNA in early development: 5-methyl cytosine content of DNA in sea urchin sperm and embryos. Nucleic Acids Res. 1978 Dec;5(12):4855–4861. doi: 10.1093/nar/5.12.4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R. J. Restriction endonucleases. CRC Crit Rev Biochem. 1976 Nov;4(2):123–164. doi: 10.3109/10409237609105456. [DOI] [PubMed] [Google Scholar]
- Schmiady H., Münke M., Sperling K. Ag-staining of nucleolus organizer regions on human prematurely condensed chromosomes from cells with different ribosomal RNA gene activity. Exp Cell Res. 1979 Jul;121(2):425–428. doi: 10.1016/0014-4827(79)90025-9. [DOI] [PubMed] [Google Scholar]
- Schreck R. R., Erlanger B. F., Miller O. J. The use of antinucleoside antibodies to probe the organization of chromosomes denatured by ultraviolet irradiation. Exp Cell Res. 1974 Sep;88(1):31–39. doi: 10.1016/0014-4827(74)90614-4. [DOI] [PubMed] [Google Scholar]
- Schreck R. R., Warburton D., Miller O. J., Beiser S. M., Erlanger B. F. Chromosome structure as revealed by a combined chemical and immunochemical procedure. Proc Natl Acad Sci U S A. 1973 Mar;70(3):804–807. doi: 10.1073/pnas.70.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shank P. R., Hughes S. H., Kung H. J., Majors J. E., Quintrell N., Guntaka R. V., Bishop J. M., Varmus H. E. Mapping unintegrated avian sarcoma virus DNA: termini of linear DNA bear 300 nucleotides present once or twice in two species of circular DNA. Cell. 1978 Dec;15(4):1383–1395. doi: 10.1016/0092-8674(78)90063-6. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stumph W. E., Wu J. R., Bonner J. Determination of the size of rat ribosomal deoxyribonucleic acid repeating units by electron microscopy. Biochemistry. 1979 Jun 26;18(13):2864–2871. doi: 10.1021/bi00580a030. [DOI] [PubMed] [Google Scholar]
- Sutter D., Doerfler W. Methylation of integrated adenovirus type 12 DNA sequences in transformed cells is inversely correlated with viral gene expression. Proc Natl Acad Sci U S A. 1980 Jan;77(1):253–256. doi: 10.1073/pnas.77.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantravahi R., Miller D. A., D'Ancona G., Croce C. M., Miller O. J. Location of rRNA genes in three inbred strains of rat and suppression of rat rRNA activity in rat-human somatic cell hybrids. Exp Cell Res. 1979 Mar 15;119(2):387–392. doi: 10.1016/0014-4827(79)90368-9. [DOI] [PubMed] [Google Scholar]
- Tantravahi R., Miller D. A., Dev V. G., Miller O. J. Detection of nucleolus organizer regions in chromosomes of human, chimpanzee, gorilla, orangutan and gibbon. Chromosoma. 1976 Jun 30;56(1):15–27. doi: 10.1007/BF00293725. [DOI] [PubMed] [Google Scholar]
- Taylor J. M., Hsu T. W., Lai M. M. Restriction enzyme sites on the avian RNA tumor virus genome. J Virol. 1978 May;26(2):479–484. doi: 10.1128/jvi.26.2.479-484.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. M., Illmensee R., Summers J. Efficeint transcription of RNA into DNA by avian sarcoma virus polymerase. Biochim Biophys Acta. 1976 Sep 6;442(3):324–330. doi: 10.1016/0005-2787(76)90307-5. [DOI] [PubMed] [Google Scholar]
- Vanyushin B. F., Tkacheva S. G., Belozersky A. N. Rare bases in animal DNA. Nature. 1970 Mar 7;225(5236):948–949. doi: 10.1038/225948a0. [DOI] [PubMed] [Google Scholar]
- Varmus H. E., Bishop J. M., Vogt P. K. Appearance of virus-specific DNA in mammalian cells following transformation by Rous sarcoma virus. J Mol Biol. 1973 Mar 15;74(4):613–626. doi: 10.1016/0022-2836(73)90052-1. [DOI] [PubMed] [Google Scholar]
- Volpe P., Eremenko T. Preferential methylation of regulatory genes in HeLa cells. FEBS Lett. 1974 Aug 25;44(2):121–126. doi: 10.1016/0014-5793(74)80708-8. [DOI] [PubMed] [Google Scholar]
- Waalwijk C., Flavell R. A. MspI, an isoschizomer of hpaII which cleaves both unmethylated and methylated hpaII sites. Nucleic Acids Res. 1978 Sep;5(9):3231–3236. doi: 10.1093/nar/5.9.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolgemuth D. J., Jagiello G. M., Henderson A. S. Quantitation of ribosomal RNA genes in fetal human oocyte nuclei using rRNA: DNA hybridization in situ. Evidence for increased multiplicity. Exp Cell Res. 1979 Jan;118(1):181–190. doi: 10.1016/0014-4827(79)90596-2. [DOI] [PubMed] [Google Scholar]
- van der Ploeg L. H., Flavell R. A. DNA methylation in the human gamma delta beta-globin locus in erythroid and nonerythroid tissues. Cell. 1980 Apr;19(4):947–958. doi: 10.1016/0092-8674(80)90086-0. [DOI] [PubMed] [Google Scholar]