Background: No study shows any negative feedback regulation of c-Myc by miRNAs.
Results: miR-185-3p is expressed in response to c-Myc and in return inhibits c-Myc expression.
Conclusion: miR-185-3p is a novel negative feedback regulator of c-Myc.
Significance: Unveiling this feedback loop of c-Myc-miR-185-3p offers new insight into c-Myc regulation and a potential anti-cancer agent.
Keywords: Cell Cycle, Cell Division, Cell Proliferation, MicroRNA, Oncogene, Transcription, Translation, c-Myc, Feedback Regulation, miR-185-3p
Abstract
The expression of the c-myc oncogene at both protein and mRNA levels is transient and begins to be turned off 3–6 h after growth stimulation of cultured cells. The exact mechanism(s) underlying this down-regulation of c-Myc remains incompletely understood. Here we report the identification of miR-185-3p as a novel feedback regulator of c-Myc. This microRNA (miRNA) was initially identified as one of the c-Myc target miRNA transcripts through analysis of RNA samples isolated from cells prior to and after serum stimulation and further verified by real-time PCR, luciferase reporter, and ChIP assays. Interestingly, overexpression of wild type, but not mutant, miR-185-3p decreased the protein, but not mRNA, level of c-Myc in a dose-dependent fashion and also drastically abated the serum induction of c-Myc level in human cancer cells by targeting the coding sequence of c-Myc mRNA, consequently suppressing c-Myc-mediated proliferation. A miR-185-3p inhibitor rescued the inhibition of c-Myc expression by endogenous miR-185-3p. Thus, our results unveil miR-185-3p as the first miRNA that monitors c-Myc levels via an autoregulatory feedback mechanism in response to serum stimulation.
Introduction
Since c-myc was first identified as the human cellular homolog of the retroviral v-myc (1), it has been intensively studied and shown to be essential for cell growth, proliferation, and animal development (2–7) because knocking it out causes embryonic lethality in mice (8). The biological importance of c-Myc is largely ascribed to its transcriptional activity. c-Myc is a nuclear transcriptional factor consisting of two major functional domains, the N-terminal regulatory and transactivational domain containing two Myc-box (MBI and MBII) motifs and the C-terminal basic helix-loop-helix leucine zipper and DNA-binding domain (2, 5, 9, 10). It forms a functional heterodimer with Max (4, 11). This transcriptional complex regulates the expression of almost 15% of human genes (3) by binding to its responsive DNA sequence element (E-box motif) (12). Most of these genes are crucial for ribosome biogenesis and protein synthesis (13–15), which are indispensable for cell growth, proliferation, and development (2–4). However, these normal functions of c-Myc are often exploited by cancer cells for their advantage because overly expressed or active c-Myc favors cell proliferation, transformation, neoplasia, and tumorigenesis in mice (16–19), and its levels are highly expressed in most human cancers (5, 20). Some of the oncogenic functions of c-Myc are also executed through its transcriptional target microRNAs (miRNAs),2 such as the miR-17-92 cluster (21). Hence, cells need to monitor c-Myc level and activity in order to grow and proliferate normally without gaining their awry transformational and tumorigenic potential.
Indeed, c-Myc is delicately regulated at transcriptional, posttranscriptional, translational, and posttranslational levels through a variety of mechanisms (3), whereas Max levels remain quite steady in cells (4). For instance, the c-Myc protein is considerably unstable with a half-life of ∼15 min due to ubiquitination-dependent proteolysis, which is mediated by E3 ubiquitin ligases, such as Fbw7, Skp2, or HectH9 (22–25). This process is also highly controlled through phosphorylation at the N-terminal Myc-box domains of c-Myc in response to Ras signaling (26, 27), leading to stabilization of c-Myc. Furthermore, both translation and stability of c-Myc mRNA are tightly regulated (3). Although a number of protein regulators have been shown to be involved in these regulations, recent studies also divulged several miRNA regulators, such as miR-24, miR-22, miR-145, or miR-let-7a (28–31). These miRNAs can inactivate c-Myc by targeting its mRNA in response to distinct signals, such as p53-responsive suppression of c-Myc by miR-145 (29).
The tight regulation of c-Myc expression can be readily detected in cultured cells, typically reflected in its bell shape-like expression pattern in response to growth signals: an immediate rise (usually peaking at 3–6 h) of c-Myc level and activity followed by a gradual descent upon serum stimulation (26, 32). The first sharp (rapid increase) phase of c-Myc level and activity after serum stimulation is chiefly attributed to the growth factor-activated Ras signaling pathway, which has been shown to induce the mRNA transcription and protein stability of c-Myc (26). By contrast, the mechanisms underlying the second (gradual decrease) phase of c-Myc response to serum remain promiscuous, although it is clear that both c-Myc protein and mRNA levels decline once they reach the induction peak (32). Our recent study demonstrates that ribosomal protein L11 (RPL11) plays a feedback role in regulating c-Myc transcriptional activity by binding to its MBII domain and excluding the binding of TRRAP, a cofactor of c-Myc (33), to this domain (32). It also suggests that RPL11 may be responsible for the second phase decrease of c-Myc activity after serum stimulation (32). Although knockdown of RPL11 resulted in the increase of both of c-Myc protein and mRNA levels (34), this role may be exerted through an indirect mechanism because overexpression of RPL11 did not simply reduce the total level of c-Myc (data not shown). Therefore, although RPL11 can suppress c-Myc activity after the peak induction in response to serum stimulation, it still remains unclear how c-Myc mRNA and protein levels are down-regulated at the late stage of the serum response.
Because none of the aforementioned miRNAs has been shown to play an autoregulatory role in monitoring c-Myc levels after serum stimulation (28–31), there might exist one or more miRNAs that could be transcriptionally activated by c-Myc and in turn regulate the expression of this transcriptional factor by either mediating its mRNA degradation or inhibiting its translation. In an attempt to identify this c-Myc-targeting miRNA(s), we performed an miRNA array analysis using RNA samples isolated from human colon cancer HCT116 p53-containing and p53-deficient cells prior to and 3 h after serum treatment. This analysis revealed several miRNAs as promising c-Myc-targeting miRNAs. Although two of them, miR-145 and miR-24, were recently reported (28, 29), we found a new miRNA, miR-185-3p, as a potential target of c-Myc. Our further studies not only confirm that miR-185-3p is an authentic transcriptional target of c-Myc but also unravel its inhibitory role in regulating c-Myc protein expression by targeting its amino acid-coding mRNA sequence without affecting its mRNA levels. In line with these results, inhibition of miR-185-3p function by its siRNA induced c-Myc level and activity. Our studies demonstrate for the first time that miR-185-3p plays an autoregulatory feedback role in monitoring the protein level of c-Myc in response to growth signals.
EXPERIMENTAL PROCEDURES
Cell Lines
Unless indicated specifically, all cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 50 units/ml penicillin, and 0.1 mg/ml streptomycin at 37 °C in 5% CO2.
Plasmids, Adenoviruses, and Antibodies
CMV-empty, CMV-β-Gal, pD40-His/V5-c-Myc, and E2F2(-E-box)-Luc plasmids were described previously (32). To generate pGL3-miR-185-3p and pGL-miR-185-3p control constructs, the E-box promoter or the sequence far away from this E-box was amplified by the primers listed in supplemental Table S1. The fragments were inserted into pGL3 with the designed cutting sites: KpnI and XhoI. To generate pSIF-miR-185-3p and pSIF-miR-185-3p mutant plasmids, the precursor and mutant sequences were synthesized and inserted into the pSIF vector at the BamHI and EcoRI sites. The pSIF vector was kindly provided and described by Yong Li (35). To make the pMIR-c-Myc and pMIR-c-Myc mutant plasmids, the targeting sequence or its mutant was synthesized and inserted into pMIR vector (Ambion, Austin, TX) by designed sites: SpeI and HindIII. Adenoviruses to overexpress full-length c-Myc or control GFP were described previously (32). The sequences of pSIF-miR-185-3p, pSIF-miR-185-3p mutant, pMIR-c-Myc, and pMIR-c-Myc mutant are summarized in supplemental Table S1. Antibodies used were anti-c-Myc (N262, Santa Cruz Biotechnology, Inc. (Santa Cruz, CA)), anti-V5 (Invitrogen), and anti-BrdU (IIB5, Santa Cruz Biotechnology, Inc.).
Transient Transfection and Immunoblot
As described previously (36), briefly, cells were transfected with the indicated plasmids as shown in each figure by using TransFectin (Bio-Rad), following the company's instructions. 48 h post-transfection, cells were harvested and lysed in lysis buffer. The total protein concentration for each sample was determined, and equal amounts of total proteins were then subjected to SDS-PAGE, followed by immunobloting.
Reverse Transcription-Polymerase Chain Reaction and Quantitative Real-time PCR Analysis
RT-PCR and quantitative real-time PCR for mature miRNA were done by using the methods described previously (37). RT-PCR and quantitative real-time PCR for other genes followed the protocol as described previously (38). Briefly, quantitative real-time PCR was performed on an ABI 7300 real-time PCR system (Applied Biosystems) using SYBR Green Mix (Applied Biosystems). Relative gene expression was calculated using the ΔCt method, following the manufacturer's instructions. All reactions were carried out in triplicate. The primer sequences used are listed in supplemental Table S1.
Luciferase Reporter Assays
Cells were transfected with pCMV-β-galactoside and the indicated plasmids (total plasmid DNA 1 μg/well) as indicated in the figures. Luciferase activity was determined and normalized by a factor of β-Gal activity in the same assay as described previously (39).
Chromatin Immunoprecipitation (ChIP)-PCR
ChIP analysis was performed as described previously (40) using anti-c-Myc (N262) for endogenous c-Myc and anti-V5 for overexpressed V5-c-Myc. Immunoprecipitated DNA fragments were analyzed by semiquantitative and/or real-time PCR amplification using primers for 5 S ribosomal DNA, miR-185-3p, and control genes. The primers are listed in supplemental Table S1.
BrdU Incorporation Assays
BrdU incorporation assays were carried out by basically following the previously described protocol (32). Cells were incubated in the presence of 10 μm BrdU for 5 h. Cells were then fixed with 95% of ethanol and 5% of acetic acid, treated with 2 m HCl containing 1% Triton X-100, and stained with the monoclonal anti-BrdU antibody, followed by staining with Alexa Fluor 546 (red) goat anti-mouse antibodies and DAPI. Stained cells were analyzed under a Zeiss Axiovert 25 fluorescent microscope.
Knockdown of the Endogenous miRNA
Anti-miRTM miRNA inhibitor and Anti-miRTM miRNA inhibitors (negative control) were purchased from Ambion. Transfections of miRNA inhibitors were performed in the same way as that of normal siRNA, as described previously (41), by using siLentFectTM lipid (Bio-Rad), following the manufacturer's protocol.
RESULTS
Identification of miR-185-3p as a Potential Transcriptional Target of c-Myc
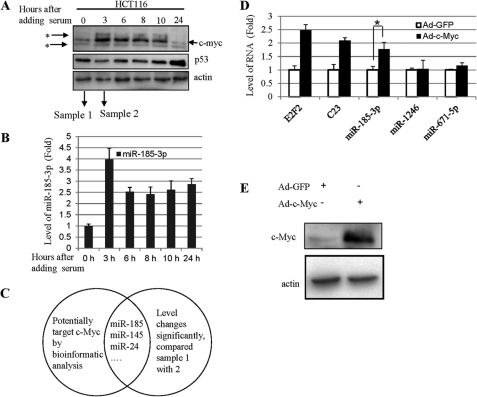
In order to identify miRNAs that may regulate the expression of c-Myc in response to serum stimulation, we isolated total RNAs from human p53-containing or p53-deficient colon cancer HCT116 cells that were treated with or without the addition of 20% of FBS after being starved in 0.2% of FBS. The RNA samples, after confirming the c-Myc induction after serum treatment (Fig. 1A), were subjected to a miRNA array analysis by LC Sciences (Houston, TX). As a result, at least three miRNAs, such as miR-24, miR-145, and miR-185-3p, were identified to potentially target c-Myc (Fig. 1C and supplemental Fig. S1). While we were conducting experiments to further study them, two of the miRNAs, miR-24 and miR-145, were later reported by other groups to suppress c-Myc expression by targeting the 3′-UTR sequence of its mRNA in response to terminal differentiation and p53 activation, respectively (28, 29). These published studies validated our procedure (Fig. 1C). Because miR-185-3p was also identified in the RNA pool isolated from p53 null HCT116 cells after serum stimulation (data not shown) and the promoter region of this miRNA-coding gene contained a potential c-Myc-responsive E-box DNA element (Fig. 2A), different from that of miR-24 and miR-145 (28, 29), we speculated that miR-185-3p might be a c-Myc transcription target and thus continued to study this miRNA.
FIGURE 1.
Bioinformatics analysis and micro-RNA array suggest that miR-185-3p and c-Myc may mutually regulate each other. A, the level of c-Myc in cells following serum stimulation. HCT116 cells were treated with 0.2% FBS for 24 h and then stimulated by 20% FBS and harvested at the indicated time points for immunoblot (IB) assays. 50 μg of protein was used for each lane (true also for the rest of the experiments). *, nonspecific band. B, the correlation of miR-185-3p and c-Myc. Cells were treated with 0.2% FBS for 24 h and then stimulated by 20% FBS. Cells were harvested at the indicated times. Values represent the mean and S.D. (error bars) of three independent experiments. C, bioinformatics analysis of microarray data shows that miR-145, 185-3p, and 24 may target c-Myc. D, c-Myc induces miR-185-3p expression in cells. H1299 infected with c-Myc-expressing adenovirus and the expression of miR-185-3p and the positive control E2F2 and C23 were determined within 48 h by quantitative real-time PCR assays. Significance was determined using an unpaired Student's t test. *, p < 0.05. Data are presented as means ± S.E. (error bars), n = 3. E, immunoblot was performed to detect the c-Myc overexpression.
FIGURE 2.
miR-185-3p is a transcriptional target of c-Myc. A, schematic representation of the E-box upstream of the miR-185-3p gene. B, c-Myc induces miR-185-3p expression in cells. H1299 cells were cotransfected with the indicated plasmids and β-Gal plasmid. 16 h after transfection, luciferase activities were measured. Luciferase activity was normalized to β-Gal, and the significance was determined using an unpaired Student's t test. *, p < 0.05. Data are presented as means ± S.E. (error bars), n = 3. C, ectopic c-Myc binds to the miR-185 promoter in 293 cells. 6 h after serum stimulation, chromatin immunoprecipitation (IP) assays were performed using 293 cells that were transfected with V5-c-Myc plasmid and with the indicated antibodies followed by PCR for miR-185 promoter sequences or negative control promoter sequences (n = 3; error bars, S.E.). Primers are listed in supplemental Table S1. D, miR-185-3p expression luciferase reporter activity is correlated with endogenous c-Myc level and in response to serum stimulation. H1299 cells were treated with 0.2% FBS for 24 h and then stimulated by 20% FBS and harvested at the indicated time points. Significance was determined using an unpaired Student's t test. *, p < 0.05. Data are presented as means ± S.E., n = 3. E, endogenous c-Myc binds to the miR-185 promoter in cells. 6 h after serum stimulation, chromatin immunoprecipitation assays were performed on U2OS cell lysates with antibodies as indicated, followed by PCR for miR-185-3p promoter sequences and negative control sequences (n = 3; error bars, S.E). Primers are listed in supplemental Table S1.
First, we verified the miRNA array data (Fig. 1C and supplemental Fig. S1) by analyzing the level of mature miR-185-3p in the RNA samples from six time points before and after serum stimulation using real-time PCR (of note, all of the primer sequences used in this study are shown in supplemental Table S1). As shown in Fig. 1B, indeed, the level of miR-185-3p reached a peak at 3 h and then declined at 6 h and stayed at a relatively level 24 h after serum stimulation, which was well correlated with the serum-induced protein level of c-Myc except at the 24 h point, when c-Myc was drastically reduced, as shown in Fig. 1A. Also, the miR-185-3p level was positively correlated with c-Myc among several cell lines tested (supplemental Fig. S4). To determine if ectopic expression of c-Myc could also induce the expression of this miRNA, we infected human lung non-small cell carcinoma H1299 cells, which are p53-deficient, with adenovirus encoding c-Myc or GFP and harvested cells for a real-time PCR analysis using primers for mature miR-185-3p, miR-1246, and miR-671-5p as well as for E2F2 and C23 mRNAs, the last two of which are c-Myc transcription targets (32) and used as positive controls. As shown in Fig. 1D and supplemental Fig. S6, miR-185-3p, E2F2, and C23, but not miR-1246 and miR-671–5p, were significantly induced when c-Myc was ectopically expressed in the cells (Fig. 1E). These results indicate that the expression of miR-185-3p can be induced by c-Myc, suggesting that this miRNA might be a transcriptional target for this transcriptional factor.
Confirmation of miR-185-3p as an Authentic Transcription Target of c-Myc
Next, we wanted to determine if miR-185-3p is a genuine transcription target of c-Myc by performing two sets of experiments. First, we identified a potential E-box DNA element at the position of −1215 nucleotides from the transcriptional starting site of the miR-185-3p-encoding gene by a bioinformatic analysis using the available University of California, Santa Cruz, online genomic database. Then we cloned this E-box DNA element sequence into a luciferase reporter plasmid as shown in Fig. 2A. Using this reporter, the c-Myc expression adenovirus, and an E2F2 luciferase reporter as a positive control, we carried out a transfection/infection coupled with a luciferase reporter assay as detailed under “Experimental Procedures.” As shown in Fig. 2B, overexpression of exogenous v5-taggged c-Myc induced the luciferase activity significantly when either the E2F2 or miR-185-3p promoter-driven luciferase reporter was used. To determine if exogenous c-Myc could bind to the promoter region of the miR-185-3p-coding gene, we conducted a ChIP assay after infecting 293 cells with adenovirus encoding v5-c-Myc. As shown in Fig. 2C and supplemental Fig. S2A, ectopic c-Myc indeed bound to the endogenous miR-185-3p promoter as well as to the 5 S ribosomal DNA promoter, which was shown as a c-Myc target as well (42), in comparison with the result using IgG control antibodies. Furthermore, the miR-185-3p promoter-driven luciferase activity was also responsive to serum stimulation in H1299 cells (Fig. 2D). Finally, endogenous c-Myc was found to bind both the miR-185-3p and 5 S ribosomal DNA promoter regions containing the E-box element as detected by ChIP assays in U2OS cells (Fig. 3E and supplemental Fig. S2B). Together with the results in Fig. 1, these results using various cell lines (of note, each of the above results was also repeated in different cells used in this study, and representative results are shown here) regardless of the p53 status demonstrate that miR-185-3p is indeed a bona fide transcriptional target of c-Myc, and miR-185-3p expression is c-Myc-dependent but p53- and cell-independent (supplemental Fig. S5).
FIGURE 3.
miR-185-3p decreased c-Myc protein, but not mRNA, levels in cells. A, the ORF sequence of c-Myc mRNA harbors putative binding sites complementary to miR-185-3p. The putative c-myc ORF sites was targeted 305–327 bp upstream from the c-myc stop codon. B and C, quantitative real-time PCR (QRT-PCR) assays were performed using RNAs isolated from H1299 cells transfected with the indicated amount of the pSIF miR-185-3p plasmid. c-Myc was induced at 6 h after changing medium. Data are presented as a function of miR-185-3p (B) and c-Myc mRNA (C) copies relative to GAPDH mRNA. The mean levels were from three independent samples. The plug-in Western blot shows the endogenous c-Myc protein level. D, miR-185-3p can decrease both endogenous and exogenous c-Myc protein levels in cells. H1299 cells were transfected with the indicated plasmids. 20 h after transfection, cells were treated with serum-free medium and stimulated by 20% FBS, and cells were collected 3 h after stimulation. Immunoblot analysis was performed with the N262 anti-c-Myc antibody. E, miR-185-3p inhibits c-Myc response to serum stimulation. H1299 or HCT116 cells were transfected with the indicated plasmids and treated with 0.2% FBS for 24 h and then stimulated by 20% FBS and harvested at the indicated time points. F, quantification of the results from E. The intensity of each band in E was quantified and graphed in this panel. The y axis represents the -fold increase of c-Myc protein levels, whereas the x axis denotes the time course after serum stimulation.
Suppression of c-Myc Level and Activity by miR-185-3p
To identify potential mRNA targets of miR-185-3p, we carried out a bioinformatic analysis based on experimentally derived rules for miRNA target recognition (43). From this analysis, we found that one of the top target candidates is c-Myc itself. However, surprisingly, the potential target sequence for miR-185-3p with a calculated energy of −46.5 kcal/mol is within the protein-coding region of c-Myc mRNA, which encodes the C-terminal domain of the c-Myc protein (Fig. 3A). This is clearly distinct from the recently reported c-Myc targeting miRNAs (28–30). This miR-185-3p-targeted sequence is highly conserved from rodent to human c-Myc mRNAs. To test if miR-185-3p could affect the level of c-Myc, we cloned a wild type or mutant miR-185-3p expression sequence, as indicated in Fig. 3A, into the pSIF miRNA expression vector (35). H1299 cells were transfected with these vectors individually and harvested 24 h post-transfection for real-time PCR and immunoblot analyses. As shown in Fig. 3, B–D, overexpression of miR-185-3p reduced the protein, but not mRNA, level of c-Myc. By contrast, the mutant miR-185-3p failed to reduce the level of both endogenous (left) and exogenous (right) c-Myc proteins, and miR-24, shown to target the 3′-UTR of c-myc (28), failed to reduce the exogenous c-Myc protein level expressed via its cDNA without 3′-UTR, as assayed by immunobloting (Fig. 3D and supplemental Fig. S3). In addition, wild type, but not mutant, miR-185-3p, when overexpressed, markedly abrogated the serum-responsive induction of c-Myc protein levels, as shown in Fig. 3, E and F. Of note, the mutant miR-185-3p did not affect the c-Myc induction pattern after serum treatment in comparison with the mock-transfected cells (data not shown; also compare Fig. 3E with Fig. 1A). These results clearly demonstrate that miR-185-3p can suppress the expression of the c-Myc protein without affecting its mRNA level and also suggest the possibility that this miRNA may target the 3′ amino acid-coding sequence of c-Myc mRNA, consequently inhibiting its translation.
Targeting the Amino Acid-coding Region of c-Myc mRNA by miR-185-3p
To test the above possibility, we constructed a luciferase reporter by cloning a wild type or mutant miR-185-3p-targeted DNA sequence derived from the c-Myc mRNA into the pMIR vector (Ambion) (Fig. 4A). H1299 cells were co-transfected with these vectors individually with or without wild type or mutant miR-185-3p expression vectors and harvested 48 h post-transfection for luciferase assays. As shown in Fig. 4B, wild type, but not mutant, miR-185-3p, when overexpressed, reduced the luciferase activity that was driven by the wild type, but not mutant, c-Myc RNA sequence in a dose-dependent fashion. This result indicates that miR-185-3p indeed binds to this amino acid-coding region of c-Myc mRNA and inhibits its translation.
FIGURE 4.
miR-185-3p targets the amino acid-coding sequence of c-Myc mRNA. A, schematic representation of the pMIR-c-Myc reporter constructs used in this set of experiment. The plasmids contain the wild type or mutant miR-185-3p-targeted site of the c-Myc mRNA. B, H1299 cells were cotransfected with the indicated plasmids. 48 h after transfection, luciferase activities were measured. Luciferase activity was normalized to β-Gal expression.
Inhibition of c-Myc-mediated Transcriptional Activity and Cell Proliferation by miR-185-3p
To investigate the functional outcome of suppression of c-Myc protein expression by miR-185-3p, we first determined if overexpression of miR-185-3p could affect c-Myc transcriptional activity by performing transfection-coupled with luciferase and real-time PCR assays using an E2F2 promoter-driven luciferase reporter and E2F2 primers. As expected, ecoptic c-Myc induced the luciferase activity driven by the wild type, but not mutant, E2F2 promoter (Fig. 5A). While titrating up the miR-185-3p expression vector, this c-Myc-induced luciferase activity was markedly inhibited in a miR-185-3p dose-dependent fashion (Fig. 5A, left graph). In addition, miR-185-3p inhibited the c-Myc-dependent expression of endogenous E2F2 mRNA (Fig. 5B, right graph). In line with these results, miR-185-3p also significantly alleviated c-Myc-induced cell proliferation as assays in BrdU-staining (Fig. 5, C and D). Taken together, these results demonstrate that miR-185-3p can repress the transcriptional activity of c-Myc by reducing its protein level, consequently inhibiting c-Myc-mediated cell proliferation.
FIGURE 5.
miR-185-3p inhibits c-Myc-dependent transcriptional activity and cell proliferation. A, miR-185-3p inhibits c-Myc-induced luciferase reporter expression. 293 cells transfected with plasmids, as indicated, were subjected to luciferase assays as described under “Experimental Procedures.” c-Myc was induced at 6 h after changing medium of cultured cells. B, miR-185-3p inhibits c-Myc-dependent transcription of the endogenous c-Myc target genes. H1299 cells were transfected with pSIF-miR-185-3p plasmid and/or infected with Ad-c-Myc, and the expression of E2F2 was measured by quantitative RT-PCR assays. C, and D, miR-185-3p reduces c-Myc-mediated cell proliferation. H1299 cells infected with Ad-c-Myc and/or transfected with pSIF-miR-185-3p were labeled with BrdU and subjected to anti-BrdU staining (red), as shown in C, and counterstained with DAPI. The percentage of BrdU-positive cells is shown in D. Significance was determined using an unpaired Student's t test. *, p < 0.05. Data are presented as means ± S.E. (error bars), n = 3.
Rescue of c-Myc Level through Inhibition of miR-185-3p
To determine if endogenous miR-185-3p plays a role in regulating c-Myc level, we employed an Anti-miRTM miRNA inhibitor specific to miR-185-3p or an Anti-miRTM miRNA control as a negative control (Ambion). Human colon cancer HCT116 or breast cancer MCF7 cells were transfected with either the miR-185-3p inhibitor or the negative control and harvested 24 h after transfection for immunblotting and real-time PCR analyses to test the level of endogenous c-Myc and the expression of its target gene E2F2 as well as miR-185-3p. As shown in Fig. 6, miR-185-3p inhibitor, but not negative control, induced the level of endogenous c-Myc proteins in both of the cell lines (top panels) as well as the expression of E2F2 mRNA (bottom panels). In accordance with these results, the level of endogenous miR-185-3p was markedly reduced by miR-185-3p inhibitor but not negative control (void columns in the bottom panels of Fig. 6, A and B). Consistent with these results, this miR-185-3p inhibitor, but not its control, further enhanced the serum induction of cell proliferation and c-Myc levels and, more remarkably, prolonged this induction period after serum stimulation of HCT116 cells by more than 2 h (Fig. 6, C and D). These results demonstrate that miR-185-3p indeed is responsible for down-regulation of c-Myc protein expression in its second descent phase after serum stimulation.
FIGURE 6.
Knocking down miR-185-3p increases c-Myc level, promotes c-Myc transcriptional activity, and thus facilitates serum-induced proliferation. Knocking down miR-185-3p increases the c-Myc level in HCT116 (A) and MCF7 (B) cells. HCT116 and MCF7 cells were double transfected with the indicated miRNA inhibitors, and cells were harvested 24 h after the second transfection and subjected to immunoblot with antibodies as indicated (top panels) and real-time PCR for the assessment of the miR-185-3p and E2F2 mRNA levels (bottom panels). c-Myc was induced at 6 h after changing medium of cultured cells. C, inhibition of miR-185-3p prolongs the serum induction of c-Myc in cells. HCT116 cells were double transfected with indicated miRNA inhibitors or negative control (CT). 6 h after the second transfection, the cells were stimulated by changing culture medium to 20% FBS fresh medium and harvested at the indicated time points. Cell lysates were subjected to Western blot (WB) analysis with the indicated antibodies. This assay was repeated three times. Bottom, quantification of the results from the assays as shown in the top as a representative graph here. The y axis represents the -fold increase of c-Myc protein levels, and the x axis denotes the time course after serum stimulation. A t test was performed with p < 0.05 for the 8 h time point (n = 3). D, miR-185-3p inhibitor, but not control inhibitor, facilitates serum-induced cell proliferation. Cells were treated the same way as in C and labeled with BrdU 2 h before harvesting and then subjected to anti-BrdU staining (red), as shown on the left. The percentage of BrdU-positive cells is shown on the right. Significance was determined using an unpaired Student's t test. *, p < 0.05. Data are presented as means ± S.E. (error bars), n = 3. E, a model for the c-Myc-miR-185-3p feedback loop.
DISCUSSION
Although several miRNAs have been recently shown to target c-Myc mRNA and suppress its expression (28–30), none of them is shown to play a negative feedback regulatory role in monitoring c-Myc level. In the current study, we identify miR-185-3p as a novel c-Myc regulator that can suppress the expression of c-Myc via an autoregulatory mechanism. First, the serum-responsive expression of miR-185-3p was well correlated with the c-Myc expression under the same condition. Also, exogenous c-Myc significantly induced the level of endogenous miR-185-3p and the E-box-containing miR-185-3p promoter-driven luciferase activity as well as bound to the endogenous miR-185-3p promoter region, as assessed by ChIP assays, confirming that miR-185-3p is a genuine transcriptional target of c-Myc. Interestingly, overexpression of wild type, not mutant, miR-185-3p led to the marked decrease of c-Myc protein, but not mRNA, levels and also alleviated the serum-stimulated induction of c-Myc, basically flattening the peak of c-Myc levels. More interestingly, unlike other known c-Myc-targeting miRNAs (28–30), miR-185-3p appeared to target the 3′ amino acid-coding region of c-Myc mRNA instead of its 3′-UTR sequence. This was further confirmed by a luciferase reporter assay. As a result of targeting c-Myc expression by miR-185-3p, this miRNA repressed the transcriptional activity and cell proliferation function of c-Myc. Conversely, miR-185-3p inhibitor, but not negative control, markedly reduced the level of endogenous miR-185-3p and consequently induced the level of endogenous c-Myc in both HCT116 and MCF7 cells. Therefore, our study uncovers miR-185-3p as the first miRNA that can act as an autoregulatory inhibitor of c-Myc by targeting the 3′ amino acid-coding sequence of c-Myc mRNA and inhibiting its translation (Fig. 6E).
It is particularly interesting to learn that miR-185-3p targets the 3′ amino acid-coding, instead of 3′-UTR, sequence of c-Myc mRNA. This is different from the previously and recently reported c-Myc targeting miRNAs, which bind to distinct seeding sequences residing in the 3′-UTR sequence of c-Myc mRNA (28–30). Although most miRNAs target a 3′-UTR sequence of mRNAs (43), targeting an open reading frame sequence of an mRNA by an miRNA molecule is not that unusual because an increasing number of miRNAs have been recently reported to target an mRNA protein-coding sequence (43, 44). For instance, a number of potential miRNA targeting sequences reside within the amino acid-coding sequences of an mRNA, as revealed by a global computer analysis of miRNA-targeted genes in human cells (45, 46). Specifically, miR-24 was recently shown to target the open reading frame region of Fas-associated factor 1 (FAF1) mRNA (47). Also, miR-206 was able to target the estrogen receptor α-coding sequence (48). Hence, it appears that targeting amino acid-coding mRNA sequences is probably a common and alternative mechanism for miRNA-mediated regulation of gene expression.
Although miR-185 was recently shown to induce cell cycle arrest (49), its molecular targets were unclear. Most recently, when we were preparing this paper, miR-185-5p,3 the miRNA processed from the 5′ stem-loop arm opposite to that for miR-185-3p, was reported to target Six1 (50). Unlike miR-185-5p, miR-185-3p specifically targets the amino acid-coding nucleotide sequence of c-Myc mRNA as a negative feedback regulator in monitoring the protein level of c-Myc, as discussed above. This finding is also physiologically significant. It has been known that both c-Myc protein and mRNA levels start to decline 3–6 h after serum stimulation (32). Although the abrogation of transcription, translation, and stability of c-Myc mRNA was believed to account for this c-Myc reduction (3), the role of miRNAs in this serum-responsive regulation of c-Myc expression has not been established. Our finding that miR-185-3p is expressed in response to serum stimulation and in turn represses the protein expression of c-Myc suggests that miR-185-3p is also responsible for the descent phase of c-Myc protein level in response to growth signals. Thus, our study offers a molecular mechanism for not only why miR-185 can lead to cell cycle arrest (49) and block cell proliferation (Fig. 5, C and D) but also why the protein level of c-Myc starts to diminish 3 h after serum treatment, although it still remains to be further investigated how c-Myc mRNA level also starts to decrease under this condition.
Identification of miR-185-3p as a suppressor of c-Myc also raises a tempting question as to whether this miRNA would play a role in suppressing c-Myc-driven tumor growth. It is likely that overexpression of miR-185-3p would eradicate the oncogenic function of c-Myc because miR-185-3p, when overexpressed in H1299 cells, significantly inhibited c-Myc-induced cell proliferation (Fig. 5). This likelihood will be further examined by using xenograft mouse models that express high level of c-Myc in the near future. Another unaddressed question is if the expression or level of miR-185-3p would be altered in human tumors. Regardless of these remaining questions, our study as presented here demonstrates that miR-185-3p is a physiological and negative feedback regulator of c-Myc.
Supplementary Material
Acknowledgments
We thank Yong Li for generously offering plasmids, Shelya Zeng and Yu Zhang for technical assistance, and the members of the Lu laboratory for active discussion.
This work was supported, in whole or in part, by National Institutes of Health, NCI, Grants CA127724, CA095441, and CA129828 (to H. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S6.
miR-185-5p and miR-185-3p are defined as the mature miRNAs processed from the 5′ and 3′ side, respectively, of the same precursor miRNA, and therefore, they share the same promoter.
- miRNA
- microRNA.
REFERENCES
- 1. Vennstrom B., Sheiness D., Zabielski J., Bishop J. M. (1982) J. Virol. 42, 773–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Adhikary S., Eilers M. (2005) Nat. Rev. Mol. Cell Biol. 6, 635–645 [DOI] [PubMed] [Google Scholar]
- 3. Secombe J., Pierce S. B., Eisenman R. N. (2004) Cell 117, 153–156 [DOI] [PubMed] [Google Scholar]
- 4. Grandori C., Cowley S. M., James L. P., Eisenman R. N. (2000) Annu. Rev. Cell Dev. Biol. 16, 653–699 [DOI] [PubMed] [Google Scholar]
- 5. Pelengaris S., Khan M., Evan G. (2002) Nat. Rev. Cancer 2, 764–776 [DOI] [PubMed] [Google Scholar]
- 6. Eisenman R. N. (2001) Genes Dev. 15, 2023–2030 [DOI] [PubMed] [Google Scholar]
- 7. Oster S. K., Ho C. S., Soucie E. L., Penn L. Z. (2002) Adv. Cancer Res. 84, 81–154 [DOI] [PubMed] [Google Scholar]
- 8. Davis A. C., Wims M., Spotts G. D., Hann S. R., Bradley A. (1993) Genes Dev. 7, 671–682 [DOI] [PubMed] [Google Scholar]
- 9. Amati B., Dalton S., Brooks M. W., Littlewood T. D., Evan G. I., Land H. (1992) Nature 359, 423–426 [DOI] [PubMed] [Google Scholar]
- 10. Lüscher B., Larsson L. G. (1999) Oncogene 18, 2955–2966 [DOI] [PubMed] [Google Scholar]
- 11. Blackwood E. M., Eisenman R. N. (1991) Science 251, 1211–1217 [DOI] [PubMed] [Google Scholar]
- 12. Roy A. L., Carruthers C., Gutjahr T., Roeder R. G. (1993) Nature 365, 359–361 [DOI] [PubMed] [Google Scholar]
- 13. Arabi A., Wu S., Ridderstråle K., Bierhoff H., Shiue C., Fatyol K., Fahlén S., Hydbring P., Söderberg O., Grummt I., Larsson L. G., Wright A. P. (2005) Nat. Cell Biol. 7, 303–310 [DOI] [PubMed] [Google Scholar]
- 14. Grandori C., Gomez-Roman N., Felton-Edkins Z. A., Ngouenet C., Galloway D. A., Eisenman R. N., White R. J. (2005) Nat. Cell Biol. 7, 311–318 [DOI] [PubMed] [Google Scholar]
- 15. Grewal S. S., Li L., Orian A., Eisenman R. N., Edgar B. A. (2005) Nat. Cell Biol. 7, 295–302 [DOI] [PubMed] [Google Scholar]
- 16. Adams J. M., Harris A. W., Pinkert C. A., Corcoran L. M., Alexander W. S., Cory S., Palmiter R. D., Brinster R. L. (1985) Nature 318, 533–538 [DOI] [PubMed] [Google Scholar]
- 17. Felsher D. W., Bishop J. M. (1999) Mol. Cell 4, 199–207 [DOI] [PubMed] [Google Scholar]
- 18. Pelengaris S., Khan M., Evan G. I. (2002) Cell 109, 321–334 [DOI] [PubMed] [Google Scholar]
- 19. Pelengaris S., Littlewood T., Khan M., Elia G., Evan G. (1999) Mol. Cell 3, 565–577 [DOI] [PubMed] [Google Scholar]
- 20. Nesbit C. E., Tersak J. M., Prochownik E. V. (1999) Oncogene 18, 3004–3016 [DOI] [PubMed] [Google Scholar]
- 21. Dews M., Homayouni A., Yu D., Murphy D., Sevignani C., Wentzel E., Furth E. E., Lee W. M., Enders G. H., Mendell J. T., Thomas-Tikhonenko A. (2006) Nat. Genet. 38, 1060–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Welcker M., Orian A., Jin J., Grim J. E., Harper J. W., Eisenman R. N., Clurman B. E. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 9085–9090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yada M., Hatakeyama S., Kamura T., Nishiyama M., Tsunematsu R., Imaki H., Ishida N., Okumura F., Nakayama K., Nakayama K. I. (2004) EMBO J. 23, 2116–2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. von der Lehr N., Johansson S., Wu S., Bahram F., Castell A., Cetinkaya C., Hydbring P., Weidung I., Nakayama K., Nakayama K. I., Söderberg O., Kerppola T. K., Larsson L. G. (2003) Mol. Cell 11, 1189–1200 [DOI] [PubMed] [Google Scholar]
- 25. Adhikary S., Marinoni F., Hock A., Hulleman E., Popov N., Beier R., Bernard S., Quarto M., Capra M., Goettig S., Kogel U., Scheffner M., Helin K., Eilers M. (2005) Cell 123, 409–421 [DOI] [PubMed] [Google Scholar]
- 26. Sears R., Nuckolls F., Haura E., Taya Y., Tamai K., Nevins J. R. (2000) Genes Dev. 14, 2501–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pulverer B. J., Fisher C., Vousden K., Littlewood T., Evan G., Woodgett J. R. (1994) Oncogene 9, 59–70 [PubMed] [Google Scholar]
- 28. Lal A., Navarro F., Maher C. A., Maliszewski L. E., Yan N., O'Day E., Chowdhury D., Dykxhoorn D. M., Tsai P., Hofmann O., Becker K. G., Gorospe M., Hide W., Lieberman J. (2009) Mol. Cell 35, 610–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sachdeva M., Zhu S., Wu F., Wu H., Walia V., Kumar S., Elble R., Watabe K., Mo Y. Y. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 3207–3212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sampson V. B., Rong N. H., Han J., Yang Q., Aris V., Soteropoulos P., Petrelli N. J., Dunn S. P., Krueger L. J. (2007) Cancer Res. 67, 9762–9770 [DOI] [PubMed] [Google Scholar]
- 31. Xiong J., Du Q., Liang Z. (2010) Oncogene 29, 4980–4988 [DOI] [PubMed] [Google Scholar]
- 32. Dai M. S., Arnold H., Sun X. X., Sears R., Lu H. (2007) EMBO J. 26, 3332–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McMahon S. B., Van Buskirk H. A., Dugan K. A., Copeland T. D., Cole M. D. (1998) Cell 94, 363–374 [DOI] [PubMed] [Google Scholar]
- 34. Dai M. S., Sears R., Lu H. (2007) Cell Cycle 6, 2735–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lu Z., Liu M., Stribinskis V., Klinge C. M., Ramos K. S., Colburn N. H., Li Y. (2008) Oncogene 27, 4373–4379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jin Y., Zeng S. X., Sun X. X., Lee H., Blattner C., Xiao Z., Lu H. (2008) Mol. Cell. Biol. 28, 1218–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tang F., Hajkova P., Barton S. C., Lao K., Surani M. A. (2006) Nucleic Acids Res. 34, e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dai M. S., Sun X. X., Lu H. (2008) Mol. Cell. Biol. 28, 4365–4376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jin Y., Dai M. S., Lu S. Z., Xu Y., Luo Z., Zhao Y., Lu H. (2006) EMBO J. 25, 1207–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zeng S. X., Dai M. S., Keller D. M., Lu H. (2002) EMBO J. 21, 5487–5497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sun X. X., Dai M. S., Lu H. (2008) J. Biol. Chem. 283, 12387–12392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dai M. S., Sun X. X., Lu H. (2010) J. Biol. Chem. 285, 12587–12594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bartel D. P. (2009) Cell 136, 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Selbach M., Schwanhäusser B., Thierfelder N., Fang Z., Khanin R., Rajewsky N. (2008) Nature 455, 58–63 [DOI] [PubMed] [Google Scholar]
- 45. Lewis B. P., Burge C. B., Bartel D. P. (2005) Cell 120, 15–20 [DOI] [PubMed] [Google Scholar]
- 46. Miranda K. C., Huynh T., Tay Y., Ang Y. S., Tam W. L., Thomson A. M., Lim B., Rigoutsos I. (2006) Cell 126, 1203–1217 [DOI] [PubMed] [Google Scholar]
- 47. Qin W., Shi Y., Zhao B., Yao C., Jin L., Ma J., Jin Y. (2010) PloS One 5, e9429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Adams B. D., Furneaux H., White B. A. (2007) Mol. Endocrinol. 21, 1132–1147 [DOI] [PubMed] [Google Scholar]
- 49. Takahashi Y., Forrest A. R., Maeno E., Hashimoto T., Daub C. O., Yasuda J. (2009) PloS One 4, e6677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Imam J. S., Buddavarapu K., Lee-Chang J. S., Ganapathy S., Camosy C., Chen Y., Rao M. K. (2010) Oncogene 29, 4971–4979 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.