Abstract
Gene looping, defined as the interaction of the promoter and the terminator regions of a gene during transcription, requires transcription factor IIB (TFIIB). We have earlier demonstrated association of TFIIB with the distal ends of a gene in an activator-dependent manner (El Kaderi, B., Medler, S., Raghunayakula, S., and Ansari, A. (2009) J. Biol. Chem. 284, 25015–25025). The presence of TFIIB at the 3′ end of a gene required its interaction with cleavage factor 1 (CF1) 3′ end processing complex subunit Rna15. Here, employing affinity chromatography and glycerol gradient centrifugation, we show that TFIIB associates with poly(A) polymerase and the entire CF1 complex in yeast cells. The factors required for general transcription such as TATA-binding protein, RNA polymerase II, and TFIIH are not a component of the TFIIB complex. This holo-TFIIB complex was resistant to MNase digestion. The complex was observed only in the looping-competent strains, but not in the looping-defective sua7-1 strain. The requirement of Rna15 in gene looping has been demonstrated earlier. Here we provide evidence that poly(A) polymerase (Pap1) as well as CF1 subunits Rna14 and Pcf11 are also required for loop formation of MET16 and INO1 genes. Accordingly, cross-linking of TFIIB to the 3′ end of genes was abolished in the mutants of Pap1, Rna14, and Pcf11. We further show that in sua7-1 cells, where holo-TFIIB complex is not formed, the kinetics of activated transcription is altered. These results suggest that a complex of TFIIB, CF1 subunits, and Pap1 exists in yeast cells. Furthermore, TFIIB interaction with the CF1 complex and Pap1 is crucial for gene looping and transcriptional regulation.
Keywords: Gene Regulation, RNA Polymerase II, Transcription, Transcription Factors, Yeast, CF1 Complex, Gene Looping, Poly(A) polymerase, TFIIB
Introduction
Transcription is regulated at multiple steps by a rich diversity of mechanisms. One such mechanism involves formation of a looped architecture due to the interaction of transcription regulatory DNA elements that are not adjacent on the linear chromosome (2, 3). Loops formed by the interaction of the enhancer with the promoter contribute, at least in part, to the transcriptional activation of genes in eukaryotes (4). Although enhancer-promoter interaction and its role in transcriptional regulation are well established, recent studies suggest that the promoter and terminator regions of a gene also cross-talk during transcription (5, 6). The interaction of 5′ and 3′ ends of a gene in a transcription-dependent manner resulting in the formation of a looped conformation is referred to as gene looping. First reported in yeast, gene looping has now been shown to occur during transcription in higher eukaryotes as well (7–9). Although the exact biological role of gene looping is not clear yet, it has been implicated in a variety of cellular functions. Transcriptional activation of genes in yeast and HIV provirus coincides with the gene assuming a looped configuration (1, 5, 6, 8, 10–12). In contrast, looping of mammalian BRCA1 gene was accompanied by its transcriptional repression in breast cancer cell lines (9), whereas human monocyte immunohistological marker gene CD68 exhibited efficient transcription-coupled splicing of its precursor mRNA upon loop formation (7). Recently, gene looping was identified as the molecular basis of transcriptional memory (10, 12). Thus, gene looping may have different regulatory roles in different cellular contexts.
The emerging ubiquity of gene looping and its potential as an important transcription regulatory mechanism necessitates understanding the mechanism of loop formation. Recent studies have implicated TFIIB4 as a major player in gene looping. TFIIB was found occupying the distal ends of a gene only when it was in looped configuration (1, 11). TFIIB has also been shown to interact with Ssu72 and Rna15 subunits of CPF and CF1 3′ end processing complexes in budding yeast (1, 11). A similar interaction of TFIIB with cleavage and polyadenylation specific factor (CPSF) and cleavage stimulatory factor (CstF) cleavage and polyadenylation complexes has recently been reported in mammalian cells (13). The overall conclusion of these results is that gene looping is primarily facilitated by the interaction of promoter-bound TFIIB with 3′ end processing/termination factors operating at the terminator region of a gene (1, 5, 10, 11). In such a scenario, Ssu72 and Rna15 may not be the only termination factors that interact with TFIIB to facilitate loop formation. Other subunits of CF1 and CPF complexes may also contribute to the protein-protein interactions that bring the promoter and the terminator together during gene looping. A thorough understanding of gene loop formation requires characterization of the macromolecular complex that serves as a bridge between the promoter and the terminator regions of a gene in looped configuration.
Here we demonstrate the existence of a complex of TFIIB, CF1 subunits, and poly(A) polymerase (Pap1) in yeast cells. The holo-TFIIB complex was observed exclusively in the looping-competent strains. In addition to Rna15, two more subunits of the complex, Rna14 and Pcf11, are required for gene looping. Pap1, which adds a poly(A) tail at the 3′ end of precursor mRNA, also physically interacts with TFIIB and is an essential looping factor. In the looping-defective sua7-1 strain, where a holo-TFIIB complex is not formed, activated transcription exhibits a kinetic lag. These results emphasize the crucial role of TFIIB interactions with the CF1 complex and Pap1 in gene looping and transcription in budding yeast.
EXPERIMENTAL PROCEDURES
Yeast Strains
The yeast strains used in this study are listed in supplemental Table S1. Strains AA1, AA2, NAH12, and NAH13, which contain a tandem affinity purification-tagged TFIIB, were constructed by transforming the temperature-sensitive mutants (rna14-1, pap1-1, pcf11-2, and hrp1-5, respectively) with DNA that was PCR-amplified from plasmid pBS1479 (TRP marker). The tandem affinity purification tags were inserted at the C terminus of TFIIB. The HA-tagged TFIIB strain (SAM56) and the HA-tagged TBP (SAM68) strain were constructed by transforming BY4733 (wild type (WT)) with DNA that was PCR-amplified from pFA6-3HA-His3MX6 (HIS marker). Strains SRR7 and SRR8, which contain C-terminal tandem affinity purification-tagged Rna14 and Pcf11, respectively, were constructed with DNA that was PCR-amplified from plasmid pBS1539.
The C-terminal Myc tagging of CF1 subunits (Rna14, Rna15, Pcf11, Hrp1, and Clp1) and poly(A) polymerase was carried out by transforming the parental strains with the PCR product amplified from pFA6-13Myc-TRP1 (TRP marker). Strains SAM50 (Myc-tagged Rna14), SAM51 (Myc-tagged Rna15), SAM52 (Myc-tagged Pcf11), SAM53 (Myc-tagged Hrp1), SAM54 (Myc-tagged Clp1), and SAM55 (Myc-tagged Pap1) were derived from a parental BY4733 (WT). Strains SAM58 (Myc-tagged Rna14), SAM59 (Myc-tagged Rna15), SAM60 (Myc-tagged Pcf11), SAM61 (Myc-tagged Hrp1), SAM62 (Myc-tagged Clp1), and SAM63 (Myc-tagged Pap1) were derived from a parental SAM56 (HA-tagged TFIIB). The looping-defective HA-tagged TFIIB strain (SAM64) was constructed by transforming YMH124 (sua7-1 mutant) with DNA that was PCR-amplified from pFA6-3HA-kanMX6 (kanamycin marker).
Cell Culture
Cultures were started from freshly streaked plates and grown overnight in 5 ml of YPD at 25 °C with shaking. The next day, the cells were diluted 0.010 into 100 ml of fresh YP-dextrose. For INO1 analysis by capture chromosome conformation (CCC) and ChIP, the wild type cells were grown to an A600 of 0.4, and the temperature-sensitive cells were grown to an A600 of 0.5 prior to induction. Cells were then transferred to 100 ml of inositol plus and inositol-depleted synthetic media and grown to an A600 of 0.7 (3 h) at 25 °C with shaking. For MET16 analysis by CCC and ChIP, the wild type cells were grown to an A600 of 0.3, and the temperature-sensitive cells were grown to an A600 of 0.45 prior to induction. Cells were transferred to 100 ml of methionine plus and methionine-depleted synthetic media and grown to an A600 of 0.7 (2 h) at 25 °C with shaking. The cells were heat-inactivated for 2 h at 37 °C and then processed for ChIP or CCC.
Primers
Primers used for CCC and ChiP experiments are listed in supplemental Table S2.
CCC
CCC experiments were performed as described previously (1).
ChIP
ChIP experiments were performed as described previously (1).
Transcription Analysis
Transcription analysis of INO1 and MET16 in wild type and sua7-1 cells was performed by the RT-PCR approach as described previously (1).
Affinity Purification
For affinity purification of HA-tagged TFIIB, TBP and Kin28 cells were grown in 1 liter of YP-dextrose to an A600 of 1.5. The cell pellet obtained from the 1-liter culture was washed with 50 ml of ice-cold 1× TBS followed by a wash with 50 ml of ice-cold double-distilled water. The pellet was resuspended in 10 ml of lysis buffer (20 mm Tris-HCl, pH 8.0, 150 mm KCl, 0.5 mm EDTA, 1 mm MgCl2, 0.1% Triton X-100, 1 mm PMSF, and 10% glycerol (v/v)). The cell suspension was flash-frozen in liquid nitrogen as described in Ansari and Schwer (14). The frozen cells were homogenized into a fine powder in a chilled mortar. The powder was then transferred to an ice-cold beaker and allowed to thaw slowly. The resulting cell lysate was then centrifuged at 16,400 rpm for 20 min in a Sorvall SS-34 rotor. The supernatant (about 10 ml) was allowed to bind to 40 μl of anti-HA-agarose beads (Sigma) in a 15-ml tube for 4 h at 4 °C with gentle shaking. The beads were washed three times with 1 ml each of ice-cold lysis buffer. Elution was performed with 100 μg of HA oligopeptides resuspended in 200 μl of lysis buffer at 25 °C for 30 min. The resulting eluate was then used for either Western blotting or glycerol gradient sedimentation.
For the MNase controls, the cell lysate was incubated with 300 units of MNase at 37 °C for 30 min prior to binding to anti-HA-agarose beads. For the high salt controls, the lysis buffer was prepared as described above using a 500 mm KCl concentration.
Glycerol Gradient Sedimentation
Affinity-purified samples were diluted with an equal volume of ice-cold lysis buffer without glycerol and incubated for 20 min on ice. The samples were then centrifuged at 30,000 rpm for 18 h at 4 °C in a 39.6-ml linear 5–30% (v/v) glycerol gradient in a Sorvall SureSpin 360 rotor. Linear gradients were prepared using a BioComp gradient master, mixing equal volumes of a 5% glycerol lysis buffer with a 30% glycerol lysis buffer. Fractions of 1.8 ml each were collected using a Beckman fraction recovery system, and 40 μl of each fraction was used for Western blotting.
Cloning and Purification of Recombinant TFIIB
The gene coding for yeast TFIIB, SUA7, was cloned into the NdeI-EcoRI sites of pET24a. Recombinant plasmid was transformed into the BL21 strain of Escherichia coli. Induction of recombinant TFIIB and preparation of cell lysate were performed as described by Ansari and Schwer (14). His-tagged TFIIB was affinity-purified on a Cobalt resin (Pierce Scientific) following the manufacturer's guidelines.
Western Blot Analysis
Anti-HA antibodies were purchased from Neomarkers. Anti-Myc antibodies were purchased from Upstate Biotech Millipore. Anti-Pap1 and Anti-Rna15 were generous gifts from Claire Moore. Anti-TFIIB antibodies were a generous gift from Michael Hampsey. Anti-TBP and anti-Rpb1 antibodies were purchased from Santa Cruz Biotechnology. Western blotting protocol was performed as described previously (1).
RESULTS
TFIIB Forms a Complex with CF1 Subunits and Pap1
TFIIB is an essential general transcription factor (15–17). Recombinant TFIIB, with a molecular mass in the range of 32–38 kDa, could complement all functions of native, biochemically purified TFIIB in an in vitro transcription assay (16–19). These results suggested that TFIIB is a single polypeptide protein that exists as a monomer in solution. There was no evidence of TFIIB being a part of a macromolecular complex containing initiation factors or termination factors or any other protein. To find proteins associated with TFIIB under physiological conditions, proteomic analysis was performed employing the tandem affinity purification approach (20–22). Neither a promoter nor a terminator-bound factor was detected in the affinity-purified TFIIB preparation in the first proteomic analysis carried out by Gavin et al. (21). However, in the second analysis, poly(A)-binding protein (Pab1), which interacts with the poly(A) tail of mRNA, was identified as the only 3′ end processing factor associated with TFIIB (20). The study carried out by Krogan et al. (22) found RNAP II subunits and two terminator-bound factors, CPF subunit Fip1 and Pab1-interacting factor Pan2, copurifying with TFIIB. The absolute requirement of TFIIB in gene looping, cross-linking of TFIIB to both the promoter and the terminator regions of a looped gene, and its functional interaction with several 3′ end processing/termination factors including CF1 subunit Rna15 (1, 11) suggested that a complex of TFIIB and termination factors exists in the cell. Apart from Rna15, the CF1 complex is composed of four more subunits namely Rna14, Pcf11, Clp1, and Hrp1. It is involved in 3′ end processing of precursor mRNA as well as termination of transcription in budding yeast (23).
We therefore investigated whether TFIIB forms a macromolecular complex with CF1 subunits and Pap1 in yeast cells. Our experimental approach involved affinity purification of TFIIB followed by detection of CF1 subunits and Pap1 in the purified preparation by Western blot. To facilitate affinity purification of TFIIB, a triple hemagglutinin (3×HA) tag was inserted at the carboxyl terminus of TFIIB. Insertion of the HA tag did not interfere with the biological activity of TFIIB as both the transcription and the gene looping of MET16 and INO1 remained unaffected in the tagged strain (data not presented). Additionally, a Myc tag was integrated at the carboxyl terminus of each of the five subunits of CF1 complex and Pap1 for their detection by Western blot. Thus, six strains were constructed, each carrying HA-tagged TFIIB and a Myc-tagged version of one of the subunits of CF1 complex and Pap1.
Cell lysates from each of the six strains described above were purified over anti-HA-agarose beads. Proteins bound to the beads were eluted with HA oligopeptides. Western blot analysis of eluates revealed the presence of Rna14, Rna15, Pcf11, Hrp1, Clp1, and Pap1 (Fig. 1A, lanes 3–8) in the affinity-purified TFIIB preparation. As a control, coimmunoprecipitation was performed from a strain carrying untagged TFIIB. Our results show that the association of CF1 subunits Rna14 and Pcf11 with TFIIB was not dependent on HA tag (supplemental Fig. S1, lanes 3 and 6). To rule out the possibility of the Myc tag of CF1 subunits and Pap1 directly interacting with anti-HA beads, we carried out purifications from strains with an untagged TFIIB. No binding of Myc-tagged CF1 subunits and Pap1 to anti-HA beads was observed in the absence of HA-tagged TFIIB (supplemental Fig. S2, lanes 3–8). MNase digestion of cell lysate prior to affinity purification did not disrupt the association of CF1 subunits and Pap1 with TFIIB (supplemental Fig. S3, lanes 3–8). These results indicate that the interaction of terminator-bound factors with TFIIB may not be mediated by DNA or RNA. To rule out the possibility that copurification of Myc-tagged CF1 subunits and Pap1 with TFIIB is not due to the interaction of the Myc tag with TFIIB, the affinity purification was performed in a strain without a Myc tag on any of the CF1 subunits or Pap1. Western blot analysis of affinity-purified TFIIB, in this case using antibodies specifically directed against Pap1 and CF1 subunit Rna15, revealed that the interaction of these factors with TFIIB is not dependent on the Myc tag (Fig. 1B, lane 1). The affinity purification of a holo-TFIIB complex described above was performed at a KCl concentration of 150 mm. To check the stability of the complex, we repeated the purification at 500 mm KCl in the lysis buffer. High ionic strength did not affect the association of TFIIB with CF1 subunits and Pap1 during affinity purification (supplemental Fig. S4).
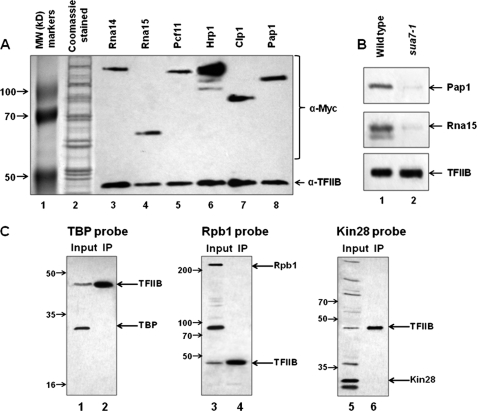
FIGURE 1.
CF1 subunits and poly(A) polymerase copurify with TFIIB on an anti-HA affinity column. A, HA-tagged TFIIB was affinity-purified from cells harboring Myc-tagged CF1 subunits or poly(A) polymerase as described under “Experimental Procedures.” Purified samples were subjected to SDS-PAGE followed by Western blot analysis using anti-HA and anti-Myc antibodies. Lane 1 displays molecular weight (MW) marker proteins, and lane 2 represents Imperial Coomassie Blue staining of the eluate from an anti-HA affinity column. B, affinity purifications were performed for HA-tagged TFIIB in the wild type and the looping-defective mutant sua7-1 strain. The eluates from the affinity columns were subjected to SDS-PAGE, and Western blotting was performed with antibodies against TFIIB, Rna15, and poly(A) polymerase. C, affinity-purified HA-tagged TFIIB was subjected to SDS-PAGE followed by Western blot analysis using anti-HA, anti-TBP, anti-Kin28, and anti-Rpb1 antibodies. IP, immunoprecipitation.
Holo-TFIIB Complex Does Not Contain General Transcription Factors
TFIIB has been shown to interact, both genetically as well as physically, with TBP and RNAP II (24–31). We therefore checked for the presence of these proteins in the affinity-purified TFIIB preparation using antibodies directed against TBP and Rpb1 subunit of polymerase. No signal for either TBP (Fig. 1C, lane 2) or Rpb1 (Fig. 1C, lane 4) was detected in the TFIIB preparation. We also did not find any evidence for the presence of another general transcription factor TFIIH in the affinity-purified TFIIB preparation. Western blot analysis using antibodies against TFIIH subunit Kin28 confirmed the absence of the factor in the TFIIB preparation (Fig. 1C, lane 6). These results suggest that the holo-TFIIB complex does not contain the factors that transiently interact with it during the transcription cycle.
Holo-TFIIB Complex Is Not Observed in Looping-defective Cells
To determine the physiological significance of TFIIB-CF1-Pap1 complex in the context of gene looping, affinity purification of TFIIB was performed in a looping-deficient mutant strain of TFIIB called sua7-1. Affinity-purified TFIIB preparation from sua7-1 cells was subjected to SDS-PAGE analysis followed by Western blotting using antibodies against Pap1 and CF1 subunit Rna15. Our results show that neither Pap1 nor Rna15 was found associated with TFIIB in sua7-1 strain (Fig. 1B, lane 2). Thus, TFIIB association with the terminator-bound factors occurred in a looping-dependent manner. These results argue in favor of a TFIIB-CF1-Pap1 complex playing a crucial role in loop formation.
Glycerol Gradient Analysis of Holo-TFIIB Complex
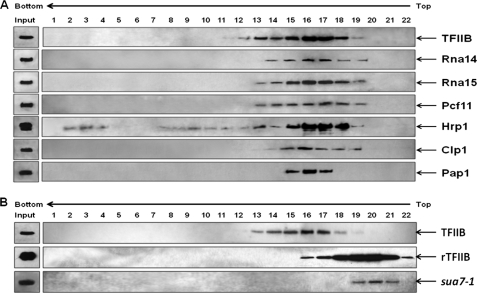
To further confirm that a holo-TFIIB complex exists in yeast cells, affinity-purified TFIIB was subjected to sedimentation analysis on a linear 5–30% (v/v) glycerol gradient in the presence of 150 mm KCl. Western blot analysis revealed that TFIIB fractionated as a single peak spanning fractions 12–19 (Fig. 2A). Pap1, as well as CF1 subunits Rna14, Rna15, Pcf11, Hrp1, and Clp1, cosedimented with TFIIB (Fig. 2A). The peak of CF1 subunits and Pap1 coincided with the TFIIB peak in fraction number 16 (Fig. 2A). To conclusively prove that TFIIB cosedimenting with CF1 subunits and Pap1 is not free TFIIB, but TFIIB in a complex with 3′ end processing/termination factors, it was important to determine the sedimentation behavior of free TFIIB. For this, we purified recombinant TFIIB from bacteria and carried out sedimentation analysis under identical conditions. Recombinant TFIIB (rTFIIB) sedimented in fractions 18–22 with the peak centered on fraction number 20 (Fig. 2B). To further confirm that TFIIB is in a complex with termination factors, we carried out sedimentation analysis of TFIIB from looping-defective sua7-1 cells that do not harbor a holo-TFIIB complex. Under identical conditions, affinity-purified TFIIB from sua7-1 cells cosedimented with rTFIIB with a peak in fraction number 20 (Fig. 2B). These results suggest that almost all TFIIB in the affinity-purified preparation is in a complex with 3′ end processing/termination factors.
FIGURE 2.
Sedimentation analysis of TFIIB. A, sedimentation analysis reveals copurification of CF1 subunits and poly(A) polymerase with TFIIB. Affinity-purified HA-tagged TFIIB was subjected to sedimentation analysis in 5–30% (v/v) glycerol gradient in 150 mm KCl. Fractions of 1.8 ml each were collected and subjected to SDS-PAGE analysis followed by Western blotting to visualize TFIIB, CF1 subunits, and poly(A) polymerase. Input bands indicate affinity-purified sample prior to sedimentation analysis. B, sedimentation analysis of affinity-purified TFIIB from wild type cells, rTFIIB, and affinity-purified TFIIB from looping-defective sua7-1 cells under the conditions as described above.
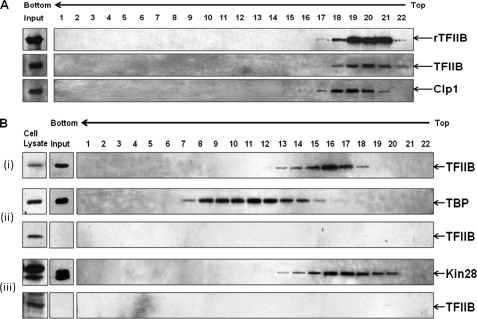
However, when sedimentation analysis of affinity-purified TFIIB was carried out in the glycerol gradient made in 500 mm KCl, TFIIB was separated from CF1 complex and sedimented at a lower rate in fractions 18–22 with the peak in fraction number 20 (Fig. 3A). This correlates with the sedimentation profile of purified TFIIB from sua7-1 cells that lack a holo-TFIIB complex in the 150 mm KCl glycerol gradient (Fig. 2B) as well as with that of recombinant TFIIB in the 500 mm KCl glycerol gradient (Fig. 3A). Because the sedimentation pattern of rTFIIB was identical in 150 and 500 mm KCl glycerol gradients (Figs. 2B and 3A), these results suggest that the reduced mobility of native affinity-purified TFIIB in the 500 mm KCl glycerol gradient is not the consequence of high ionic strength but due to the separation of TFIIB from poly(A) polymerase and CF1 subunits (as indicated by a shift in Clp1 peak in Fig. 3A). Thus, holo-TFIIB complex is not stable upon prolonged exposure to high salt in a centrifugal field, although it is able to withstand high ionic strength for a short period of time during affinity purification.
FIGURE 3.
Holo-TFIIB complex is susceptible to high ionic strength and sediments between TFIID and TFIIH complexes. A, affinity-purified HA-tagged TFIIB and rTFIIB were subjected to sedimentation analysis in 5–30% (v/v) glycerol gradient in 500 mm KCl. Fractions were collected and processed as in Fig. 2. B, affinity-purified preparations of TFIIB (panel i), TBP (panel ii), and Kin28 (panel iii) were subjected to sedimentation analysis in 5–30% (v/v) glycerol gradient in 150 mm KCl as described previously. Fractions were collected, processed as above, and probed for TFIIB and TBP or TFIIB and Kin28.
We then compared the sedimentation profile of the holo-TFIIB complex with the sedimentation profiles of the TFIID and TFIIH complexes. The affinity-purified TFIIB, TFIID, and TFIIH were subjected to sedimentation analysis under identical conditions (Fig. 3B). The presence of TFIID and TFIIH in the gradient fractions was detected by Western blot analysis using antibodies against the TBP and Kin28 subunits of TFIID and TFIIH, respectively. TFIID, which has a molecular mass of about 750 kDa (32), sedimented in fractions 7–15 with the peak in fraction number 11 (Fig. 3B, panel ii), whereas TFIIH with an approximate molecular mass of 500 kDa (33) sedimented in fractions 13–20 (Fig. 3B, panel iii). Thus, the sedimentation coefficient of the holo-TFIIB complex is intermediate between that of the TFIID and TFIIH complexes. We also looked for the presence of TFIIB in the gradient-purified TFIID and TFIIH preparations. No signal for TFIIB was detected in either the TFIID (Fig. 3B, panel ii) or the TFIIH (Fig. 3B, panel iii) glycerol gradient fractions. This corroborated our earlier results that TFIIB is not in a complex with general transcription factors.
Pap1, Rna14, and Pcf11 Are Required for Gene Looping
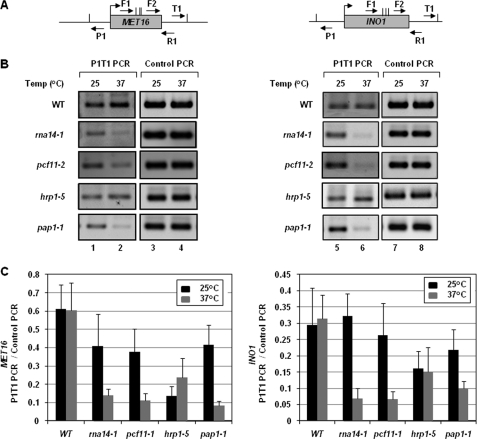
The presence of a TFIIB-CF1-Pap1 complex exclusively in the looping-competent strain supports the notion that the interaction of TFIIB with the terminator-bound factors is critical for gene looping. In such a scenario, TFIIB-associated termination factors are also expected to be required for loop formation. We therefore examined the role of components of a holo-TFIIB complex in gene looping employing the CCC approach. We have previously demonstrated the interaction of the promoter region of MET16 and INO1 with their terminator site using this approach (1). A PCR product obtained using divergent primers P1 and T1 was taken as a measure of gene looping in these experiments (Fig. 4A) (34). Using this protocol, we demonstrated the requirement of the Rna15 component of CF1 complex in gene looping (1). Here we examine the role of TFIIB-associated CF1 subunits other than Rna15, namely Rna14, Pcf11, and Hrp1 as well as poly(A) polymerase in gene looping. CCC analysis was therefore carried out for MET16 and INO1 under transcriptionally inductive conditions in the mutant strain of the factor under investigation. As a CCC control, complete digestion of chromatin by restriction enzyme and ligation dependence of P1-T1 PCR products was routinely monitored.
FIGURE 4.
Gene looping requires the CF1 complex and poly(A) polymerase. A, schematic representation of MET16 and INO1 indicating the positions of AluI restriction sites (vertical lines) and PCR primers (arrows) used in CCC analysis. B, CCC analysis of MET16 and INO1 to detect gene looping in W303-1a (wild type) and mutant strains of Rna14 (rna14-1), Pcf11 (pcf11-2), Hrp1 (hrp1-5), and poly(A) polymerase (pap1-1) following 120 min of induction followed by incubation at either permissive (25 °C) or non-permissive (37 °C) temperatures. P1T1 PCR reflects the looping signal, whereas Control PCR represents the loading control indicating that equal amount of template DNA was present in each of the CCC reactions. C, quantification of the CCC results shown in B, representing ChIP signal/Input signal. Error bars indicate one standard deviation.
During termination of transcription, Rna14 and Pcf11 play a role analogous to that of Rna15 (35). To determine whether these subunits also have a role in loop formation, we performed CCC analysis in rna14-1 and pcf11-2 mutant strains under induced conditions. A distinct P1-T1 PCR signal was obtained for both MET16 and INO1 when the mutants were grown at permissive temperature (25 °C) (Fig. 4B, lanes 1 and 5, and 4C). However, upon shifting to non-permissive temperature (37 °C), P1-T1 looping signal decreased by about 6–8-fold for both MET16 and INO1 (Fig. 4B, lanes 2 and 6, and 4C). No such diminution in looping signal was detected in the isogenic wild type strain following the temperature shift (Fig. 4B, lanes 2 and 6, and 4C). Thus, both Rna14 and Pcf11, like Rna15, are essential for gene looping in yeast.
Hrp1 is another subunit of CF1 complex, which has been implicated in maintaining the length of poly(A) tail (36). We next asked whether Hrp1, like other components of CF1 complex, is required for loop formation. CCC analysis was carried out in the hrp1-5 strain as described above. We did not observe any decrease in P1-T1 PCR signal for either MET16 or INO1 in hrp1-5 cells following the temperature shift to 37 °C (Fig. 4B, lanes 2 and 6, and 4C). Like Hrp1, poly(A) polymerase is required at the polyadenylation step of 3′ end processing. It is, however, not a CF1 subunit but is a component of CPF complex (37). A marked diminution (3–5-fold) of the P1-T1 PCR product for MET16 and INO1 was observed in pap1-1 strain following the temperature shift from 25 to 37 °C (Fig. 4B, lanes 2 and 6, and 4C). Thus, of the two factors operating at the polyadenylation step of 3′ end processing, our results show that only Pap1 is essential for gene looping.
In conclusion, CF1 components Rna14, Rna15, and Pcf11 are required for gene looping. In addition, polyadenylating enzyme, poly(A) polymerase, is also essential for loop formation. Although we did not observe any decrease in looping signal of genes in hrp1-5 strain at elevated temperature, we cannot make a conclusion regarding the role of Hrp1 in loop formation. To confirm the role of Hrp1 in gene looping, we need to check other temperature-sensitive mutant alleles of Hrp1. We could not examine the role of the remaining CF1 subunit Clp1 in gene looping due to non-availability of a good conditional mutant allele of the factor. Thus, gene looping exhibits an obligate requirement for CF1 complex and poly(A) polymerase.
TFIIB Localization on the Terminator Region Requires Pap1, Rna14, and Pcf11
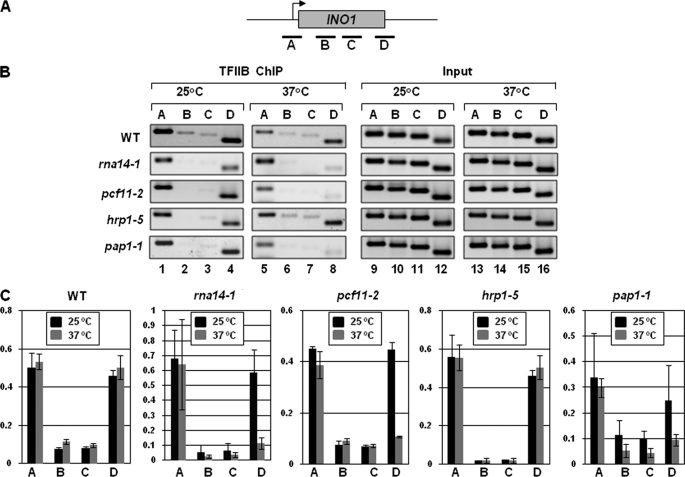
Our hypothesis is that gene looping is facilitated by the interaction of promoter-bound TFIIB with the terminator-bound factors including CF1 complex subunits and Pap1 (5). Cross-linking of TFIIB to the promoter and the terminator regions of a looped gene (1, 11), the presence of a TFIIB-CF1-Pap1 complex in looping-competent strains, and loss of gene looping in the mutants of TFIIB, CF1 subunits, and Pap1 provide support to our hypothesis. Accordingly, in a previous study, we have demonstrated that cross-linking of TFIIB to the 3′ end of a gene is compromised in a mutant of Rna15 that is deficient in gene looping (1). We therefore asked whether TFIIB association with the 3′ end of genes is also dependent on other components of holo-TFIIB complex such as Rna14, Pcf11, Hrp1, and Pap1. TFIIB ChIP was therefore performed for MET16 and INO1 genes in the mutants of Rna14 (rna14-1), Pcf11 (pcf11-2), Hrp1 (hrp1-5), and Pap1 (pap1-1) and isogenic wild type strains grown under transcriptionally inductive conditions at the permissive (25 °C) and non-permissive (37 °C) temperatures. TFIIB cross-links to the 3′ end of INO1, in the wild type strain, at 25 and 37 °C during induced conditions (Fig. 5B, lanes 4 and 8, and 5C). In contrast, TFIIB cross-linking to the terminator was abolished in rna14-1, pcf11-2, and pap1-1 strains at restrictive temperature (37 °C) (Fig. 5B, lane 8, and 5C), whereas the cross-linking to the promoter remained intact (Fig. 5B, lane 5, and 5C). TFIIB occupancy of the terminator region of INO1 remained unaffected in hrp1-5 strain following a temperature shift to 37 °C (Fig. 5B, lanes 4 and 8, and 5C). Identical results were obtained with MET16 (supplemental Fig. S5). This is in accordance with CCC results where no decrease in looping signal was observed in hrp1-5 strain at non-permissive temperature.
FIGURE 5.
TFIIB cross-linking to the terminator region is dependent upon a functional CF1 complex and poly(A) polymerase. A, schematic depiction of INO1 indicating the positions of ChIP primer pairs A, B, C, and D. B, ChIP analysis showing cross-linking of TFIIB to different regions of INO1 in W303-1a (wild type) and mutant strains of Rna14 (rna14-1), Pcf11 (pcf11-2), Hrp1 (hrp1-5), and poly(A) polymerase (pap1-1) following 120 min of induction followed by incubation at either permissive (25 °C) or non-permissive (37 °C) temperatures. The Input signal represents DNA prior to immunoprecipitation. C, quantification of the data shown in B, representing ChIP signal/input signal. Error bars indicate one standard deviation.
Thus, Rna14, Pcf11, and Pap1, like Rna15, are required for interaction of TFIIB with the 3′ end of MET16 and INO1. To comment on the role of Hrp1 in TFIIB cross-linking to the 3′ end of genes, we need to perform TFIIB-ChIP in other temperature-sensitive mutants of hrp1.
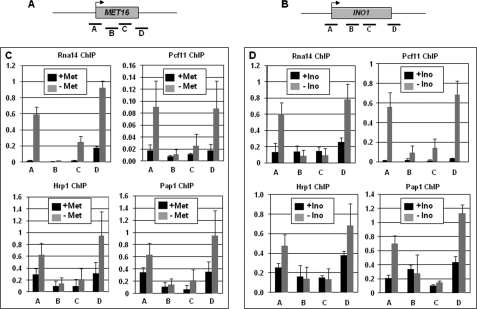
TFIIB-interacting Termination Factors Occupy the Distal Ends of a Gene in Looped Configuration
If the presence of TFIIB on the terminator region is due to its interaction with components of CF1 subunits and Pap1, we expected TFIIB-interacting termination factors to be present on both the terminator and the promoter regions of a transcriptionally activated gene. ChIP analysis was therefore performed to determine the presence of CF1 subunits on different regions of MET16 and INO1 during the repressed and activated transcriptional state of genes. Our results show that CF1 subunits Rna14, Pcf11, and Hrp1 as well as poly(A) polymerase indeed cross-link to both the terminator and the promoter regions of MET16 during activated transcription (Fig. 6C, gray bars). Identical results were obtained with INO1 (Fig. 6D, gray bars). These results provide support for the existence of a complex of TFIIB-CF1 subunits and Pap1 at the promoter-terminator junction. A similar TFIIB-dependent localization of CPSF and CstF 3′ end processing complexes on the distal ends of a gene was recently demonstrated in mammalian cells as well (13).
FIGURE 6.
The CF1 complex and poly(A) polymerase cross-link to the promoter and the terminator regions of MET16 and INO1 during activated transcription. A and B, schematic depiction of MET16 and INO1 indicating the positions of ChIP primer pairs A, B, C, and D. C and D, quantification of the ChIP analysis data showing cross-linking of Rna14, Pcf11, Hrp1, and Pap1 to different regions of MET16 and INO1 during repressed (black bars) and activated (gray bars) transcription. Met, methionine; Ino, inositol. Error bars indicate one standard deviation.
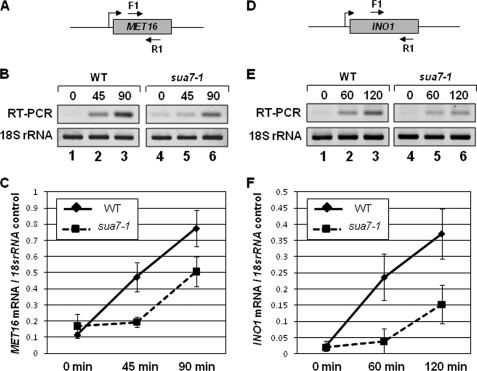
Kinetics of Activated Transcription Is Compromised in the Absence of Gene Looping
We have earlier demonstrated that gene looping is conferred by the activator-dependent interaction of the promoter-bound proteins with the terminator-bound factors (1). Here we provide evidence for the existence of a complex of promoter-bound TFIIB with the terminator-associated factors in yeast cells. This complex could be the molecular basis of gene looping as it exists only in the looping-competent strains, but not in the looping-defective strain. Gene looping has been proposed to enhance transcription efficiency of a gene by coupling termination to reinitiation (5). In such a scenario, efficiency of transcription is expected to decrease in the absence of gene looping. We therefore compared kinetics of activated transcription of MET16 and INO1 in wild type cells that harbor the holo-TFIIB complex and in the looping-defective sua7-1 strain. Our results suggest that although both MET16 and INO1 exhibited induced transcription in the sua7-1 strain, activated transcription exhibited a kinetic lag in the looping-defective strain (Fig. 7). The level of MET16 RNA in sua7-1 cells was about 1.5 times less than in isogenic wild type strain following 90 min after transfer of cells to inducing conditions (Fig. 7B, lanes 3 and 6, and 7C). Similarly, the INO1 RNA level in sua7-1 cells was ∼2.5 times less than in wild type cells at 120 min after induction of transcription (Fig. 7E, lanes 3 and 6, and 7F). A possible interpretation of these results is that looped configuration helps a gene to achieve higher transcription efficiency within a short period of time following exposure of the cells to induction signal.
FIGURE 7.
Induced transcription in the looping-defective mutant of TFIIB exhibits a kinetic lag. A and D, schematic depiction of MET16 and INO1 indicating the positions of RT-PCR primer pairs. B and E, RT-PCR analysis of MET16 and INO1 in WT and looping-defective sua7-1 strains following transfer of cells to transcription-inducing conditions at the indicated time points. C and F, quantification of the data shown in B and E. Error bars indicate one standard deviation.
DISCUSSION
The results presented here indicate that TFIIB associates with Pap1 and the CF1 3′ end processing complex in yeast cells. We provide several lines of evidence in support of the existence of a complex of TFIIB and termination factors. First, affinity purification of HA-tagged TFIIB yielded a complex composed of Pap1 as well as CF1 subunits Rna14, Rna15, Pcf11, Clp1, and Hrp1. Second, holo-TFIIB complex is devoid of known TFIIB-interacting proteins such as RNAP II and TFIID. Thus, a TFIIB complex is not formed by transiently interacting proteins. Third, a glycerol gradient sedimentation profile of affinity-purified TFIIB showed a TFIIB peak cosedimenting with CF1 subunits and Pap1. Fourth, the sedimentation rate of affinity-purified TFIIB is more than that of free TFIIB, thereby suggesting that it is in a complex. These results provide strong support in favor of the existence of a macromolecular complex composed of TFIIB and 3′ end processing/termination factors in yeast cells.
Our results indicate that the association of TFIIB with Pap1 and the CF1 complex occurs only when the conditions are favorable for gene looping. First, Pap1 and CF1 subunit, Rna15, associate with TFIIB in a looping-competent strain. No such association was observed in looping-deficient sua7-1 strain. Second, cross-linking of TFIIB to the 3′ end of the gene, which is essential for loop formation, was abolished in looping-defective temperature-sensitive mutants of Pap1 and the CF1 subunits Rna14 and Pcf11 at non-permissive temperatures. Third, CF1 subunits and Pap1 were found localized to the 5′ end of a gene only when it was in a looped configuration. These results suggest that a complex of TFIIB, Pap1, and CF1 is formed at the promoter-terminator junction to facilitate loop formation.
We expected at least two populations of TFIIB in a cell: 1) free TFIIB that is not engaged in transcription; 2) TFIIB in association with the terminator-bound factors on genes that are in looped configuration. Contrary to our expectation, we did not find any low sedimentation coefficient peak of free TFIIB in the glycerol gradient. One possible reason for this could be that our affinity purification step is selectively purifying the holo-TFIIB complex. Following elution of TFIIB from affinity beads with oligopeptides, there was still a substantial amount of TFIIB bound to the beads that could be eluted with 0.5% SDS (data not presented). This tightly bound TFIIB could be free TFIIB that cannot be eluted with anti-HA oligopeptides. The earlier attempt to purify native TFIIB from yeast did not observe a holo-TFIIB complex (17). A possible explanation for this is that the holo-TFIIB complex is not stable upon prolonged exposure to high ionic strength. When we performed sedimentation analysis of affinity-purified TFIIB at 500 mm KCl, TFIIB dissociated from the complex and sedimented at the position of free TFIIB. During the purification of native TFIIB from budding yeast by Tschochner et al. (17), at several steps in the purification protocol, ionic strength equivalent to or higher than 500 mm potassium acetate was used. This may have resulted in separation of TFIIB from the termination factors, and consequently, a holo-TFIIB complex was not observed.
Holo-TFIIB complex may also include factors other than CF1 subunits and Pap1. It is likely that some components of CPF complex are present in the TFIIB macromolecular assembly. Ssu72, which is a subunit of CPF complex (21, 38), exhibits a genetic as well as a physical interaction with TFIIB, and is a strong constituent candidate of the TFIIB complex (39–41). The presence of Pab1, Pab1-binding protein Pan1, and CPF subunit Fip1 in the tandem affinity-purified preparation of TFIIB makes them likely components of the TFIIB complex. The presence of some promoter-bound factors in TFIIB preparation also cannot be ruled out.
A similar interaction of mammalian TFIIB with CPSF and CstF, which are homologues of yeast CPF and CF1 cleavage and polyadenylation complexes, has been observed recently (13). TFIIB exhibited a physical interaction with CstF-64 and mSsu72 subunits of CstF and CPSF complexes, respectively. CstF-64 and mSsu72 were also found cross-linked to the distal ends of a gene in a manner analogous to their yeast counterparts. Furthermore, TFIIB phosphorylation was required for recruitment of CstF-64 and mSsu72 to the promoter region of a gene. Whether association of TFIIB with CstF and CPSF complexes facilitates juxtaposition of the promoter and terminator regions to form a gene loop in higher eukaryotes remains to be elucidated.
TFIIB is absolutely required for initiation of transcription. The interactions of TFIIB with the promoter-bound factors are well established. The essential role of TFIIB in gene looping, its interaction with the terminator-bound factors, and the kinetic delay in induced transcription in the strain lacking a holo-TFIIB complex suggests a novel role of TFIIB in looping-mediated transcriptional regulation. It has been proposed that gene looping, which juxtaposes the terminator with the promoter, brings about transcriptional activation by coupling termination with reinitiation (4, 5). Although such a termination-reinitiation link is yet to be demonstrated for RNAP II-transcribed genes, it has been demonstrated for RNAP III and mitochondrial RNA polymerase-transcribed genes (42, 43). If RNAP II-transcribed genes also exhibit a similar termination-reinitiation coupling, the TFIIB-termination factor complex is likely to play a key role in the process.
Supplementary Material
Acknowledgments
We are grateful to Dr. Michael Hampsey (University of Medicine and Dentistry of New Jersey), Dr. John Lopes (University of Massachusetts), Dr. Lori Pile (Wayne State University), and Dr. Phillip Cunningham (Wayne State University) for critical reading of the manuscript. We thank Sara Stanley for cloning and purification of recombinant TFIIB. The anti-TFIIB antibodies and the sua7-1 strain were generous gifts from Dr. Michael Hampsey. The anti-Rna15 and anti-Pap1 antibodies were generous gifts from Dr. Claire Moore. We are thankful to Dr. David Brow for providing us with the hrp1-5 strain. We also thank the Dr. Phillip Cunningham laboratory, especially Jennifer Jasinksi, for help with the instruments to perform the glycerol gradient sedimentation.
This work was supported by National Science Foundation (NSF) Grant MCB1020911.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S5.
- TFIIB
- transcription factor IIB
- rTFIIB
- recombinant TFIIB
- CPSF
- cleavage and polyadenylation specific factor
- CstF
- cleavage stimulatory factor
- CCC
- capture chromosome conformation
- Pab1
- poly(A)-binding protein
- Pap1
- poly(A) polymerase
- RNAP
- RNA polymerase
- TBP
- TATA-binding protein
- CPF
- cleavage and polyadenylation factor.
REFERENCES
- 1. El Kaderi B., Medler S., Raghunayakula S., Ansari A. (2009) J. Biol. Chem. 284, 25015–25025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Papantonis A., Cook P. R. (2010) Curr. Opin. Cell Biol. 22, 271–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vilar J. M., Saiz L. (2005) Curr. Opin. Genet. Dev. 15, 136–144 [DOI] [PubMed] [Google Scholar]
- 4. Dieci G., Sentenac A. (2003) Trends Biochem. Sci. 28, 202–209 [DOI] [PubMed] [Google Scholar]
- 5. Ansari A., Hampsey M. (2005) Genes Dev. 19, 2969–2978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. O'Sullivan J. M., Tan-Wong S. M., Morillon A., Lee B., Coles J., Mellor J., Proudfoot N. J. (2004) Nat. Genet. 36, 1014–1018 [DOI] [PubMed] [Google Scholar]
- 7. O'Reilly D., Greaves D. R. (2007) Genomics 90, 407–415 [DOI] [PubMed] [Google Scholar]
- 8. Perkins K. J., Lusic M., Mitar I., Giacca M., Proudfoot N. J. (2008) Mol. Cell 29, 56–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tan-Wong S. M., French J. D., Proudfoot N. J., Brown M. A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 5160–5165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lainé J. P., Singh B. N., Krishnamurthy S., Hampsey M. (2009) Genes Dev. 23, 2604–2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Singh B. N., Hampsey M. (2007) Mol. Cell 27, 806–816 [DOI] [PubMed] [Google Scholar]
- 12. Tan-Wong S. M., Wijayatilake H. D., Proudfoot N. J. (2009) Genes Dev. 23, 2610–2624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang Y., Fairley J. A., Roberts S. G. (2010) Curr. Biol. 20, 548–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ansari A., Schwer B. (1995) EMBO J. 14, 4001–4009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Deng W., Roberts S. G. (2007) Chromosoma 116, 417–429 [DOI] [PubMed] [Google Scholar]
- 16. Pinto I., Ware D. E., Hampsey M. (1992) Cell 68, 977–988 [DOI] [PubMed] [Google Scholar]
- 17. Tschochner H., Sayre M. H., Flanagan P. M., Feaver W. J., Kornberg R. D. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 11292–11296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ha I., Lane W. S., Reinberg D. (1991) Nature 352, 689–695 [DOI] [PubMed] [Google Scholar]
- 19. Malik S., Hisatake K., Sumimoto H., Horikoshi M., Roeder R. G. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 9553–9557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gavin A. C., Aloy P., Grandi P., Krause R., Boesche M., Marzioch M., Rau C., Jensen L. J., Bastuck S., Dümpelfeld B., Edelmann A., Heurtier M. A., Hoffman V., Hoefert C., Klein K., Hudak M., Michon A. M., Schelder M., Schirle M., Remor M., Rudi T., Hooper S., Bauer A., Bouwmeester T., Casari G., Drewes G., Neubauer G., Rick J. M., Kuster B., Bork P., Russell R. B., Superti-Furga G. (2006) Nature 440, 631–636 [DOI] [PubMed] [Google Scholar]
- 21. Gavin A. C., Bösche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J. M., Michon A. M., Cruciat C. M., Remor M., Höfert C., Schelder M., Brajenovic M., Ruffner H., Merino A., Klein K., Hudak M., Dickson D., Rudi T., Gnau V., Bauch A., Bastuck S., Huhse B., Leutwein C., Heurtier M. A., Copley R. R., Edelmann A., Querfurth E., Rybin V., Drewes G., Raida M., Bouwmeester T., Bork P., Seraphin B., Kuster B., Neubauer G., Superti-Furga G. (2002) Nature 415, 141–147 [DOI] [PubMed] [Google Scholar]
- 22. Krogan N. J., Cagney G., Yu H., Zhong G., Guo X., Ignatchenko A., Li J., Pu S., Datta N., Tikuisis A. P., Punna T., Peregrín-Alvarez J. M., Shales M., Zhang X., Davey M., Robinson M. D., Paccanaro A., Bray J. E., Sheung A., Beattie B., Richards D. P., Canadien V., Lalev A., Mena F., Wong P., Starostine A., Canete M. M., Vlasblom J., Wu S., Orsi C., Collins S. R., Chandran S., Haw R., Rilstone J. J., Gandi K., Thompson N. J., Musso G., St Onge P., Ghanny S., Lam M. H., Butland G., Altaf-Ul A. M., Kanaya S., Shilatifard A., O'Shea E., Weissman J. S., Ingles C. J., Hughes T. R., Parkinson J., Gerstein M., Wodak S. J., Emili A., Greenblatt J. F. (2006) Nature 440, 637–643 [DOI] [PubMed] [Google Scholar]
- 23. Mandel C. R., Bai Y., Tong L. (2008) Cell. Mol. Life Sci. 65, 1099–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bagby S., Kim S., Maldonado E., Tong K. I., Reinberg D., Ikura M. (1995) Cell 82, 857–867 [DOI] [PubMed] [Google Scholar]
- 25. Chen H. T., Hahn S. (2003) Mol. Cell 12, 437–447 [DOI] [PubMed] [Google Scholar]
- 26. Nikolov D. B., Chen H., Halay E. D., Usheva A. A., Hisatake K., Lee D. K., Roeder R. G., Burley S. K. (1995) Nature 377, 119–128 [DOI] [PubMed] [Google Scholar]
- 27. Buratowski S., Zhou H. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 5633–5637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chew B. S., Lehming N. (2007) Biochem. J. 406, 265–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cho E. J., Buratowski S. (1999) J. Biol. Chem. 274, 25807–25813 [DOI] [PubMed] [Google Scholar]
- 30. Pardee T. S., Bangur C. S., Ponticelli A. S. (1998) J. Biol. Chem. 273, 17859–17864 [DOI] [PubMed] [Google Scholar]
- 31. Zhang D. Y., Dorsey M. J., Voth W. P., Carson D. J., Zeng X., Stillman D. J., Ma J. (2000) Nucleic Acids Res. 28, 1913–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Auty R., Steen H., Myers L. C., Persinger J., Bartholomew B., Gygi S. P., Buratowski S. (2004) J. Biol. Chem. 279, 49973–49981 [DOI] [PubMed] [Google Scholar]
- 33. Takagi Y., Komori H., Chang W. H., Hudmon A., Erdjument-Bromage H., Tempst P., Kornberg R. D. (2003) J. Biol. Chem. 278, 43897–43900 [DOI] [PubMed] [Google Scholar]
- 34. Singh B. N., Ansari A., Hampsey M. (2009) Methods 48, 361–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Birse C. E., Minvielle-Sebastia L., Lee B. A., Keller W., Proudfoot N. J. (1998) Science 280, 298–301 [DOI] [PubMed] [Google Scholar]
- 36. Kessler M. M., Henry M. F., Shen E., Zhao J., Gross S., Silver P. A., Moore C. L. (1997) Genes Dev. 11, 2545–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Millevoi S., Vagner S. (2010) Nucleic Acids Res. 38, 2757–2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. He X., Khan A. U., Cheng H., Pappas D. L., Jr., Hampsey M., Moore C. L. (2003) Genes Dev. 17, 1030–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dichtl B., Blank D., Sadowski M., Hübner W., Weiser S., Keller W. (2002) EMBO J. 21, 4125–4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sun Z. W., Hampsey M. (1996) Mol. Cell Biol. 16, 1557–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wu W. H., Hampsey M. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 2764–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dieci G., Sentenac A. (1996) Cell 84, 245–252 [DOI] [PubMed] [Google Scholar]
- 43. Martin M., Cho J., Cesare A. J., Griffith J. D., Attardi G. (2005) Cell 123, 1227–1240 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.