Abstract
The semaphorins are a family of secreted or membrane-bound proteins that are known to guide axons in the developing nervous system. Genetic evidence revealed that a class III semaphorin, semaphorin 3E (Sema3E), and its receptor Plexin-D1 also control the vascular patterning during development. At the molecular level, we have recently shown that Sema3E acts on Plexin-D1 expressed in endothelial cells, thus initiating a novel antiangiogenic signaling pathway that results in the retraction of filopodia in endothelial tip cells. Sema3E induces the rapid disassembly of integrin-mediated adhesive structures, thereby inhibiting endothelial cell adhesion to the extracellular matrix. This process requires the activation of small GTPase Arf6 (ADP-ribosylation factor 6), which regulates intracellular trafficking of β1 integrin. However, the molecular mechanisms by which Sema3E-Plexin-D1 activates Arf6 remained to be identified. Here we show that GEP100 (guanine nucleotide exchange protein 100)/Brag2, a guanine nucleotide exchange factor for Arf6, mediates Sema3E-induced Arf6 activation in endothelial cells. We provide evidence that upon activation by Sema3E, Plexin-D1 recruits phosphatidylinositol-4-phosphate 5-kinase, and its enzymatic lipid product, phosphatidylinositol 4,5-bisphosphate, binds to the pleckstrin homology domain of GEP100. Phosphatidylinositol 4,5-bisphosphate binding to GEP100 enhances its guanine nucleotide exchange factor activity toward Arf6, thus resulting in the disassembly of integrin-mediated focal adhesions and endothelial cell collapse. Our present study reveals a novel phospholipid-regulated antiangiogenic signaling pathway whereby Sema3E activates Arf6 through Plexin-D1 and consequently controls integrin-mediated endothelial cell attachment to the extracellular matrix and migration.
Keywords: Cell Adhesion, Cell Motility, Endothelium, Integrin, Semaphorin, Arf6, Antiangiogenesis
Introduction
Angiogenesis, the formation of new blood capillaries from pre-existing ones, is an essential process during embryonic development. In adults, angiogenesis is required for tissue repair during physiological wound healing, whereas dysregulated angiogenesis contributes to a variety of pathological conditions, such as diabetic retinopathy, age-related macular degeneration, rheumatoid arthritis, and cancer (1). Hence, the identification of molecular mechanisms controlling normal and aberrant angiogenesis may afford new therapeutic opportunities for multiple human diseases (2). In this regard, recent studies revealed that axon guidance molecules, including netrin, slit, eph, and semaphorins, play a key role in developmental and postnatal angiogenesis (3). Among them, multiple secreted class III semaphorins (Sema3s)4 are reported to control endothelial cell migration in vitro and in vivo (4–7). Sema3s signal through A-type and D-type Plexin family proteins (Plexin-A1, -A2, and -A3 and Plexin-D1) and utilize their co-receptor neuropilins (Nrp1 and Nrp2) to tightly control pro- and antiangiogenic responses (8). However, the downstream signaling pathways initiated by these semaphorin receptors are complex and not fully understood, because Nrps are also co-receptors for multiple VEGF receptors (9). Hence, semaphorins can also antagonize the potent pro-angiogenic biochemical routes activated by VEGF family members (8).
Whereas most Sema3s require Nrp as a ligand-binding subunit, Sema3E binds directly to its receptor Plexin-D1 and controls vascular patterning independently of Nrps (5). In line with these findings, we have recently shown that Sema3E acts on Plexin-D1 in endothelial cells to initiate a novel antiangiogenic signaling pathway (10). Specifically, activation of Plexin-D1 by Sema3E causes the rapid disassembly of integrin-mediated focal adhesions, thereby inhibiting endothelial cell adhesion to the extracellular matrix and causing the retraction of filopodia in endothelial tip cells in growing blood vessels. This process requires Sema3E-induced activation of small GTPase Arf6 (ADP-ribosylation factor 6), which regulates intracellular trafficking of β1 integrin (11, 12). However, the molecular mechanisms by which Sema3E-Plexin-D1 activates Arf6 remained to be identified.
Like other small GTPases, Arf6 cycles between an active GTP-bound form and an inactive GDP-bound form, and this GTPase cycle is regulated by guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (13). GEFs facilitate the dissociation of GDP from small GTPases, which is the rate-limiting step in the activation of most small GTPases. The human genome encodes 15 Arf GEFs, which are divided into five subfamilies. Among them, three families of Arf GEFs, BRAG (brefeldin-resistant Arf GEF), ARNO (Arf nucleotide binding site opener)/cytohesin, and EFA6 (exchange factor for Arf6), can all activate Arf6 (14). By the use of dominant negative approaches and RNA interference techniques, we now show that guanine nucleotide exchange protein 100 (GEP100), also known as Brag2a, a GEF that preferentially activates Arf6 (15), mediates Sema3E-induced Arf6 activation in endothelial cells. At the biochemical level, we provide evidence that upon Sema3E activation, Plexin-D1 recruits phosphatidylinositol-4-phosphate 5-kinase (PIP5K) and that its enzymatic lipid product, PI(4,5)P2, binds to the pleckstrin homology (PH) domain of GEP100, thus resulting in its increased GEF activity toward Arf6. Overall, our results reveal a novel phospholipid-regulated antiangiogenic signaling pathway linking Plexin-D1 to Arf6 and endothelial cell integrin function and cell adhesion.
EXPERIMENTAL PROCEDURES
Cell Culture
Primary human umbilical vascular endothelial cells (HUVECs) were grown in endothelial cell medium EGM-2 BulletKit (Lonza). Simian fibroblasts COS-7 and HEK-293T cells were grown in DMEM (Sigma), plus 10% fetal bovine serum (Sigma).
Expression Vectors, siRNA, and Transfection
pCMV-Sport6-Sema3E-HisMyc and pCEFL-Plexin-D1 were generated as described previously (10). Expression vectors for HA-tagged GEP100 wild type, HA-tagged [E498K]Brag2, and Myc-tagged [E156K]ARNO were obtained from J. Casanova. HA-GEP100-ΔPH in which the PH domain of GEP100 was deleted (amino acids 769–862) was generated by standard PCR techniques. pFLAG-EFA6B E651K mutant and membrane-targeted red fluorescent protein (RFP) expression vector (pRFP-CAAX) were provided by J. G. Donaldson. Zyxin-RFP was provided by A. Kobielak. PH domain of GEP100 was amplified and subcloned into pEGFP-C1 vector as an EcoRI-BamHI fragment, thus generating EGFP-GEP100-PH expressing plasmid. pGEX4T3-GEP100-Sec7-PH, an expression vector for GST-tagged GEP100 (amino acids 499–863), was generated by a standard PCR method. pCS2-hPIP5K1β was provided by D. Wu. pcDNA3-Plexin-D1-V5 was obtained from J. Epstein. TurboFect reagent (Fermentas) and Lipofectamine reagent (Invitrogen) were used for transfection of HEK-293T and COS-7 cells, respectively. The following predesigned siRNAs were used: AllStars negative control siRNA, human GEP100, human PIP5KIβ (Qiagen), and human Plexin-D1 (Invitrogen). The cells were transfected with control or with two independent siRNAs targeting different sequences of the molecule using Lipofectamine RNAiMAX transfection reagent (Invitrogen). After incubation for 4–5 days, the cells were used for the experiments. All of the results are representative of two independent siRNA sequences per target gene.
Reagents and Antibodies
Recombinant Sema3E/Fc chimeric protein was from R&D Systems. Sema3E conditioned medium was prepared as described previously (10). Alexa Fluor 488-labeled phalloidin and Alexa Fluor-conjugated secondary antibodies were from Invitrogen. LY294002 was purchased from EMD Biosciences. Recombinant epidermal growth factor was purchased from R&D Systems. Glass coverslips or cell culture surfaces were coated with 10 μg/ml of type I rat tail collagen (BD Biosciences) or 5 μg/ml of human plasma fibronectin (Invitrogen). The following antibodies were used: rabbit anti-Plexin-D1 was generated as described previously (10). Rabbit anti-Arf6 was provided by J. G. Donaldson. Rabbit anti-GEP100/Brag2 was provided by J. Casanova. Other antibodies were mouse anti-FLAG (Sigma); mouse anti-Myc, mouse anti-HA, and mouse anti-GFP (Covance); mouse anti-α-tubulin, rabbit anti-Akt, and rabbit anti-phospho-Akt T308 (Cell Signaling Technology); mouse anti-GAPDH (StressGen); rabbit anti-PIP5KIβ (Thermo Scientific); and mouse anti-V5 (Invitrogen).
COS-7 Collapse Assay
COS-7 cells were grown on collagen-coated 6-well tissue culture plate, co-transfected with 0.5 μg of GFP- or RFP-CAAX and 1.5 μg of pCEFL-Plexin-D1, with or without dominant negative Arf6 GEF constructs. The cells were serum-starved for 6 h and treated with Sema3E-containing conditioned medium for 30 min. The cells were fixed with 3.7% formaldehyde and visualized under a fluorescence microscope. Cell collapse was scored for about 150 fluorescence-positive cells for each experimental condition in triplicate wells, and the percentage of collapsed cells was calculated.
Immunoblotting and Immunoprecipitation
Cells were lysed in lysis buffer containing 50 mm Tris-HCl, pH 7.5, 100 mm NaCl, 1% Triton X-100, and protease inhibitor mixture (Sigma). Precleared lysates were subjected to SDS-PAGE, and the proteins were transferred onto a polyvinylidene fluoride membrane (Millipore), blocked with 5% skim milk in Tris-buffered saline, and then incubated with primary antibodies. The primary antibodies were detected with horseradish peroxidase-conjugated secondary antibodies or Alexa Fluor 680-conjugated secondary antibodies, using Pierce ECL Western blotting Substrate (Thermo Scientific) or Odyssey Infrared Imaging System (LI-COR Biosciences), respectively. To detect the interaction between Plexin-D1 and PIP5K1β, HEK-293T cells were transfected with V5-tagged Plexin-D1 and PIP5K1β, serum-starved for 6 h, and stimulated with Sema3E. Plexin-D1 was immunoprecipitated with anti-V5 antibody from the precleared lysates. Immunoprecipitated Plexin-D1 and aliquots of cell lysate were subjected to SDS-PAGE, and immunoblotted with anti-PIP5K1β and anti-V5 antibodies, respectively.
Immunofluorescent Microscopy
The cells cultured on fibronectin-coated coverslips were serum-starved and treated with Sema3E (100 ng/ml) for 30 min, fixed with 3.7% formaldehyde in PBS for 30 min, and permeabilized with 0.05% Triton X-100 for 10 min. The cells were blocked with 3% FBS-containing PBS for 30 min and incubated with indicated antibodies for 1 h at room temperature. The immunopositive reaction was visualized with Alexa488-labeled secondary antibody (Invitrogen). Actin was visualized using Alexa Fluor 488 Phalloidin (Invitrogen). The samples were mounted in Vectashield propidium iodide-containing mounting medium (Vector Labs) and visualized under a LSM700 confocal microscope controlled by Zen software (Carl Zeiss).
mRNA Extraction and qPCR
Total RNA was extracted from GEP100 siRNA-transfected cells using TRIzol reagent (Invitrogen). Reverse transcription was conducted using SuperScript III RT system (Invitrogen) according to manufacturer's instructions. GEP100 mRNA levels were measured by qPCR with specific gene primers as follows: GEP100 variant 1 forward, 5′-CTGTGGCGGTCGTCATGTGGTG-3′, and reverse, 5′-CGCCCTCAACACCGCTGTCAAG-3′; and GEP100 variant 2 forward, 5′-TCAGAAGCGGGGAAGCGAGG-3′, and reverse, 5′-TGGTGGCTGGAAGGGCTGAAATTC-3′. iCycler iQ real time PCR detection system and SYBR green PCR mix (Bio-Rad) were used to carry out the qPCR. The relative abundance of GEP100 transcript was quantified using the comparative Ct method with GAPDH as an internal control. The data were analyzed from three independent experiments and statistical significance validated by Student's t test.
GST Pulldown Assay
Arf6 activation was monitored by GST pulldowns by using GST-GGA3 (Arf-binding domain of GGA3) recombinant protein bound to glutathione slurry resin (Amersham Biosciences). The cells were lysed in buffer containing 50 mm Tris-HCl, pH 7.5, 100 mm NaCl, 2 mm MgCl2, 0.1% SDS, 0.5% sodium deoxycholate, 1% Triton X-100, 10% glycerol, 1 mm Na3VO4, and protease inhibitor mixture (Sigma). Precleared lysates were incubated with slurry resin. The resin was washed three times in lysis buffer, and the proteins collected on the resin were subjected to SDS-PAGE followed by immunoblotting with anti-Arf6 antibody.
Lipid Binding Assay
Recombinant GST-GEP100 were purified using glutathione-Sepharose 4B (GE Healthcare) according to manufacturer's instructions. Large unilamellar vesicles (LUVs) containing various phospholipids were prepared by extrusion with lipids purchased from Avanti Polar Lipids and Echelon Biosciences as described previously (16). They contained molar ratios of 40% phosphatidylcholine, 25% phosphatidylethanolamine, 15% phosphatidylserine, 10% cholesterol, and 10% phosphatidylinositol (PI), or 9.5% PI plus 0.5% of various phosphatidylinositol phosphate (PIP), including PI 3-phosphate, PI 4-phosphate, PI 5-phosphate, PI 3,4-bisphosphate, PI 3,5-bisphosphate, PI(4,5)P2, and PI 3,4,5-trisphosphate. For the PIP titration experiments, PIP amount varied from 0%, 0.1%, 0.25%, and 0.5% to 1%. The amount of PI was changed accordingly to 10%, 9.9%, 9.75%, 9.5%, and 9%. The purified recombinant protein (500 nm) was incubated with sucrose-loaded LUVs containing 500 μm of total phospholipids at 30 °C for 5 min. The LUVs were precipitated by ultracentrifugation at 75,000 rpm for 15 min at 4 °C, and the precipitated proteins with LUV were separated by SDS-PAGE and visualized by Coomassie Blue staining. The signal was quantified by densitometry using Image J software. The Kd for the GST-GEP100-PI complex was determined by titrating each PI into the binding reaction.
GTPγS Binding Assay
Myristoylated Arf6 (myrArf6) was purified as described previously (17). GST-GEP 100 catalyzed GTPγS binding to myrArf6 were performed in the nucleotide exchange buffer (25 mm HEPES, pH 7.4, 100 mm NaCl, 1 mm dithiothreitol, 2 mm MgCl2, 1 mm EDTA, 1 mm ATP, 5 μm GTPγS, trace amount of [35S]GTPγS) as described (18, 19). The high MgCl2 was used in this reaction to slow down the spontaneous nucleotide exchange. The reactions also contained 0.5 mm LUVs and 1 μm myrArf6, with or without 100 or 20 nm GST-GEP100, for testing effects of different PIP or PIP titration experiments, respectively. The reactions were incubated at 30 °C for 3 min and terminated with 2 ml of ice-cold 20 mm Tris, pH 8.0, 100 mm NaCl, 10 mm MgCl2, and 1 mm dithiothreitol. Protein-bound nucleotide was trapped on nitrocellulose, and the bound radioactivity was counted by liquid scintillation.
PIP5K Activity Assay
PIP5K activity was determined as described previously (20) with some modifications. Briefly, serum-starved cells left untreated or stimulated with Sema3E were lysed in 50 mm Tris-HCl, pH 7.5, 100 mm NaCl, 1% Triton X-100, and protease inhibitor mixture (Sigma). Precleared lysates were immunoprecipitated with an anti-Plexin-D1 polyclonal antibody for 2 h at 4 °C, and immunocomplexes were recovered with protein G-Sepharose (GE Healthcare). Pellets were then washed twice with PBS containing 1% Nonidet P-40 and 1 mm sodium vanadate; washed twice with 0.5 m LiCl in 100 mm Tris-HCl, pH 7.5; washed once with TNE buffer (10 mm Tris-HCl, pH 7.5, 100 mm NaCl, 1 mm EDTA); washed once with 20 mm HEPES, pH 7.6; and resuspended with kinase reaction buffer (20 mm Tris-HCl, pH 7.6, 100 mm NaCl, 0.5 mm EGTA). Reactions were performed in a 50-μl volume of kinase reaction buffer containing 5 μCi of [γ-32P]ATP/reaction, 10 μm MgCl2, 67 μm PI 4-phosphate, and 133 μm phosphoserine at 30 °C for 30 min. The reactions were stopped by the addition of 100 μl of 1 n HCl, and the lipid products were extracted using 200 μl of chloroform-methanol (1:1). The organic phase was then spotted on thin layer chromatography plates and separated. Phosphorylated PI 4-phosphate was visualized by autoradiography and quantified with Image J software.
RESULTS
GEP100 Mediates Sema3E-induced Cell Collapse
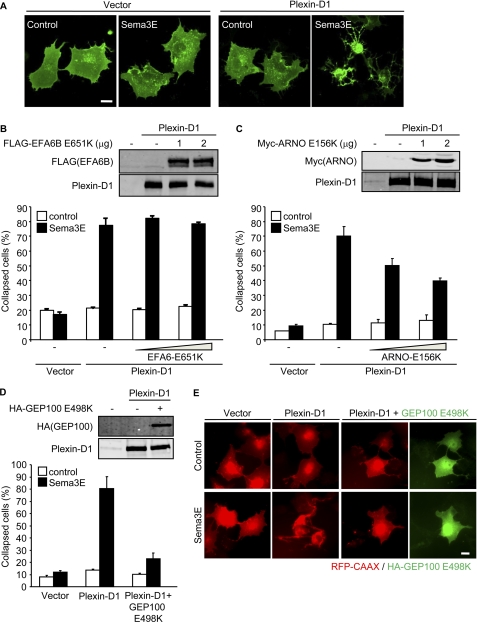
Three families of Arf GEFs can activate Arf6, BRAG (brefeldin-resistant Arf GEF), ARNO (Arf nucleotide binding site opener)/cytohesin, and EFA6 (exchange factor for Arf6), many of which exhibit multiple splice variants and isoforms (14). To begin exploring which of these Arf-GEF families is responsible for Sema3E-induced Arf6 activation, we initially took advantage of a dominant negative approach. Expression of Plexin-D1 in COS-7 cells induces cytoskeletal collapse in response to Sema3E stimulation, which can mimic Sema3E-mediated signaling in endothelial cells (10). Indeed, when COS-7 cells were co-transfected with control or Plexin-D1-expressing plasmids and membrane-targeted EGFP (EGFP-CAAX) and were stimulated with Sema3E, only the cells expressing Plexin-D1 showed a cytoskeletal collapse phenotype (Fig. 1A). Using this cellular reconstitution system, we first examined the effect of dominant negative Arf6 GEFs including EFA6, ARNO, and GEP100/Brag2. The dominant negative form of EFA6B, which displays an ubiquitous tissue distribution among EFA6 family proteins (21), had no effect on Sema3E-induced cell collapse (Fig. 1B). Dominant negative ARNO (ARNO E156K) had a partial response, rescuing some of the Sema3E-induced cell collapse at very high expression levels (Fig. 1C). However, co-transfection of dominant negative GEP100 (GEP100 E498K) exerted a much more potent effect, virtually eliminating the cell collapse phenotype induced by Sema3E (Fig. 1, D and E). These results and the limited activity of ARNO on Arf6 in vivo (22) suggested that GEP100 may represent the most likely potential candidate to initiate the Sema3E-Plexin-D1-Arf6 signaling pathway.
FIGURE 1.
Dominant negative GEP100 inhibits Sema3E-induced cell collapse. A, COS-7 cells were transfected with control or Plexin-D1-expressing plasmids and EGFP-CAAX, serum-starved, and stimulated with Sema3E-containing conditioned media for 30 min. B, COS-7 cells were transfected with control or Plexin-D1-expressing plasmids and RFP-CAAX, with or without FLAG-tagged EFA6B E651K. The protein expression was analyzed by Western blotting with the antibodies indicated (top panel). The cells were serum-starved and stimulated with Sema3E-containing conditioned medium. RFP-positive cells exhibiting a collapse phenotype were scored as a percentage of the total number of transfected cells. C, cell collapse assay was performed as described for B with Myc-tagged ARNO E156K. D, cell collapse assay was performed as described for B with HA-tagged GEP100 E498K. E, expression of HA-GEP100 E498A was visualized by immunofluorescence using anti-HA antibody (green). The graphs represent the means ± S.E. of three independent experiments. Scale bars, 20 μm.
GEP100 Is Required for Sema3E-induced Arf6 Activation and Focal Adhesion Disassembly in Endothelial Cells
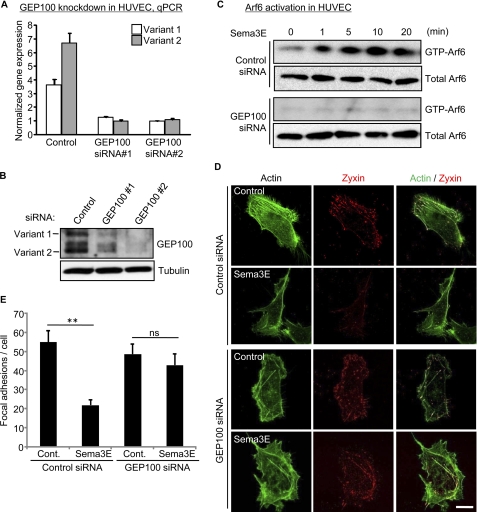
To further investigate the involvement of GEP100 in Sema3E-induced Arf6 activation, we silenced GEP100 by siRNA-mediated knockdown in endothelial cells. GEP100 exists in two isoforms, Brag2A and Brag2B, both of which can activate Arf6 (23). qPCR analysis with primers specific for each isoform showed that the expression of both isoforms was markedly reduced by siRNA transfection (Fig. 2A). Reduced GEP100 protein expression was also confirmed by Western blotting using anti-GEP100-specific antibodies (Fig. 2B). As we have previously shown, Sema3E activates endogenous Arf6 as fast as 1 min in endothelial cells (10). However, this Arf6 activation was completely abolished in GEP100-silenced cells (Fig. 2C). These results demonstrated the importance of GEP100 in Arf6 activation upon Sema3E stimulation. Sema3E inhibits endothelial cell adhesion by reducing focal adhesions (FAs) (10), which are dynamic cell-to-extracellular matrix adhesive structures containing integrin receptors, signaling and scaffolding proteins (24). To examine the effect of GEP100 depletion on Sema3E-induced FA disassembly, we next visualized FAs by using RFP-tagged zyxin (25) as a probe. Control or GEP100 siRNA-transfected endothelial cells were plated on fibronectin-coated dishes, transfected with zyxin-RFP, and stimulated with Sema3E. In control cells, Sema3E treatment significantly reduced the number of FAs, concomitant with decreased actin stress fibers visualized by phalloidin staining. However, in GEP100-silenced cells, this FA disassembly phenotype upon Sema3E stimulation was nearly completely rescued (Fig. 2D). We found that, compared with control cells, GEP100-silenced cells retained more FAs after Sema3E stimulation (Fig. 2E). Taken together, these results support the possibility that GEP100 mediates Sema3E-induced Arf6 activation, which results in FA and stress fiber disassembly, thereby inhibiting endothelial cell adhesion and migration.
FIGURE 2.
Knockdown of GEP100 inhibits Sema3E-induced Arf6 activation and focal adhesion disassembly. A and B, siRNA-mediated knockdown of GEP100 in HUVECs. The cells were transfected with control or GEP100 siRNAs. GEP100 mRNA levels (A) and protein levels (B) were assessed 5 days later by qPCR and Western blotting, respectively. C, control or GEP100 siRNA-transfected HUVECs were serum-starved and stimulated with 100 ng/ml Sema3E, and Arf6 activity was measured as described under “Experimental Procedures.” D, control or GEP100 siRNA-transfected HUVECs were plated on fibronectin-coated glass coverslips and transfected with Zyxin-RFP. The cells were serum-starved, treated with 100 ng/ml Sema3E for 15 min, and stained for F-actin (green). RFP signals were visualized directly (red). Scale bar, 20 μm. E, quantitative analysis of focal adhesion numbers from D. The number of focal adhesions in each cell was evaluated from images of Zyxin signal. The number represents the mean ± S.E. from ∼50 cells in each group. The data shown are representative of three independent experiments. ns, no significant difference; **, p < 0.01; Cont., control.
A Role for the GEP100 PH Domain in Sema3E Signaling
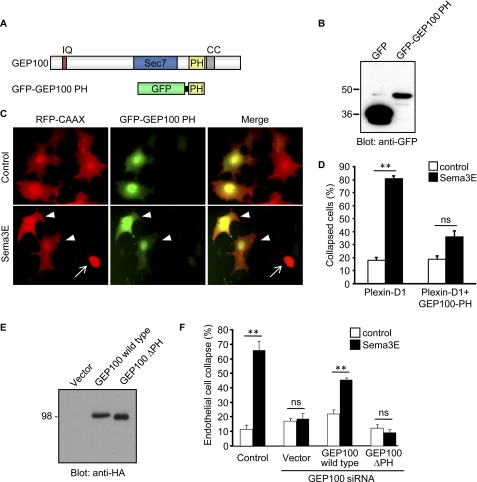
These findings prompted us to investigate the mechanism by which Sema3E activates GEP100. It was recently reported that GEP100 associates with the ligand-activated EGF receptor (26). This association is mediated via PH domain of GEP100, which binds to phopshorylated Tyr-1068 or Tyr-1086 on the EGF receptor. However, we did not detect any phosphorylation of tyrosine residues in Plexin-D1 upon Sema3E stimulation nor GEP100 associated with Plexin-D1 under multiple experimental conditions (data not shown). Thus, we speculated that Sema3E may regulate GEP100 by alternative molecular intermediates. Because PH domains are present in various GEFs and GTPase-activating proteins and play pivotal roles in controlling the biological activities of these small GTPase regulating molecules, we hypothesized that the PH domain of GEP100 may be involved in Sema3E-induced GEP100 activation. To test this hypothesis, we generated GFP-tagged GEP100 PH domain construct (Fig. 3, A and B) and tested the effect of overexpression of GFP-GEP100 PH on Sema3E-induced cell collapse. COS-7 cells were transfected either with Plexin-D1 and RFP-CAAX or with Plexin-D1, RFP-CAAX, and GFP-GEP100-PH, and stimulated with Sema3E. The cells expressing Plexin-D1 and RFP-CAAX showed a collapse phenotype (Fig. 3C, arrow); however, the cells co-transfected with GFP-GEP100-PH showed only a slight change in their morphology but did not collapse (Fig. 3, C, arrowheads, and D). These results suggested that the GFP-GEP100-PH functions as a dominant negative construct and that the PH domain of GEP100 might participate in Sema3E-induced GEP100 activation and its consequent downstream signal transduction. To further confirm the role of PH domain in Sema3E-induced signal transduction, next we generated a GEP100 mutant construct lacking the PH domain (GEP100 ΔPH) (Fig. 3E). GEP100 silencing inhibited Sema3E-induced stress fiber disassembly in endothelial cells, and it was rescued by reintroducing wild type GEP100, but not GEP100 ΔPH, although both were expressed at comparable levels (Fig. 3, E and F). These findings indicated a critical role for GEP100 PH domain in Sema3E-induced antiangiogenic phenotype.
FIGURE 3.
PH domain of GEP100 is involved in Sema3E-induced cell collapse. A, domain structure of GEP100 and schematic representation of chimeric protein encoding PH domain of GEP100 fused to GFP. IQ, IQ motif; Sec7, Sec7 domain; PH, pleckstrin homology domain; CC, coiled-coil domain. B, expression of GFP-GEP100 PH in COS-7 cells. C and D, COS-7 cells were transfected with Plexin-D1-expressing plasmids, RFP-CAAX, and GFP-GEP100 PH. The cells were serum-starved and subjected to collapse assay as described for Fig. 1B. GFP-negative cell collapsed upon Sema3E stimulation (arrow), whereas GFP-GEP100 PH-expressing cells did not collapse (arrowheads). E, expression of HA-tagged GEP100 constructs in COS-7 cells. ΔPH, mutant GEP100 lacking PH domain. F, GEP100 siRNA-transfected HUVECs were plated on fibronectin-coated glass coverslips. The cells were transfected with GEP100 wild type or GEP100 ΔPH, serum-starved, treated with 100 ng/ml Sema3E for 15 min, and stained for F-actin. The endothelial cells exhibiting collapse phenotype were scored as a percentage of the total number of cells. The graph represents the means ± S.E. of three independent experiments. ns, no significant difference; **, p < 0.01. Scale bars, 20 μm.
The PH Domain of GEP100 Binds to PI(4,5)P2 and PIP3, Which Enhances Its GEF Activity
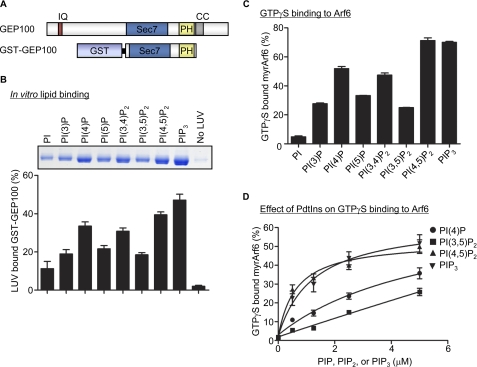
Although it was reported that nucleotide exchange activity of GEP100 is not affected by phosphoinositides (15), the direct interaction of phosphoinositides with PH domain of GEP100 has not been determined. To examine PI binding to GEP100, we expressed and purified GST-tagged GEP100 recombinant protein, which is composed of the Sec7 (catalytic domain) and PH domain of GEP100 (Fig. 4A), and performed in vitro lipid binding assay. LUVs containing various phospholipids were prepared as described under “Experimental Procedures,” then incubated with GST-GEP100, and separated from bulk solution by ultracentrifugation. The precipitated proteins with LUV were separated by SDS-PAGE and visualized (Fig. 4B, top panel). Interestingly, GEP100 showed high affinity for LUVs containing phosphoinositides with a phosphate in position 4 of the inositol ring (Fig. 4B and Table 1). The Kd for the GEP100-PI complex was determined by titrating each PI into the binding reaction, and we found that PI(4,5)P2 has the highest affinity for GEP100 compared with the other PI species (Table 1). Next we tested the effect of phosphoinositides on guanine nucleotide exchange activity of GEP100 using GTPγS binding assay. Recombinant GEP100 enhanced GTPγS binding to Arf6 as previously reported (15), and the activity of GEP100 was significantly enhanced by the addition of PI(4,5)P2 and PIP3 (Fig. 4C). Indeed, the activity of GEP100 was increased in a PI concentration-dependent manner (Fig. 4D). Taken together, these results indicate that PI binding to PH domain of GEP100, specifically PI(4,5)P2 and PIP3, may contribute to the regulation of the GEF activity of GEP100 toward Arf6.
FIGURE 4.
PH domain of GEP100 has high affinity to PI(4,5)P2 and PIP3. A, schematic representation of GEP100 protein encoding Sec7 and PH domain of GEP100 fused to GST. IQ, IQ motif; Sec7, Sec7 domain; PH, pleckstrin homology domain; CC, coiled-coil domain. B, GEP100 phosphoinositide binding specificity. 500 nm GEP100 recombinant protein was incubated with sucrose-loaded LUVs containing phospholipids indicated in the figure. Vesicles were precipitated by ultracentrifugation, and associated proteins were separated by SDS-PAGE. The amount of bound protein was determined by densitometry of the Coomassie Blue staining. A representative image of a gel is shown on top. C, phosphoinositide dependence of GEF activity of GEP100. 1 μm of myrArf6 was incubated for 3 min at 30 °C with 100 nm GST-GEP100, 5 μm GTPγS, and 0.5 mm LUVs containing the indicated phospholipids. D, effect of phospholipids on GTPγS binding to Arf6. 1 μm of myrArf6 was incubated with 20 nm GST-GEP100, 5 μm GTPγS, and the indicated amount of phospholipids. The graphs represent average ± S.E. of three independent experiments.
TABLE 1.
Phospholipid association with GEP100
GEP100-PH recombinant protein was incubated with LUVs containing PI 4-phosphate, PI(4,5)P2, or PIP3, and lipid binding assay was performed as described for Fig. 4B. The Kd for the GST-GEP100-PI complex was determined by titrating each PI into the binding reaction.
| Phospholipid | Kd |
|---|---|
| μm | |
| PI(4)P | 6.52 ± 4.13 |
| PI(4,5)P2 | 0.97 ± 0.25 |
| PIP3 | 1.53 ± 0.25 |
PI(4,5)P2 Is Required for Sema3E-induced Arf6 Activation
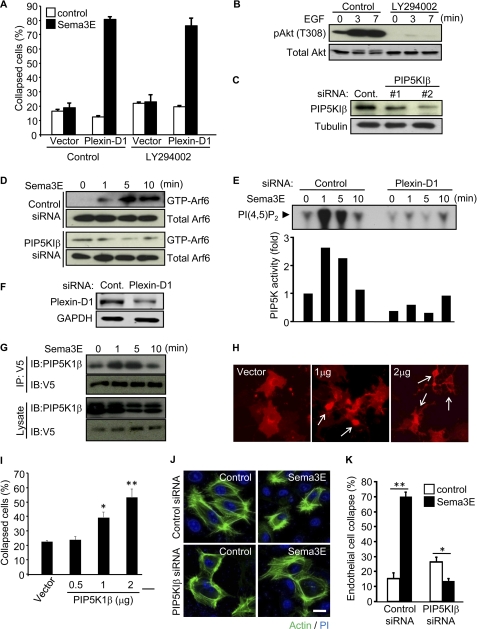
To determine the PI participating in Sema3E-induced signaling pathway, first we treated the cells with the pan-phosphoinositide 3-kinase (PI3K) inhibitor LY294002 (27) to prevent PIP3 production and performed cell collapse assay. However, inhibition of PI3Ks failed to block Sema3E-induced cytoskeletal collapse (Fig. 5A), suggesting that PIP3 is not involved in this pathway. The inhibitory effect of LY294002 in COS-7 cells was confirmed by reduced phosphorylation of Akt, a major downstream effector of PI3K (Fig. 5B). Next, to reduce the cellular PI(4,5)P2 level, we silenced type I PIP5K (PIP5KI), which is a key regulator of PI(4,5)P2 production. Three isoforms of PIP5KI have been identified, and among them, we focused on PIP5KIβ, which was recently reported to regulate focal adhesion disassembly by controlling β1 integrin trafficking (28). Transfection of PIP5KIβ siRNAs successfully reduced PIP5KIβ protein expression in endothelial cells (Fig. 5C). In these PIP5KIβ knockdown cells, Arf6 activation induced by Sema3E was nearly abolished (Fig. 5D). These results prompted us to hypothesize that Sema3E activates PIP5KIβ through Plexin-D1. To test this possibility, we incubated HUVECs with Sema3E and performed in vitro kinase assays with immunoprecipitated Plexin-D1 using PI 4-phosphate as a substrate. Sema3E treatment significantly enhanced PIP5K activity within 1 min, as judged from the amount of generated phosphorylated lipid product, PI(4,5)P2 (Fig. 5E). Although we used multiple controls, this rapid activation of PIP5K was not observed in Plexin-D1 knockdown HUVECs, supporting the specificity of the Plexin-D1/PIP5K interactions (Fig. 5, E and F). Consistent with these findings, co-expression and co-immunoprecipitation analysis showed the interaction of Plexin-D1 and PIP5K1β upon Sema3E stimulation (Fig. 5G). On the other hand, overexpression of PIP5K1β was itself sufficient to induce a cell collapse phenotype, indicating that the up-regulation of cellular PI(4,5)P2 may initiate FA disassembly (Fig. 5, H and I). Furthermore, PIP5KIβ silencing inhibited Sema3E-induced stress fiber disassembly in endothelial cells (Fig. 5, J and K). Taken together, these results suggest that Sema3E activates PIP5K through Plexin-D1, and the local accumulation of PI(4,5)P2 may play an important role as a second messenger in Sema3E-induced antiangiogenic signaling by enabling the GEP100-mediated activation of Arf6.
FIGURE 5.
PI(4,5)P2 is required for Sema3E-induced Arf6 activation. A, COS-7 cells were transfected with control or Plexin-D1-expressing plasmids together with RFP-CAAX. The cells were serum-starved, treated with 10 μm of LY294002 for 30 min, and subjected to collapse assay as described for Fig. 1B. B, COS-7 cells treated as in A were stimulated with 50 ng/ml EGF instead of Sema3E and analyzed for the phosphorylation of Akt with anti-phospho-Akt (threonine 308) antibody. C, siRNA-mediated knockdown of PIP5K1β in HUVECs. The cells were transfected with control or PIP5K1β siRNAs, and the PIP5K1β protein level was assessed 5 days later by Western blotting. D, control or PIP5K1β siRNA-transfected HUVECs were serum-starved and stimulated with 100 ng/ml Sema3E, and Arf6 activity was measured as described under “Experimental Procedures.” E, control or PIP5K1β siRNA-transfected HUVECs were serum-starved and stimulated with 100 ng/ml Sema3E, and the total cell lysates were subjected to immunoprecipitation using anti-Plexin-D1. The immunoprecipitated Plexin-D1 complexes were assayed to phosphorylate PI 4-phosphate. The data are representative of three independent experiments. F, Plexin-D1 knockdown was confirmed by Western blotting. G, HEK-293T cells were transfected with PIP5K1β- and V5-tagged Plexin-D1-expressing plasmids, serum-starved, and stimulated with Sema3E. The cell lysates were subjected to immunoprecipitation (IP) with anti-V5 antibody and immunoblotted (IB) with the indicated antibodies. H and I, COS-7 cells were transfected with RFP-CAAX and the indicated amount of PIP5K1β plasmid and then subjected to collapse assay as described for Fig. 1B. Overexpression of PIP5K1β itself induces cell collapse phenotype (arrows). J, control or PIP5K1β siRNA-transfected HUVECs were plated on fibronectin-coated glass coverslips. The cells were serum-starved, treated with 100 ng/ml Sema3E for 15 min, and stained for F-actin (green) and nuclei (blue). Scale bar, 20 μm. K, endothelial cell exhibiting collapse phenotype were scored as a percentage of the total number of cells. The graphs represent the means ± S.E. of three independent experiments. *, p < 0.05; **, p < 0.01; Cont., control.
DISCUSSION
Recent genetic evidence in model organisms and in multiple in vitro study systems revealed that Plexin-D1 transduces the potent antiangiogenic signals initiated by Sema3E (29). Indeed, Sema3E acts as an endothelial guidance molecule, controlling vascular patterning during development (5, 30) and causing the retraction of filopodia in endothelial tip cells in growing blood vessels under physiological and pathological conditions (10). At the molecular level, we have recently observed that upon stimulation by Sema3E, Plexin-D1 activates Arf6 potently, and that in turn the ability of Arf6 to regulate the intracellular trafficking of β1 integrin results in the disassembly of integrin-mediated adhesive structures and the inability of endothelial cells to adhere to the extracellular matrix, thus causing endothelial cell collapse (10). In this study, we investigated the molecular mechanisms by which Sema3E binding to Plexin-D1 leads to Arf6 activation. We now provide evidence that upon Sema3E stimulation, Plexin-D1 associates with PIP5KIβ and that the enzymatic product of this lipid kinase, PI(4,5)P2, binds to the PH domain of GEP100, thus increasing its GEF activity toward Arf6 and resulting in Arf6 activation and the disassembly of integrin-mediated FA. These findings suggest that PIP5Ks and GEP100 are integral components of a novel antiangiogenic pathway linking Sema3E-Plexin-D1 to Arf6 in endothelial cells, thereby controlling endothelial integrin-mediated cell adhesion.
The BRAG family Arf GEF consists of three isoforms: BRAG1/IQsec2, BRAG2/IQsec1/GEP100, and BRAG2/IQsec3/synArfGEF. In mammals, BRAG1 and BRAG3 are primarily expressed in the brain; however, the expression of BRAG2/GEP100 is more ubiquitous (15, 31, 32). GEP100 acts preferentially on Arf6 and partially colocalizes with this GTPase at the cell periphery (15). Several biological functions for GEP100 have been reported. In HeLa cells, GEP100 controls endocytosis and recycling of β1 integrins, thus regulating cell adhesion (23). GEP100 also plays a role in actin cytoskeleton remodeling, which involves direct interaction with α-catenin (33). Furthermore, GEP100 was recently reported to be involved in breast cancer invasion (26) and phagocytosis (34). Although these findings suggest an important function for GEP100 in cell adhesion and migration, how GEP100 is activated remains unclear. GEP100 contains an IQ-like domain, a catalytic Sec7 domain, a PH domain, and a coiled-coil domain, and it was shown in one report that GEP100 can bind to ligand-activated EGF receptor through its PH domain, thus activating Arf6 (26). In this regard, we did not detect the association of GEP100 with Plexin-D1 nor tyrosine phosphorylation of GEP100 upon Sema3E stimulation under multiple experimental conditions (data not shown). However, the strict requirement for GEP100 for Sema3E-induced Arf6 activation and focal adhesion disassembly prompted us to explore alternative mechanisms regulating GEP100. In this regard, we found that overexpression of GEP100 PH domain is sufficient to block Sema3E-induced cytoskeletal collapse phenotype, suggesting the role of PH domain in the activation of GEP100. Under the conditions of our assays, which included phospholipids presented as large unilamellar vesicles and which used appropriately myristoylate Arfs as substrates (both different than in previous work (15)), we found that PI(4,5)P2 bound to and activated GEP100, leading to increased nucleotide exchange on Arf6. These results together with our results examining PIP kinases and PIP2 formation support the conclusion that GEP100 is a downstream target of phosphoinositides in a pathway critical to the antiangiogenic effects of Sema3E.
Phosphoinositides are key regulators of a large variety of cellular functions. Among them, PI(4,5)P2 has been well studied because of its role as a precursor of inositol(1,4,5)-triphosphate, diacylglycerol, and PIP3, all of which affect multiple downstream targets, ranging from calcium and calcium-regulated processes, calcium/calmodulin-regulated kinases, the large family of PKCs, and numerous growth and survival signaling molecules that are controlled by the accumulation of PIP3 (35, 36). However, recent reports revealed that PI(4,5)P2 can itself act as a second messenger, controlling focal adhesion dynamics and actin cytoskeleton (37, 38), both of which are crucial to cell migration. Considering the fact that cellular PI(4,5)P2 levels are relatively high, overall concentration undergoes limited changes upon stimulation of molecules increasing its synthesis or promoting its turnover or phosphorylation to PIP3. However, the localized generation of PI(4,5)P2 at specific cellular microdomains may underlie its function as a signaling molecule. PI(4,5)P2 is generated mainly by phosphorylation of PI 4-phosphate by PIP5KIs and, to a lower extent, by phosphorylation of PI 5-phosphate by type II phosphatidylinositol-5-phosphate 4-kinases (37). Of interest, there are three isoforms of PIP5KIs, α, β, and γ (39), which are known to play a regulatory function on the actin cytoskeleton (40–42). Specifically, a recent report showed that knockdown of one PIP5K isoform, PIP5KIβ, blocks the internalization of β1 integrin and impairs FA turnover and cell migration (28), albeit by a yet to be identified mechanism. Indeed, our results indicate that PIP5KIβ is required for Sema3E-induced Arf6 activation and FA disassembly, supporting the idea that the local production of PI(4,5)P2 through PIP5KIβ may contribute to the control of integrin-dependent cellular structures and cell migration. This effect seems to be specific for PIP5KIβ, because PIP5KIα knockdown did not rescue the Sema3E-induced endothelial cell collapse phenotype (data not shown), and PIP5KIγ-knockdown endothelial cells were not able to attach to the extracellular matrix (data not shown) as reported previously (42, 43), suggesting that PIP5KIγ regulates the basal formation of FA. Overall, our results support that PIP5KIβ plays a key role in promoting Arf6 activation, which may in turn explain its ability to control integrin internalization and therefore FA turnover.
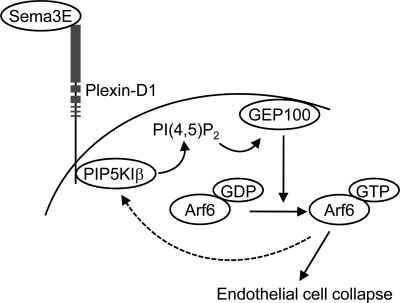
The mechanism by which Plexin-D1 activates PIP5Ks still remains to be fully elucidated. PIP5K activity can be detected in anti-Plexin-D1 immunoprecipitates, and co-immunoprecipitation analysis showed that Plexin-D1 and PIP5K1β can interact upon Sema3E stimulation. Thus, we can speculate that ligand-activated Plexin-D1 binds and recruits PIP5KIβ directly or that Plexin-D1 forms protein complexes including PIP5KIβ, thus resulting in local PI(4,5)P2 production and the activation of the GEP100-Arf6 pathway. Of direct relevance to our study, it has been reported that myristoylated membrane-bound Arf6 can itself activate PIP5Ks (44). This raises the possibility of the existence of a feed-forward mechanism linking Plexin-D1 to the rapid and localized activation of Arf6 and PIP5Ks. As represented graphically in Fig. 6, our present findings support a model in which Sema3E binding to Plexin-D1 initiates PI(4,5)P2 production via PIP5KIβ, thus generating PI(4,5)P2 locally. PI(4,5)P2 then binds to the PH domain of an Arf6 GEF, GEP100, resulting in Arf6 activation, and active Arf6 can itself recruit and activate PIP5K1β even further, likely resulting in increased Arf6 stimulation. In this scenario, the ability PIP5K1β to act downstream from Plexin-D1 and both upstream and downstream of GEP100-Arf6 may help explain the rapid and potent activation of Arf6 by Plexin-D1 and its dramatic impact on FA disassembly, integrin internalization, and cellular collapse caused by Sema3E on endothelial cells (10). Although the existence of this proposed positive feedback loop may warrant further investigation, given the remarkable antiangiogenic activity of Sema3E (10, 30, 45), we can also speculate that the activation of PIP5K1β-GEP100-Arf6 signaling circuitry may ultimately provide new biochemical targets for pharmacological intervention in many disease conditions characterized by aberrant angiogenesis.
FIGURE 6.
Proposed model for Sema3E-induced Arf6 activation. Sema3E binding to Plexin-D1 induces PIP5KIβ activation, which generates PI(4,5)P2 locally. PI(4,5)P2 then binds to the PH domain of an Arf6 GEF, GEP100, resulting in Arf6 activation. Active Arf6 itself recruits and further activates PIP5KIβ, likely resulting in increased Arf6 activity. Sema3E-induced Arf6 activation induces rapid FA disassembly and endothelial cell collapse, thereby inhibiting angiogenesis.
Acknowledgments
We thank J. Cassanova (University of Virginia School of Medicine), A. Kobielak (The University of Southern California), D. Wu (Yale University, School of Medicine), and J. Epstein (University of Pennsylvania) for providing reagents and D. Martin and M. Samaan (National Institute of Dental and Craniofacial Research, National Institutes of Health) for comments and discussions.
This work was supported, in whole or in part, by a National Institutes of Health grant from Intramural Research Program of the National Institute of Dental and Craniofacial Research.
- Sema3
- class III semaphorin
- GEF
- guanine nucleotide exchange factor
- PIP5K
- phosphatidylinositol-4-phosphate 5-kinase
- PI(4,5)P2
- phosphatidylinositol 4,5-bisphosphate
- PH
- pleckstrin homology
- HUVEC
- human umbilical vascular endothelial cell
- RFP
- red fluorescent protein
- qPCR
- quantitative PCR
- LUV
- large unilamellar vesicle
- PI
- phosphatidylinositol
- PIP
- phosphatidylinositol phosphate
- PIP3
- PI 3,4,5-trisphosphate
- GTPγS
- guanosine 5′-O-(thiotriphosphate)
- myrArf6
- myristoylated Arf6
- PIP5KI
- type I PIP5K.
REFERENCES
- 1. Carmeliet P. (2005) Nature 438, 932–936 [DOI] [PubMed] [Google Scholar]
- 2. Folkman J. (2007) Nat. Rev. Drug Discov 6, 273–286 [DOI] [PubMed] [Google Scholar]
- 3. Carmeliet P., Tessier-Lavigne M. (2005) Nature 436, 193–200 [DOI] [PubMed] [Google Scholar]
- 4. Serini G., Valdembri D., Zanivan S., Morterra G., Burkhardt C., Caccavari F., Zammataro L., Primo L., Tamagnone L., Logan M., Tessier-Lavigne M., Taniguchi M., Püschel A. W., Bussolino F. (2003) Nature 424, 391–397 [DOI] [PubMed] [Google Scholar]
- 5. Gu C., Yoshida Y., Livet J., Reimert D. V., Mann F., Merte J., Henderson C. E., Jessell T. M., Kolodkin A. L., Ginty D. D. (2005) Science 307, 265–268 [DOI] [PubMed] [Google Scholar]
- 6. Acevedo L. M., Barillas S., Weis S. M., Göthert J. R., Cheresh D. A. (2008) Blood 111, 2674–2680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kessler O., Shraga-Heled N., Lange T., Gutmann-Raviv N., Sabo E., Baruch L., Machluf M., Neufeld G. (2004) Cancer Res. 64, 1008–1015 [DOI] [PubMed] [Google Scholar]
- 8. Neufeld G., Kessler O. (2008) Nat. Rev. Cancer 8, 632–645 [DOI] [PubMed] [Google Scholar]
- 9. Staton C. A., Kumar I., Reed M. W., Brown N. J. (2007) J. Pathol. 212, 237–248 [DOI] [PubMed] [Google Scholar]
- 10. Sakurai A., Gavard J., Annas-Linhares Y., Basile J. R., Amornphimoltham P., Palmby T. R., Yagi H., Zhang F., Randazzo P. A., Li X., Weigert R., Gutkind J. S. (2010) Mol. Cell Biol. 30, 3086–3098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brown F. D., Rozelle A. L., Yin H. L., Balla T., Donaldson J. G. (2001) J. Cell Biol. 154, 1007–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Powelka A. M., Sun J., Li J., Gao M., Shaw L. M., Sonnenberg A., Hsu V. W. (2004) Traffic 5, 20–36 [DOI] [PubMed] [Google Scholar]
- 13. Donaldson J. G., Jackson C. L. (2000) Curr. Opin. Cell Biol. 12, 475–482 [DOI] [PubMed] [Google Scholar]
- 14. Casanova J. E. (2007) Traffic 8, 1476–1485 [DOI] [PubMed] [Google Scholar]
- 15. Someya A., Sata M., Takeda K., Pacheco-Rodriguez G., Ferrans V. J., Moss J., Vaughan M. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 2413–2418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Campa F., Yoon H. Y., Ha V. L., Szentpetery Z., Balla T., Randazzo P. A. (2009) J. Biol. Chem. 284, 28069–28083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ha V. L., Thomas G. M., Stauffer S., Randazzo P. A. (2005) Methods Enzymol. 404, 164–174 [DOI] [PubMed] [Google Scholar]
- 18. Randazzo P. A., Terui T., Sturch S., Fales H. M., Ferrige A. G., Kahn R. A. (1995) J. Biol. Chem. 270, 14809–14815 [DOI] [PubMed] [Google Scholar]
- 19. Jian X., Cavenagh M., Gruschus J. M., Randazzo P. A., Kahn R. A. (2010) Traffic 11, 732–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tolias K. F., Rameh L. E., Ishihara H., Shibasaki Y., Chen J., Prestwich G. D., Cantley L. C., Carpenter C. L. (1998) J. Biol. Chem. 273, 18040–18046 [DOI] [PubMed] [Google Scholar]
- 21. Derrien V., Couillault C., Franco M., Martineau S., Montcourrier P., Houlgatte R., Chavrier P. (2002) J. Cell Sci. 115, 2867–2879 [DOI] [PubMed] [Google Scholar]
- 22. Cohen L. A., Honda A., Varnai P., Brown F. D., Balla T., Donaldson J. G. (2007) Mol. Biol. Cell 18, 2244–2253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dunphy J. L., Moravec R., Ly K., Lasell T. K., Melancon P., Casanova J. E. (2006) Curr. Biol. 16, 315–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Broussard J. A., Webb D. J., Kaverina I. (2008) Curr. Opin. Cell Biol. 20, 85–90 [DOI] [PubMed] [Google Scholar]
- 25. Bhatt A., Kaverina I., Otey C., Huttenlocher A. (2002) J. Cell Sci. 115, 3415–3425 [DOI] [PubMed] [Google Scholar]
- 26. Morishige M., Hashimoto S., Ogawa E., Toda Y., Kotani H., Hirose M., Wei S., Hashimoto A., Yamada A., Yano H., Mazaki Y., Kodama H., Nio Y., Manabe T., Wada H., Kobayashi H., Sabe H. (2008) Nat. Cell Biol. 10, 85–92 [DOI] [PubMed] [Google Scholar]
- 27. Vlahos C. J., Matter W. F., Hui K. Y., Brown R. F. (1994) J. Biol. Chem. 269, 5241–5248 [PubMed] [Google Scholar]
- 28. Chao W. T., Ashcroft F., Daquinag A. C., Vadakkan T., Wei Z., Zhang P., Dickinson M. E., Kunz J. (2010) Mol. Cell Biol. 30, 4463–4479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gay C. M., Zygmunt T., Torres-Vázquez J. (2011) Dev. Biol. 349, 1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Torres-Vázquez J., Gitler A. D., Fraser S. D., Berk J. D., Van N. P., Fishman M. C., Childs S., Epstein J. A., Weinstein B. M. (2004) Dev. Cell 7, 117–123 [DOI] [PubMed] [Google Scholar]
- 31. Murphy J. A., Jensen O. N., Walikonis R. S. (2006) Brain Res. 1120, 35–45 [DOI] [PubMed] [Google Scholar]
- 32. Inaba Y., Tian Q. B., Okano A., Zhang J. P., Sakagami H., Miyazawa S., Li W., Komiyama A., Inokuchi K., Kondo H., Suzuki T. (2004) J. Neurochem. 89, 1347–1357 [DOI] [PubMed] [Google Scholar]
- 33. Hiroi T., Someya A., Thompson W., Moss J., Vaughan M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 10672–10677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Someya A., Moss J., Nagaoka I. (2010) J. Biol. Chem. 285, 30698–30707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Di Paolo G., De Camilli P. (2006) Nature 443, 651–657 [DOI] [PubMed] [Google Scholar]
- 36. Manning B. D., Cantley L. C. (2007) Cell 129, 1261–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kwiatkowska K. (2010) Cell Mol. Life Sci. 67, 3927–3946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yin H. L., Janmey P. A. (2003) Annu. Rev. Physiol. 65, 761–789 [DOI] [PubMed] [Google Scholar]
- 39. van den Bout I., Divecha N. (2009) J. Cell Sci. 122, 3837–3850 [DOI] [PubMed] [Google Scholar]
- 40. Tolias K. F., Hartwig J. H., Ishihara H., Shibasaki Y., Cantley L. C., Carpenter C. L. (2000) Curr. Biol. 10, 153–156 [DOI] [PubMed] [Google Scholar]
- 41. Rozelle A. L., Machesky L. M., Yamamoto M., Driessens M. H., Insall R. H., Roth M. G., Luby-Phelps K., Marriott G., Hall A., Yin H. L. (2000) Curr. Biol. 10, 311–320 [DOI] [PubMed] [Google Scholar]
- 42. Ling K., Doughman R. L., Firestone A. J., Bunce M. W., Anderson R. A. (2002) Nature 420, 89–93 [DOI] [PubMed] [Google Scholar]
- 43. Wang Y., Litvinov R. I., Chen X., Bach T. L., Lian L., Petrich B. G., Monkley S. J., Kanaho Y., Critchley D. R., Sasaki T., Birnbaum M. J., Weisel J. W., Hartwig J., Abrams C. S. (2008) J. Clin. Invest. 118, 812–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Honda A., Nogami M., Yokozeki T., Yamazaki M., Nakamura H., Watanabe H., Kawamoto K., Nakayama K., Morris A. J., Frohman M. A., Kanaho Y. (1999) Cell 99, 521–532 [DOI] [PubMed] [Google Scholar]
- 45. Moriya J., Minamino T., Tateno K., Okada S., Uemura A., Shimizu I., Yokoyama M., Nojima A., Okada M., Koga H., Komuro I. (2010) Circ. Res. 106, 391–398 [DOI] [PubMed] [Google Scholar]