Abstract
The mechanism by which misfolded proteins in the endoplasmic reticulum (ER) are retrotranslocated to the cytosol for proteasomal degradation is still poorly understood. Here, we show that importin β, a well established nucleocytoplasmic transport protein, interacts with components of the retrotranslocation complex and promotes ER-associated degradation (ERAD). Knockdown of importin β specifically inhibited the degradation of misfolded ERAD substrates but did not affect turnover of non-ERAD proteasome substrates. Genetic studies and in vitro reconstitution assays demonstrate that importin β is critically required for ubiquitination of mutant α1-antitrypsin, a luminal ERAD substrate. Furthermore, we show that importin β cooperates with Ran GTPase to promote ubiquitination and proteasomal degradation of mutant α1-antitrypsin. These results establish an unanticipated role for importin β in ER protein quality control.
Keywords: E3 Ubiquitin Ligase, Endoplasmic Reticulum (ER), ER Quality Control, Nuclear Transport, Ubiquitin
Introduction
Many newly synthesized proteins in the endoplasmic reticulum (ER)2 fail to fold properly because of transcriptional and translational errors or imbalanced production of accessory subunits (1, 2). Additionally, pathogenic conditions, such as genetic mutations, hypoxia, oxidative stress, ischemia, and disturbance of calcium homeostasis, can also cause proteins to misfold in the ER (3–5). Fortunately, cells have evolved protein quality control systems that can efficiently eliminate misfolded proteins from the ER before they wreak havoc on cells. A major mechanism that recognizes and degrades misfolded and unassembled ER proteins at the ER is the ER-associated degradation (ERAD) pathway (1, 2, 6–8), which exports misfolded ER proteins into the cytosol for degradation by the ubiquitin proteasome system.
ER luminal chaperones and lectins recognize and deliver ERAD substrates to membrane-anchored protein complexes that form putative protein-conducting channels from which the substrates are subsequently retrotranslocated into the cytosol (8, 9). Each of these retrotranslocation complexes usually contains one or more membrane-bound ubiquitin ligases (E3s), which ubiquitinate ERAD substrates en route to the cytosol. In budding yeast, the Hrd1p E3 complex degrades substrates whose lesions reside in either the transmembrane domain or lumen of the ER, whereas the other complex containing the Doa10p E3 disposes of substrates with lesions in their cytosolic domains (10, 11). These ERAD complexes are conserved in mammalian cells, but as expected, the repertoire of E3s for mammalian ERAD is more complex. In addition to the Hrd1p and Doa10p orthologs (Hrd1 and gp78 for Hrd1p and TEB4 for Doa10p) (12–16), several other membrane-bound E3s, such as RMA1/RNF5 (ring finger protein 5), RFP2 (Ret finger protein 2), and TRC8 (translocation in renal cancer from chromosome 8) have been implicated in ERAD (17, 18). In addition, cytosolic E3s, including CHIP (C terminus of Hsc70-interacting protein) (19, 20), Parkin (21), and the SCF (Skpl-Cullin-F-box protein family) multisubunit E3 (22), can also be recruited to the cytosolic surface of the ER to act on ERAD substrates.
ERAD substrates have to be fully retrotranslocated or dislocated from the ER for elimination (1, 2, 6–8), which is mediated by a conserved cytosolic AAA ATPase termed p97 in mammals or Cdc48p in yeast (6, 23). The mechanism underlying protein retrotranslocation is poorly defined. It is generally believed that substrates are retrotranslocated from the ER through one or a few proteineous channels (1), although the identity of such channel has not been revealed. One channel candidate protein is the multispanning membrane protein, Derlin-1, based on its involvement in retrotranslocation of nascent major histocompatibility complex class I heavy chains (24, 25), pαF (26), simian virus 40 (27), and cholera toxin (28). Alternative candidates include the multispanning E3 ligases such as gp78, Hrd1, and TEB4 (29–31). Upon emerging into the cytosol, substrates are ubiquitinated by membrane-associated E3s and subsequently dislocated into the cytosol by the p97/Cdc48 complex (32, 33). The ER membrane-anchored Ubx domain proteins, Ubx2p in yeast and erasin/UbxD2 and/or UbxD8 in mammalian cells, have been shown to recruit p97/Cdc48 to the ER for retrotranslocation (34–36). In addition, several additional ER membrane proteins, including Hrd1, gp78, VIMP (p97/valosin-containing protein-interacting membrane protein), Derlin-1, and Herp, also interact with p97 in mammalian cells (25, 37–39), suggesting that recruitment of p97 to the ER might occur through a concerted effort of multiple proteins. ERAD substrates retrotranslocated by p97/Cdc48p are then subjected to deglycosylation and ubiquitin chain editing before being shuttled to the proteasome for degradation (40–43).
To identify additional factors involved in ERAD, we searched for proteins that bind to the p97 receptor VIMP. Through this approach, we identified importin β as a new protein involved in ERAD. Importin β (also known as karyopherin β1 or kap95 in budding yeast) is a multifunctional protein that is known to mediate nuclear import of numerous proteins, act as a cytoplasmic chaperone that prevents aggregation for some proteins, and negatively regulate mitotic spindle assembly, nuclear membrane fusion, and nuclear pore complex formation (44). During nuclear import, importin β binds cargo directly or utilizes importin α as an adaptor to bind its cargo proteins bearing a classical nuclear localization signal. The importin β-cargo complex is transported through the nuclear pore complex into the nucleus where RanGTP binds to importin β to induce cargo release. The importin β-RanGTP complex is then recycled back to the cytoplasm where RanGTP is dissociated from importin β and converted to RanGDP (45). RanGDP is imported into the nucleus by NTF2 (nuclear transport factor 2) and then converted to RanGTP by RCC1 (regulator of chromosome condensation 1) (46, 47). The process results in RanGTP predominantly in the nucleus and RanGDP mainly in the cytoplasm (45). Here we present evidence for a novel function of importin β in promoting ERAD. We show that importin β is an important component of a retrotranslocation machinery involved in ERAD and that it cooperates with Ran GTPase to promote ubiquitination and degradation of the luminal ERAD substrate, the null Hong Kong variant of α-1-antitrypsin (NHK).
EXPERIMENTAL PROCEDURES
Plasmid Constructs
pGEX-VIMP was constructed by inserting the ORF of VIMP (NcoI/XhoI fragment) from pcDNA3.1-hVIMP (25) into pGEX-4T-3. To construct pCIneo-VIMP-FLAG, the PCR fragment encoding VIMP with a C-terminal FLAG tag was inserted into the EcoRI/SalI sites of pCIneo. pQE-importin β is a kind gift from Dr. Dirk Görlich (48). pQE-importin β-N603 (amino acids 1–603) and pQE-importin β-N297 (amino acids 1–297) were made by self-ligation of the largest fragments of pQE-impβ cut with BglII (blunted with Klenow) or HindIII, respectively. pQE-importin βΔN32 (amino acids 33–871), ΔN128 (amino acids 129–871), and ΔN169 (amino acids 170–871) were constructed by PCR. To construct pCIneo-importin β, importin β ORF was cloned into pCIneo vector via XbaI and NotI. To construct pCIneo-importin β-N297, HindIII (blunted with Klenow) and XbaI fragment encoding the N-terminal 297 amino acids of importin β was excised from pCIneo-importin β and then inserted into pCIneo cut by XbaI (blunted with Klenow) and NheI. To make pCIneo-importin β-N603, the BglII (Klenow) and XbaI fragment encoding importin β (amino acids 1–603) was excised from pCIneo-importin β and then inserted into XbaI/SmaI sites of pCIneo. pcDNA3.1-Derlin-2-HA was constructed by inserting the PCR fragment encoding Derlin-2 with a C-terminal HA tag into Hind III/XhoI sites of pcDNA3.1. pKW356 expressing His-tagged Ran was provided by Dr. Karsten Weis (49). hATM/pcDNA3.1Zeo(+) and hATNHK/pcDNA3.1Zeo(+) were provided by Dr. Richard Sifers (50). pCIneo-NHK-HA was constructed by inserting the PCR fragment encoding NHK with a C-terminal HA tag to NheI/SmaI sites of pCIneo vector. Plasmid encoding importin α1-GST was provided by Dr. Reinhard Depping (University of Luebeck, Luebeck, Germany). pcDNA3-HA-Cdc24 was provided by Dr. Yun Qiu (University of Maryland, Baltimore, MD). The GFP-p53 construct was provided by Dr. Wei Gu (51). Ub-R-GFP plasmid was provided by Dr. Maria G. Masucci (52). pcDNA3.1-Derlin-1-HA, pCI-HA-CD3δ, pFLAG-gp78, and pCI-Hrd1-FLAG were previously reported (12, 25, 30).
Antibodies
Monoclonal anti-gp78 (2G5), polyclonal anti-Derlin-1, and anti-Hrd1 antibodies have been previously described (38, 53). Rabbit polyclonal antibodies against the importin β C-terminal region were purchased from Calbiochem. Rabbit polyclonal antibodies against importin β (amino acids 1–300), and mouse monoclonal anti-Ran, anti-ubiquitin, anti-NTF2 antibodies were purchased from Santa Cruz Biotechnology Inc. Mouse anti-p97 and anti-calnexin antibodies were purchased from Affinity BioReagents. Goat anti-α1-antitrypsin antibodies were purchased from Bethyl Laboratories. Mouse monoclonal anti-GAPDH antibody was purchased from Ambion. Mouse monoclonal anti-importin α, anti-β-actin (AC-74), anti-HA (HA-7), HRP-conjugated anti-FLAG, and rabbit anti-VIMP and anti-FLAG antibodies were purchased from Sigma. Mouse monoclonal anti-importin β antibody (3E9) was purchased from Abcam Inc. Mouse monoclonal antibody against calreticulin was purchase from BD Bioscience. Anti-GFP-agarose was purchased from MBL International Corporation. HRP-conjugated anti-GFP antibody was purchased from Rockland Inc.
Recombinant Protein Expression
Generally, bacteria expressing recombinant proteins were induced with 0.1 mm isopropyl β-d-thiogalactopyranoside either at 37 °C for 1 h or at 23 °C for 3 h. All His-tagged proteins were purified using Ni-NTA-agarose (Qiagen) following the user's manual. GST fusion proteins were expressed and lysed as previously described (54). Then the lysates were incubated with glutathione-Sepharose beads (Amersham Biosciences). After washing, GST fusion proteins were eluted from the beads with 10 mm reduced glutathione in 50 mm Tris/HCl (pH 8.0). All His-tagged and GST-tagged proteins were dialyzed in PBS to remove imidazole or glutathione from the proteins.
Nucleotide Loading of Ran
20 μm recombinant Ran was incubated with 1 mm of the GTP or GDP in 50 mm Hepes, pH 7.3, 1 mm magnesium acetate, 10 mm EDTA, 2.5 mm DTT, 1 mm ATP at 20 °C for 30 min (55). After the addition of magnesium acetate to 5 mm, free nucleotides were removed by dialysis in PBS.
GST Pulldown
GST fusion proteins were immobilized on glutathione-Sepharose beads and incubated with indicated proteins for 2 h at 4 °C in in vitro binding buffer (25 mm Tris/HCl, pH 8.0, 200 mm KCl, 2 mm MgCl2, 1 mm ATP, 1 mm DTT, 5% glycerol, and 1% Triton X-100) (56). After washing three times, the beads were boiled in SDS sample buffer followed by SDS-PAGE.
Immunofluorescence and Microscopy
HeLa cells grown on slide glass were transfected with pCIneo-VIMP-FLAG. 24 h after transfection, the cells were washed with PBS and permeabilized with 0.05% saponin in PBS for 5 min to deplete the cytosol. Then the cells were fixed in 4% paraformaldehyde for 30 min at 4 °C and blocked in 0.1% saponin, 0.1% human serum albumin. The cells were labeled with mouse monoclonal anti-importin β antibody (3E9) and rabbit polyclonal anti-FLAG antibody for 1 h followed by labeling with Alexa® Fluor 594-conjugated goat anti-mouse IgG (H+L) and Alexa® Fluor 488 conjugated goat anti-rabbit IgG (H+L) for 1 h. Fluorescence microscopy was performed using a Zeiss 510 laser scanning confocal microscope.
Immunoprecipitation
The cells were lysed in cell lysis buffer (150 mm NaCl, 10 mm Tris/HCl, pH 7.4, 1 mm EDTA, 1 mm EGTA, 0.2% Nonidet P-40, and protease inhibitor mixture). Normally, 500 μg of total proteins were used for IP in a total volume of 750 μl containing 4% glycerol. Depending on experimental requirements, Trueblot anti-mouse IgG or anti-rabbit IgG beads (eBiosciences), protein A-Sepharose (Zymed Laboratories Inc.), or protein G plus-Sepharose (Calbiochem) was used to precipitate antibodies. Anti-FLAG® M2 Affinity Gel (Sigma A2220) was used for FLAG-tagged protein IP. After incubation for 2 h at 4 °C, the beads were washed three times and processed for immunoblotting (IB).
RNA Interference
NTF2 siRNAs were purchased from Santa Cruz Biotecnologies. All other siRNAs were ordered from Ambion, including Silencer® Negative Control #1, importin β and p97 siRNAs. Three importin β siRNAs were designed according to a previously reported study (57). p97 siRNA was as previously reported (58). siRNAs were transfected with Lipofectamine 2000 (Invitrogen). 48 h after transfection, the cells were processed for IB, IP, or pulse-chase experiments.
Pulse-Chase
Pulse-chase was done as previously described (38). In brief, transfected HEK293 cells were starved for methionine and cysteine for 1 h and then pulse-labeled with l-[35S]methionine and l-[35S]cysteine (RedivueTM Pro-mix l-[35S] in vitro cell labeling mix; GE HealthCare) (150 μCi/ml) for 30 min at 37 °C. The cells were then washed with PBS and chased in complete DMEM with excess amount of methionine and cysteine for the indicated chase times. Following the chase, the cells were lysed in cell lysis buffer (150 mm NaCl, 10 mm Tris/HCl, pH 7.4, 1 mm EDTA, 1 mm EGTA, 0.2% Nonidet P-40, 0.5% Triton X-100, and protease inhibitor mixture) with 0.1% SDS. The lysates were immunoprecipitated with antibodies as indicated, and the immunoprecipitates were subjected to SDS-PAGE and autoradiography.
In Vivo Ubiquitination
HEK293 cells were transfected with indicated plasmids. 6 h after transfection, the cells were reseeded in 100-mm dishes at 5 × 105 cells/dish. siRNAs were transfected with Lipofectamine 2000 on the second day after the reseeding as indicated. 48 h after siRNA transfection, the cells were treated with 10 μm MG132 for 3–6 h to inhibit proteasomal degradation. Then the cells were harvested and lysed in 2% SDS. After incubation at 37 °C for 30 min, the lysates were diluted 20 times in cell lysis buffer. The cell debris was removed by centrifugation. 200 μg of cleared lysates were used for IP with antibody as indicated at 4 °C overnight. The precipitates were processed for IB.
In Vitro Ubiquitination
In vitro ubiquitination was performed as described (33, 59) with minor modifications. Microsomes were prepared from HEK293 cells expressing HA-tagged NHK. The microsomes (pellet) were washed in PBS containing 0.5 m NaCl and centrifuged again at 105,000 × g for 1 h. To prepare cytosol, HEK293 cells were lysed by freezing and thawing and centrifuged at 105,000 × g for 1h. The supernatant (cytosol) was used in in vitro ubiquitination assays. Importin β-depleted cytosol was prepared by incubating the cytosol with monoclonal anti-importin β antibody (3E9) and protein A-Sepharose. Ran and importin α were depleted from the cytosol by incubating cytosol with Ni-NTA-agarose prebound with importin β-N603 or -ΔN169, respectively. Control cytosol was obtained by incubation with protein A-Sepharose or Ni-NTA-agarose. An in vitro ubiquitination reaction contains ∼40 μg of microsomes, 400 μg of cytosol, 1 μg of ubiquitin, 5 mm ATP, 1 mm DTT, 300 μm MG132, protease inhibitor mixture, and recombinant proteins as indicated in a total volume of 40 μl. The reactions were incubated at 37 °C or on ice for 1 h and stop by adding of 10 μl of 250 mm N-ethylmaleimide and 40 μl of 2% SDS. The samples were boiled for 20 min. Then they were diluted with 500 μl of cell lysis buffer. The samples were cleared by spinning at 14,000 rpm for 10 min, and then HA-tagged NHK was immunoprecipitated with anti-HA antibody at 4 °C overnight.
RESULTS
Importin β Is a Novel Component of the ERAD Complexes
VIMP is a component of the mammalian gp78 and Hrd1 ERAD complexes and has no ortholog in yeast. It is a type III transmembrane protein with a sizable cytosolic domain, which recruits p97 to the ER membrane to promote ERAD (25, 37). To identify additional factors specifically involved in mammalian ERAD, we searched for proteins that interact with VIMP using a GST pulldown assay. We incubated GST-tagged VIMP with HEK293 cell lysates and found two VIMP-binding proteins (supplemental Fig. S1). Mass spectrometry identified these two proteins as p97, a known VIMP-interacting protein, and importin β.
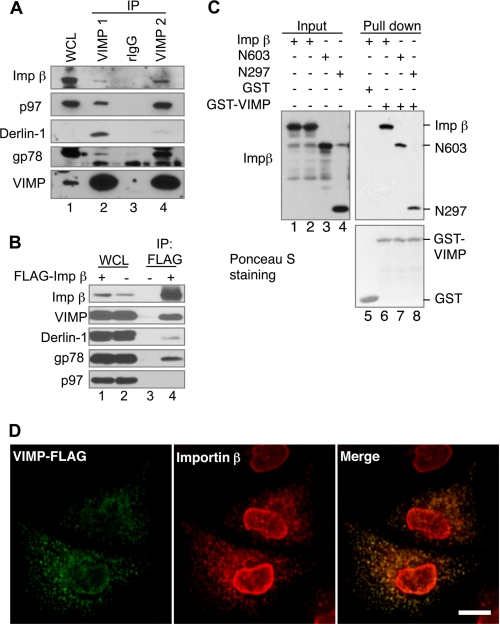
Importin β is a well known carrier protein that mediates nuclear transport of numerous proteins. In this regard, it is unanticipated to see an interaction of importin β with VIMP. We therefore first confirmed that endogenous VIMP and importin β could interact in cells by IP using two different VIMP antibodies. Importin β along with known VIMP-interacting proteins, including gp78, Derlin-1, and p97, were all coimmunoprecipitated with VIMP (Fig. 1A). IP with an anti-FLAG antibody showed that endogenous VIMP along with gp78 and Derlin-1 but not p97 were coprecipitated with ectopically expressed FLAG-tagged importin β (Fig. 1B). GST pulldown assay using GST-VIMP incubated with purified recombinant importin β, and its truncation mutants revealed that VIMP can bind directly to the N-terminal region of importin β (Fig. 1C). Together, these results demonstrate that importin β interacts with VIMP, Derlin1, and gp78 but not p97.
FIGURE 1.
Importin β is a novel VIMP interacting protein. A, interaction of endogenous VIMP and importin β. HEK293 cell lysate was subject to IP using two different rabbit polyclonal anti-VIMP antibodies (VIMP1 and VIMP2) or rabbit IgG (rIgG) as a negative control. WCL, whole cell lysate. B, FLAG-tagged importin β binds VIMP. HEK293 cells transfected with the indicated plasmids were processed for IP with anti-FLAG antibody. C, VIMP binds directly to the N-terminal region of importin β. GST pulldown assay was conducted by incubating recombinant importin β or its truncation mutants with GST and GST-VIMP as indicated. The association of importin β or its mutants with VIMP was detected by IB. D, importin β colocalizes with VIMP. HeLa cells transiently expressing FLAG-tagged VIMP were permeabilized with 0.05% saponin to release the cytosol before fixation. Then the cells were immunostained with mouse monoclonal anti-importin β antibody (3E9) and rabbit anti-FLAG antibody. Alexa® Fluor 594-conjugated goat anti-mouse IgG and Alexa® Fluor 488-conjugated goat anti-rabbit IgG were used as secondary antibodies. Bar, 10 μm.
Next, we determined whether VIMP and importin β colocalize in cells using laser scanning confocal microscopy. HeLa cells expressing FLAG-tagged VIMP were treated with saponin to release cytosolic contents prior to fixation with paraformaldehyde. The cells were stained for VIMP-FLAG and endogenous importin β. Our results revealed that importin β was predominantly localized to the nucleus in saponin-treated cells. However, a significant amount of importin β was detected as discrete foci that overlap with the VIMP-FLAG signal (Fig. 1D). These foci may represent special functional domains in the ER membrane where these proteins interact to regulate ERAD.
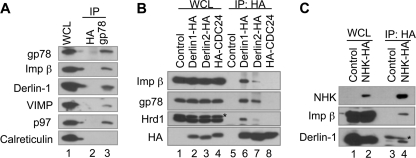
Furthermore, we demonstrated that importin β was coimmunoprecipitated with endogenous gp78 (Fig. 2A), as did the ectopically expressed gp78, Hrd1, Derlin-1, and Derlin-2 (Fig. 2B and supplemental Fig. S2A). Interestingly, overexpression of gp78 or Hrd1 in HEK293 cells resulted in increased association of importin β with ER-enriched microsomes (supplemental Fig. S2B), indicating that gp78 and Hrd1 may be involved in the recruitment of importin β to the ER. In addition, IP of the well established luminal ERAD substrate, NHK, also coprecipitated importin β (Fig. 2C). These results suggest that importin β is an integral component of the ERAD complex that contains gp78, Hrd1, VIMP, and Derlin-1.
FIGURE 2.
Importin β is a novel component of the ERAD complex. A, importin β interacts with gp78. HEK293 cell lysates were subject to IP with monoclonal anti-gp78 antibody (2G5) or monoclonal anti-HA antibody as a control followed by IB for the indicated proteins. B, importin β interacts with Derlin-1 and Derlin-2. HEK293 cells transfected with plasmids encoding Derlin-1-HA, Derlin-2-HA, or HA-CDC24 (as a negative control) were processed for IP with anti-HA antibody followed by IB for the indicated proteins. The asterisk indicates an additional band that is reactive with anti-Hrd1 antibodies. C, importin β interacts with the luminal ERAD substrate NHK. HEK293 cells transfected with plasmids encoding NHK-HA were processed for IP with anti-HA antibody followed by IB for the indicated proteins. The asterisk indicates the light chain of anti-HA antibody used in IP. WCL, whole cell lysate.
Importin β Knockdown Stabilizes ERAD Substrates
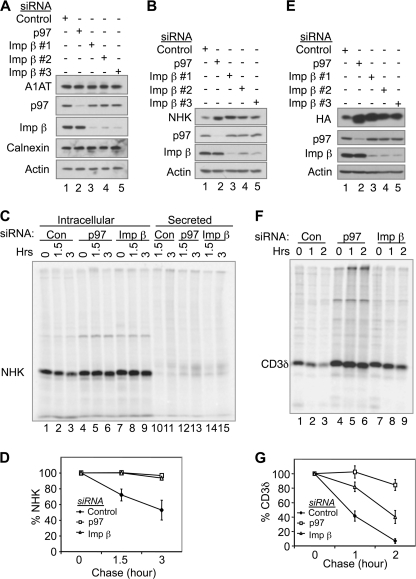
To examine whether importin β regulates ERAD, we characterized the effect of RNAi-mediated knockdown of importin β on the degradation of NHK. To avoid potential off target activity of siRNA transfection, we tested the effects of three previously reported importin β siRNAs (57). WT α-1-antitrypsin (A1AT) was used as a negative control because it folds properly and is therefore not an ERAD substrate. All three siRNAs had little effect on WT A1AT but caused accumulation of NHK to a level comparable with that of p97 depletion (Fig. 3, A and B). To more accurately measure the stability of NHK, pulse-chase experiments were conducted in control and importin β knockdown cells. The results showed that newly synthesized NHK was rapidly degraded in control cells, consistent with NHK being a short-lived protein (Fig. 3, C and D). In contrast, in importin β knockdown cells, NHK degradation was significantly inhibited, similar to that seen in p97 knockdown cells (Fig. 3, C and D). Knockdown of importin β also stabilized CD3δ, a membrane-bound ERAD substrate, as was showed by both steady-state levels and pulse-chase results (Fig. 3, E–G), although the stabilization effect was less efficient than p97 depletion. These results suggest that importin β is involved in degradation of some ERAD substrates. Notably, knockdown of importin β did not affect the degradation of Ub-R-GFP, a proteasomal substrate of the N-end rule pathway (52) (supplemental Fig. S3A), suggesting that the effect of importin β knockdown on ERAD is not due to a general compromisation of proteasomal degradation.
FIGURE 3.
Importin β knockdown stabilizes ERAD substrates. A, HEK293 cells expressing wild type A1AT were transfected with siRNA targeting p97 or importin β as indicated. 48 h after transfection, the cells were processed for IB. Three previously reported siRNAs targeting different regions of the importin β transcript (57) were used as indicated. B, as in A, except that HEK293 cells expressing NHK were transfected with siRNAs. C, HEK293 cells expressing NHK were transfected with siRNAs targeting p97 or importin β as indicated. 48 h after transfection, the cells were pulse-labeled with l-[35S]methionine and l-[35S]cysteine (150 μCi/ml) for 30 min at 37 °C followed by chase for the indicated times. The cells were then processed for IP with anti-A1AT antibodies (reactive with NHK). Immunoprecipitates were resolved by SDS-PAGE, and the image was acquired on a Typhoon Scanner. D, quantification of NHK degradation in C. The data represent the means ± S.E. of three independent experiments. The relative band intensity of NHK at chase 0 h was set to 100%. E, as in A, except that HEK293 cells expressing HA-tagged CD3δ were transfected with siRNAs. F and G, as in C and D, except that HEK293 cells expressing HA-tagged CD3δ were transfected with siRNAs, and the lysates were immunoprecipitated with anti-HA antibody. Image was acquired on a Typhoon Scanner. Imp, importin; Con, control.
Importin β Is Required for Ubiquitination of NHK
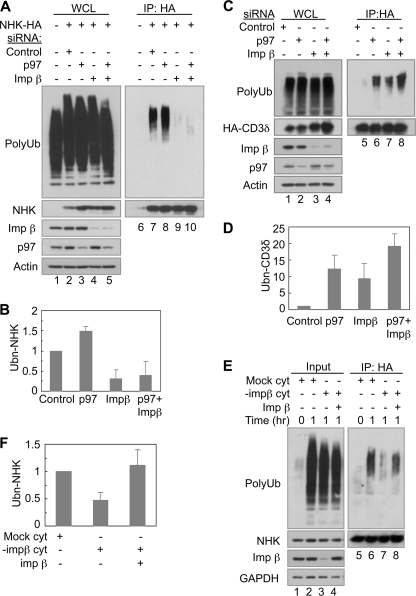
To determine how importin β might function in ERAD, we tested whether importin β is required for ubiquitination of NHK. To this end, cells transfected with various siRNAs were treated with a proteasome inhibitor to block the degradation of ubiquitinated NHK. NHK was immunoprecipitated from detergent extracts under denaturing conditions, and its ubiquitination was examined by IB. Polyubiquitin immunoreactivity was detected in NHK-expressing cells treated with a control siRNA, but not in cells lacking NHK expression (Fig. 4A, lane 7 versus lane 6), indicating that the polyubiquitin signal was derived from the polyubiquitinated NHK. Consistent with the notion that p97 acts at a post-ubiquitination step during ERAD (60), knockdown of p97 increased ubiquitinated NHK levels (Fig. 4, A and B). In contrast, knockdown of importin β consistently reduced the amount of ubiquitinated NHK by ∼70% (Fig. 4, A, lane 9 versus lane 7, and B). Importantly, simultaneously knockdown of importin β and p97 abolished the accumulation of ubiquitinated NHK observed in p97 knockdown cells (Fig. 4, A, lane 10 versus lane 8, and B), indicating that importin β is essential for NHK ubiquitination in cells.
FIGURE 4.
Importin β is required for ubiquitination of NHK. A, importin β knockdown inhibits NHK ubiquitination. HEK293 cells expressing HA-tagged NHK were transfected with siRNAs as indicated. 48 h after transfection, the cells were treated with MG132 (10 μm) for 6 h and then lysed in SDS. Lysates were used for IP with an anti-HA antibody followed by IB for ubiquitin. B, quantification of NHK ubiquitination in A. The data represent the means ± S.E. of three independent experiments. The relative intensity of the polyubiquitin smear (Ubn-NHK) from cells transfected with control siRNA was set to 1. C and D, importin β knockdown increases CD3δ ubiquitination; as in A and B, except that HEK293 cells expressing HA-tagged CD3δ were transfected with siRNAs. E, importin β is required for NHK ubiquitination in vitro. Microsomes were prepared from HEK293 cells expressing HA-tagged NHK. Cytosol was prepared from HEK293 cells. Importin β in the cytosol was depleted (−impβ cyt) by incubating the cytosol with anti-importin β antibody (3E9) and protein A-Sepharose beads at 4 °C for 2 h. Mock control was incubated with protein A-Sepharose beads only. 40 μg of microsomes were incubated together with cytosol (∼10 μg/μl) and recombinant importin β (1 μg) as indicated for 1 h at 37 °C. The reactions were stopped by adding denaturing buffer. HA-tagged NHK was immunoprecipitated with anti-HA antibody. A fraction of each lysate (input) was analyzed directly by IB. F, quantification of NHK ubiquitination in E. The data represent the means ± S.E. of three independent experiments. The relative intensity of the polyubiquitin smear (Ubn-NHK) in the reaction with mock cytosol for 1 h (lane 6) was set to 1. WCL, whole cell lysate; Imp, importin.
In contrast to the results seen with NHK, knockdown of importin β increased accumulation of ubiquitinated CD3δ, as did knockdown of p97 (Fig. 4, C and D). Simultaneously, knockdown of importin β and p97 resulted in even greater accumulation of ubiquitinated CD3δ (Fig. 4, C and D). Thus, importin β knockdown stabilized both NHK and CD3δ but had opposite effects on their ubiquitination. In addition, importin β knockdown did not affect the ubiquitination of the non-ERAD substrate Ub-R-GFP, suggesting that the ubiquitin-proteasome activity is not generally compromised (supplemental Fig. S3B). These results suggest that importin β plays an essential role in regulating degradation of both luminal (NHK) and membrane (CD3δ) ERAD substrates but appears to play a more critical role in regulating ubiquitination of luminal substrates.
To see whether importin β directly regulates ERAD, we set up a cell-free ubiquitination assay to monitor the ubiquitination of NHK in vitro (33, 59). Microsomes were isolated from HEK293 cells expressing HA-tagged NHK and incubated with cytosol in the presence of ATP and the proteasome inhibitor MG132. The reactions were stopped by the addition of a denaturing buffer, and the protein extracts were subject to IP for NHK with an antibody to its HA tag. The immunoprecipitated materials were analyzed by IB with an anti-ubiquitin antibody. No or very little signal was detected in sample using microsomes isolated from control cells (without NHK expression) and in sample not incubated at 37 °C, indicating that the polyubiquitin signal is derived from ubiquitinated NHK (supplemental Fig. S4). Intriguingly, when microsomes were incubated with cytosol depleted of importin β, ubiquitination of NHK was diminished (Fig. 4, E, lane 7 versus lane 6, and F). Importantly, NHK ubiquitination was recovered when recombinant importin β was added to the importin β-depleted cytosol (Fig. 4, E, lane 8 versus lane 7, and F). These results suggest that importin β is required for NHK ubiquitination in vitro.
A Mutant of Importin β Inhibits ERAD of NHK
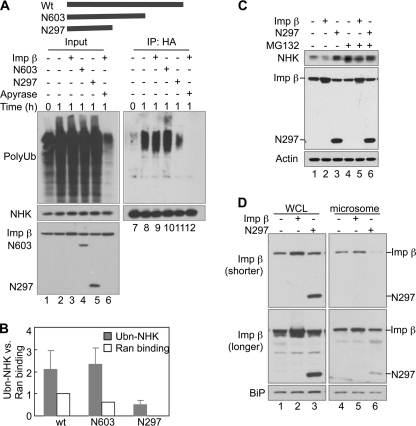
To further characterize the function of importin β, we tested the effects of two truncation mutants of importin β on ubiquitination of NHK in vitro (Fig. 5A). We found that the N-terminal 603-amino acid fragment of importin β (N603) was sufficient to promote NHK ubiquitination in vitro, whereas the N-terminal 297-amino acid fragment (N297) inhibited NHK ubiquitination (Fig. 5, A and B). The addition of apyrase, an ATP diphosphohydrolase, completely diminished NHK ubiquitination, indicating that the in vitro ubiquitination is ATP-dependent (Fig. 5A). Consistent with the in vitro findings, overexpression of N297 in NHK-expressing cells resulted in NHK accumulation, whereas overexpression of WT importin β slightly decreased NHK levels (Fig. 5C). MG132 blunted these effects (Fig. 5C), indicating that changes in the steady-state levels of NHK are largely controlled by proteasomal degradation. Overexpression of WT importin β and N297 did not alter the levels of the non-ERAD substrate Ub-R-GFP (supplemental Fig. S5A), suggesting that they do not cause a general alteration of proteins degraded by the ubiquitin-proteasome system. In addition, N297 overexpression did not affect the nuclear import of p53, suggesting that N297 may not block the nuclear transport (supplemental Fig. S5B). Thus, the inhibitory effect of N297 on ERAD is unlikely to be caused by a defect in nuclear transport.
FIGURE 5.
The N-terminal fragment of importin β containing 297 amino acids (N297) inhibits ERAD of NHK. A, effects of C-terminal truncation mutants of importin β on NHK ubiquitination in vitro. The diagram represents recombinant proteins used in this experiment. WT importin β (1 μg) or its truncation mutants N603 (0.7 μg) or N297 (0.35 μg) was used as indicated. Apyrase was added to deplete ATP. B, quantification of NHK ubiquitination in A. The data represent the means ± S.E. of three independent experiments. The relative intensity of polyubiquitin smear (Ubn-NHK) in reaction with control cytosol for 1 h (lane 8) was set to 1. Ran binding was quantified from supplemental Fig. S6B. C, N297 overexpression causes accumulation of NHK in cells. HEK293 cells steadily expressing NHK-HA were transfected with plasmids encoding WT importin β or its mutant N297 as indicated. 24 h later, the cells were treated with MG132 as indicated for 6 h and then analyzed by IB. D, effect of N297 overexpression on association of importin β with ER. Microsomes were prepared from HEK293 cells transfected with plasmids encoding WT importin β or its mutant N297 as indicated. A shorter exposure (shorter) and a longer exposure (longer) images were shown for importin β blotting. Imp, importin.
We have shown that N297 bound directly to VIMP, as did WT importin β (Fig. 1C), suggesting that N297 retains some binding abilities of importin β. To see whether N297 can compete with importin β to bind to ER, we overexpressed N297 in HEK293 cells and prepared the ER-enriched microsomes from these cells. All of the microsomes were washed with excess volume of fractionation buffer to minimize the contamination of cytosol. We blotted for importin β to see whether N297 overexpression has any effect on the level of association of importin β with microsomes. Less importin β was detected in microsomes prepared from cells overexpressing N297, suggesting that N297 somehow inhibits importin β binding to ER (Fig. 5D). These results further illustrate the importance of importin β binding to the ER for ERAD. Interestingly, in these assays, only low amounts of N297 was detected in microsomes. We speculate that this arises because N297 has weaker binding than WT importin β to proteins in the ER. For example, we found that N297 bound weaker than WT importin β to VIMP (Fig. 1C). The weaker binding would make it easier for N297 to be washed from microsomes.
Importin β and Ran GTPase Cooperate to Facilitate NHK Ubiquitination in Vitro
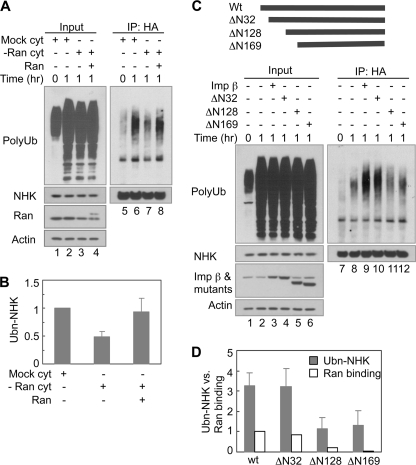
Ran is a small GTPase that interacts with importin β and regulates its function in nuclear import (45). We next asked whether Ran also participates in ERAD using the cell-free ubiquitination assay. Similar to our results for importin β, depletion of Ran from the cytosol also inhibited NHK ubiquitination (Fig. 6, A, lane 7 versus lane 6, and B). NHK ubiquitination increased significantly upon supplementation of the Ran-depleted cytosol with recombinant His-tagged Ran (Fig. 6, A, lane 8 versus lane 7, and B). These results suggest that Ran GTPase is also required to promote NHK ubiquitination in vitro.
FIGURE 6.
Importin β cooperates with Ran to promote NHK ubiquitination. A, Ran is required for NHK ubiquitination in vitro. Ran in the cytosol was depleted (−Ran cyt) by incubating the cytosol with Ni-NTA-agarose prebound with His-tagged importin β-N603 at 4 °C for 2 h. Mock control was only incubated with Ni-NTA-agarose. His-tagged recombinant Ran is ∼1 kDa larger than endogenous Ran. B, quantification of NHK ubiquitination in A. The data represent the means ± S.E. of three independent experiments. The relative intensity of the polyubiquitin smear (Ubn-NHK) in the reaction with mock cytosol for 1 h (lane 6) was set to 1. C, effects of N-terminal truncation mutants of importin β on NHK ubiquitination in vitro. The diagram represents recombinant proteins used in this experiment. 1 μg of each recombinant protein was used as indicated. D, quantification of NHK ubiquitination in C. The data represent the means ± S.E. of three independent experiments. The relative intensity of polyubiquitin smear (Ubn-NHK) in reaction with control cytosol for 1 h (lane 8) was set to 1. Ran binding was quantified from supplemental Fig. S6A. The relative band intensity of Ran bound to WT importin β was set to 1. Imp, importin.
We next determined whether Ran cooperates with importin β to promote NHK ubiquitination through their interaction. Previous studies indicate that the N-terminal HEAT repeats of importin β are important for Ran binding (48, 61). Removal of the N-terminal 71 amino acids was reported to abolish the Ran binding activity of importin β (62). If Ran binding is important for importin β to act in ERAD, removal of the N-terminal sequence should affect the activity of importin β in ERAD. We tested three importin β truncation mutants ΔN32, ΔN128, and ΔN169 that were deleted of HEAT repeat 1, HEAT repeats 1–3, and HEAT repeats 1–4, respectively (Fig. 6C). IP results showed that the ΔN32 mutant bound to Ran comparably with WT importin β, whereas the ΔN128 and ΔN169 mutants could only bind weakly to Ran (supplemental Fig. S6A). Correlating with their Ran binding activity, the ΔN32 mutant promoted NHK ubiquitination similarly to WT importin β, whereas the ΔN128 and ΔN169 mutants displayed significantly reduced ubiquitination activity (Fig. 6, C and D). In addition, N603 that was sufficient to enhance NHK ubiquitination in vitro bound Ran efficiently, whereas the dominant negative inhibitor N297 had no Ran binding activity (supplemental Fig. S6B). These results suggest that the interaction between importin β and Ran is required to promote NHK ubiquitination in ERAD.
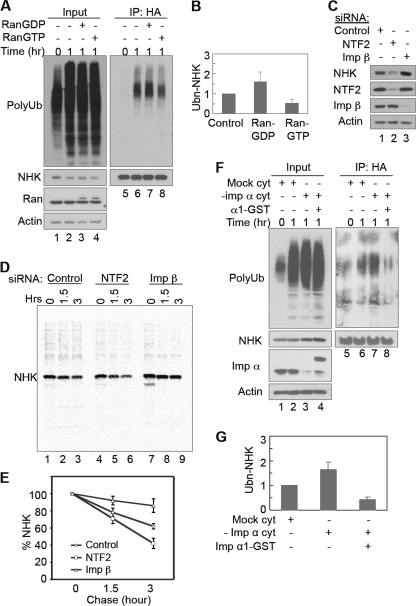
RanGDP but Not RanGTP Enhances ERAD of NHK
Ran exists in GTP- or GDP-bound form. RanGTP is mainly in the nucleus, whereas RanGDP is predominantly in the cytoplasm. To determine which form of Ran is the active species in ERAD, we first studied of the effects of Ran preloaded with GTP or GDP on NHK ubiquitination using the cell-free assay. The addition of RanGDP enhanced NHK ubiquitination, whereas the addition of RanGTP inhibited the ubiquitination (Fig. 7, A and B), suggesting that RanGDP but not RanGTP is required to promote NHK ubiquitination in vitro. We next examined how modulation of RanGDP in cells affects ERAD. It is known that cytoplasmic RanGDP is imported into the nucleus by NTF2 (46). Therefore, knockdown of NTF2 expression by RNAi is expected to slow down the nuclear import of RanGDP and result in an increase in the cytoplasmic level of RanGDP. We studied the effect of NTF2 knockdown on NHK degradation in cells. The steady-state level of NHK was significantly decreased in the cells transfected with NTF2 siRNAs (Fig. 7C). This decrease is due to the increased degradation of NHK, as was revealed by pulse-chase experiments (Fig. 7, D and E). These results support the notion that cytoplasmic RanGDP cooperates with importin β to facilitate NHK degradation.
FIGURE 7.
RanGDP but not RanGTP promotes ERAD of NHK. A, RanGDP promotes, whereas RanGTP inhibits, NHK ubiquitination in vitro. 1 μg of Ran preloaded with GDP or GTP was used in in vitro NHK ubiquitination as indicated. His-tagged recombinant Ran is ∼1 kDa larger than endogenous Ran. B, quantification of NHK ubiquitination in A. The data represent the means ± S.E. of three independent experiments. The relative intensity of polyubiquitin smear (Ubn-NHK) in reaction with control cytosol for 1 h (lane 6) was set to 1. C and D, HEK293 cells stably expressing NHK were transfected with siRNA targeting NTF2 or importin β as indicated. 48 h after transfection, the cells were processed for IB (C), or cells were pulse-labeled and chased for the indicated times before being processed for IP with anti-A1AT antibodies (D). E, quantification of NHK degradation in D. The data represent the means ± S.E. of three independent experiments. The relative band intensity of NHK at chase 0 h was set to 100%. F, importin α inhibits NHK ubiquitination in vitro. Importin α in the cytosol was depleted (−imp α cyt) by incubating the cytosol with Ni-NTA-agarose prebound with His-tagged importin β-ΔN169 at 4 °C for 2 h. Mock control was incubated with Ni-NTA-agarose only. 1 μg of importin α1-GST was used as indicated. G, quantification of NHK ubiquitination in F. The data represent the means ± S.E. of three independent experiments. The relative intensity of the polyubiquitin smear (Ubn-NHK) in the reaction with mock cytosol for 1 h (lane 6) was set to 1. Imp, importin.
It is generally believed that the affinity of RanGDP with importin β is much lower than that of RanGTP, leading to suggestions that the formation of a stable complex between RanGDP and importin β in the cytosol is unfavorable. However, interaction between importin β and RanGDP has been demonstrated in vitro and in the cytoplasm of cells (61–65). We have shown that importin β requires its Ran binding activity to promote ERAD (Fig. 6 and supplemental Fig. S6). To see whether the weak interaction between RanGDP and importin β is important to their function in ERAD, we studied the effect of importin α on NHK ubiquitination using the in vitro assay. The rationale behind this experiment is that importin α is able to dissociate RanGDP but not RanGTP or nucleotide-free Ran from importin β (65, 66). Accordingly, we predicted that an elevation of importin α levels should inhibit ERAD. As shown in Fig. 7 (F and G), cytosol depleted of importin α produced increased NHK ubiquitination compared with mock-depleted cytosol. Supplementation of the depleted cytosol with recombinant importin α1-GST protein reduced NHK ubiquitination. Taken together, our results suggest that RanGDP is likely to be the active species that cooperates with importin β to promote ubiquitination and degradation of NHK.
DISCUSSION
In this study, we demonstrate a surprising connection between ERAD and importin β, a well known regulator of nucleocytoplasmic transport. Our in vivo and in vitro studies support the conclusion that importin β is required for ubiquitination of the luminal soluble ERAD substrate NHK. We demonstrated that siRNA-mediated knockdown of importin β reduces NHK ubiquitination. This phenotype can be recapitulated in an in vitro assay in which importin β is biochemically depleted from the cytosol and readdition of recombinant importin β to importin β-depleted cytosol restores the ubiquitination of NHK. By contrast, ubiquitination of the membrane ERAD substrate CD3δ increased in importin β knockdown cells. Importin β knockdown stabilizes both NHK and CD3δ. Therefore, diminished NHK ubiquitination is not likely to be resulted from a defect in the ubiquitin conjugating/deconjugating systems associated with the ER membrane. Otherwise, both luminal and membrane substrates should be affected in a similar way. Importin β is likely to act in a step required for the degradation of both substrates but only required for the ubiquitination of NHK. As a luminal ERAD substrate, NHK must first be retrotranslocated to the cytosolic side of the ER for ubiquitination because all known E3s involved in ERAD have their catalytic domains located in the cytosolic side. Thus, the ubiquitination levels of an ER luminal substrate, such as NHK, can be a faithful indicator of retrotranslocation when proteasomal degradation is inhibited. On the other hand, the membrane-bound substrates that contain cytosolically exposed domains, such as CD3δ, may be ubiquitinated directly on their cytosolic residues without the need of retrotranslocation. We propose that importin β may have a function in NHK retrotranslocation. Importin β may act first to promote extrusion of NHK into the cytosol for ubiquitination, after which p97 binds to the ubiquitin conjugates on NHK and extracts it from the translocation site for subsequent proteasomal degradation. Our data showed that importin β does not interact with p97, suggesting that they may not need physical interaction to cooperate in ERAD. However, we cannot exclude the possibility that importin β and p97 have weak or transient interaction during ERAD. Our study reveals association of importin β with several ERAD components in a retrotranslocation complex, including VIMP, Derlin-1, gp78, and Hrd1. Among these proteins, at least VIMP binds importin β directly. These interactions may recruit importin β to the ER and enable the latter to directly participate in retrotranslocation. In support of this possibility, a mutant of importin β (N297) that inhibits importin β association with the ER-enriched microsomes also inhibits ERAD of NHK.
Structural studies show that in the absence of protein binding, importin β adopts an extended S-shaped superhelical architecture formed by HEAT repeats that consist of pairs of anti-parallel α-helices. The spring-like superhelical structure enables importin β to adapt its geometry to fit cargos of different sizes and shapes. When importin β binds to its cargo or the regulators Ran and importin α, it undergoes a drastic conformational change, which converts it to a compact spring-like form. This compact conformation stores energy, which is used for nuclear transport (67). By analogy, importin β might alter between the two different energy states at the site of retrotranslocation, which could provide the necessary energy for protein retrotranslocation. Our results suggest that importin β interacts with RanGDP to facilitate ERAD, whereas RanGTP and importin α inhibit ERAD. We speculate that RanGDP is easier to be dissociated from importin β because of its weak affinity to the latter, and hence importin β in complex with RanGDP is easier to switch from the compact conformation to the extended conformation and release energy. It is conceivable that the conformational change in importin β, if it occurs within a retrotranslocation complex, may generate a localized force to regulate the gating of a retrotranslocation channel. By contrast, the high affinity binding of RanGTP or importin α to importin β may keep importin β in the compact conformation, which results in a blockage of retrotranslocation.
Cytosolic localization of RanGDP and importin α may be pivotal to regulate the ERAD capacity. Previous studies reported that a significant amount of Ran was distributed to the cytoplasm from the nucleus in response to oxidative stress induced by H2O2 (68, 69). In contrast, H2O2-induced stress accumulates importin α, a potential inhibitor of ERAD, in the nucleus (68). Oxidative stress is known to cause overproduction of misfolded proteins in the ER. Therefore, the relocation of Ran and importin α under oxidative stress induced by H2O2 may represent a novel strategy to cope with cellular stress. Clearly, the unexpected interaction of importin β with ERAD machinery reveals a new layer of complexity for the regulation of ER quality control system in mammalian cells.
Supplementary Material
Acknowledgments
We thank Drs. Dirk Görlich, Richard Sifers, Karsten Weis, Reinhard Depping, Yun Qiu, Maria G. Masucci, and Wei Gu for providing plasmid constructs. We are grateful to Drs. Gong Li and Joe Kao for providing expert help in confocal microscopy.
This work was supported, in whole or in part, by National Institutes of Health Grants GM06696 (to S. F.) and GM066287 (to M. J. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6.
- ER
- endoplasmic reticulum
- ERAD
- ER-associated degradation
- A1AT
- α-1-antitrypsin
- NHK
- null Hong Kong variant of α-1-antitrypsin
- IP
- immunoprecipitation
- IB
- immunoblotting
- VIMP
- p97/valosin-containing protein-interacting membrane protein
- NTF2
- nuclear transport factor 2
- Ni-NTA
- nickel-nitrilotriacetic acid.
REFERENCES
- 1. Vembar S. S., Brodsky J. L. (2008) Nat. Rev. Mol. Cell Biol. 9, 944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hirsch C., Gauss R., Horn S. C., Neuber O., Sommer T. (2009) Nature 458, 453–460 [DOI] [PubMed] [Google Scholar]
- 3. Kim I., Xu W., Reed J. C. (2008) Nat. Rev. Drug Discov. 7, 1013–1030 [DOI] [PubMed] [Google Scholar]
- 4. Zhang K., Kaufman R. J. (2008) Nature 454, 455–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sato B. K., Schulz D., Do P. H., Hampton R. Y. (2009) Mol. Cell 34, 212–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tsai B., Ye Y., Rapoport T. A. (2002) Nat. Rev. Mol. Cell Biol. 3, 246–255 [DOI] [PubMed] [Google Scholar]
- 7. Hampton R. Y. (2002) Curr. Opin. Cell Biol. 14, 476–482 [DOI] [PubMed] [Google Scholar]
- 8. Hebert D. N., Bernasconi R., Molinari M. (2010) Semin. Cell Dev. Biol. 21, 526–532 [DOI] [PubMed] [Google Scholar]
- 9. Wang S., Ng D. T. (2008) Nat. Cell Biol. 10, 251–253 [DOI] [PubMed] [Google Scholar]
- 10. Carvalho P., Goder V., Rapoport T. A. (2006) Cell 126, 361–373 [DOI] [PubMed] [Google Scholar]
- 11. Denic V., Quan E. M., Weissman J. S. (2006) Cell 126, 349–359 [DOI] [PubMed] [Google Scholar]
- 12. Fang S., Ferrone M., Yang C., Jensen J. P., Tiwari S., Weissman A. M. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 14422–14427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nadav E., Shmueli A., Barr H., Gonen H., Ciechanover A., Reiss Y. (2003) Biochem. Biophys. Res. Commun. 303, 91–97 [DOI] [PubMed] [Google Scholar]
- 14. Amano T., Yamasaki S., Yagishita N., Tsuchimochi K., Shin H., Kawahara K., Aratani S., Fujita H., Zhang L., Ikeda R., Fujii R., Miura N., Komiya S., Nishioka K., Maruyama I., Fukamizu A., Nakajima T. (2003) Genes Dev. 17, 2436–2449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kikkert M., Doolman R., Dai M., Avner R., Hassink G., van Voorden S., Thanedar S., Roitelman J., Chau V., Wiertz E. (2004) J. Biol. Chem. 279, 3525–3534 [DOI] [PubMed] [Google Scholar]
- 16. Hassink G., Kikkert M., van Voorden S., Lee S. J., Spaapen R., van Laar T., Coleman C. S., Bartee E., Früh K., Chau V., Wiertz E. (2005) Biochem. J. 388, 647–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kostova Z., Tsai Y. C., Weissman A. M. (2007) Semin. Cell Dev. Biol. 18, 770–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mehnert M., Sommer T., Jarosch E. (2010) Bioessays 32, 905–913 [DOI] [PubMed] [Google Scholar]
- 19. Meacham G. C., Patterson C., Zhang W., Younger J. M., Cyr D. M. (2001) Nat. Cell Biol. 3, 100–105 [DOI] [PubMed] [Google Scholar]
- 20. Connell P., Ballinger C. A., Jiang J., Wu Y., Thompson L. J., Höhfeld J., Patterson C. (2001) Nat. Cell Biol. 3, 93–96 [DOI] [PubMed] [Google Scholar]
- 21. Imai Y., Soda M., Inoue H., Hattori N., Mizuno Y., Takahashi R. (2001) Cell 105, 891–902 [DOI] [PubMed] [Google Scholar]
- 22. Yoshida Y., Chiba T., Tokunaga F., Kawasaki H., Iwai K., Suzuki T., Ito Y., Matsuoka K., Yoshida M., Tanaka K., Tai T. (2002) Nature 418, 438–442 [DOI] [PubMed] [Google Scholar]
- 23. Bays N. W., Hampton R. Y. (2002) Curr. Biol. 12, R366–71 [DOI] [PubMed] [Google Scholar]
- 24. Lilley B. N., Ploegh H. L. (2004) Nature 429, 834–840 [DOI] [PubMed] [Google Scholar]
- 25. Ye Y., Shibata Y., Yun C., Ron D., Rapoport T. A. (2004) Nature 429, 841–847 [DOI] [PubMed] [Google Scholar]
- 26. Wahlman J., DeMartino G. N., Skach W. R., Bulleid N. J., Brodsky J. L., Johnson A. E. (2007) Cell 129, 943–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schelhaas M., Malmström J., Pelkmans L., Haugstetter J., Ellgaard L., Grünewald K., Helenius A. (2007) Cell 131, 516–529 [DOI] [PubMed] [Google Scholar]
- 28. Bernardi K. M., Forster M. L., Lencer W. I., Tsai B. (2008) Mol. Biol. Cell 19, 877–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Meusser B., Hirsch C., Jarosch E., Sommer T. (2005) Nat. Cell Biol. 7, 766–772 [DOI] [PubMed] [Google Scholar]
- 30. Zhong X., Shen Y., Ballar P., Apostolou A., Agami R., Fang S. (2004) J. Biol. Chem. 279, 45676–45684 [DOI] [PubMed] [Google Scholar]
- 31. Carvalho P., Stanley A. M., Rapoport T. A. (2010) Cell 143, 579–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ye Y., Meyer H. H., Rapoport T. A. (2003) J. Cell Biol. 162, 71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nakatsukasa K., Huyer G., Michaelis S., Brodsky J. L. (2008) Cell 132, 101–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Neuber O., Jarosch E., Volkwein C., Walter J., Sommer T. (2005) Nat. Cell Biol. 7, 993–998 [DOI] [PubMed] [Google Scholar]
- 35. Lim P. J., Danner R., Liang J., Doong H., Harman C., Srinivasan D., Rothenberg C., Wang H., Ye Y., Fang S., Monteiro M. J. (2009) J. Cell Biol. 187, 201–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mueller B., Klemm E. J., Spooner E., Claessen J. H., Ploegh H. L. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 12325–12330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ye Y., Shibata Y., Kikkert M., van Voorden S., Wiertz E., Rapoport T. A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 14132–14138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ballar P., Shen Y., Yang H., Fang S. (2006) J. Biol. Chem. 281, 35359–35368 [DOI] [PubMed] [Google Scholar]
- 39. Schulze A., Standera S., Buerger E., Kikkert M., van Voorden S., Wiertz E., Koning F., Kloetzel P. M., Seeger M. (2005) J. Mol. Biol. 354, 1021–1027 [DOI] [PubMed] [Google Scholar]
- 40. Park H., Suzuki T., Lennarz W. J. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 11163–11168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang Q., Li L., Ye Y. (2006) J. Cell Biol. 174, 963–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ernst R., Mueller B., Ploegh H. L., Schlieker C. (2009) Mol. Cell 36, 28–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ernst R., Claessen J. H., Mueller B., Sanyal S., Spooner E., van der Veen A. G., Kirak O., Schlieker C. D., Weihofen W. A., Ploegh H. L. (2011) PLoS Biol. 8, e1000605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Harel A., Forbes D. J. (2004) Mol. Cell 16, 319–330 [DOI] [PubMed] [Google Scholar]
- 45. Cook A., Bono F., Jinek M., Conti E. (2007) Annu. Rev. Biochem. 76, 647–671 [DOI] [PubMed] [Google Scholar]
- 46. Ribbeck K., Lipowsky G., Kent H. M., Stewart M., Görlich D. (1998) EMBO J. 17, 6587–6598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Görlich D., Seewald M. J., Ribbeck K. (2003) EMBO J. 22, 1088–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kutay U., Izaurralde E., Bischoff F. R., Mattaj I. W., Görlich D. (1997) EMBO J. 16, 1153–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nachury M. V., Maresca T. J., Salmon W. C., Waterman-Storer C. M., Heald R., Weis K. (2001) Cell 104, 95–106 [DOI] [PubMed] [Google Scholar]
- 50. Wu Y., Swulius M. T., Moremen K. W., Sifers R. N. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 8229–8234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li M., Brooks C. L., Wu-Baer F., Chen D., Baer R., Gu W. (2003) Science 302, 1972–1975 [DOI] [PubMed] [Google Scholar]
- 52. Dantuma N. P., Lindsten K., Glas R., Jellne M., Masucci M. G. (2000) Nat. Biotechnol 18, 538–543 [DOI] [PubMed] [Google Scholar]
- 53. Yang H., Zhong X., Ballar P., Luo S., Shen Y., Rubinsztein D. C., Monteiro M. J., Fang S. (2007) Exp. Cell Res. 313, 538–550 [DOI] [PubMed] [Google Scholar]
- 54. Fang S., Jensen J. P., Ludwig R. L., Vousden K. H., Weissman A. M. (2000) J. Biol. Chem. 275, 8945–8951 [DOI] [PubMed] [Google Scholar]
- 55. Melchior F., Guan T., Yokoyama N., Nishimoto T., Gerace L. (1995) J. Cell Biol. 131, 571–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Meyer H. H., Shorter J. G., Seemann J., Pappin D., Warren G. (2000) EMBO J. 19, 2181–2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Willingham A. T., Orth A. P., Batalov S., Peters E. C., Wen B. G., Aza-Blanc P., Hogenesch J. B., Schultz P. G. (2005) Science 309, 1570–1573 [DOI] [PubMed] [Google Scholar]
- 58. Ballar P., Zhong Y., Nagahama M., Tagaya M., Shen Y., Fang S. (2007) J. Biol. Chem. 282, 33908–33914 [DOI] [PubMed] [Google Scholar]
- 59. Garza R. M., Sato B. K., Hampton R. Y. (2009) J. Biol. Chem. 284, 14710–14722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ye Y., Meyer H. H., Rapoport T. A. (2001) Nature 414, 652–656 [DOI] [PubMed] [Google Scholar]
- 61. Lee S. J., Matsuura Y., Liu S. M., Stewart M. (2005) Nature 435, 693–696 [DOI] [PubMed] [Google Scholar]
- 62. Chi N. C., Adam E. J., Adam S. A. (1997) J. Biol. Chem. 272, 6818–6822 [DOI] [PubMed] [Google Scholar]
- 63. Chi N. C., Adam E. J., Visser G. D., Adam S. A. (1996) J. Cell Biol. 135, 559–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Plafker K., Macara I. G. (2002) J. Biol. Chem. 277, 30121–30127 [DOI] [PubMed] [Google Scholar]
- 65. Forwood J. K., Lonhienne T. G., Marfori M., Robin G., Meng W., Guncar G., Liu S. M., Stewart M., Carroll B. J., Kobe B. (2008) J. Mol. Biol. 383, 772–782 [DOI] [PubMed] [Google Scholar]
- 66. Lonhienne T. G., Forwood J. K., Marfori M., Robin G., Kobe B., Carroll B. J. (2009) J. Biol. Chem. 284, 22549–22558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zachariae U., Grubmüller H. (2008) Structure 16, 906–915 [DOI] [PubMed] [Google Scholar]
- 68. Miyamoto Y., Saiwaki T., Yamashita J., Yasuda Y., Kotera I., Shibata S., Shigeta M., Hiraoka Y., Haraguchi T., Yoneda Y. (2004) J. Cell Biol. 165, 617–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yasuda Y., Miyamoto Y., Saiwaki T., Yoneda Y. (2006) Exp. Cell Res. 312, 512–520 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.