Background: The ESCRT (endosomal sorting complex required for transport) machinery governs the formation of multivesicular endosomes.
Results: A complex formed by ESCRT-II and Vps20 directs ESCRT-III polymerization specifically to membranes of elevated curvature.
Conclusion: Curvature sensing by components of the ESCRT machinery spatially restricts the scission activity of ESCRT-III.
Significance: These findings highlight a new regulatory mechanism that controls ESCRT function.
Keywords: Atomic Force Microscopy, Endosomes, Membrane Fusion, Membrane Trafficking, Protein Structure, ESCRT, Membrane Curvature
Abstract
The scission of membranes necessary for vesicle biogenesis and cytokinesis is mediated by cytoplasmic proteins, which include members of the ESCRT (endosomal sorting complex required for transport) machinery. During the formation of intralumenal vesicles that bud into multivesicular endosomes, the ESCRT-II complex initiates polymerization of ESCRT-III subunits essential for membrane fission. However, mechanisms underlying the spatial and temporal regulation of this process remain unclear. Here, we show that purified ESCRT-II binds to the ESCRT-III subunit Vps20 on chemically defined membranes in a curvature-dependent manner. Using a combination of liposome co-flotation assays, fluorescence-based liposome interaction studies, and high-resolution atomic force microscopy, we found that the interaction between ESCRT-II and Vps20 decreases the affinity of ESCRT-II for flat lipid bilayers. We additionally demonstrate that ESCRT-II and Vps20 nucleate flexible filaments of Vps32 that polymerize specifically along highly curved membranes as a single string of monomers. Strikingly, Vps32 filaments are shown to modulate membrane dynamics in vitro, a prerequisite for membrane scission events in cells. We propose that a curvature-dependent assembly pathway provides the spatial regulation of ESCRT-III to fuse juxtaposed bilayers of elevated curvature.
Introduction
Formation of multivesicular endosomes (MVEs),3 which is mediated by a set of protein complexes collectively known as the ESCRT (endosomal sorting complex required for transport) machinery, is critical for the down-regulation of activated cell surface receptors and thereby exhibits properties of a tumor suppressor pathway (1, 2). Although endocytosis from the plasma membrane restricts receptors from further access to extracellular ligands, most activated receptors continue signaling from internal membrane compartments (3). However, sequestration of receptors within endosomes terminates their signaling potential by prohibiting access to cytoplasmic effector molecules. Unlike endocytosis, ESCRT-mediated vesicle biogenesis involves a budding event away from the cytoplasm. In a topologically similar fashion, components of the ESCRT machinery also function in the budding of several enveloped viruses from the plasma membrane and in cytokinesis (4, 5).
Early acting ESCRT complexes (ESCRT-0, -I, and -II) function in cargo recognition (6). Each complex contains one or more ubiquitin-interacting domains that can recruit and concentrate ubiquitinylated cargoes of the MVE pathway. Additionally, each complex exhibits membrane-binding activity. ESCRT-0 associates avidly with the endosomally enriched lipid phosphatidylinositol 3-phosphate (PI-3-P), and under steady-state conditions, the majority of ESCRT-0 localizes to endosomes (7). In contrast, ESCRT-I and ESCRT-II are predominantly cytoplasmic in most cell types and are transiently recruited onto the membrane during MVE biogenesis (8, 9). Recent studies have implicated both complexes in membrane deformation and the generation of nascent buds on giant unilamellar vesicles (GUVs). Addition of the ESCRT-III subunits Vps20 and Vps32 is sufficient to release nascent vesicles into the GUV lumen, indicating a specific role for ESCRT-III in membrane cleavage (10, 11). Surprisingly, in the absence of other ESCRT machinery, Vps32 and an activated form of Vps20 (lacking its regulatory C terminus) are also sufficient to generate lumenal vesicles in a GUV-based assay (11). Although high concentrations of Vps32 are necessary, these studies suggest that ESCRT-III may also function in vesicle formation. Consistent with this idea, overexpression of human Vps32 in mammalian cells results in the formation of tubules that extend away from the cytoplasm, and the yeast ESCRT-III subunits mediate the formation of inward invaginations on small unilamellar vesicles (12, 13).
The ESCRT-III subunits exist in an autoinhibited state in solution (14). ESCRT-III assembly requires Vps20 activation, which is mediated by ESCRT-II in the case of MVE biogenesis (15). Fluorescence spectroscopy measurements suggest that Vps20 undergoes a conformational change when bound to ESCRT-II on liposomes, which may foster an interaction between Vps20 and Vps32 that takes place only on membranes (13). Additionally, when mixed together, ESCRT-II and Vps20 associate with liposomes more tightly than either alone (16). Although this phenomenon has not been mechanistically explained, a molecular model predicts that ESCRT-II bound to Vps20 would exhibit a convex membrane-binding surface (17), which may explain their increased binding to the curved surfaces of liposomes. Here, we demonstrate that association of ESCRT-II with Vps20 generates a curvature-sensitive complex that is capable of nucleating two filaments of Vps32. Our data further indicate that Vps32 filaments assemble specifically on membranes of high curvature and enable membrane remodeling necessary for scission. By sensing membrane curvature, ESCRT-III function is both spatially and temporally restricted to fission events during vesicle biogenesis and cytokinesis.
EXPERIMENTAL PROCEDURES
Protein Purification and Hydrodynamic Studies
Recombinant protein expression was performed using Escherichia coli BL21(DE3) cells. For ESCRT-II, all subunits were cloned into the polycistronic expression vector pST39 (18), and a single tag was appended onto Vps25 to enable purification (described briefly below). Purifications were conducted using glutathione-agarose beads (for GST-Vps20) or nickel affinity resin (for intact ESCRT-II and monomeric Vps32). The GST moiety was removed from Vps20 using PreScission protease. For size exclusion chromatography, samples (2 ml) were applied to a Superose 6 gel filtration column (GE Healthcare), and 1-ml fractions were collected. The Stokes radius of each protein or protein complex was calculated from its elution volume based on the elution profiles of characterized globular standards (19). 4-ml glycerol gradients (10–30%) were poured using a Gradient Master and fractionated (200 μl) from the top by hand. Sedimentation values were calculated by comparing the position of the peak with that of characterized standards run on a separate gradient in parallel (20). To fluorescently label Vps20, a 20-fold molar excess of BODIPY-FL-labeled maleimide was incubated with Vps20 overnight with rotation. The reaction was quenched using an excess of glutathione. Unbound dye was removed by gel filtration chromatography. Based on the amount of protein recovered and its absorbance, stoichiometry (BODIPY-FL:Vps20) was determined to be ∼1:1.
Production of Liposomes and Co-flotation Assays
Liposomes (36.5% phosphatidylcholine, 30% phosphatidylethanolamine (PE), 30% phosphatidylserine, 3% PI-3-P, and 0.5% rhodamine-labeled PE) were prepared by extrusion through polycarbonate filters with pore sizes of 30 and 200 nm (Avanti Polar Lipids). Dynamic light scattering measurements were conducted to determine the actual size of liposomes generated. For co-flotation assays, liposomes were incubated with protein in buffer (50 mm Hepes (pH 7.6) and 100 mm NaCl) prior to mixing with Accudenz density medium. Mixtures were overlaid with decreasing concentrations of Accudenz (0–40%) and centrifuged for 2 h at 280,000 × g. During this period, liposomes and associated proteins floated to the buffer/Accudenz interface and were harvested by hand. Recovery of liposomes was normalized based on the fluorescence intensity of the sample, and equivalent fractions (when comparing flotation experiments that used liposomes of differing sizes) were separated by SDS-PAGE and stained with Coomassie Blue to determine the relative amount of protein that bound (21). Similar results were obtained with liposomes composed of 51.5% phosphatidylcholine, 30% PE, 15% phosphatidylserine, 3% PI-3-P, and 0.5% rhodamine-PE.
Fluorescence Confocal Microscopy and Analysis
Fluorescent images of BODIPY-FL-Vps20 and rhodamine-labeled liposomes were acquired on a swept-field confocal microscope (Nikon Ti-E) equipped with a Roper CoolSNAP HQ2 CCD camera using a Nikon ×60, 1.4 numerical aperture Plan Apo oil objective lens. Acquisition parameters were controlled by Nikon Elements software. To image immobilized liposomes, coverslips were first cleaned and coated with a solution of PEG and biotin-PEG (40:1 ratio). The PEG-coated glass was then incubated with 1 μm avidin for 10 min and washed several times with buffer (50 mm Hepes (pH 7.6) and 100 mm NaCl). 100 μl of liposomes (15 mm; 35.9% phosphatidylcholine, 30% PE, 30% phosphatidylserine, 3% PI-3-P, 1% biotinyl-PE, and 0.1% rhodamine-PE) was mixed with ESCRT-II (250 nm) and BODIPY-FL-Vps20 (250 nm) and incubated on avidin-coated coverslips for 30 min. Unbound liposomes and protein were aspirated and replaced with 3 μl of 50 mm Hepes (pH 7.6) and 100 mm NaCl prior to inversion onto a depression slide for imaging. Image analysis was conducted using MetaMorph software. Based on the plausible assumptions that the fluorescence intensity of each liposome is proportional to its surface area and that the ratio of BODIPY-FL to rhodamine fluorescence is proportional to the concentration of labeled Vps20 bound to each liposome, we were able to determine the relationship between Vps20 membrane binding and membrane curvature in the presence of ESCRT-II. Specifically, we plotted the fluorescence intensity of each liposome (following background subtraction) on the x axis and the relative concentration of Vps20 that bound to each liposome on the y axis and determined the best fit curve to be a power series described by y = kx−0.993. Because the surface area of a liposome is proportional to the square of its radius, [Vps20] bound ∝ 1/radius2. Furthermore, because the curvature of a membrane is defined as the inverse of its radius, [Vps20] bound ∝ curvature2.
Atomic Force Microscopy (AFM)
AFM imaging was performed using a Veeco Digital Instruments Multimode instrument controlled by a Nanoscope IIIa controller. All imaging was conducted under fluid using NSC-18 cantilevers with a chromium-gold back-side coating (MikroMasch). Their resonant frequencies under fluid were 30–35 kHz, and the actual scanning frequencies were ∼5% below the maximal resonance peak. The root mean square voltage was maintained at 2 V. Lipid mixtures containing phosphatidylcholine (54%), PE (30%), phosphatidylserine (15%), and PI-3-P (1%) were dried under nitrogen and hydrated in Biotechnology Performance Certified (BPC) water overnight. Suspensions were probe-sonicated at an amplitude of 10 μA until the mixture became transparent. Liposomes were incubated in the presence or absence of proteins for 30 min and then placed on freshly cleaved mica. In each case, 40 μl of the liposome mixture and an equal volume of Hepes-buffered saline (50 mm Hepes (pH 7.6), 150 mm NaCl, and 1 mm Ca2+) were applied to the mica surface. The mica was washed twice with Hepes-buffered saline and placed in the fluid cell of the atomic force microscope. The assembled lipid bilayer was immersed in 150 μl of Hepes-buffered saline, and all imaging was performed at room temperature. AFM images were plane-fitted to remove tilt, and each scan line was fitted to a first-order equation. Particles were identified and their dimensions were measured manually using the section tool in the Nanoscope software. The height and radius of each particle were used to calculate its molecular volume using the following equation: Vm = (πh/6)(3r2 + h2), where h is the particle height, and r is the radius (20, 22). Each particle was measured twice in both dimensions, and an average was taken for the calculation. Widths and heights of filaments were determined by taking cross-sections at three points along the filaments and taking the average. Exposed mica surface area measurements were conducted using the Scanning Probe Image Processor software SPIP. Exposed mica was detected using the “pore detection” function to isolate areas with heights below a specified threshold in relation to the bilayer height. The surface areas of these regions were summed, and percent changes over time were calculated relative to the maximum area of exposed mica in each independent experiment.
RESULTS
A Complex of ESCRT-II Bound to Vps20 Binds Preferentially to Membranes of High Curvature
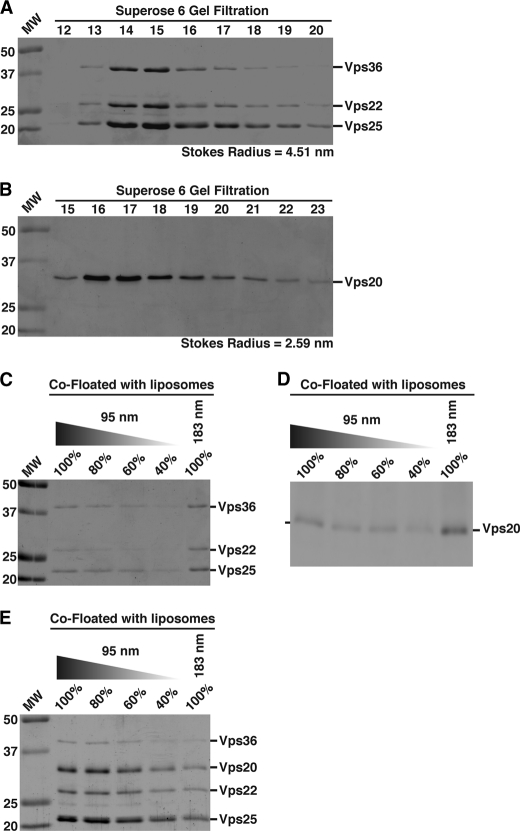
To study the role of membrane curvature in the regulation of ESCRT assembly, we first examined the binding properties of ESCRT-II and Vps20 to synthetic liposomes of two different diameters (supplemental Fig. S1A). We used recombinant Caenorhabditis elegans proteins due to their robust expression in E. coli and high degree of purity following affinity and size exclusion chromatography (Fig. 1, A and B). The hydrodynamic properties of C. elegans ESCRT-II and Vps20 were nearly identical to those of human ESCRT-II and CHMP6, respectively, consistent with their high level of amino acid sequence similarity (Fig. 1, A and B, and supplemental Fig. S1, B and C) (23). Using an assay in which proteins are mixed with liposomes and then floated through a gradient (supplemental Fig. S1D), we found that ESCRT-II and Vps20 did not individually exhibit a preference for 183-nm vesicles compared with 95-nm vesicles (Fig. 1, C and D). In contrast, a mixture of ESCRT-II and Vps20 bound significantly more tightly to the smaller, more highly curved vesicles (by ∼2-fold) (Fig. 1E), suggesting that the assembled ESCRT-II·Vps20 “supercomplex” senses elevated membrane curvature.
FIGURE 1.
ESCRT-II bound to Vps20 binds preferentially to liposomes of higher curvature. A, ESCRT-II initially purified using nickel affinity chromatography was subjected to size exclusion chromatography, and the peak fractions eluted were separated by SDS-PAGE and stained using Coomassie Blue. A Stokes radius was calculated based on the elution profile of characterized standards. Data shown are representative of at least three independent experiments. B, Vps20 was purified initially as a GST fusion protein on glutathione-agarose. The GST moiety was removed using PreScission protease, and untagged Vps20 was analyzed as described for A. Data shown are representative of at least three independent experiments. C–E, a co-flotation assay was used to analyze the binding of ESCRT-II, Vps20, or a mixture of ESCRT-II and Vps20 to liposomes of different diameters (95 and 183 nm). Fractions were resolved by SDS-PAGE and stained using Coomassie Blue (C and E) or immunoblotted using anti-Vps20 antibodies (D). A dilution series of each protein or protein mixture that co-floated with 95-nm liposomes was loaded to quantify the relative amount that co-floated with 183-nm vesicles. Data shown are representative of at least three independent experiments. On the basis of densitometry measurements performed on each Coomassie Blue-stained band, we found that 101 ± 17% of the ESCRT-II complex co-floated with 183-nm vesicles relative to 95-nm vesicles. For Vps20, 108 ± 13% of the protein co-floated with 183-nm vesicles relative to 95-nm vesicles. For a mixture of ESCRT-II and Vps20, 45 ± 13.1% of the complex co-floated with 183-nm vesicles compared with 95-nm vesicles.
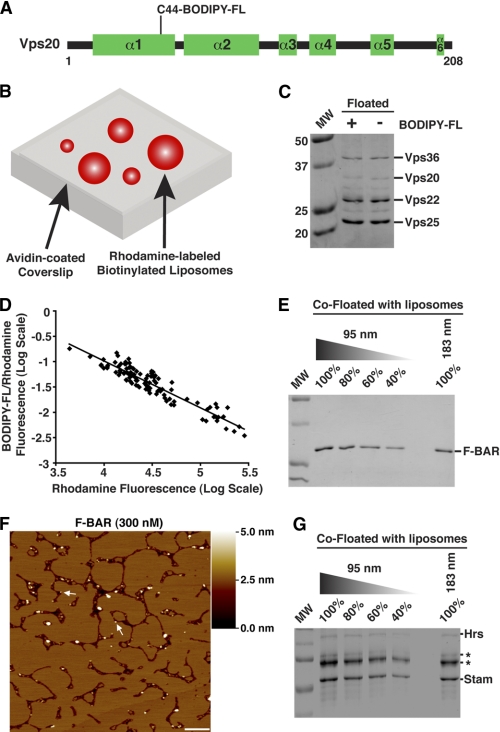
To confirm a curvature-sensitive association between membranes and ESCRT-II·Vps20, we used a fluorescence-based assay. We labeled the only endogenous cysteine residue in Vps20 with BODIPY-FL and measured its binding to liposomes of various sizes that contained rhodamine-PE in the presence of unlabeled ESCRT-II (Fig. 2, A and B, and supplemental Fig. S2A). The BODIPY-FL modification did not affect the interaction between Vps20 and ESCRT-II (Fig. 2C). Ratiometric confocal imaging showed a reciprocal relationship between the intensities of the fluorophores (Fig. 2D), indicating that ESCRT-II·Vps20 binds more avidly to smaller liposomes of higher curvature. The best fit curve through the data was essentially described by a power series in the form of y ∝ x−1, demonstrating that membrane binding of ESCRT-II·Vps20 varied as the multiplicative inverse of the liposome surface area (supplemental Fig. S2B; see “Experimental Procedures” for additional details). Thus, ESCRT-II·Vps20 membrane binding is proportional to the square of the membrane curvature. On the basis of these findings, we conclude that ESCRT-II bound to Vps20 senses the curvature of lipid bilayers. In contrast, we were unable to detect the association of BODIPY-FL-Vps20 with liposomes in the absence of ESCRT-II (supplemental Fig. S2C), consistent with a necessity to perform immunoblot analysis to detect Vps20 in co-flotation assays (see Fig. 1D).
FIGURE 2.
Complex of ESCRT-II bound to Vps20 senses membrane curvature. A, schematic illustrating the secondary structure organization of Vps20, highlighting the position of the BODIPY-FL modification. α-Helical domains are predicted based on sequence alignment with CHMP6. B, schematic representation of a fluorescence-based liposome binding assay. C, a co-flotation assay was used to analyze the association of ESCRT-II with either unlabeled Vps20 or Vps20 conjugated to BODIPY-FL. Labeled Vps20 exhibited a slightly reduced mobility during SDS-PAGE compared with unlabeled Vps20. D, the log of the BODIPY-FL:rhodamine fluorescence ratio for individual liposomes was plotted against the log of liposome fluorescence intensity. More than 200 immobilized liposomes were analyzed in three independent experiments. E, the FCHo F-BAR domain was analyzed using the co-flotation assay described in the legend to Fig. 1. Data shown are representative of at least three independent experiments. Based on densitometry measurements, 80 ± 3.5% of the FCHo F-BAR domain co-floated with 183-nm vesicles relative to 95-nm vesicles. F, representative AFM images of bilayers assembled in the presence of the FCHo F-BAR domain (300 nm). Arrows highlight the presence of F-BAR particles at the bilayer edges. Based on hydrodynamic studies, the F-BAR domain forms a homodimer in solution. A shade-height scale bar is shown to the right. White scale bar = 500 nm. G, the ESCRT-0 complex was analyzed using the co-flotation assay. Because Hrs stability during flotation was compromised (asterisks highlight breakdown products of Hrs based on immunoblot analysis, which co-migrated with bacterial heat shock proteins that co-purified with recombinant ESCRT-0 during SDS-PAGE), the intensity of STAM was used to determine that ESCRT-0 binds to liposomes in a curvature-independent fashion. Data shown are representative of at least three independent experiments. Based on densitometry measurements, 95 ± 13% of ESCRT-0 co-floated with 183-nm vesicles relative to 95-nm vesicles.
Vps20 Regulates ESCRT-II Distribution on Supported Lipid Bilayers
Methodology to visualize ESCRT complex assembly in vitro is currently limited. Although the development of a fluorescence microscopy-based assay using GUVs has been instrumental in defining distinct roles for each ESCRT complex, the relatively low resolution of the light microscope prohibits the acquisition of nanometer scale structural information regarding ESCRT complex assembly on the membrane surface (10, 11). Moreover, we have found GUVs to be highly dynamic, capable of undergoing spontaneous membrane deformations, including lumenal vesicle formation, in a protein-independent manner (supplemental Movies S1 and S2). Although the addition of ESCRT proteins increases the frequency at which lumenal vesicles form within GUVs, these caveats have prevented us from reproducibly interpreting data acquired using this assay. To study the assembly of ESCRT components on membranes, we took advantage of an alternative approach that uses AFM. With this method, components of the ESCRT machinery can be visualized at nanometer resolution on supported lipid bilayers (SLBs) that exhibit a thickness of ∼4 nm using label-free recombinant proteins. During assembly of SLBs, we observed that several gaps formed throughout the surface, where the underlying mica could be visualized (supplemental Fig. S2D). Based on molecular dynamics simulations, the edges of SLBs are predicted to be highly curved surfaces (supplemental Fig. S2E) (24) with a radius (∼2 nm) comparable with that of a constricted vesicle bud neck (∼10–15 nm) (25–29). Although direct experimental data to describe the bilayer edge are lacking, the energy cost to maintain exposed hydrocarbon tails in an aqueous solution greatly exceeds the energy necessary for migration of polar headgroups around a bilayer edge (30–34). Thus, the use of SLBs afforded us the unique opportunity to study the association of ESCRT components with both flat and highly curved membrane surfaces simultaneously.
To validate the system, we examined the well characterized, curvature-sensitive F-BAR domain derived from FCHo (35) on SLBs. Although particles corresponding to the F-BAR domain varied in size, the majority (89.9% of particles within the volume range of a F-BAR homodimer) associated with edges of SLBs, consistent with its elevated affinity for curved membranes (Fig. 2, E and F). In contrast, the ESCRT-0 complex, composed of Hrs and STAM, is curvature-insensitive (Fig. 2G) and exhibits a uniform distribution throughout the bilayer (20). Together, these data illustrate the utility of our SLB system in characterizing the membrane-binding properties of proteins.
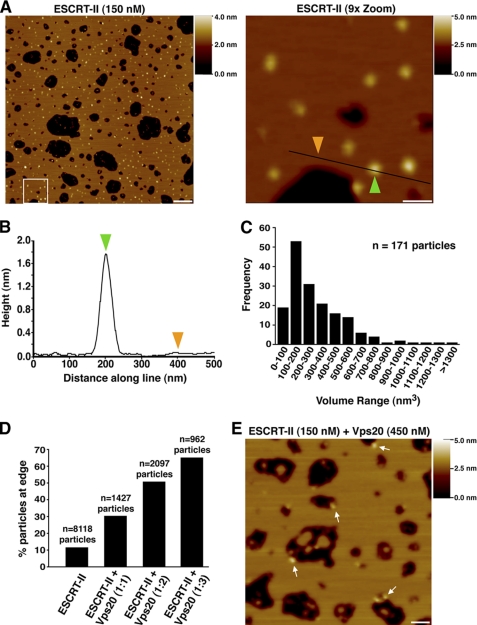
ESCRT-II bound to membranes as individual particles that were distributed evenly across the bilayer (Fig. 3A). Analysis of the particles produced a volume distribution with a peak in the region of 100–200 nm3, similar to that expected of a single ESCRT-II complex (140 nm3) based on amino acid composition (Fig. 3, B and C). We conclude that ESCRT-II on membranes behaves as an individual heterotetrameric complex that lacks curvature sensitivity.
FIGURE 3.
Vps20 targets ESCRT-II to membranes of high curvature. A, left panel, representative AFM image of a bilayer assembled in the presence of ESCRT-II (150 nm). Right panel, 9-fold magnification of the boxed region in the left panel. A shade-height scale bar is shown to the right of each panel. White scale bar = 500 nm (left panel) and 100 nm (right panel). B, analysis of the height distribution along the line drawn in the right panel in A. Colored arrowheads highlight the heights at two positions along the line as shown in A. C, frequency distribution of molecular volumes for the ESCRT-II complex bound to the bilayer surface. The total number of particles analyzed is indicated. D, bar graph showing the percentage of particles that appeared at the edges of bilayers following the addition of various amounts of Vps20 to ESCRT-II. The total number of particles analyzed is indicated above each bar. E, representative AFM image of a bilayer with an identical composition as described for A assembled in the presence of ESCRT-II (150 nm) and Vps20 (450 nm). Arrows highlight the presence of ESCRT-II·Vps20 particles at the bilayer edges. A shade-height scale bar is shown to the right. White scale bar = 125 nm.
When ESCRT-II and Vps20 were co-incubated with liposomes at a 1:1 molar ratio and analyzed by AFM, the total number of particles observed on the membrane was decreased by ∼4-fold compared with ESCRT-II alone (supplemental Fig. S2F). Specifically, we observed a decrease in particle association with flat regions of the SLB. These data suggested that a complex of ESCRT-II bound to Vps20 binds less efficiently to flat lipid bilayers. Additionally, the particles were no longer distributed evenly across the membrane. Instead, ∼30% were concentrated at the periphery of the gaps that had formed during bilayer assembly (Fig. 3D). Because Vps20 can associate with both copies of Vps25 present in ESCRT-II (15), we increased the molar ratio of ESCRT-II to Vps20 to 1:2. Under these conditions, we observed a further shift in particle distribution, with ∼50% at the edges of gaps (Fig. 3D). Although binding of Vps20 to ESCRT-II does not alter the volume of particles observed by AFM sufficiently to allow us to detect this interaction, we suspected that the remaining particles that bound throughout the bilayer were ESCRT-II complexes that were not bound to Vps20. We therefore added a 3-fold molar excess of Vps20 to increase the likelihood of ESCRT-II·Vps20 supercomplex formation. Under these conditions, we found that a clear majority (∼65%) of particles targeted to the highly curved edges of SLBs (Fig. 3, D and E). Similar results were obtained using SLBs that lacked PI-3-P (supplemental Fig. S2G), indicating that recruitment of ESCRT-II·Vps20 to bilayer edges is independent of phosphoinositides. We conclude that the association of ESCRT-II with Vps20 dramatically increases the relative affinity of the proteins for highly curved surfaces, providing a mechanism for spatially restricting Vps20-mediated ESCRT-III assembly to the neck of nascent, inward-budding vesicles.
The ESCRT-II·Vps20 Complex Directs ESCRT-III Filament Assembly Specifically on Highly Curved Membranes
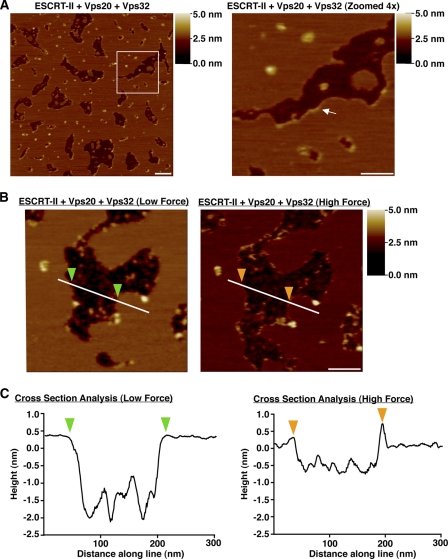
To directly examine ESCRT-III polymer formation, we first purified recombinant Vps32, an ESCRT-III subunit known to function downstream of Vps20. Based on gel filtration and glycerol gradient studies, Vps32 exists in a monomeric conformation in solution, exhibiting nearly identical hydrodynamic properties to carbonic anhydrase, a well characterized globular monomer (supplemental Fig. S3, A and B). In contrast to Vps20, which failed to detectably associate with SLBs (supplemental Fig. S3C), Vps32 bound to membranes as individual particles that were scattered throughout the bilayer (supplemental Fig. S3D). However, when Vps32 was co-incubated with ESCRT-II and Vps20, we observed the formation of flexible filaments that lined the edges of the bilayer gaps (Fig. 4A). The size and shape of the gap made no significant impact on filament assembly, nor did the addition of Vps24 and Vps2 (data not shown). To further define how the Vps32 filaments associate with the bilayer, we modulated the force applied by the AFM tip (supplemental Fig. S3E). With force that was sufficient to compress or penetrate the bilayer, we were able to clearly visualize the filaments bordering the gap. However, when the force exerted by the AFM tip was lowered, filaments were no longer visible (Fig. 4, B and C), suggesting that the filament height was equal to or below the surface of the bilayer. On the basis of these findings, we conclude that Vps32 filaments bind specifically to the edges of highly curved membranes. In agreement with this idea, we failed to observe cases in which ESCRT-III filaments extended onto the flat surface of the SLB, suggesting that similar to ESCRT-II·Vps20, Vps32 filaments are also curvature-sensitive during polymerization.
FIGURE 4.
ESCRT-II and Vps20 nucleate a strand of Vps32 monomers that bind specifically to highly curved bilayers. A, left panel, representative AFM images of a bilayer assembled in the presence of ESCRT-II (100 nm), Vps20 (150 nm), and Vps32 (450 nm). Right panel, 4-fold magnification of the boxed region in the left panel. The arrow highlights the presence of a filament along the bilayer edge. A shade-height scale bar is shown to the right of each panel. White scale bar = 200 nm (left panel) and 100 nm (right panel). B, representative AFM images of an identical region of a bilayer formed in the presence of ESCRT-II (100 nm), Vps20 (150 nm), and Vps32 (450 nm) following the application of low force (left panel) and high force (right panel) with the AFM tip. A shade-height scale bar is shown to the right. White scale bar = 100 nm. C, the height relative to the bilayer is plotted along the lines shown in B. Color-coded arrowheads highlight the bilayer edges.
The Vps32 filaments often emerged from one or both sides of a central particle of ESCRT-II·Vps20 (supplemental Fig. S3, F and G). Using the height and width of each filament, we calculated the volume of a spherical monomeric subunit to range from 20 to 40 nm3, similar to the volume of a single Vps32 molecule (30 nm3) based on its amino acid composition (supplemental Fig. S3H). Furthermore, on the basis of the hydrodynamic properties of Vps32 (supplemental Fig. S3, A and B) and volume calculations using AFM, we determined that each 100-nm filament (in length) contains approximately eight subunits of Vps32 arranged in an end-to-end configuration and is on average 13.4 ± 2.5 nm wide, similar to the diameter of endogenous ESCRT-III filaments that assemble during abscission (36).
ESCRT-III Promotes Membrane Remodeling
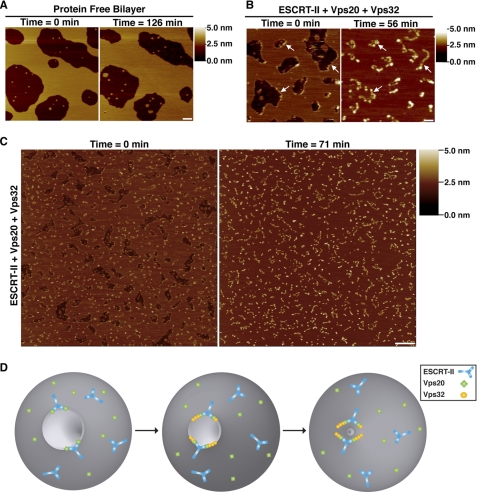
Imaging of SLBs in the absence of protein by AFM revealed that some gaps filled with membrane over time (Fig. 5A), indicating that the bilayer is mobile on the mica surface. We took advantage of this phenomenon to study the effect of Vps32 filament assembly on membrane dynamics and found that membrane closure occurred more rapidly and uniformly in the presence of ESCRT proteins (Fig. 5, A–C, and supplemental Fig. S4). This process is unlikely to depend on Vps32 molecules that fail to incorporate into filaments, as monomers of Vps32 did not concentrate at the gap edges (supplemental Fig. S3D). In some cases, the ESCRT-III filaments remained relatively immobile during the closure process (Fig. 5B). However, such occurrences were in the minority, as indicated by the overall redistribution of filaments following membrane sealing (Fig. 5C). Strikingly, in the majority of cases, we found that Vps32 filaments remained associated with the bilayer surface after membrane closure (Fig. 5, B and C). These data indicate that the filaments permit the passive movement of membrane but cannot disassemble spontaneously, consistent with previous work demonstrating that ESCRT-III removal from membranes requires an energy-dependent step that involves the Vps4 ATPase (37). Together, these data demonstrate that individual ESCRT-III filaments are sufficient to drive the remodeling of lipid bilayers.
FIGURE 5.
ESCRT-III filaments alter the dynamics of highly curved membranes. A, representative AFM images of protein-free bilayers taken 126 min apart. A shade-height scale bar is shown to the right. White scale bar = 200 nm. B, representative AFM images of bilayers assembled in the presence of ESCRT-II (150 nm), Vps20 (150 nm), and Vps32 (450 nm) taken 56 min apart. Arrows highlight the presence of the same filaments at each time point. A shade-height scale bar is shown to the right. White scale bar = 200 nm. C, representative AFM images of a bilayer formed in the presence of ESCRT-II (100 nm), Vps20 (150 nm), and Vps32 (450 nm) taken 71 min apart. A shade-height scale bar is shown to the right. White scale bar = 500 nm. D, diagram illustrating a model for ESCRT-III-mediated membrane scission. The association of ESCRT-II and Vps20 (step 1) generates a curvature-sensitive protein assembly that is capable of nucleating strings of Vps32 monomers (step 2) that bind to the highly curved surface of a vesicle bud neck. The binding energy between polymerized ESCRT-III and the membrane is sufficient to overcome the energy barrier to membrane fusion (step 3). Also shown on the membrane surface is the presence of free ESCRT-II and Vps20, which do not exhibit enhanced binding to regions of elevated curvature. Notably, Vps20 is myristoylated in vivo, suggesting that it may bind constitutively to the endosomal membrane as depicted in the model (6).
DISCUSSION
Viral budding, cytokinesis, and lumenal vesicle formation within endosomes all share a common requirement for ESCRT-mediated membrane fission, which necessitates a significant input of free energy. This barrier is overcome by electrostatic interactions between components of the ESCRT machinery and the membrane (38). ESCRT-III subunits are ideally suited to mediate membrane fission, as each component harbors an electrically polarized core that contains N-terminal basic residues that bind strongly to acidic phospholipids. In contrast, upstream ESCRT complexes are unlikely to participate in the scission process but instead function as adaptors to target ESCRT-III to various cellular locations. In the case of HIV-1 infection, virally encoded Gag binds two upstream ESCRT factors, Tsg101 and Alix, to drive ESCRT-III polymerization on the plasma membrane (39). Although HIV-1 bud formation is ESCRT-independent, bud release strictly requires ESCRT function. Similarly during cytokinesis, the midbody-associated protein Cep55 recruits Tsg101 and Alix to allow the assembly of ESCRT-III on the intracellular bridge (5, 40).
The ESCRT Complexes Possess Distinct Activities during MVE Biogenesis
ESCRT-III activity enables membrane scission and daughter cell separation, but initial steps of cleavage furrow formation do not require the ESCRT machinery. At the endosome, ESCRT-0, -I, and -II cooperate to properly localize ESCRT-III to act at the final scission step of lumenal vesicle formation (1, 6). However, the formation of nascent buds does not appear to require ESCRT-III (10). Together, these findings strongly support a model in which ESCRT-III functions after the membrane-bending steps that occur prior to fission. Consistent with this idea, we found that ESCRT-III polymerizes specifically on highly curved membranes. By sensing curvature, ESCRT-III function is spatially restricted to unique areas of a lipid bilayer and may thereby prevent nonspecific ESCRT-mediated membrane remodeling. Importantly, we have also demonstrated that ESCRT-II·Vps20 membrane binding is proportional to the square of the curvature, which is directly related to the bending energy of a membrane. Thus, as membrane bending increases, so does the membrane binding of ESCRT-II·Vps20, which may ultimately trigger a conformational switch enabling Vps32 filament assembly. On the basis of our data, we speculate that Vps20 associates with ESCRT-II to generate a mechanosensitive complex, which specifically nucleates ESCRT-III polymerization subsequent to membrane-bending events (Fig. 5D).
Requirements for ESCRT-mediated Membrane Scission
Several models that share common features currently exist to explain ESCRT-mediated membrane scission. The “purse-string” model suggests that the AAA ATPase Vps4 mediates disassembly of ESCRT-III circular arrays to draw opposing membranes together and allow fission (13). Although Vps4 is required for ESCRT-III disassembly and recycling, recent evidence indicates that the ATPase is dispensable for vesicle budding (10, 11, 41). Consistent with this idea, we found that ESCRT-III disassembly is not required to alter membrane dynamics, which would be necessary during fission. Alternative models suggest that ESCRT-III spirals or dome-like structures are sufficient to draw opposing membranes together (11, 12, 38, 41). Although our data do not preclude the possibility that such polymers are actually generated in vivo, we found no evidence for their formation using purified recombinant proteins. A limitation of our study is the use of SLBs assembled on a mica surface, which may not be conducive to the formation of Vps32 flat spirals. Nevertheless, our data indicate that the binding of single ESCRT-III filaments to membranes can generate sufficient energy to modulate membrane dynamics. In particular, we found that the association of a Vps32 polymer with the edge of a lipid bilayer promoted its movement on an artificial surface and resulted in membrane sealing. Although additional studies are required to decipher the mechanisms by which ESCRT-III mediates SLB gap closure, this process may be akin to fission events, which similarly involve the reorganization and movement of lipid bilayers to ultimately seal a membrane. In the future, it will be essential to establish tractable assays in vivo, which permit high-resolution analysis of normal ESCRT assembly on endosomes, to validate our findings and those obtained from other artificial systems that are currently in use.
On the basis of our data examining ESCRT-III assembly on a lipid bilayer that closely resembles a constricting bud neck, we offer a new model for ESCRT-mediated fission (Fig. 5D). We propose that ESCRT-III filaments assemble specifically on highly curved membrane surfaces, contributing sufficient binding energy to promote membrane fission. Such a model is consistent with the wide range of ESCRT-mediated fission events, including the release of nascent vesicles (∼40–50 nm in diameter) and sealing of the relatively large intracellular bridge during cytokinesis (∼200 nm in diameter).
Supplementary Material
Acknowledgments
We thank E. Chapman, M. Jackson, C. Johnson, and E. Hui for advice and helpful discussions; R. Murphy for use of dynamic light scattering instrumentation; N. George for technical assistance; and members of the Audhya laboratory for critically reading this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant 1R01GM088151-01A1 (to A. A.). This work was also supported by a Shaw Scientist award (Greater Milwaukee Foundation; to A. A.) and a Biotechnology and Biological Sciences Research Council doctoral training award (to I. F.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and Movies S1 and S2.
- MVE
- multivesicular endosome
- PI-3-P
- phosphatidylinositol 3-phosphate
- GUV
- giant unilamellar vesicle
- PE
- phosphatidylethanolamine
- AFM
- atomic force microscopy
- SLB
- supported lipid bilayer.
REFERENCES
- 1. Raiborg C., Stenmark H. (2009) Nature 458, 445–452 [DOI] [PubMed] [Google Scholar]
- 2. Saksena S., Emr S. D. (2009) Biochem. Soc. Trans. 37, 167–172 [DOI] [PubMed] [Google Scholar]
- 3. Sorkin A., Von Zastrow M. (2002) Nat. Rev. Mol. Cell Biol. 3, 600–614 [DOI] [PubMed] [Google Scholar]
- 4. Garrus J. E., von Schwedler U. K., Pornillos O. W., Morham S. G., Zavitz K. H., Wang H. E., Wettstein D. A., Stray K. M., Côté M., Rich R. L., Myszka D. G., Sundquist W. I. (2001) Cell 107, 55–65 [DOI] [PubMed] [Google Scholar]
- 5. Carlton J. G., Martin-Serrano J. (2007) Science 316, 1908–1912 [DOI] [PubMed] [Google Scholar]
- 6. Hurley J. H., Emr S. D. (2006) Annu. Rev. Biophys. Biomol. Struct. 35, 277–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gaullier J. M., Simonsen A., D'Arrigo A., Bremnes B., Stenmark H., Aasland R. (1998) Nature 394, 432–433 [DOI] [PubMed] [Google Scholar]
- 8. Xie W., Li L., Cohen S. N. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 1595–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Langelier C., von Schwedler U. K., Fisher R. D., De Domenico I., White P. L., Hill C. P., Kaplan J., Ward D., Sundquist W. I. (2006) J. Virol. 80, 9465–9480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wollert T., Hurley J. H. (2010) Nature 464, 864–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wollert T., Wunder C., Lippincott-Schwartz J., Hurley J. H. (2009) Nature 458, 172–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hanson P. I., Roth R., Lin Y., Heuser J. E. (2008) J. Cell Biol. 180, 389–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Saksena S., Wahlman J., Teis D., Johnson A. E., Emr S. D. (2009) Cell 136, 97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zamborlini A., Usami Y., Radoshitzky S. R., Popova E., Palu G., Göttlinger H. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 19140–19145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Teis D., Saksena S., Judson B. L., Emr S. D. (2010) EMBO J. 29, 871–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Teo H., Perisic O., González B., Williams R. L. (2004) Dev. Cell 7, 559–569 [DOI] [PubMed] [Google Scholar]
- 17. Im Y. J., Wollert T., Boura E., Hurley J. H. (2009) Dev. Cell 17, 234–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tan S. (2001) Protein Expr. Purif. 21, 224–234 [DOI] [PubMed] [Google Scholar]
- 19. Audhya A., McLeod I. X., Yates J. R., Oegema K. (2007) PLoS ONE 2, e956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mayers J. R., Fyfe I., Schuh A. L., Chapman E. R., Edwardson J. M., Audhya A. (2011) J. Biol. Chem. 286, 9636–9645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tucker W. C., Weber. T., Chapman E. R. (2004) Science 304, 435–438 [DOI] [PubMed] [Google Scholar]
- 22. Barrera N. P., Ormond S. J., Henderson R. M., Murrell-Lagnado R. D., Edwardson J. M. (2005) J. Biol. Chem. 280, 10759–10765 [DOI] [PubMed] [Google Scholar]
- 23. Im Y. J., Hurley J. H. (2008) Dev. Cell 14, 902–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kasson P. M., Pande V. S. (2004) Biophys. J. 86, 3744–3749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kozlovsky Y., Kozlov M. M. (2003) Biophys. J. 85, 85–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jackson M. B. (2009) J. Membr. Biol. 231, 101–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee M. C., Schekman R. (2004) Science 23, 479–480 [DOI] [PubMed] [Google Scholar]
- 28. Liu J., Kaksonen M., Drubin D. G., Oster G. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 10277–10282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu J., Sun Y., Oster G. F., Drubin D. G. (2010) Curr. Opin. Cell Biol. 22, 36–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang H., de Joannis J., Jiang Y., Gaulding J. C., Albrecht B., Yin F., Khanna K., Kindt J. T. (2008) Biophys. J. 95, 2647–2657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lister J. D. (1975) Phys. Lett. A 53, 193–194 [Google Scholar]
- 32. Jiang F. Y., Bouret Y., Kindt J. T. (2004) Biophys. J. 87, 182–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. de Joannis J., Jiang F. Y., Kindt J. T. (2006) Langmuir 22, 998–1005 [DOI] [PubMed] [Google Scholar]
- 34. Jiang Y., Kindt J. T. (2007) J. Chem. Phys. 126, 045105 [DOI] [PubMed] [Google Scholar]
- 35. Henne W. M., Kent H. M., Ford M. G., Hegde B. G., Daumke O., Butler P. J., Mittal R., Langen R., Evans P. R., McMahon H. T. (2007) Structure 15, 839–852 [DOI] [PubMed] [Google Scholar]
- 36. Guizetti J., Schermelleh L., Mäntler J., Maar S., Poser I., Leonhardt H., Müller-Reichert T., Gerlich D. W. (2011) Science 331, 1616–1620 [DOI] [PubMed] [Google Scholar]
- 37. Babst M., Wendland B., Estepa E. J., Emr S. D. (1998) EMBO J. 17, 2982–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fabrikant G., Lata S., Riches J. D., Briggs J. A., Weissenhorn W., Kozlov M. M. (2009) PLoS Comput. Biol. 5, e1000575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fujii K., Hurley J. H., Freed E. O. (2007) Nat. Rev. Microbiol. 5, 912–916 [DOI] [PubMed] [Google Scholar]
- 40. Lee H. H., Elia N., Ghirlando R., Lippincott-Schwartz J., Hurley J. H. (2008) Science 322, 576–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lata S., Schoehn G., Jain A., Pires R., Piehler J., Gottlinger H. G., Weissenhorn W. (2008) Science 321, 1354–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.