Background: It is important to understand the pathophysiology of Huntington disease (HD).
Results: Huntingtin altered AMPAR-mediated synaptic transmission via a mechanism depending on the microtubule (MT) motor KIF5. The AMPAR/KIF5/MT complex was disrupted in a HD mouse model.
Conclusion: AMPAR trafficking and function is impaired by mutant huntingtin.
Significance: It could underlie the deficits in movement control and cognitive processes in HD conditions.
Keywords: Glutamate Receptors Ionotropic (AMPA, NMDA); Kinesin; Microtubules; Neurons; Trafficking; HAP1; Huntingtin; Huntington Disease
Abstract
Emerging evidence from studies of Huntington disease (HD) pathophysiology suggests that huntingtin (htt) and its associated protein HAP1 participate in intracellular trafficking and synaptic function. However, it is largely unknown whether AMPA receptor trafficking, which is crucial for controlling the efficacy of synaptic excitation, is affected by the mutant huntingtin with polyglutamine expansion (polyQ-htt). In this study, we found that expressing polyQ-htt in neuronal cultures significantly decreased the amplitude and frequency of AMPAR-mediated miniature excitatory postsynaptic current (mEPSC), while expressing wild-type huntingtin (WT-htt) increased mEPSC. AMPAR-mediated synaptic transmission was also impaired in a transgenic mouse model of HD expressing polyQ-htt. The effect of polyQ-htt on mEPSC was mimicked by knockdown of HAP1 and occluded by the dominant negative HAP1. Moreover, we found that huntingtin affected mESPC via a mechanism depending on the kinesin motor protein, KIF5, which controls the transport of GluR2-containing AMPARs along microtubules in dendrites. The GluR2/KIF5/HAP1 complex was disrupted and dissociated from microtubules in the HD mouse model. Together, these data suggest that AMPAR trafficking and function is impaired by mutant huntingtin, presumably due to the interference of KIF5-mediated microtubule-based transport of AMPA receptors. The diminished strength of glutamatergic transmission could contribute to the deficits in movement control and cognitive processes in HD conditions.
Introduction
Because the discovery that Huntington disease (HD)2 is caused by an abnormally elongated polyglutamine (polyQ) repeat in the N-terminal region of the large protein huntingtin (htt) (1), extensive studies have been performed to explore the normal function of huntingtin in neurons and the molecular mechanism by which polyQ-htt causes selective neuronal dysfunction and degeneration. It is thought that huntingtin is a scaffold protein that facilitates the assembly of various protein complexes (2). Huntingtin-associated protein 1 (HAP1) is the first identified htt-binding partner (3). HAP1 is enriched in the brain and it interacts more tightly with polyQ-htt than WT-htt (3). It has been suggested that huntingtin and HAP1 participate in intracellular trafficking and synaptic function, and polyglutamine expansion affects vesicular transport (4, 5).
HAP1 associates with kinesin or dynein microtubule motor proteins (6–9), which play a critical role in the anterograde or retrograde transport of neuronal cargos between cell bodies and dendritic/axonal terminals (10, 11). Growing evidence suggests that deficits in these neuronal transport systems underlie the pathogenesis of a number of neurodegenerative diseases (12). It has been found that mutant huntingtin impairs the HAP1-dependent vesicular transport of brain-derived neurotrophic factor (BDNF) along microtubules, which is concomitant with its altered interactions with HAP1 and the dynein-associated protein dynactin p150 (8). Mutant huntingtin also reduces the association of HAP1 with dynactin p150 and kinesin light chain and thereby decreases the intracellular level of receptor tyrosine kinase (TrkA), a nerve growth factor receptor whose internalization and trafficking are required for neurite outgrowth (13). Moreover, suppressing the expression of HAP1 inhibits the kinesin-dependent GABAAR trafficking and synaptic inhibition (14, 15). These findings suggest that HD may be associated with disrupted HAP1 transport of cargos along microtubules that are critical for maintaining neuronal functions.
AMPA receptor, the major mediator of excitatory transmission in the CNS, is highly involved in synaptic plasticity and cognitive functions. Biochemical studies have found that the kinesin motor protein 5 (KIF5) is important for the microtubule-based transport of GluR2-containing AMPA receptors (16). Because HAP1 interacts with KIF5 (9, 15), it prompts us to speculate that the abnormal interaction between mutant huntingtin and HAP1 will cause the disruption of the HAP1/KIF5/GluR2 complex, leading to impaired transport of AMPA receptors along dendritic microtubules. The loss of glutamatergic synaptic transmission could underlie some of the defects in information processing occurring in HD.
MATERIALS AND METHODS
Primary Neuronal Culture and Transfection
Rat cortical or striatal cultures were prepared as described previously (17, 18). Briefly, cells from 18d rat embryos were dissociated using trypsin and trituration through a Pasteur pipette. When neurons were attached to the coverslip within 24 h, the medium was changed to Neurobasal with B27 supplement. Cultured neurons (12–14 DIV) were transfected with various constructs or siRNA using Lipofectamine 2000 method, which included EGFP-tagged N-terminal fragment of huntingtin containing the first 480 amino acids with 17Q (WT-htt) or 68Q (polyQ-htt), HAP1 siRNA (15), and KLC1 (Kinesin Light Chain 1) siRNA (Santa Cruz Biotechnology). To knockdown the expression of KIF17, the antisense oligonucleotide (1 μm) (19, 20) was added directly to the culture medium. Electrophysiological recordings were performed 2–3 days after transfection.
Electrophysiological Recording of Synaptic AMPAR Currents
Recording of miniature excitatory postsynaptic currents (mEPSC) in cultured neurons used the whole-cell patch clamp technique (18). Electrodes (5–9 MΩ) were filled with the internal solution (in mm): 130 Cs-methanesulfonate, 10 CsCl, 4 NaCl, 10 HEPES, 1 MgCl2, 5 EGTA, 2.2 QX-314, 12 phosphocreatine, 5 MgATP, 0.2 Na2GTP, 0.1 leupeptin, pH 7.2–7.3, 265–270 mOsm. The external solution consisted of (in mm): 127 NaCl, 5 KCl, 10 HEPES, 2 MgCl2, 2 CaCl2, 2 glucose, pH 7.3–7.4, 300–305 mOsm. TTX (0.5 μm), bicuculline (10 μm) and D-APV (25 μm) were added to block action potentials, GABAA receptors and NMDA receptors, respectively. The cell membrane potential was held at −70 mV and all recordings were performed at room temperature.
For evoked EPSC recording (21) in striatal slices, the internal solution contained: 130 Cs-methanesulfonate, 10 CsCl, 4 NaCl, 10 HEPES, 1 MgCl2, 5 EGTA, 2.2 QX-314, 12 phosphocreatine, 5 MgATP, 0.2 Na3GTP, and 0.1 leupeptin, pH 7.2–7.3, 265–270 mOsm. Slices (300 μm) were placed in a perfusion chamber attached to the fixed stage of an upright microscope (Olympus) and submerged in continuously flowing oxygenated artificial CSF (ACSF) containing bicuculline (10 μm) and D-APV (25 μm). Cells were visualized with a 40X water-immersion lens and illuminated with near infrared (IR) light, and the image was detected with an IR-sensitive CCD camera. A Multiclamp 700A amplifier was used for these recordings. Tight seals (2–10 GΩ) from visualized pyramidal neurons were obtained by applying negative pressure. The membrane was disrupted with additional suction, and the whole-cell configuration was obtained. The access resistances ranged from 13–18 MΩ. Evoked currents were generated with a pulse from a stimulation isolation unit controlled by a S48 pulse generator (Astro Med, West Warwick, RI). A bipolar stimulating electrode (FHC, Bowdoinham, ME) was positioned ∼100 μm from the neuron under recording. Membrane potential was held at −70mV during the recording.
Mini Analysis Program (Synaptosoft, Leonia, NJ) was used to analyze the spontaneous synaptic events. Statistical comparisons of mEPSC were made using Kolmogorov-Smirnov (K-S) test. Clampfit (Axon instruments) was used to analyze evoked synaptic currents. ANOVA, followed by post hoc Tukey tests, was performed to detect statistical significance between groups subjected to different treatments.
Knockdown Measurement
The full-length open reading frame of KLC1 was amplified from rat brain cDNA by PCR, and a Flag tag was added to the N-terminal in frame. The PCR product was cloned to T/A vector, and then subcloned to pcDNA3.1 expression vector. The construct was verified by DNA sequencing. To test the knockdown effect of siRNA, the plasmid FlagKLC1 was transfected to HEK293 cells with a KLC1 siRNA (Santa Cruz Biotechnology, 20 nm) or a scrambled control siRNA. Two days after transfection, the cells were harvested and subjected to Western blotting with anti-Flag. Actin was used as a loading control.
Co-immunoprecipitation
Slices were homogenized in 0.5% Nonidet P-40 or 1% Triton X-100 lysis buffer, then lysates were ultracentrifuged (200,000 × g) at 4 °C for 60 min. Supernatant fractions were incubated with anti-α-tubulin (15 μg; Sigma, T6199) or anti-GluR2 (15 μg, Millipore, MAB397) for overnight at 4 °C, followed by incubation with 50 μl of protein A/G plus agarose (Santa Cruz Biotechnology) for 1 h at 4 °C. Immunoprecipitates were washed three times with lysis buffer containing 0.2 m NaCl, then boiled in 2× SDS loading buffer for 5 min, and separated on 7.5% SDS-polyacrylamide gels. Western blotting experiments were performed with anti-KIF5 heavy chain (1:500, SUK4) (15), anti-GluR2 (1:500, Millipore, MAB397) or anti-HAP1 (1:200, Santa Cruz Biotechnology, sc-30125). To compare GluR1 and GluR2 in KIF5 immunoprecipitates, the antibody (4 μg) against KIF5 (SUK4), GluR1 (Santa Cruz Biotechnology, sc-13152) or GluR2 (Millipore, MAB397) was used.
Biochemical Measurement of Surface-expressed Receptors
The surface AMPA receptors were detected as previously described (18, 21). Briefly, striatal slices were incubated with ACSF containing 1 mg/ml Sulfo-NHS-LC-Biotin (Pierce) for 20 min on ice. The slices were then rinsed three times in TBS to quench the biotin reaction, followed by homogenization in 300 μl of modified radioimmunoprecipitation assay (RIPA) buffer (1% Triton X-100, 0.1% SDS, 0.5% deoxycholic acid, 50 mm NaPO4, 150 mm NaCl, 2 mm EDTA, 50 mm NaF, 10 mm sodium pyrophosphate, 1 mm sodium orthovanadate, 1 mm PMSF, and 1 mg/ml leupeptin). The homogenates were centrifuged at 14,000 × g for 15 min at 4 °C. To measure total protein, 15 μg of protein were removed. For surface protein, 150 μg of protein were incubated with 100 μl 50% Neutravidin-agarose (Pierce) for 2 h at 4 °C, and bound proteins were resuspended in 25 μl of SDS sample buffer and boiled. Quantitative Western blots were performed on both total and biotinylated (surface) proteins using antibody against GluR1 (1:200, Santa Cruz Biotechnology) and GluR2 (1:500, Millipore).
RESULTS
Huntingtin Regulates Synaptic AMPAR Responses
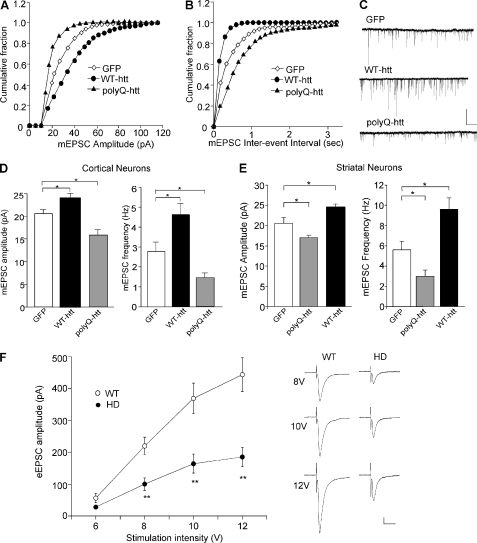
To determine the role of huntingtin in the regulation of AMPARs, we examined the effect of WT-htt and polyQ-htt on miniature EPSC (mEPSC), a synaptic response to the quantal release of single glutamate vesicles, in cultured neurons. As shown in Fig. 1, A and C, expression of WT-htt in cortical pyramidal neurons caused a significant increase in the mEPSC amplitude (indicated by a rightward shift in the cumulative distribution plot of amplitudes), while expression of polyQ-htt caused a significant decrease in the mEPSC amplitude (GFP: 20.2 ± 0.8 pA, n = 32; WT-htt: 24.3 ± 0.8 pA, n = 20; polyQ-htt: 16.4 ± 0.9 pA, n = 20, p < 0.05, ANOVA, Fig. 1D). The frequency of mEPSC was also significantly increased in WT-htt-expressing cortical neurons (indicated by a leftward shift in the cumulative distribution plot of inter-event intervals), and decreased in polyQ-htt-expressing cortical neurons (Fig. 1, B and C, GFP: 2.8 ± 0.5 Hz, n = 32; WT-htt: 4.6 ± 0.6 Hz, n = 20; polyQ-htt: 1.4 ± 0.3 Hz, n = 20, p < 0.05, ANOVA, Fig. 1D). Similar effects were observed in cultured striatal medium spiny neurons (GFP: 20.5 ± 1.4 pA, 5.6 ± 0.8 Hz, n = 9; polyQ-htt: 17.0 ± 0.6 pA, 3.0 ± 0.6 Hz, n = 15; WT-htt: 24.7 ± 0.7 pA, 9.6 ± 1.1 Hz, n = 24, p < 0.05, ANOVA, Fig. 1E). These results indicate that AMPAR-mediated synaptic responses is enhanced by WT-htt, and impaired by the mutant htt containing polyQ expansion.
FIGURE 1.
Synaptic AMPAR responses are enhanced by WT-htt and impaired by polyQ-htt. A and B, cumulative distribution plot of the mEPSC amplitude and inter-event interval in cortical neurons transfected with GFP, WT-htt (17Q) or polyQ-htt (68Q). C, representative mEPSC traces. Scale bar: 25 pA, 2 s. D and E, summary data (mean ± S.E.) of mEPSC amplitude and frequency in cortical or striatal neurons with different transfections. *: p < 0.05, ANOVA. F, summarized input-output curves of AMPAR-EPSC evoked by a series of stimulus intensities in striatal MSNs taken from WT versus N171–82Q mice (4.5-month-old). Inset, representative AMPAR-EPSC traces. Scale bars: 50 pA, 20 ms. **, p < 0.01, ANOVA.
To verify results from the cellular model of HD, we also examined AMPAR-mediated synaptic transmission in an animal model of HD. Transgenic mice, N171–82Q, which express a mutant N-terminal fragment of huntingtin (the first 171 amino acids of human htt with 82Q), develop behavioral abnormalities resembling HD, including loss of coordination, tremors, hypokinesis, and abnormal gait (22). We compared the synaptic strength by measuring input/output curves of evoked AMPAR-EPSC in striatal medium spiny neurons (MSNs) from WT versus N171–82Q mice. As shown in Fig. 1F, AMPAR-mediated excitatory synaptic responses induced by a series of stimulus intensities were markedly reduced in striatal MSNs from N171–82Q mice (8V: WT: 219.9 ± 27.2 pA, n = 15, HD: 100.4 ± 19.6 pA, n = 11; 10V: WT: 368.9 ± 47.7 pA, HD: 164.2 ± 29.6 pA; 12V: WT: 443.2 ± 53.0 pA, HD: 185.3 ± 30.6 pA, p < 0.01, ANOVA). It suggests that AMPAR-mediated corticostriatal synaptic transmission is significantly compromised in the HD model.
The PolyQ-htt-induced Impairment of Synaptic AMPAR Responses Requires HAP1
Next, we examined whether the effect of polyQ-htt on AMPARs involved Huntingtin-associated protein-1 (HAP1). HAP1 binds to htt in a glutamine repeat length-dependent manner (3, 6). HAP1 is enriched in the brain (23, 24) and transports together with htt bidirectionally in neurons (25). We first used RNA interference to knock down HAP1 expression, and examined its impact on mEPSC. A small interfering RNA (siRNA) that has been proven to be highly effective in suppressing HAP1 expression (15) was used. As shown in Fig. 2, A–D, the mEPSC amplitude and frequency were significantly smaller in HAP1 siRNA-transfected neurons, compared with neurons transfected with a control siRNA (control siRNA: 21.0 ± 1.2 pA, 3.5 ± 0.6 Hz, n = 8; HAP1 siRNA: 15.3 ± 1.1 pA, 1.7 ± 0.4 Hz, n = 10, p < 0.05, ANOVA), similar to the effect of polyQ-htt.
FIGURE 2.
Knockdown of HAP1 causes a loss of synaptic AMPAR responses. A and B, cumulative distribution plot of the mEPSC amplitude and inter-event interval in cortical neurons transfected with GFP, HAP1 siRNA, or a scrambled control siRNA. C, representative mEPSC traces. Scale bar: 25 pA, 2 s. D, summary data (mean ± S.E.) of mEPSC amplitude and frequency in cortical neurons with different transfections. *, p < 0.05, ANOVA.
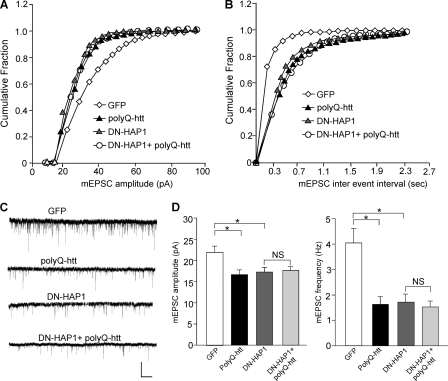
To confirm that polyQ-htt affects AMPARs via a HAP1-depdenent mechanism, we transfected neurons with the dominant negative HAP1 (DN-HAP1) (15) in conjugation with polyQ-htt. As shown in Fig. 3, A–D, DN-HAP1 caused a significant reduction of mEPSC amplitude and frequency, mimicking the effect of polyQ-htt (GFP: 21.3 ± 1.4 pA, 4.0 ± 0.6 Hz, n = 9; polyQ-htt: 16.6 ± 1.1 pA, 1.6 ± 0.3 Hz, n = 10; DN-HAP1: 17.2 ± 1.1 pA, 1.7 ± 0.3 Hz, n = 10, p < 0.05, ANOVA). In the presence of DN-HAP1, the effect of polyQ-htt on mEPSC was occluded (17.5 ± 1.2 pA, 1.5 ± 0.3 Hz, n = 10). Taken together, these results suggest that HAP1 is required for the polyQ-htt-induced loss of synaptic AMPAR responses.
FIGURE 3.
Dominant negative HAP1 (DN-HAP1) occludes the reducing effect of polyQ-htt on synaptic AMPAR responses. A and B, cumulative distribution plot of the mEPSC amplitude and inter-event interval in cortical neurons transfected with GFP, polyQ-htt, DN-HAP1, or DN-HAP1 plus polyQ-htt. C, representative mEPSC traces. Scale bar: 25 pA, 2 s. D, summary data (mean ± S.E.) of mEPSC amplitude and frequency in cortical neurons with different transfections. *, p < 0.05, ANOVA.
KIF5-mediated Microtubule-based Transport of GluR2-containing AMPARs Is Involved in Huntingtin Regulation of Synaptic AMPAR Responses
How does polyQ-htt decrease and WT-htt increase mEPSC? One possibility is through the regulation of AMPAR trafficking. Biochemical evidence shows that KIF5 is the kinesin motor protein to transport GluR2-containing AMPARs along microtubule tracks in dendrites (16). The conventional kinesin complex consists of two heavy chains (KHC, 110–120 kDa) and two light chains (KLC1 and KLC2, 60–70 kDa) (26). Kinesin heavy chain has a conserved N-terminal motor domain, which is responsible for microtubule binding, and a diverse C-terminal non-motor domain containing KLC binding domain and cargo binding domain. It has been found that HAP1 directly interacts with KIF5 light chain (9) and heavy chain (15). We hypothesize that htt/HAP1 might alter glutamatergic transmission by interfering with KIF5-dependent AMPAR transport along microtubules.
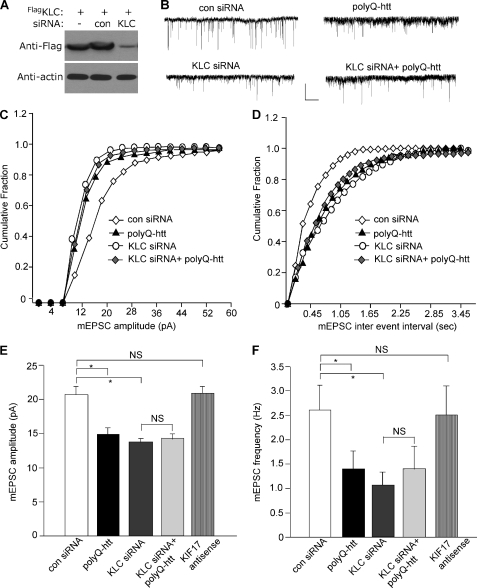
To test this, we knocked down KIF5 light chain, which is essential for the proper function of KIF5 heavy chains (27), and examined its impact on mEPSC. As illustrated in Fig. 4A, the KLC1 siRNA, but not a control siRNA, caused an effective suppression of KLC1 expression. KLC1 knockdown resulted in a significant decrease in the mEPSC amplitude and frequency (Fig. 4, B–F, control siRNA: 20.7 ± 1.2 pA, 2.6 ± 0.5 Hz, n = 11; KLC1 siRNA, 13.7 ± 0.6 pA, 1.0 ± 0.3 Hz, n = 11, p < 0.05, ANOVA), similar to the effect of polyQ-htt (14.8 ± 0.9 pA, 1.4 ± 0.4 Hz, n = 10). Moreover, in neurons with KLC1 knockdown, the effect of polyQ-htt on mEPSC was occluded (14.3 ± 0.8 pA, 1.4 ± 0.5 Hz, n = 9). To test the specificity of KIF5 involvement, we also examined the potential involvement of another kinesin motor protein, KIF17, which transports NMDA receptors along microtubules (19, 28). As shown in Fig. 4, E and F, cellular knockdown of KIF17 with antisense oligonucleotides did not induce any significant changes in mEPSC (20.8 ± 1.0 pA, 2.5 ± 0.6 Hz, n = 9).
FIGURE 4.
The polyQ-htt-induced impairment of synaptic AMPAR responses is occluded by knockdown of KIF5, but not KIF-17. A, representative Western blots in HEK293 cells transfected with FLAG-tagged rat KLC1 in the absence or presence of a control siRNA or a KLC1 siRNA. B–D, representative mEPSC traces (B) and cumulative distribution plot of the mEPSC amplitude (C) and inter-event interval (D) from cortical neurons transfected with control siRNA, polyQ-htt, KLC1 siRNA, or KLC1 siRNA plus polyQ-htt. Scale bar: 25 pA, 1 s. E and F, summary data (mean ± S.E.) of mEPSC amplitude and frequency in cortical neurons with different transfections. *, p < 0.05, ANOVA. NS, no significance.
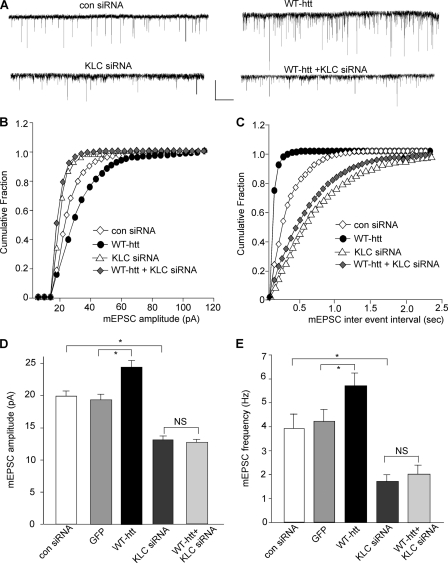
We further tested whether the WT-htt-induced enhancement of mEPSC also requires KIF5 motor protein. Neurons were transfected with KLC1 siRNA and WT-htt together. As shown in Fig. 5, A–E, the enhancing effect of WT-htt on mEPSC was prevented by KLC1 knockdown (KLC1 siRNA: 13.1 ± 0.6 pA, 1.7 ± 0.3 Hz, n = 11; KLC1 siRNA+WT-htt: 12.6 ± 0.5 pA, 2.0 ± 0.4 Hz, n = 12). Taken together, these results suggest that polyQ-htt-induced impairment and WT-htt-induced facilitation of synaptic AMPAR responses are both mediated by a KIF5-dependent mechanism.
FIGURE 5.
The WT-htt-induced enhancement of synaptic AMPAR responses is blocked by knockdown of KIF5. A–C, representative mEPSC traces (A) and cumulative distribution plot of the mEPSC amplitude (B) and inter-event interval (C) from cortical neurons transfected with control siRNA, WT-htt, KLC1 siRNA, or KLC1 siRNA plus WT-htt. Scale bar: 25 pA, 1 s. D and E, summary data (mean ± S.E.) of mEPSC amplitude and frequency in cortical neurons with different transfections. *, p < 0.05, ANOVA. NS, no significance.
Because GABAA receptors are also affected by HAP1, KIF5 and polyQ-htt (15, 29), we examined whether the results seen here could be a direct effect on AMPARs or an indirect effect caused by the homeostatic changes following the loss of GABA receptors. To do so, we blocked GABAARs in cultures with bicuculline throughout the polyQ-htt experssion, and then examined the impact of polyQ-htt on mEPSC. As shown in Fig. 6, A and B, polyQ-htt produced a significant reduction of mEPSC in the presence of bicuculline (GFP: 22.9 ± 1.0 pA, 6.7 ± 1.6 Hz, n = 11; polyQ-htt: 16.4 ± 1.1 pA, 2.5 ± 0.7 Hz, n = 6, p < 0.05). It suggests that the effect of polyQ-htt on AMPARs is independent of its effect on GABAARs.
FIGURE 6.
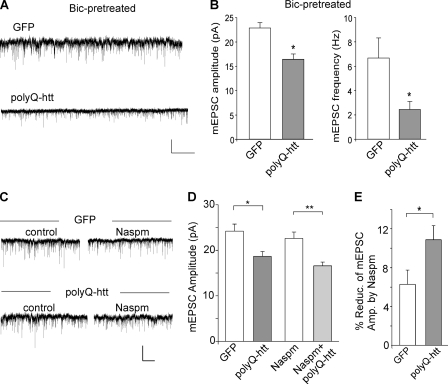
PolyQ-htt impairs synaptic AMPAR responses independent of GABAARs, and mainly targets GluR2-containing AMPARs. A and B, representative mEPSC traces and summary data (mean ± S.E.) of mEPSC amplitude and frequency from bicuculline-treated cortical neurons transfected with GFP versus polyQ-htt. Bicuculline (10 μm) was added 2 h before transfection and remained throughout the course of expressing polyQ-htt. Scale bar: 10 pA, 2 s. *, p < 0.05, t test. C and D, representative mEPSC traces and summary data (mean ± S.E.) of mEPSC amplitude from cortical neurons transfected with GFP versus polyQ-htt in the absence or presence of Naspm. *, p < 0.05; **, p < 0.005, t test. Scale bar: 10 pA, 10 s. E, summary data (mean ± S.E.) of the percentage block of mEPSC amplitude by Naspm in GFP- versus polyQ-htt-transfected cortical neurons. *, p < 0.05; t test.
To determine the subunit of AMPARs that is affected by polyQ-htt, we applied Naspm (10 μm), a selective blocker of GluR2-lacking AMPARs. As shown in Fig. 6, C–E, polyQ-htt decreased mEPSC amplitude to a bigger extent in the presence of Naspm when only GluR2-containing channels remained (GFP: 22.6 ± 1.4 pA, n = 8; polyQ-htt: 16.6 ± 0.8 pA, n = 8, ∼27% decrease, p < 0.005), compared with the control condition when all AMPAR channels were present (GFP: 24.2 ± 1.5 pA, n = 8; polyQ-htt: 18.7 ± 1.0 pA, n = 8, ∼23% decrease, p < 0.05). Moreover, polyQ-htt-transfected neurons had more Naspm-sensitive component (GluR1/1) than GFP-transfected neurons (GFP: 6.3 ± 1.5%, n = 8; polyQ-htt: 10.9 ± 1.4%, n = 8, p < 0.05). These results suggest that the majority of AMPARs contain the GluR2 subunit, and GluR2 is the major target that is suppressed by polyQ-htt.
The HAP1/GluR2/KIF5/Microtubule Complex Is Disrupted in HD
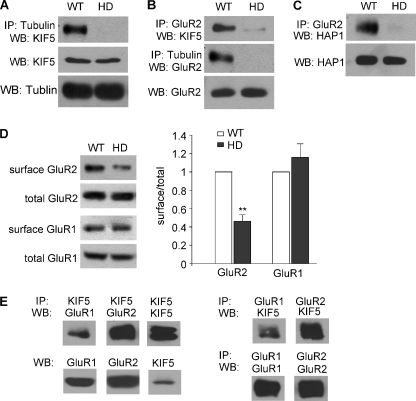
Previous biochemical studies have found that polyQ-htt binds to HAP1 with a higher affinity than WT-htt (3), and this abnormal interaction impairs anterograde transport of cargos (5). We hypothesize that the HAP1/KIF5/GluR2 complex may be altered in HD, which underlies the polyQ-htt-induced impairment of synaptic AMPAR responses. To test this, co-immunoprecipitation assays from WT versus N171–82Q mice were carried out to compare the HAP1/KIF5/GluR2 complex in vivo. As shown in Fig. 7A, the association of KIF5 heavy chain (HC) to microtubules was severely compromised in HD mice. Furthermore, a strong decrease was found with the GluR2 bound to KIF5 molecular motors or microtubules in HD mice (Fig. 7B). The GluR2/HAP1 complex was also significantly reduced in HD mice (Fig. 7C). None of these proteins show significant changes in their expression. It suggests that polyQ-htt causes a dissociation of the KIF5/GluR2 complex from microtubules, and a dissociation of the cargo GluR2 from KIF5 motor proteins. These changes in the HAP1/KIF5/GluR2 multiprotein complex are likely to cause the disrupted transport of AMPARs along microtubules in HD.
FIGURE 7.
HAP1/GluR2/KIF5/microtubule complex is disrupted in the mouse model of HD. A–C, co-immunoprecipitation assays showing the association between tubulin and KIF5 (A), GluR2 and KIF5 (B), tubulin and GluR2 (B), GluR2 and HAP1 (C) from striatal slices of WT versus HD mice (4-month-old). Each experiment was repeated in 3–5 pairs of mice. D, immunoblots and quantification analysis showing the surface and total GluR2 and GluR1 subunits in lysates of striatal slices taken from WT versus HD mice (4-month-old). *, p < 0.01, ANOVA. E, co-immunoprecipitation assays showing the association between KIF5 and GluR1 or GluR2 in brain lysates.
The loss of MT-based transport of GluR2 suggests that GluR2-containing AMPARs at the plasma membrane are reduced in HD mice. To test this, surface biotinylation assays in striatal slices from WT versus N171–82Q mice were performed to detect the surface and total level of AMPAR subunits. As shown in Fig. 7D, HD mice showed a significant decrease in the surface GluR2 subunit (46.2 ± 7.1% of control, n = 7, p < 0.01, ANOVA), while the surface GluR1 subunit was largely unchanged (115.9 ± 14.8% of control, n = 4). The total level of these receptor subunits remained similar in WT versus HD mice. These results suggest that the polyQ-htt-induced impairment of glutamatergic transmission could be attributable to the diminished surface delivery of GluR2-containing AMPARs.
Because GluR1/2 heteromers account for a major portion of synaptic AMPAR channels in cortical neurons, the specific impairment of GluR2 transport and not GluR1 in HD mice is surprising. To explain it, we compared GluR1 and GluR2 in KIF5 immunoprecipitates. As shown in Fig. 7E, while the majority of GluR2 bound to KIF5, much less GluR1 was found in the KIF5 complex. It suggests that a big portion of GluR1/2 heteromers is independent of the microtubule motor KIF5. Consistently, whole-cell recordings have found that blocking KIF5 function only leads to 35–40% reduction of AMPAR-EPSC (30, 31), which may attributable to the loss of GluR2/3 channel transport on microtubules. The lack of effect on surface GluR1 in HD mice may be due to the small portion of GluR1 (in GluR1/2 and GluR1/1 channels) that is dependent on the MT/KIF5-based transport.
DISCUSSION
The cellular function of WT-htt and the pathogenic mechanisms induced by polyQ-htt are hot topics in the HD field. Wt-htt, together with HAP1, is known to affect vital cellular function like vesicular transport (6, 8, 25) and neurite outgrowth (13, 32). Several studies have identified an altered microtubule (MT)-dependent axonal transport of organelles in Drosophila or mammalian neurons of HD (8, 33–36), which is thought to contribute to the neuronal toxicity. There is also evidence suggesting that the neuronal damage in HD arises as a result of excessive activation of NMDA receptors (37–42), which may be due to the increased numbers of surface NMDARs resulting from their accelerated forward trafficking (43).
In this study, we have provided evidence showing that htt/HAP1 plays a critical role in regulating dendritic transport of AMPARs and excitatory synaptic transmission. In cultured neurons, overexpression of WT-htt causes a strong facilitation, while polyQ-htt causes a significant impairment, of AMPAR-mediated synaptic currents, an effect dependent on HAP1. In the transgenic mouse model of HD expressing an 82Q-containing N-terminal fragment of htt, the AMPAR-mediated synaptic strength is also markedly reduced, with a concomitant decrease of surface GluR2 subunits. Consistently, AMPA currents are smaller in isolated cortical pyramidal neurons from the R6/2 transgenic mice that express exon 1 of the human HD gene with ∼150 CAG repeats (44). Moreover, the evoked and spontaneous AMPAR-EPSC have smaller amplitudes in striatal MSNs from the most severe model of HD, the YAC transgenic mice expressing full-length human HD gene with 128 CAG repeats (YAC128) (45). In addition, spontaneous EPSC frequency in striatal MSNs is significantly decreased in symptomatic R6/2 mice (46). Decreased glutamate transmission is also found specifically in dopamine D1 receptor-containing MSNs from the fully symptomatic YAC128 mice (47). The compromised glutamatergic transmission could contribute to the loss of movement control and cognitive processes in HD conditions (48, 49). Interestingly, a recent study has found that treatment with ampakine, a positive modulator of AMPARs, rescues synaptic plasticity and memory in HD mice (50).
While presynaptic dysfunction (45–47) could contribute to the diminished mEPSC or sEPSC frequency and eEPSC amplitude in neurons expressing polyQ-htt, the significantly altered mEPSC amplitude by wild-type or mutant htt suggests that htt/HAP1 may also regulate AMPAR function by changing postsynaptic AMPAR trafficking. The polyQ-htt-induced decrease in mEPSC frequency can be explained, at least partially, by a reduction of the number of postsynaptic AMPA receptors, which leads to fewer synaptic events beyond the detection threshold. AMPA receptors are transported along microtubules on dendrites before being delivered to synapses via actin cytoskeletons. This MT-based transport of AMPARs is controlled by the kinesin motor protein KIF5, which binds to GluR2-containing vesicles via the adaptor protein GRIP1 (16). Because HAP1 also interacts with KIF5 (9, 15), the HAP1/htt complex could potentially influence the association of AMPARs with KIF5 motors, therefore affecting the MT-based transport of AMPARs. Consistently, we show that inhibiting KIF5 function by knocking down its light chain prevents the impairing effect of polyQ-htt and the facilitating effect of WT-htt on AMPAR synaptic currents. It suggests that WT-htt enhances the KIF5-mediated transport of AMPAR-containing vesicles along MTs on dendrites. This function is altered when htt contains the polyQ expansion.
In agreement with an important role of KIF5 and HAP1 in regulating AMPAR trafficking and function, we show that the association of the kinesin motor complex to microtubules is diminished in the HD mouse model. It could be due to the reduced tubulin acetylation (51) or caused by the polyQ-htt-induced activation of JNK3 and phosphorylation of KIF5 (52). The AMPAR/KIF5/HAP1 complex is also disrupted in HD mice, suggesting that polyQ-htt may impair microtubule-based AMPAR trafficking by sequestering HAP1 and KIF5 motors away from AMPAR-containing vesicles.
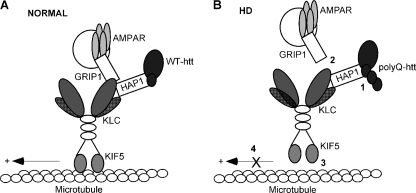
Based on these findings, we propose a model (Fig. 8) that schematically represents the potential mechanism by which htt influences AMPAR trafficking and function. In normal conditions, HAP1, which is complexed with wild-type htt, interacts with the kinesin motor protein KIF5, and facilitates the transport of GluR2-containing vesicles along microtubule tracks. In HD conditions, the HAP1 binding to htt is enhanced by expanded polyQ tracts (3). When this happens, KIF5/GluR2 complex falls apart and KIF5 motor loses the binding to microtubules. Thus, KIF5 is unable to transport GluR2-containing vesicles along the microtubule rails. Since the delivery of GluR2 subunits is an important factor to determine the number of functional heteromeric AMPA receptors at the synaptic membrane (53), the polyQ-htt-induced impairment of GluR2 transport causes the loss of AMPAR synaptic responses. The selective impairment of GluR2 transport and surface expression by polyQ-htt found in this study also suggests that remaining AMPARs in HD has a bigger portion of the Ca2+-permeable GluR1 homomers, thus the calcium permeability may be higher than normal in HD.
FIGURE 8.
A schematic model demonstrating the potential mechanism for huntingtin regulation of AMPAR trafficking. A, in normal conditions, WT-htt binds to HAP1 and enables the binding of HAP1 to KIF5 motor protein. KIF5 heavy chain interacts with GRIP1/GluR2-containing vesicles, and the whole complex is anterogradely transported along the microtubule (MT) rails. B, in HD conditions, polyQ-htt interacts abnormally with HAP1 (Step 1), thereby interrupting the binding of GluR2/KIF5 (Step 2), and KIF5/MT (Step 3). Consequently, KIF5 is unable to transport the GluR2 cargo along MTs (Step 4), leading to insufficient synaptic delivery of AMPAR channels.
Acknowledgment
We thank Xiaoqing Chen for technical support.
This work was supported, in whole or in part, by National Institutes of Health Grant NS69929 (to Z. Y.).
- HD
- Huntington Disease
- htt
- huntingtin
- MT
- microtubule
- AMPA
- α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid
- AMPAR
- AMPA receptor
- HAP
- huntingtin-associated protein
- BDNF
- brain-derived neurotrophic factor
- mEPSC
- miniature excitatory postsynaptic currents.
REFERENCES
- 1. Mangiarini L., Sathasivam K., Seller M., Cozens B., Harper A., Hetherington C., Lawton M., Trottier Y., Lehrach H., Davies S. W., Bates G. P. (1996) Cell 87, 493–506 [DOI] [PubMed] [Google Scholar]
- 2. Harjes P., Wanker E. E. (2003) Trends Biochem. Sci 28, 425–433 [DOI] [PubMed] [Google Scholar]
- 3. Li X. J., Li S. H., Sharp A. H., Nucifora F. C., Jr., Schilling G., Lanahan A., Worley P., Snyder S. H., Ross C. A. (1995) Nature 378, 398–402 [DOI] [PubMed] [Google Scholar]
- 4. Smith R., Brundin P., Li J. Y. (2005) Cell Mol. Life Sci. 62, 1901–1912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Li X. J., Li S. H. (2005) Trends Pharmacol. Sci. 26, 1–3 [DOI] [PubMed] [Google Scholar]
- 6. Engelender S., Sharp A. H., Colomer V., Tokito M. K., Lanahan A., Worley P., Holzbaur E. L., Ross C. A. (1997) Hum. Mol. Genet. 6, 2205–2212 [DOI] [PubMed] [Google Scholar]
- 7. Li S. H., Gutekunst C. A., Hersch S. M., Li X. J. (1998) J. Neurosci. 18, 1261–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gauthier L. R., Charrin B. C., Borrell-Pagès M., Dompierre J. P., Rangone H., Cordelières F. P., De Mey J., MacDonald M. E., Lessmann V., Humbert S., Saudou F. (2004) Cell 118, 127–138 [DOI] [PubMed] [Google Scholar]
- 9. McGuire J. R., Rong J., Li S. H., Li X. J. (2006) J. Biol. Chem. 281, 3552–3559 [DOI] [PubMed] [Google Scholar]
- 10. Goldstein L. S., Yang Z. (2000) Annu. Rev. Neurosci. 23, 39–71 [DOI] [PubMed] [Google Scholar]
- 11. Hirokawa N., Noda Y. (2008) Physiol. Rev. 88, 1089–1118 [DOI] [PubMed] [Google Scholar]
- 12. Goldstein L. S. (2003) Neuron 40, 415–425 [DOI] [PubMed] [Google Scholar]
- 13. Rong J., McGuire J. R., Fang Z. H., Sheng G., Shin J. Y., Li S. H., Li X. J. (2006) J. Neurosci. 26, 6019–6030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kittler J. T., Thomas P., Tretter V., Bogdanov Y. D., Haucke V., Smart T. G., Moss S. J. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 12736–12741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Twelvetrees A. E., Yuen E. Y., Arancibia-Carcamo I. L., MacAskill A. F., Rostaing P., Lumb M. J., Humbert S., Triller A., Saudou F., Yan Z., Kittler J. T. (2010) Neuron 65, 53–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Setou M., Seog D. H., Tanaka Y., Kanai Y., Takei Y., Kawagishi M., Hirokawa N. (2002) Nature 417, 83–87 [DOI] [PubMed] [Google Scholar]
- 17. Gu Z., Liu W., Yan Z. (2009) J. Biol. Chem. 284, 10639–10649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wei J., Liu W., Yan Z. (2010) J. Biol. Chem. 285, 26369–26376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guillaud L., Setou M., Hirokawa N. (2003) J. Neurosci. 23, 131–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yuen E. Y., Jiang Q., Chen P., Gu Z., Feng J., Yan Z. (2005) J. Neurosci. 25, 5488–5501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yuen E. Y., Liu W., Karatsoreos I. N., Feng J., McEwen B. S., Yan Z. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 14075–14079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schilling G., Becher M. W., Sharp A. H., Jinnah H. A., Duan K., Kotzuk J. A., Slunt H. H., Ratovitski T., Cooper J. K., Jenkins N. A., Copeland N. G., Price D. L., Ross C. A., Borchelt D. R. (1999) Hum. Mol. Genet. 8, 397–407 [DOI] [PubMed] [Google Scholar]
- 23. Gutekunst C. A., Li S. H., Yi H., Ferrante R. J., Li X. J., Hersch S. M. (1998) J. Neurosci. 18, 7674–7686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Page K. J., Potter L., Aronni S., Everitt B. J., Dunnett S. B. (1998) Eur. J. Neurosci. 10, 1835–1845 [DOI] [PubMed] [Google Scholar]
- 25. Block-Galarza J., Chase K. O., Sapp E., Vaughn K. T., Vallee R. B., DiFiglia M., Aronin N. (1997) Neuroreport 8, 2247–2251 [DOI] [PubMed] [Google Scholar]
- 26. Gyoeva F. K., Sarkisov D. V., Khodjakov A. L., Minin A. A. (2004) Biochemistry 43, 13525–13531 [DOI] [PubMed] [Google Scholar]
- 27. Rahman A., Kamal A., Roberts E. A., Goldstein L. S. (1999) J. Cell Biol. 146, 1277–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Setou M., Nakagawa T., Seog D. H., Hirokawa N. (2000) Science 288, 1796–1802 [DOI] [PubMed] [Google Scholar]
- 29. Cummings D. M., André V. M., Uzgil B. O., Gee S. M., Fisher Y. E., Cepeda C., Levine M. S. (2009) J. Neurosci. 29, 10371–10386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim C. H., Lisman J. E. (2001) J. Neurosci. 21, 4188–4194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yuen E. Y., Yan Z. (2011) J. Biol. Chem. 286, 24957–24965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li Y., Chin L. S., Levey A. I., Li L. (2002) J. Biol. Chem. 277, 28212–28221 [DOI] [PubMed] [Google Scholar]
- 33. Gunawardena S., Her L. S., Brusch R. G., Laymon R. A., Niesman I. R., Gordesky-Gold B., Sintasath L., Bonini N. M., Goldstein L. S. (2003) Neuron 40, 25–40 [DOI] [PubMed] [Google Scholar]
- 34. Szebenyi G., Morfini G. A., Babcock A., Gould M., Selkoe K., Stenoien D. L., Young M., Faber P. W., MacDonald M. E., McPhaul M. J., Brady S. T. (2003) Neuron 40, 41–52 [DOI] [PubMed] [Google Scholar]
- 35. Lee W. C., Yoshihara M., Littleton J. T. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 3224–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Trushina E., Dyer R. B., Badger J. D., 2nd, Ure D., Eide L., Tran D. D., Vrieze B. T., Legendre-Guillemin V., McPherson P. S., Mandavilli B. S., Van Houten B., Zeitlin S., McNiven M., Aebersold R., Hayden M., Parisi J. E., Seeberg E., Dragatsis I., Doyle K., Bender A., Chacko C., McMurray C. T. (2004) Mol. Cell. Biol. 18, 8195–8209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cepeda C., Ariano M. A., Calvert C. R., Flores-Hernández J., Chandler S. H., Leavitt B. R., Hayden M. R., Levine M. S. (2001) J. Neurosci. Res. 66, 525–539 [DOI] [PubMed] [Google Scholar]
- 38. Trettel F., Rigamonti D., Hilditch-Maguire P., Wheeler V. C., Sharp A. H., Persichetti F., Cattaneo E., MacDonald M. E. (2000) Hum. Mol. Genet. 9, 2799–2809 [DOI] [PubMed] [Google Scholar]
- 39. Laforet G. A., Sapp E., Chase K., McIntyre C., Boyce F. M., Campbell M., Cadigan B. A., Warzecki L., Tagle D. A., Reddy P. H., Cepeda C., Calvert C. R., Jokel E. S., Klapstein G. J., Ariano M. A., Levine M. S., DiFiglia M., Aronin N. (2001) J. Neurosci. 21, 9112–9123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zeron M. M., Hansson O., Chen N., Wellington C. L., Leavitt B. R., Brundin P., Hayden M. R., Raymond L. A. (2002) Neuron 33, 849–860 [DOI] [PubMed] [Google Scholar]
- 41. Li L., Murphy T. H., Hayden M. R., Raymond L. A. (2004) J. Neurophysiol. 92, 2738–2746 [DOI] [PubMed] [Google Scholar]
- 42. Fan M. M., Raymond L. A. (2007) Prog. Neurobiol. 81, 272–293 [DOI] [PubMed] [Google Scholar]
- 43. Fan M. M., Fernandes H. B., Zhang L. Y., Hayden M. R., Raymond L. A. (2007) J. Neurosci. 27, 3768–3779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. André V. M., Cepeda C., Venegas A., Gomez Y., Levine M. S. (2006) J. Neurophysiol. 95, 2108–2119 [DOI] [PubMed] [Google Scholar]
- 45. Milnerwood A. J., Raymond L. A. (2007) J. Physiol. 585, 817–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cepeda C., Hurst R. S., Calvert C. R., Hernández-Echeagaray E., Nguyen O. K., Jocoy E., Christian L. J., Ariano M. A., Levine M. S. (2003) J. Neurosci. 23, 961–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. André V. M., Cepeda C., Fisher Y. E., Huynh M., Bardakjian N., Singh S., Yang X. W., Levine M. S. (2011) J. Neurosci. 31, 1170–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lione L. A., Carter R. J., Hunt M. J., Bates G. P., Morton A. J., Dunnett S. B. (1999) J. Neurosci. 19, 10428–10437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Smith M. A., Brandt J., Shadmehr R. (2000) Nature 403, 544–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Simmons D. A., Rex C. S., Palmer L., Pandyarajan V., Fedulov V., Gall C. M., Lynch G. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 4906–4911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dompierre J. P., Godin J. D., Charrin B. C., Cordelières F. P., King S. J., Humbert S., Saudou F. (2007) J. Neurosci. 27, 3571–3583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Morfini G. A., You Y. M., Pollema S. L., Kaminska A., Liu K., Yoshioka K., Björkblom B., Coffey E. T., Bagnato C., Han D., Huang C. F., Banker G., Pigino G., Brady S. T. (2009) Nat. Neurosci. 12, 864–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Malinow R., Malenka R. C. (2002) Annu. Rev. Neurosci. 25, 103–126 [DOI] [PubMed] [Google Scholar]