Abstract
Prognosis for patients with early stage kidney cancer has improved, but the treatment options for patients with locally advanced disease and metastasis remain few. Understanding the molecular mechanisms that regulate invasion and metastasis is critical for developing successful therapies to treat these patients. Proinflammatory prostaglandin E2 plays an important role in cancer initiation and progression via activation of cognate EP receptors that belong to the superfamily of G protein-coupled receptors. Here we report that prostaglandin E2 promotes renal cancer cell invasion through a signal transduction pathway that encompasses EP4 and small GTPase Rap. Inactivation of Rap signaling with Rap1GAP, like inhibition of EP4 signaling with ligand antagonist or knockdown with shRNA, reduces the kidney cancer cell invasion. Human kidney cells evidence increased EP4 and decreased Rap1GAP expression levels in the malignant compared with benign samples. These results support the idea that targeted inhibition of EP4 signaling and restoration of Rap1GAP expression constitute a new strategy to control kidney cancer progression.
Keywords: Epac, Cell Invasion
Introduction
Kidney cancer is increasingly common, and the number of patients dying of this disease has also increased over the past several years (1). Kidney cancer accounts for roughly 4% of all cancer cases, and it occurs more often in males than in females. Common causes of kidney cancer have largely been attributed to genetic (e.g. inactivation of the von Hippel-Lindau gene product), environmental, and behavioral (e.g. smoking and diet) factors. Renal cell carcinoma (RCC)2 accounts for the majority (90%) of kidney cancer cases (2) and mostly originates in proximal renal tubules. RCC comprises several distinct histological subtypes that are traditionally classified by light microscopy, including clear cell (80% of all RCC cases), papillary, chromophobe, and oncocytoma (3). Although RCC presents as a localized disease in the majority of cases, more than one-third of patients exhibit metastatic lesions (4) that produce the highest mortality of any adult urological cancer (5).
Kidney tumor mass is sustained by the release of circulating and locally produced factors acting through cellular receptors that can switch the susceptible quiescent kidney cells to an activated state. Prostaglandins are naturally occurring lipids that are produced from cyclooxygenase (COX)-mediated metabolism of arachidonic acid (6, 7). The prostaglandins are abundantly expressed in the kidney and act locally to regulate renal function and systemic blood pressure (6). Notably, COX-2 expression is up-regulated in many human malignancies, including RCC (8–10) and correlates with poor prognosis. Clinical practice-based outcomes reveal a drawback to the safe use of drugs that target COX-2 as a result of increasing renal and cardiovascular risk (11, 12).
PGE2 is the predominant prostaglandin in the kidney, and a large body of evidence demonstrates that its levels are increased in patients diagnosed with cancer (13–16). PGE2 exerts its effects on target cells through activation of cognate receptors named EP1, EP2, EP3, and EP4 (6, 17) that belong to the superfamily of G protein-coupled receptors. Stimulation with PGE2 activates at least three distinct subfamilies of heterotrimeric G proteins, namely Gq, Gi, and Gs. In most cells, PGE2-bound EP1 couples to Gq and induces the activation of protein kinase C through the release of Ca2+ ions from intracellular stores (18). EP3 couples predominantly to Gi and inhibits the accumulation of second messenger cAMP (19, 20). Stimulated EP2 and EP4 couple to Gs leading to synthesis of cAMP and activation of the cAMP-dependent protein kinase (PKA) (21, 22). Hence, PGE2 transduces the multiple receptor-specific signaling events in target cells.
Emerging evidence implicates prostaglandins in cancer cell migration (23, 24). In this study, we explored the possible involvement of EPs and their downstream effectors in kidney cancer cell invasion. The results show that PGE2 promotes kidney cancer cell invasion through activation of EP4 and small GTPase Rap proteins. Interference of EP4-to-Rap signaling with complementary pharmacologic and biologic reagents reduces invasion of the kidney cancer cells. EP4 protein expression is increased in malignant compared with benign human kidney cells and inversely correlates with Rap1GAP protein expression. These studies identify EP4 and Rap1GAP proteins as positive and negative regulators, respectively, of kidney cancer cell invasion, and suggest their utility as prognostic markers and therapeutic targets to limit patient morbidity and mortality.
MATERIALS AND METHODS
Reagents
The mammalian expression plasmids were obtained as follows: FLAG epitope-tagged Rap1GAP from L. Quilliam (Indiana University) and YFP-Epac1-CFP from V. Nikolaev (University of Wuerzburg). Bacterial GST-RalGDS plasmid was obtained from J. Bos (University Medical Center Utrecht). HA epitope-tagged GAP domains of Rap1GAP were cloned by PCR amplification using the FLAG-Rap1GAP cDNA as a template, and all cDNA clones were verified by sequencing. Antibodies were obtained as follows: anti-EP1, anti-EP2, anti-EP3, and anti-EP4 from Cayman Chemical; anti-VASP from Millipore; anti-HA from Sigma; anti-Rap and anti-Rap1GAP from Santa Cruz; anti-GAPDH from Chemicon; and secondary antibodies from Jackson ImmunoResearch Laboratories. Reagents were obtained as follows: PGE2, AH23848, GW627368, H89, and anti-EP4 antibody (C terminus) blocking peptide from Cayman Chemical. Human embryonic kidney (HEK)-293 cells stably overexpressing EP4 were kind gift of J. Regan (University of Arizona).
Cell Culture
Human RCC7 and Caki-1 cells were maintained in RPMI1640, and HEK-293 cells in DMEM supplemented with 10% fetal bovine serum (FBS), 1% penicillin/streptomycin, and 10 mm HEPES buffer. The HK-2 cells were maintained in keratinocyte/SFM medium with EGF, bovine pituitary extract, and 1% penicillin/streptomycin. Cells were transfected with the appropriate cDNA and Lipofectamine 2000 (Invitrogen), and experiments were performed 1–2 days after transfection. For stable overexpression of individual HA-tagged wild-type (GAP-WT) or mutated (GAP-K194A, GAP-K285A, and GAP-N290A) GAP domain of Rap1GAP, transfected RCC7 cells were generated and cultured in the presence of G418 (500 μg/ml). Control RCC7 cells (EV) were similarly generated using pcDNA3.1 empty vector. EP4 knockdown with shRNA was peformed exactly as described in an earlier publication (24). Knockdown of endogenous Epac1 and -2 was achieved with Dharmacon ON-TARGETplus SMARTpool. Cells grown on 6-well plates to 40% confluence were transfected with 50–100 nm siRNA and 5 μl of DharmaFECT 2 reagent. For all assays, pooled transfected cells were equally divided to ensure the identical cell populations, and the agonist-regulated cell proliferation was determined by counting cells using trypan blue and hemocytometer.
Reverse Transcription Quantitative PCR Assay
Total RNA was isolated using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. Reverse transcription was conducted using SuperScriptTM III First-strand Synthesis System (Invitrogen). Gene levels were determined by two-step quantitative real-time PCR (SYBR Green I) with gene-specific primers amplification followed by melting curve analysis. The primers designed for PTGER1–4 were as follows: PTGER1 (sense), 5′-ACC TTC TTT GGC GGC TCT C-3′, PTGER1 (antisense), 5′-GCA CGA CAC CAC CAT GAT AC-3′; PTGER2 (sense), 5′-CAG TCT CCC TGC TCT TCT GC-3′, PTGER2 (antisense), 5′-GCA CCG AGA CAA TGA GAA GC-3′; PTGER3 (sense), 5′-TCA TCG TCG TGT ACC TGT CC-3′, PTGER3 (antisense), 5′-CGA TGA ACA ACG AGG AGA GC-3′; and PTGER4 (sense), 5′-TGC TCT TCT TCA GCC TGT CC-3′; PTGER4 (antisense), 5′-AGA CTG CAA AGA GCG TGA GG-3′. GAPDH primers were (sense) 5′-GGT CAT GAG TCC TTC CAC GAT-3′ and (antisense) 5′-CAT GGG TGT GAA CCA TGA GAA-3′. Calculation of relative mRNA of the probed genes were carried out by taking the normalized threshold cycle value (ΔCt) for each sample (ΔCt = Ct of Queried gene − Ct GAPDH) − the ΔCt of the control samples (ΔΔCt), and converting the difference to fold-expression using the following equation: fold = 2∧(−ΔΔCt). Experiments were repeated at least three times, each in triplicate.
FRET Assay
Cells were transfected with a cDNA encoding CFP-Epac1-YFP fusion protein and seeded onto glass coverslips. After 48 h, cells were inspected with a SP2 scanning confocal microscope (Leica) and imaged using a 63 × 1.4 NA oil immersion objective. Cells were excited at 425 nm, and emission of CFP and YFP was detected simultaneously through 470 ± 20- and 530 ± 25-nm bandpass filters. Cells were continuously perfused with a solution (150 mm NaCl, 5 mm KCl, 10 mm HEPES, 10 mm glucose, 1.5 mm CaCl2, and 2.5 mm MgCl2) containing PGE2 in the absence or presence of AH23848 (5 μm). Solution changes were made by using a multiport attachment and perfusion capillary positioned directly in front of the cell under study. Exposure time was 200–500 s and images were taken every 10–30 s. Fluorescent images were background corrected by subtracting autofluorescence intensities of background with no cells. Data were digitized and the ratio of YFP/CFP emissions were calculated at different time points and normalized by dividing all ratios by the emission ratio just before stimulation.
Western Blot Analysis
Appropriately treated cells were lysed in RIPA buffer (150 mm NaCl, 50 mm Tris-HCl, pH 8, 1 mm EDTA, 0.25% (w/v) sodium deoxycholate, 0.1% (v/v) Nonidet P-40, 1 mm NaF, 1 mm sodium pyrophosphate, 100 μm Na3VO4, 1 mm phenylmethylsulfonyl fluoride, 10 μg/ml of leupeptin, 10 μg/ml of aprotinin, and 0.7 μg/ml of pepstatin) and analyzed by SDS-PAGE and Western blotting. All primary antibodies were used at a dilution of 1:1,000 except Rap1GAP, which was used at a dilution of 1:500. For anti-EP4 antibody neutralization, blocking peptides (that target the C terminus of EP4) were pre-mixed with the anti-EP4 antibodies in blocking solution for 1 h prior to incubation with filter. Peroxidase-conjugated secondary antibodies were used at a dilution of 1:10,000, and membranes were developed using an ECL plus Western blotting Detection System (GE Healthcare).
Rap Activation Assay
Rap activation was determined using an established pulldown method based on the specific binding of a GST fusion protein containing the Ras-binding domain of RalGDS (GST-RalGDS-RBD) to the active GTP-bound form of Rap (25, 26). Briefly, whole cell lysates were centrifuged at 14,000 × g for 10 min at 4 °C, and the supernatant was removed and assayed for protein concentration. The GST-tagged RalGDS-RBD protein was expressed in BL21 cells and purified using glutathione-Sepharose 4B beads and an equal amount was added to 500 μg of cell extracts. Mixtures were incubated for 1 h at 4 °C, washed with PBS, and boiled in Laemmli sample buffer. The precipitated proteins were resolved by SDS-PAGE and analyzed by Western blotting to detect Rap1.
Cell Adhesion Assay
Cells were seeded in fibronectin-coated 96-well plates at 2.0 × 104 cells/well and treated, or not, with PGE2 or FBS for 1 h at 37 °C. Wells were washed three times with PBS to remove non-adherent cells, followed by addition of ice-cold methanol for 10 min. Fixed cells were incubated with crystal violet (0.5%) solution for additional 10 min at ambient temperature, followed by repeated washing with water. Wells were allowed to dry and adherent stained cells were solubilized with SDS (1%) solution. Absorbance was determined at a wavelength of 570 nm.
Cell Invasion Assay
Cells were starved overnight in RPMI1640 containing 0.1% FBS, washed with PBS, and detached. A total of 2 × 105 cells in 100 μl were placed into a transwell chamber containing collagen-coated filters, placed on the feeder tray that contained RPMI1640 supplemented, or not, with PGE2 at 37 °C in a humidified atmosphere. For experiments using EP4 antagonist AH23848 (1 and 5 μm) or PKA inhibitor H89 (5–20 μm), reagent was added to cells 10 and 30 min, respectively, prior to stimulation with PGE2. Cells in the upper well were removed with cotton swabs. The membranes were then fixed with ethanol and stained with crystal violet. Invading cells were counted using a phase-contrast microscope and for each membrane, five randomly selected fields were counted.
Statistical Analysis
The significance of agonist-induced cellular response was analyzed by one-way analysis of variance with Tukey post-test or two-way analysis of variance with Bonferroni post-test and implied at p < 0.05. For quantitative PCR experiments, mean ± S.E. for final gene pool analysis was calculated by propagation of error (addition). All statistical analyses were done, and all graphs were generated, using GraphPad Prism 5.0 software (GraphPad). The x and y labels of all presented data were prepared using Adobe Illustrator CS5 suite (Adobe).
RESULTS
EP4 Is Expressed in Renal Cancer Cells
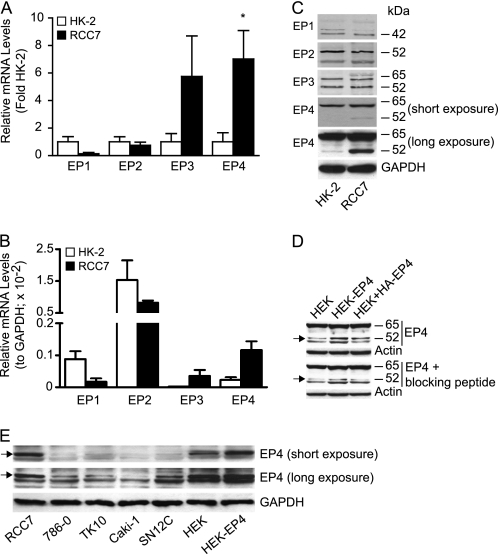
G protein-coupled receptor signaling arrays were used to compare the gene expression profile in renal cancer RCC7 cells to that in benign human kidney HK-2 cells. The results evidenced a distinct gene expression pattern between the malignant RCC7 and normal HK-2 cells. The RCC7 cells expressed elevated gene levels for prostaglandin E and D receptors, in comparison to the HK-2 cells. Four distinct genes encode for the prostaglandin E receptors (PTGER) and reverse transcription followed by real time quantitative PCR analysis confirmed expression of all four PTGER subtypes in both the RCC7 and HK-2 cells (Fig. 1A). The expression results also revealed a significant increase only in the PTGER4 gene level in the RCC7, compared with the HK-2 cells (Fig. 1A). The PTGER3 gene levels appeared to be more in the RCC7 compared with HK-2 cells, but the increase did not reach statistical significance. In comparison to GAPDH gene expression, PTGER2 gene levels were expressed most in HK-2 and RCC7 cells (Fig. 1B). Western blot analysis evidenced the expression of bands corresponding to the reported EP1 (44 kDa), EP2 (53 kDa), EP3 (52 and 62 kDa), and EP4 (52 kDa) protein masses (Fig. 1C). The results also confirmed the conclusion that EP4 protein expression is elevated in cancer RCC7, compared with benign HK-2 cells (Fig. 1C). To ascertain specificity of the anti-EP4 antibodies, EP4 antibody blocking peptides were pre-mixed with the antibodies followed by Western blot analysis. Results show that neutralization of anti-EP4 antibodies yielded a specific decrease in the intensity of the expected EP4 (52 kDa) protein band (Fig. 1D, compare arrow pointing band in upper and lower panels). Increased intensity of protein bands that migrated with apparent molecular masses predicted for EP4 in HEK-EP4 cells that stably overexpress the EP4 gene or in HEK + HA-EP4 cells that were transiently transfected with HA-EP4 cDNA, in comparison to control HEK cells (Fig. 1D, upper panel), further suggested specificity of the EP4 antibody used. Because EP4 has been implicated in transduction of mitogenic signals, we determined its expression in four additional human kidney cancer cell lines (obtained from the NCI) and found it to be expressed at high levels in aggressive SN12C and lower in less aggressive 786-O, TK10, and Caki-1 (27–30) cells (Fig. 1E). These findings support the idea that EP4 expression is increased in human renal cancer compared with benign cells.
FIGURE 1.
Expression of EPs in kidney cancer cells. A and B, expression of PTGER genes in HK-2 and RCC7 cells. Isolated RNA was reverse transcribed and equal amounts of cDNA was subjected to real-time PCR amplification to establish relative expression of the PTGER1 (EP1), PTGER2 (EP2), PTGER3 (EP3), or PTGER4 (EP4) genes. The gene expression was calculated as described, and the data were expressed as RNA levels in RCC7 relative to HK-2 cells (A) and EP relative to GAPDH in RCC7 and HK-2 cells (B). Each point represents the mean ± S.E. of values obtained from three experiments each performed in triplicate. *, p < 0.05 versus corresponding HK-2 samples. C, expression of the EP4 protein is increased in RCC7 compared with HK-2 cells. Equal amounts of cell lysates were analyzed by immunoblotting using specific anti-EP1, -EP2, -EP3, -EP4, or -GAPDH antibodies. GAPDH was used to calibrate total protein loading. D, EP4 antibody blocking peptides decrease the intensity of the detected EP4 protein band. Two sets of equal amounts of cell lysate from HEK, HEK-EP4, and HEK + HA-EP4 were fractionated on SDS-PAGE and transferred to filters. After blocking, one filter was incubated with anti-EP4 antibodies alone and the other with anti-EP4 antibodies that were pre-mixed with blocking peptides. The filters were analyzed by immunoblotting to detect EP4 and Actin (used as loading control) proteins. HEK-EP4, HEK293 cells stably overexpressing EP4; HEK + HA-EP4, HEK293 cells transiently transfected with HA-EP4 cDNA. Arrows on left indicate EP4 protein. E, expression of EP4 protein in kidney cancer cell lines. Equal amounts of cell lysates were analyzed by immunoblotting to detect EP4 and GAPDH (used as loading control) proteins. Arrows on left indicate EP4 protein. For C and E, contemporaneous short and long exposures of the same filter are shown to provide visual evidence for the relative expression of the EP4 protein.
EP4 Mediates the PGE2-induced RCC7 Cell Invasion
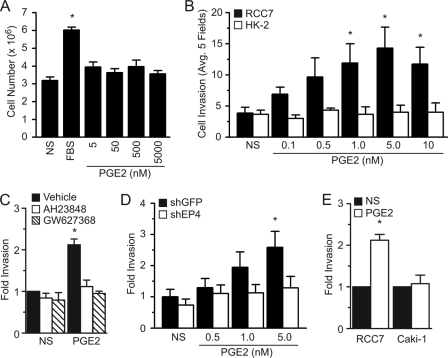
Stimulation of non-kidney cancer cells with PGE2 promotes cell growth and survival (15, 31), and we tested whether activation of endogenous EPs with PGE2 could also impact the RCC7 cell proliferation. Results shown in Fig. 2A demonstrate that, whereas treatment with serum caused a significant increase in the RCC7 cell number, exposure to various concentrations of PGE2 had no such effect.
FIGURE 2.
EP4 mediates the PGE2-induced RCC7 cell invasion. A, effect of PGE2 on RCC7 cell proliferation. Cells were stimulated with PGE2 (or 10% FBS used as a control) for 2 days at 37 °C, harvested, and incubated with 0.1% trypan blue stain. Cells excluding the dye were counted under light microscopy with hemocytometer. Each point represents the mean ± S.E. of values obtained from four experiments. *, p < 0.05 versus non-stimulated (NS) samples. B, PGE2 promotes RCC7 cell invasion. Equal number of RCC7 or HK-2 cells were starved overnight and allowed to invade collagen-coated transwell filters in the presence or absence of PGE2 (in nm). Cells that migrated to the bottom of the filter were stained with crystal violet and five fields were randomly selected and counted using a phase-contrast microscope. Each point represents the mean ± S.E. of values obtained from five experiments. *, p < 0.05 versus not stimulated (NS) samples. C, effect of EP4 antagonists AH23848 and GW627368 on RCC7 cell invasion. Invasion assays were done using cells pre-treated for 10 min, or not, with AH23848 (5 μm) or GW627368 (1 μm) and stimulated with PGE2 (5 nm). Data represent the fold-increase relative to nonstimulated values and *, p < 0.05. D, knockdown of endogenous EP4 expression attenuates the PGE2-induced RCC7 invasion. Cells stably expressing shGFP (control) or shEP4 were treated as in B. Five fields were randomly selected and counted and each point represents the mean ± S.E. of values obtained from three experiments. *, p < 0.05 versus corresponding nonstimulated samples. E, effect of PGE2 (5 nm) on the invasion of Caki-1 cells. Cells were treated and analyzed exactly as in B. *, p < 0.05 versus corresponding NS samples.
Morbidity and mortality of patients diagnosed with renal cell carcinomas are primarily caused by cancer cell metastasis to distal organs, and available evidence shows that the PGE2-EP4 axis may regulate the colon cancer cell migration (23). We examined impact of PGE2 stimulation on RCC7 cell invasion using a transwell invasion assay (32). Stimulation with PGE2 induced a dose-dependent increase in RCC7 cell invasion of a collagen barrier reconstituted in a transwell migration chamber (Fig. 2B). Maximal cell invasion was achieved with a PGE2 concentration of 5 nm, which is attainable in the kidney and falls within the Kd range for the EP4 (6, 19, 22); the EPs bind most potently to PGE2 with Kd values in the range of 1–40 nm, and PGE2 has high affinity to EP4 with a Kd value of 2 nm. Distinctly, the stimulation of normal HK-2 kidney cells with PGE2 did not increase their invasion (Fig. 2B), suggesting an EP4-mdiated response.
We used two EP4 antagonists, namely AH23848 and GW627368, as diagnostic tools to estimate the contribution of EP4 to the PGE2-induced RCC7 cell invasion. Exposure to AH23848 or GW627368 abrogated the PGE2-mediated RCC7 cell invasion (Fig. 2C). In addition to exerting an antagonist effect on EP4, the AH23848 acts as antagonist on the thromboxane A2 TP receptor. Treatment of RCC7 cells with the TP receptor agonist I-BOP showed no effect on VASP phosphorylation or cell invasion (data not shown). Nonetheless, to confirm the selective role of EP4 in PGE2-mediated cell invasion, RCC7 cells stably expressing shRNA targeting the PTGER4 gene were established and exhibited ∼60% reduction in EP4 protein expression. The RCC7-shEP4 cells failed to invade following stimulation with PGE2 (Fig. 2D). To add confidence to the conclusion that EP4 mediates the RCC cell invasion, we compared the PGE2-induced invasiveness of RCC7 (that express EP4; Fig. 1D) and Caki-1 (that do not express EP4; Fig. 1D) cells. Results show that although PGE2 promoted the RCC7 cell invasion, it failed to do so in the Caki-1 cells (Fig. 2E). Together, these results support the notion that PGE2-mediated RCC7 cell invasion occurs via EP4.
PGE2 Promotes Rap Activation
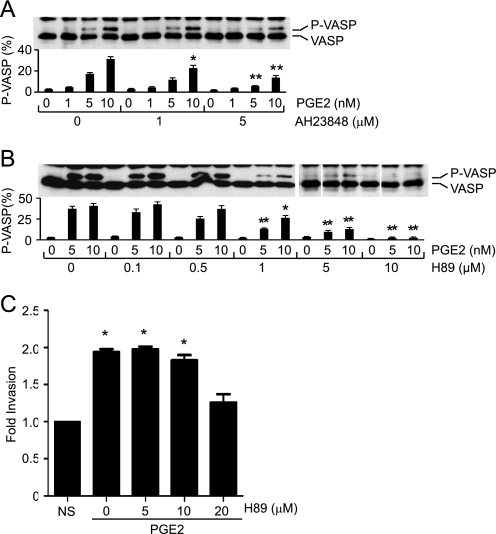
In most cell types, activated EP4 couples to heterotrimeric Gs proteins leading to the synthesis of second messenger cAMP through activation of adenylyl cyclases (19, 22). Stimulation of the RCC7 cells with PGE2 induced the dose-dependent accumulation of cAMP that was inhibited by the ligand antagonist AH23848 (data not shown), demonstrating a principally EP4-mediated signal. The treatment with AH23848 had no measurable effect on the PGE2-induced and Gq-coupled EP1-mediated Ca2+ mobilization. The cAMP acts as a master regulator of several effectors, including PKA, ion channels, and Epac, a guanine nucleotide exchange factor for small GTPase Rap proteins (33, 34). To begin to elucidate the signaling pathway(s) connecting the PGE2-EP4 module to RCC7 cell invasion, we first established the PGE2-mediated PKA activation. Stimulation of RCC7 cells with PGE2 induced a dose-dependent and EP4-mediated (i.e. signal is sensitive to AH23848) VASP phosphorylation (Fig. 3A). VASP is a phosphoprotein, and both PKA and PKG have been implicated as responsible kinases. Treatment with cAMP mimetic dibutyryl-cAMP increased VASP phosphorylation, whereas treatment with the cAMP antagonist (Rp)-cAMPS blocked the PGE2- and PGE1-OH-induced VASP phosphorylation (data not shown). Moreover, treatment with H89, a selective PKA inhibitor (35), decreased VASP phosphorylation in a dose-dependent manner (Fig. 3B), demonstrating the PGE2-dependent PKA activation. Remarkably, similar treatment with H89 concentrations capable of obliterating the PGE2-induced VASP phosphorylation (Fig. 3B) had no significant effects on the PGE2-mediated cell invasion (Fig. 3C), suggesting PKA exerts little impact on the PGE2-mediated RCC7 cell invasion.
FIGURE 3.
Role of PKA in the PGE2-induced RCC7 cell invasion. A, effect of AH23848 on VASP phosphorylation. RCC7 cells were pretreated, or not, for 10 min with AH23848 (1 μm, 5 μm) and stimulated with PGE2 (in nm) for 5 min. Cell monolayers were lysed and subjected to Western blot analysis using anti-VASP antibodies. Note that the phosphorylation of VASP retards its migration on SDS-PAGE. Data are expressed as the intensity ratio of phosphorylated (upper band) to total (upper and lower bands) VASP protein. Values shown represent mean ± S.E. from three separate experiments. *, p < 0.05 and **, p < 0.01 versus PGE2-treated, but AH23848-untreated samples. B, PKA phosphorylates VASP. RCC7 cells were treated, or not, for 30 min with the PKA inhibitor H89 (0.1 μm to 10 μm) and stimulated with PGE2 (in nm) for 5 min. Cells were analyzed for VASP phosphorylation by immunoblotting with anti-VASP antibodies. Data are expressed as the intensity ratio of phosphorylated (upper band) to total (upper and lower bands) VASP protein. Values shown represent mean ± S.E. from three separate experiments. *, p < 0.05 and **, p < 0.01 versus PGE2-treated, but H89-untreated samples. C, treatment with H89 does not impact the PGE2-induced RCC7 cell invasion. H89 (5–20 μm) was added, or not, to the top and bottom compartments and cells were allowed to invade collagen-coated transwell filters in response to PGE2 (5 nm) stimulation. Each point represents the fold-increase relative to the vehicle alone sample and *, p < 0.05 versus not stimulated (NS) samples.
In addition to PKA, cAMP binds to and activates the Rap guanine nucleotide exchange factor Epac (33, 36). Using Forster resonance energy transfer (FRET) to measure activation-dependent conformational changes in response to cAMP binding (37), we are able to show that PGE2 induces Epac activation (Fig. 4A) in RCC7 cells ectopically expressing Epac1 (fused to CFP at the amino terminus and to YFP at the carboxyl terminus). The PGE2-induced FRET signal was mediated by EP4; antagonism with AH23848 inhibited the FRET response, and a second challenge with PGE2 (after washing cells) elicited a robust FRET signal, excluding the possibility of EP4 desensitization throughout the duration of the experiment (Fig. 4A). PGE2 also induced a significant increase in the levels of GTP-bound Rap1 (Fig. 4, B and C), as determined with a GST pulldown assay using the Ras-binding domain (RBD) of Ral GDP-dissociation stimulator (Ral-GDS). Remarkably, the PGE2-mediated Rap1·GTP accumulation was inhibited when cells were pretreated with the EP4 antagonist AH23848 (Fig. 4, B and C), evidencing an EP4-dependent signal. To directly implicate Epac in PGE2-induced Rap1·GTP accumulation, we knocked down expression of endogenous Epac1 and -2 and examined Rap·GTP levels. Results show that treatment with Epac siRNA abrogated the Rap1·GTP accumulation following stimulation with PGE2 (Fig. 4D), providing support to the conclusion that PGE2 activates Rap, at least in part, through Epac.
FIGURE 4.
PGE2 promotes the Rap activation in RCC7 cells. A, PGE2 induces the EP4-dependent conformational change in Epac1. A representative time course of the YFP:CFP emission ratio in RCC7 cells expressing the cAMP biosensor CFP-Epac1-YFP. Cells were treated with PGE2 (10 nm) alone or together with AH23848 (5 μm), as described. B and C, PGE2 activates Rap. RCC7 cells were treated, or not, with AH23848 (5 μm) for 10 min, then with PGE2 (in nm) for an additional 5 min. Cell lysates were subjected to pulldown assays using GST fusion of the RBD domain of RalGDS protein. Levels of activated Rap1 (Rap1·GTP) and total Rap1 proteins were determined by immunoblotting using anti-Rap1 antibodies. Representative blots are shown in B and data summary are shown in C. Data are presented as fold-increase of basal Rap1·GTP, where the basal amount of Rap1·GTP in untreated cells is assigned a value of 1.0. Data shown represent the mean ± S.E. values from five separate experiments. *, p < 0.05 versus non-stimulated control values. D, knockdown of Epac abrogates the PGE2-induced Rap1·GTP accumulation. RCC7 cells transiently transfected with scrambled siRNA (siCon) or siRNA targeting Epca1 and -2 genes (siEpac) were subjected to Rap1·GTP pulldown assay after treatment with PGE2 (5 nm) for 5 min, as described in B. Data represent the fold-increase relative to nonstimulated (NS) values and *, p < 0.05.
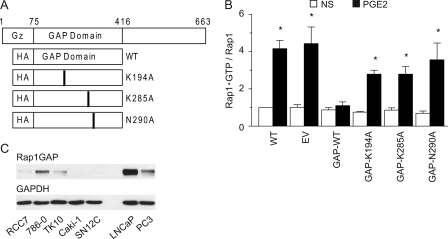
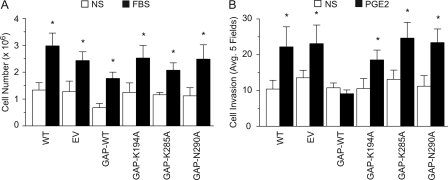
Rap1·GTP accumulation reached maximal levels at PGE2 concentrations (0.5–5 nm) that parallel doses required to induce the RCC7 cell invasion (Fig. 2B). To directly link the PGE2-EP4-mediated RCC7 cell invasion to Rap activation, we created a series of polyclonal cell lines that express either the wild-type or enzymatically inactive GTPase activating protein (GAP) domain only (34, 38, 39) of the Rap inactivator Rap1GAP (Fig. 5A). Rap1GAP acts as a Rap inactivator by stimulating the low intrinsic GTPase activity of the Rap and promoting the GTP hydrolysis. Highly invariant lysine 194 and 285 and asparagine 290 residues are essential for the GAP activity of Rap1GAP: mutation of Lys-194 and Lys-285 reduces Rap1GAP activity by 25- and 100-fold, respectively, and mutation of Asn-290 eliminates any measurable activity (38, 39). Consistent with these results, we find that expression of the wild-type GAP domain only of Rap1GAP obliterates the Rap1 activation (Fig. 5B), whereas expression of the GAP-impaired domains of Rap1GAP showed little effect on the Rap1 activation (Fig. 5B).
FIGURE 5.
Expression of Rap1GAP in human cancer cells. A, schematic presentation of the Rap1GAP protein structure. The GAP domain was cloned as a fusion protein with hemagglutinin (HA) tag. Lysine residues 194 and 285 and asparagine residue 290 were changed to alanine by site-directed mutagenesis. Gz, binding domain to Gαz. B, Rap activation is inhibited by the forced overexpression of wild-type GAP domain of Rap1GAP. RCC7 cells stably expressing GAP domain (wild-type, K194A, K285A, or N290A) of Rap1GAP were stimulated with PGE2 (5 nm), lysed, and mixed with agarose beads conjugated to GST-RalGDS-RBD to capture the Rap1·GTP. Precipitated proteins were analyzed by immunoblotting using anti-Rap1 antibodies. Data are presented as fold-increase of basal Rap1·GTP, where the basal amount of Rap1·GTP in untreated cells is assigned a value of 1.0. Data shown represent the mean ± S.E. values from three separate experiments. *, p < 0.05 versus nonstimulated (NS) control values. WT, wild type RCC7 cells; EV, RCC7 cells stably expressing empty vector. C, expression of Rap1GAP protein in human kidney cancer cell lines. Cell lysates were obtained from the NCI and analyzed by Western blotting for expression of Rap1GAP (upper panel) and GAPDH (lower panel) proteins.
Signaling by activated EP4 and Rap is interdicted by the Rap inactivator Rap1GAP. Protein expression analysis revealed that the invasive (Fig. 2) and metastatic RCC7 (data not shown), like aggressive prostate cancer PC3 cells (40), express less Rap1GAP protein (Fig. 5C). In contrast, the weakly tumorigenic and not metastatic prostate cancer LNCaP cells (40) expressed higher levels of the Rap1GAP protein (Fig. 5C), in agreement with a recent report (41), and supporting the proposition that Rap1GAP expression is lost/decreased in the aggressive cancer cells. To provide evidence for this possibility, we screened the human kidney cancer cell lines (obtained from NCI) for expression of the Rap1GAP and found 60% of the cell lines (3 of 5) do not express detectable Rap1GAP protein (Fig. 5C). Hence, Rap1GAP expression (in both prostate and kidney cancer cell lines) coincides with weak metastatic character, and absence of Rap1GAP expression correlates with the metastatic potential.
Rap Mediates the PGE2-induced RCC7 Cell Invasion
Taking advantage of the reagents that we generated (i.e. the RCC7 cell lines that stably express wild-type or mutated GAP domains of Rap1GAP), we sought to implicate Rap signaling in the PGE2-mediated RCC7 cell invasion. First, we determined the effect of expressing the various GAP domains of Rap1GAP on the RCC7 cell proliferation in serum. The RCC7 cell clones exhibited similar fold-increases in their growth rates, suggesting Rap signaling does not impact proliferation of the RCC7 cells (Fig. 6A). However, PGE2-induced cell invasion was dramatically impaired in the RCC7 cells that express wild-type GAP domain of Rap1GAP (GAP-WT), but not in the cells that express the inactive forms of the GAP domain of Rap1GAP (Fig. 6B). Together, these results demonstrate that PGE2-mediated RCC7 cell invasion is controlled, at least in part, by a EP4 → Rap signal.
FIGURE 6.
Rap mediates the PGE2-induced RCC7 cell invasion. A, role of Rap signaling in RCC7 cell proliferation. Cells stably expressing wild-type or mutated GAP domains of Rap1GAP were grown in starvation medium, or in medium containing 10% fetal bovine serum. The cells were cultured for 3 days, harvested, and stained with trypan blue. The cells excluding the dye were counted under light microscopy with a hemocytometer. Each point represents the mean ± S.E. of values obtained from three experiments. *, p < 0.05 versus same cell type in starvation medium. NS, non-stimulated; WT, control wild-type RCC7 cells; EV, RCC7 cells transfected with empty vector. B, PGE2 induces the Rap-mediated RCC7 cell invasion. RCC7 cells stably expressing the GAP domain (wild-type, K194A, K285A, or N290A) of Rap1GAP were allowed to invade collagen-coated transwell filters in response to stimulation with PGE2 (5 nm). Cells that migrated to the bottom side of the filter were stained and inspected using a phase-contrast microscope. Cells in randomly selected five fields were counted and each experiment was repeated three times. *, p < 0.05 versus not stimulated (NS) samples. WT, wild-type RCC7 cells; EV, RCC7 cells stably expressing empty vector.
DISCUSSION
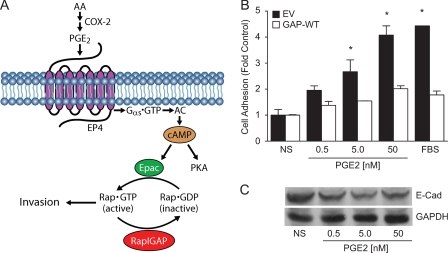
Majority of kidney cancer-related deaths result from cancer metastasis to distal organs and the prognosis of patients with metastatic disease is poor with a median survival of 10 months (4, 42, 43). Although surgery is highly effective for the treatment of localized low-grade RCC (43), current management options of patients diagnosed with locally advanced or metastatic kidney cancer are not curative, reinforcing the need to identify mechanisms involved in kidney cancer initiation, survival, and metastasis as a prerequisite for discovering effective therapeutics. The major finding of this study is that PGE2 promotes clear cell RCC cell invasion by activating cognate receptor EP4, thereby leading to the activation of small GTPase Rap. As depicted schematically in Fig. 7A, the signaling cascade connecting stimulated EP4 to Rap may include heterotrimeric Gαs protein, cAMP, and the Rap activator Epac. Under physiologic conditions, this proinvasive signal is counterbalanced by the Rap inactivator Rap1GAP, whose expression is lost in the clear cell RCC.
FIGURE 7.
Rap mediates the cell adhesion. A, schematic presentation of the signal relay from activated EP4 to Rap and cell invasion. AA, arachidonic acid; AC, adenylyl cyclase. B, effect of Rap activity on cell adhesion. RCC7 cells stably expressing empty vector (EV) or the GAP domain (GAP-WT) of Rap1GAP were suspended in starvation medium supplemented, or not, with PGE2 or FBS. Adherent cells were stained with crystal violet and dye absorbance was measured at a wavelength of 570 nm. Data are presented as fold-increase above basal, where the basal absorbance in untreated cells is assigned a value of 1.0. Data shown represent the mean ± S.E. from three separate experiments. *, p < 0.05 versus nonstimulated (NS) control values. EV and GAP-WT denote, respectively, RCC7 cells stably expressing empty vector and GAP domain of Rap1GAP. C, effect of PGE2 on E-cadherin expression. RCC7 cells were treated, or not, with PGE2 for 24 h and lysates were analyzed by Western blotting for expression of E-cadherin (upper panel) and GAPDH (lower panel) proteins.
PGE2 is a product of the COX-2 enzyme that is overexpressed under pathophysiologic conditions, including kidney cancer (8–10), and is associated with poor prognosis and reduced survival time. Specific COX-2 inhibitors have been tried as therapeutics to treat cancer patients but unwanted cardiovascular and renal (11, 12) side effects limited their application and emphasized a need to identify COX-2 effectors for therapeutic intervention. Expression of the PGE2 receptor EP4 is elevated in RCC cells, and it mediates RCC7 cell invasion. Hence, the targeting of EP4 with specific ligand antagonists or neutralizing antibodies may harness the benefits to interfere with the RCC cell invasion, whereas circumventing the health safety concerns associated with specific COX-2 inhibitors.
Activated EP4 initiates multiple signaling pathways that are transduced by activated Gαs (Fig. 7A), and our gene array results demonstrated the increased expression of Gαs in malignant RCC7 compared with benign HK-2 kidney epithelial cells (data not shown). Activating mutations of GNAS (referred to as gsp oncogene) have been detected in a number of endocrine malignancies, including pituitary and thyroid adenomas (44, 45) and Leydig cell tumors (46). More recent work has revealed activating Gαs mutations in kidney (47) and colorectal and breast (48–50) cancers, and increased Gαs expression in breast cancer associates with poor prognosis (49). Somatic (tumor specific) activating mutations of the GNAS were found in a significant portion of clear cell RCC cases, and it was reported that 16% of the patients (5 of 30) diagnosed with clear cell RCC expressed mutations of the GNAS (47), rendering the enzyme constitutively active. These results link Gαs signaling to kidney carcinogenesis and are consistent with the conclusion that activated EP4 (that signals through Gαs) associates with kidney cancer.
Activated Gαs transduces signals mainly through adenylyl cyclases that produce cAMP (Fig. 7A). The best studied effector of cAMP is PKA, which has been demonstrated to exert cell type- and context-dependent responses ranging from induction of cell proliferation to inhibition of cell survival. Our results show that PKA does not significantly impact the PGE2-regulated RCC7 cell invasion. Rather, we find that Rap mediates the effects of EP4-induced RCC7 cell invasion. Data from several in vitro and in vivo cell-based and mouse models provide evidence that aberrant Rap1 activation, either through activating mutations in its exchange factors or by inactivating mutations in its GAPs, contribute to several types of malignancies (51). Of note, Rap may be activated by several guanine exchange factors, including Epac, C3G, PDZ-GEF, RasGRP, phospholipase Cϵ, and DOCK4 (52).
Although many Rap1 functions are attributed to its ability to regulate integrins and cell adhesion dynamics (53, 54), the responsible mechanisms involved in Rap-mediated cancer progression remain to be elucidated. Our results show that stimulation with PGE2 promotes the RCC7 cell adhesion to collagen (data not shown) and fibronectin (Fig. 7B) and increases the number of focal adhesions (data not shown). In addition, E-cadherin has been suggested as another Rap target, and our data show that stimulation with PGE2 decreases the E-cadherin expression in the RCC7 cells (Fig. 7C). Hence, PGE2-induced RCC7 cell invasion and metastasis may proceed via the activation of multiple Rap effectors that converge to activate the integrins and focal adhesions.
Rap signaling is negatively impacted by the inactivator Rap1GAP (Fig. 7A) and previous studies suggest Rap1GAP and its family members Rap1GAPII, Spa1/SIPA1, and E6TP1 possess tumor metastasis suppressor activities. Rap1GAP protein levels are decreased in several types of cancer, including pancreatic adenocarcinoma (55), papillary thyroid carcinoma (56, 57), colorectal carcinoma (58), and melanoma (59). The implication of Rap1GAP as a tumor suppressor was largely deduced based upon experimental results using forced overexpression of full-length Rap1GAP (60, 61). In addition to its GAP domain, the Rap1GAP protein encompasses regulatory N terminus and C terminus domains (34, 38), rendering it capable of exerting other functions, like regulation of heterotrimeric Gz protein signaling (62) or serving as a docking site for binding partner proteins due to phosphorylation-dependent modification of its C terminus (63, 64). In these experiments, we employed only the GAP domain of Rap1GAP, allowing us to confidently conclude that Rap signals mediate the PGE2-regulated RCC cell invasion.
In summary, we have uncovered a PGE2-controlled signaling pathway that regulates the clear cell RCC cell invasion. The responsible signal pathway contains drug targetable intermediates, including EP4, Epac, and Rap1GAP that may be used to benefit patients with progressive disease. The absence of Rap1GAP together with increased EP4 protein expression in localized tumor cells may also serve as early markers for the development of a more aggressive, invasive phenotype. EP4 signaling may be interdicted with specific ligand antagonists as G protein-coupled receptors have proven to be viable drug targets, accounting for 40–60% of all therapeutic drugs. Hence, the combined targeted inhibition of EP4 activation and rescued expression of Rap1GAP may improve prognosis of patients diagnosed with advanced kidney cancer.
Acknowledgments
We thank our colleagues for providing valuable reagents and the NCI for providing the NCI-60 cancer cell lysate. We also thank Drs. N. Lambert, Z. Nie, P. Arora, and T. Polascik for technical assistance and advice.
This work was supported, in whole or in part, by National Institutes of Health Public Health Service Grant CA129155 from the NCI.
- RCC
- renal cell carcinoma
- PGE
- prostaglandin E
- PKA
- protein kinase A
- EP
- prostaglandin E receptors
- RBD
- Ras-binding domain
- GAP
- GTPase activating protein
- EV
- empty vector
- VASP
- Vasodilator-stimulated phosphoprotein.
REFERENCES
- 1. Jemal A., Siegel R., Ward E., Hao Y., Xu J., Thun M. J. (2009) CA Cancer J. Clin. 59, 225–249 [DOI] [PubMed] [Google Scholar]
- 2. Biswas S., Eisen T. (2009) Nat. Rev. Clin. Oncol. 6, 478–487 [DOI] [PubMed] [Google Scholar]
- 3. Young A. N., Dale J., Yin-Goen Q., Harris W. B., Petros J. A., Datta M. W., Wang M. D., Marshall F. F., Amin M. B. (2006) Urology 67, 873–880 [DOI] [PubMed] [Google Scholar]
- 4. Schlesinger-Raab A., Treiber U., Zaak D., Hölzel D., Engel J. (2008) Eur. J. Cancer 44, 2485–2495 [DOI] [PubMed] [Google Scholar]
- 5. Murai M., Oya M. (2004) Curr. Opin. Urol. 14, 229–233 [DOI] [PubMed] [Google Scholar]
- 6. Hao C. M., Breyer M. D. (2008) Annu. Rev. Physiol. 70, 357–377 [DOI] [PubMed] [Google Scholar]
- 7. Zeldin D. C. (2001) J. Biol. Chem. 276, 36059–36062 [DOI] [PubMed] [Google Scholar]
- 8. Chen Q., Shinohara N., Abe T., Harabayashi T., Nonomura K. (2004) J. Urol. 172, 2153–2157 [DOI] [PubMed] [Google Scholar]
- 9. Mungan M. U., Gurel D., Canda A. E., Tuna B., Yorukoglu K., Kirkali Z. (2006) Eur. Urol. 50, 92–97 [DOI] [PubMed] [Google Scholar]
- 10. Tawfik O. W., Kramer B., Shideler B., Danley M., Kimler B. F., Holzbeierlein J. (2007) Arch. Pathol. Lab. Med. 131, 261–267 [DOI] [PubMed] [Google Scholar]
- 11. Cheng H. F., Harris R. C. (2005) Curr. Pharm. Des. 11, 1795–1804 [DOI] [PubMed] [Google Scholar]
- 12. Lanas A., Hunt R. (2006) Ann. Med. 38, 415–428 [DOI] [PubMed] [Google Scholar]
- 13. Asano T., Shoda J., Ueda T., Kawamoto T., Todoroki T., Shimonishi M., Tanabe T., Sugimoto Y., Ichikawa A., Mutoh M., Tanaka N., Miwa M. (2002) Clin. Cancer Res. 8, 1157–1167 [PubMed] [Google Scholar]
- 14. Pugh S., Thomas G. A. O. (1994) Gut 35, 675–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang D., Wang H., Brown J., Daikoku T., Ning W., Shi Q., Richmond A., Strieter R., Dey S. K., DuBois R. N. (2006) J. Exp. Med. 203, 941–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Muller M., Sales K. J., Katz A. A., Jabbour H. N. (2006) Endocrinology 147, 3356–3365 [DOI] [PubMed] [Google Scholar]
- 17. Sugimoto Y., Narumiya S. (2007) J. Biol. Chem. 282, 11613–11617 [DOI] [PubMed] [Google Scholar]
- 18. Funk C. D., Furci L., FitzGerald G. A., Grygorczyk R., Rochette C., Bayne M. A., Abramovitz M., Adam M., Metters K. M. (1993) J. Biol. Chem. 268, 26767–26772 [PubMed] [Google Scholar]
- 19. Namba T., Sugimoto Y., Negishi M., Irie A., Ushikubi F., Kakizuka A., Ito S., Ichikawa A., Narumiya S. (1993) Nature 365, 166–170 [DOI] [PubMed] [Google Scholar]
- 20. Sonnenburg W. K., Zhu J. H., Smith W. L. (1990) J. Biol. Chem. 265, 8479–8483 [PubMed] [Google Scholar]
- 21. Bastien L., Sawyer N., Grygorczyk R., Metters K. M., Adam M. (1994) J. Biol. Chem. 269, 11873–11877 [PubMed] [Google Scholar]
- 22. Regan J. W., Bailey T. J., Pepperl D. J., Pierce K. L., Bogardus A. M., Donello J. E., Fairbairn C. E., Kedzie K. M., Woodward D. F., Gil D. W. (1994) Mol. Pharmacol. 46, 213–220 [PubMed] [Google Scholar]
- 23. Buchanan F. G., Gorden D. L., Matta P., Shi Q., Matrisian L. M., DuBois R. N. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 1492–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim J. I., Lakshmikanthan V., Frilot N., Daaka Y. (2010) Mol. Cancer Res. 8, 569–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Herrmann C., Horn G., Spaargaren M., Wittinghofer A. (1996) J. Biol. Chem. 271, 6794–6800 [DOI] [PubMed] [Google Scholar]
- 26. Franke B., Akkerman J. W., Bos J. L. (1997) EMBO J. 16, 252–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bisignani G. J., McLaughlin P. J., Ordille S. D., Beltz M. S., Jarowenko M. V., Zagon I. S. (1999) J. Urol. 162, 2186–2191 [DOI] [PubMed] [Google Scholar]
- 28. Nomura T., Huang W. C., Seo S., Zhau H. E., Mimata H., Chung L. W. (2007) J. Urol. 178, 292–300 [DOI] [PubMed] [Google Scholar]
- 29. An Z., Jiang P., Wang X., Moossa A. R., Hoffman R. M. (1999) Clin. Exp. Metastasis 17, 265–270 [DOI] [PubMed] [Google Scholar]
- 30. Kadhim S. A., Bowlin T. L., Waud W. R., Angers E. G., Bibeau L., DeMuys J. M., Bednarski K., Cimpoia A., Attardo G. (1997) Cancer Res. 57, 4803–4810 [PubMed] [Google Scholar]
- 31. Castellone M. D., Teramoto H., Williams B. O., Druey K. M., Gutkind J. S. (2005) Science 310, 1504–1510 [DOI] [PubMed] [Google Scholar]
- 32. Kelly P., Moeller B. J., Juneja J., Booden M. A., Der C. J., Daaka Y., Dewhirst M. W., Fields T. A., Casey P. J. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 8173–8178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bos J. L., de Rooij J., Reedquist K. A. (2001) Nat. Rev. Mol. Cell Biol. 2, 369–377 [DOI] [PubMed] [Google Scholar]
- 34. de Rooij J., Zwartkruis F. J., Verheijen M. H., Cool R. H., Nijman S. M., Wittinghofer A., Bos J. L. (1998) Nature 396, 474–477 [DOI] [PubMed] [Google Scholar]
- 35. Chijiwa T., Mishima A., Hagiwara M., Sano M., Hayashi K., Inoue T., Naito K., Toshioka T., Hidaka H. (1990) J. Biol. Chem. 265, 5267–5272 [PubMed] [Google Scholar]
- 36. Bos J. L. (2007) Trends Biochem. Sci. 31, 680–686 [DOI] [PubMed] [Google Scholar]
- 37. Ponsioen B., Zhao J., Riedl J., Zwartkruis F., van der Krogt G., Zaccolo M., Moolenaar W. H., Bos J. L., Jalink K. (2004) EMBO Rep. 5, 1176–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Brinkmann T., Daumke O., Herbrand U., Kühlmann D., Stege P., Ahmadian M. R., Wittinghofer A. (2002) J. Biol. Chem. 277, 12525–12531 [DOI] [PubMed] [Google Scholar]
- 39. Daumke O., Weyand M., Chakrabarti P. P., Vetter I. R., Wittinghofer A. (2004) Nature 429, 197–201 [DOI] [PubMed] [Google Scholar]
- 40. Sobel R. E., Sadar M. D. (2005) J. Urol. 173, 342–359 [DOI] [PubMed] [Google Scholar]
- 41. Bailey C. L., Kelly P., Casey P. J. (2009) Cancer Res. 69, 4962–4968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Atkins M. B., Regan M., McDermott D. (2004) Clin. Cancer Res. 10, 6342S–6346S [DOI] [PubMed] [Google Scholar]
- 43. Belldegrun A. S., Klatte T., Shuch B., LaRochelle J. C., Miller D. C., Said J. W., Riggs S. B., Zomorodian N., Kabbinavar F. F., Dekernion J. B., Pantuck A. J. (2008) Cancer 113, 2457–2463 [DOI] [PubMed] [Google Scholar]
- 44. Landis C. A., Masters S. B., Spada A., Pace A. M., Bourne H. R., Vallar L. (1989) Nature 340, 692–696 [DOI] [PubMed] [Google Scholar]
- 45. Vallar L., Spada A., Giannattasio G. (1987) Nature 330, 566–568 [DOI] [PubMed] [Google Scholar]
- 46. Fragoso M. C., Latronico A. C., Carvalho F. M., Zerbini M. C., Marcondes J. A., Araujo L. M., Lando V. S., Frazzatto E. T., Mendonca B. B., Villares S. M. (1998) J. Clin. Endocrinol. Metab. 83, 2074–2078 [DOI] [PubMed] [Google Scholar]
- 47. Kalfa N., Lumbroso S., Boulle N., Guiter J., Soustelle L., Costa P., Chapuis H., Baldet P., Sultan C. (2006) J. Urol. 176, 891–895 [DOI] [PubMed] [Google Scholar]
- 48. Chin K., DeVries S., Fridlyand J., Spellman P. T., Roydasgupta R., Kuo W. L., Lapuk A., Neve R. M., Qian Z., Ryder T., Chen F., Feiler H., Tokuyasu T., Kingsley C., Dairkee S., Meng Z., Chew K., Pinkel D., Jain A., Ljung B. M., Esserman L., Albertson D. G., Waldman F. M., Gray J. W. (2006) Cancer Cell 10, 529–541 [DOI] [PubMed] [Google Scholar]
- 49. Neve R. M., Chin K., Fridlyand J., Yeh J., Baehner F. L., Fevr T., Clark L., Bayani N., Coppe J. P., Tong F., Speed T., Spellman P. T., DeVries S., Lapuk A., Wang N. J., Kuo W. L., Stilwell J. L., Pinkel D., Albertson D. G., Waldman F. M., McCormick F., Dickson R. B., Johnson M. D., Lippman M., Ethier S., Gazdar A., Gray J. W. (2006) Cancer Cell 10, 515–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sjöblom T., Jones S., Wood L. D., Parsons D. W., Lin J., Barber T. D., Mandelker D., Leary R. J., Ptak J., Silliman N., Szabo S., Buckhaults P., Farrell C., Meeh P., Markowitz S. D., Willis J., Dawson D., Willson J. K., Gazdar A. F., Hartigan J., Wu L., Liu C., Parmigiani G., Park B. H., Bachman K. E., Papadopoulos N., Vogelstein B., Kinzler K. W., Velculescu V. E. (2006) Science 314, 268–274 [DOI] [PubMed] [Google Scholar]
- 51. Hattori M., Minato N. (2003) J. Biochem. 134, 479–484 [DOI] [PubMed] [Google Scholar]
- 52. Pannekoek W. J., Kooistra M. R., Zwartkruis F. J., Bos J. L. (2009) Biochim. Biophys. Acta 1788, 790–796 [DOI] [PubMed] [Google Scholar]
- 53. Katagiri K., Ohnishi N., Kabashima K., Iyoda T., Takeda N., Shinkai Y., Inaba K., Kinashi T. (2004) Nat. Immunol. 5, 1045–1051 [DOI] [PubMed] [Google Scholar]
- 54. Boettner B., Van Aelst L. (2009) Curr. Opin. Cell Biol. 21, 684–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhang L., Chenwei L., Mahmood R., van Golen K., Greenson J., Li G., D'Silva N. J., Li X., Burant C. F., Logsdon C. D., Simeone D. M. (2006) Cancer Res. 66, 898–906 [DOI] [PubMed] [Google Scholar]
- 56. Zuo H., Gandhi M., Edreira M. M., Hochbaum D., Nimgaonkar V. L., Zhang P., Dipaola J., Evdokimova V., Altschuler D. L., Nikiforov Y. E. (2010) Cancer Res. 70, 1389–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nellore A., Paziana K., Ma C., Tsygankova O. M., Wang Y., Puttaswamy K., Iqbal A. U., Franks S. R., Lv Y., Troxel A. B., Feldman M. D., Meinkoth J. L., Brose M. S. (2009) J. Clin. Endocrinol. Metab. 94, 1026–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tsygankova O. M., Ma C., Tang W., Korch C., Feldman M. D., Lv Y., Brose M. S., Meinkoth J. L. (2010) Mol. Cell Biol. 30, 3262–3274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zheng H., Gao L., Feng Y., Yuan L., Zhao H., Cornelius L. A. (2009) Cancer Res. 69, 449–457 [DOI] [PubMed] [Google Scholar]
- 60. Mitra R. S., Goto M., Lee J. S., Maldonado D., Taylor J. M., Pan Q., Carey T. E., Bradford C. R., Prince M. E., Cordell K. G., Kirkwood K. L., D'Silva N. J. (2008) Cancer Res. 68, 3959–3969 [DOI] [PubMed] [Google Scholar]
- 61. Zhang Z., Mitra R. S., Henson B. S., Datta N. S., McCauley L. K., Kumar P., Lee J. S., Carey T. E., D'Silva N. J. (2006) Am. J. Pathol. 168, 585–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Meng J., Glick J. L., Polakis P., Casey P. J. (1999) J. Biol. Chem. 274, 36663–36669 [DOI] [PubMed] [Google Scholar]
- 63. Polakis P., Rubinfeld B., McCormick F. (1992) J. Biol. Chem. 267, 10780–10785 [PubMed] [Google Scholar]
- 64. McAvoy T., Zhou M. M., Greengard P., Nairn A. C. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 3531–3536 [DOI] [PMC free article] [PubMed] [Google Scholar]