Abstract
Caspase-2, the most evolutionarily conserved member in the human caspase family, may play important roles in stress-induced apoptosis, cell cycle regulation, and tumor suppression. In biochemical assays, caspase-2 uniquely prefers a pentapeptide (such as VDVAD) rather than a tetrapeptide, as required for efficient cleavage by other caspases. We investigated the molecular basis for pentapeptide specificity using peptide analog inhibitors and substrates that vary at the P5 position. We determined the crystal structures of apo caspase-2, caspase-2 in complex with peptide inhibitors VDVAD-CHO, ADVAD-CHO, and DVAD-CHO, and a T380A mutant of caspase-2 in complex with VDVAD-CHO. Two residues, Thr-380 and Tyr-420, are identified to be critical for the P5 residue recognition; mutation of the two residues reduces the catalytic efficiency by about 4- and 40-fold, respectively. The structures also provide a series of snapshots of caspase-2 in different catalytic states, shedding light on the mechanism of capase-2 activation, substrate binding, and catalysis. By comparing the apo and inhibited caspase-2 structures, we propose that the disruption of a non-conserved salt bridge between Glu-217 and the invariant Arg-378 is important for the activation of caspase-2. These findings broaden our understanding of caspase-2 substrate specificity and catalysis.
Keywords: Apoptosis, Caspase, Enzyme Inhibitors, Enzyme Structure, Huntington Disease, Peptide Interactions, X-ray Crystallography
Introduction
Caspases are a family of cysteine-dependent aspartate-specific proteases that have essential roles in various cellular processes, such as apoptosis, proliferation, and differentiation, as well as in pathological conditions including cancer and inflammation (1). Caspases are synthesized and stored as inactive zymogens, and their activities are strictly regulated by protein-protein interactions and by proteolysis. Based on function and prodomain structure, caspases are grouped into inflammatory and apoptotic caspases (2). In the apoptotic cascade, caspases are further divided into initiator caspases and executioner caspases. Initiator caspases (such as caspase-8 and -9) are activated when they bind to multiprotein complexes that oligomerize and orient the caspases to cleave themselves. Activated initiator caspases then cleave executioner caspases such as caspase-3 and -7, which further cleave cellular proteins, leading to cell death (3). Although the roles of these four caspases have been characterized in detail, the function of some of the other caspases, including caspase-2, is not well understood (4).
Caspase-2 is the first identified mammalian member and the most conserved member of the caspase family; nevertheless, its physiological role is enigmatic. Caspase-2 possesses features of both initiator and executioner caspases (5). The predicted substrate specificity of caspase-2 is more like that of caspase-3 and -7, linking it to executioner caspases, whereas its long N-terminal caspase recruitment domain (CARD)2 suggests a potential role as an initiator caspase. Moreover, the lack of an obvious phenotype in gene knock-out mice makes its essential role in development unclear (6). Caspase-2 may have important functions in stress-induced cell death pathways (7). For instance, the expression level of caspase-2 has been shown to be elevated in the brains of patients with some neurodegenerative diseases (8). In addition, evidence for the potential roles of caspase-2 in non-apoptotic pathways, including cell cycle regulation and DNA repair, is emerging (9, 10). Despite this progress, many questions about caspase-2 remain. For instance, exactly how and under what conditions caspase-2 becomes activated is not well known. Whether caspase-2 is mandatory for apoptosis under specific circumstances or whether it primarily functions in cell cycle regulation is yet to be determined (7). To more fully investigate the true roles of caspase-2, it would be very helpful to develop caspase-2-specific probes.
In this study, we investigated the substrate specificity and catalysis of caspase-2 biochemically and structurally. Previously, the substrate specificity of caspases has been studied with short peptide substrates; in general, most caspases recognize four-residue sequences on the N-terminal side of the scissile bond, and there is little prime-side sequence selectivity beyond a preference for small residues. Interestingly, caspase-2 is the only family member to show a strong preference for pentapeptides instead of tetrapeptides, with VDVAD being its optimal substrate sequence (11–13). Thus, there may be opportunities to build more selective caspase-2 inhibitors by understanding the importance of the P5 position. By using fluorogenic peptide substrates with the sequences VDVAD, ADVAD, and DVAD, we quantitatively dissect the impact of the side chain and backbone of the P5 residue on caspase-2 catalytic efficiency. Based on the x-ray crystal structures, enzymatic measurements, and mutational analysis, we identify Thr-380 and Tyr-420 as the key residues in the P5 recognition. Additionally, the five structures described here serve as snapshots along the catalytic pathway. Based on these images, we propose that the breaking of a salt bridge between Glu-217 and Arg-378 is important in the activation of caspase-2.
MATERIALS AND METHODS
Reagents
Acetyl (Ac)-VDVAD-CHO and Ac-VDVAD-AFC (AFC = 7-amino-4-trifluoromethylcoumarin) were purchased from AnaSpec, Inc. (Fremont, CA). Ac-ADVAD-CHO, Ac-ADVAD-AFC, Ac-DVAD-CHO, and Ac-DVAD-AFC were custom-synthesized by Elim Biopharmaceuticals Inc. (Hayward, CA).
Cloning, Expression, and Purification of ΔCARD Caspase-2
The gene encoding caspase-2 (residues 170–452) was amplified from full-length human caspase-2 cDNA (OriGene) by PCR using the primers 5′-AACATATGGGTCCTGTCTGCCTTCAGGTG-3′ and 5′-AACTCGAGTGTGGGAGGGTGTCC-3′ (restriction sites are underlined). The amplified insert was digested with NdeI and XhoI and cloned into the pET23b expression vector (Novagen), yielding a construct encoding caspase-2 with a C-terminal His6 tag. Escherichia coli BL21(DE3) pLysS cells were transformed with the resulting plasmid, and the transformant was grown in LB medium at 37 °C to an A600 of 0.6. The culture was then cooled to 16 °C, induced with 0.2 mm isopropyl β-d-galactopyranoside, and incubated for an additional 14 h at 16 °C. The cells were centrifuged at 5000 rpm for 15 min and resuspended in lysis/wash buffer (100 mm NaCl, 100 mm Tris, pH 8.0). The resuspended cells were passed through a Microfluidizer (microfluidics) five times to ensure lysis. The cell debris was removed by centrifugation, and the supernatant was applied to a 1-ml HisTrap FF column (GE Healthcare) equilibrated with 100% buffer A (100 mm NaCl, 100 mm Tris, pH 8.0). Untagged protein was removed by washing with 20% buffer B (100 mm NaCl, 100 mm Tris, pH 8.0, 200 mm imidazole). Caspase-2 was eluted with 100% buffer B into 40 ml of 20 mm Tris, pH 8. Final purification was achieved on a Hi-Trap Q anion exchange column (GE Healthcare) with an increasing linear gradient of 0–1 m NaCl. Caspase-2 eluted at ∼200 mm NaCl. The purified protein was concentrated to ∼10 mg/ml using a Millipore Ultrafree device with a molecular mass cutoff of 10,000 Da. Typical yields of purified protein, as determined by Bradford assay (14), were 2–4.5 mg/liter of culture. Protein was characterized by active-site titration using Z-VAD-FMK and found to be 100% active.
Caspase-2 Assay
Enzymatic activity of caspase-2 was determined using the fluorescent caspase-2 substrate Ac-VDVAD-AFC (AnaSpec, Inc.). AFC released by substrate cleavage was detected at λex = 390 nm and λem = 490 nm using an Analyst HT plate reader (Molecular Devices, Sunnyvale, CA). GraphPad Prism was used to obtain Km and kcat values by fitting reaction velocities into the Michaelis-Menten equation. For the measurement of IC50 of the peptide aldehyde inhibitors, caspase-2 was preincubated with the peptide aldehyde in assay buffer (50 mm Mes, pH 6.5, 150 mm NaCl, 1.5% sucrose, 10 mm DTT, and 0.05% Chaps) at room temperature for 10 min. The reaction was quenched by 20 μl of 4 m acetic acid 1 h after the substrate was added. IC50 values were determined by a dose-response curve using GraphPad Prism.
Construction, Expression, and Purification of Caspase-2 Mutants T380A, Y420A, and T380A/Y420A
Mutagenesis was performed with the QuikChange site-directed mutagenesis kit (Stratagene). Primers 5′-CTGCCGCCATGCGGAACGCCAAACGAGGTTC-3′, 5′-CAAGGATCGGGAAGGTGCTGCTCCTGGCACAG-3′, and their complementary oligonucleotides were used to introduce T380A and Y420A mutations, respectively, and the double mutant T380A/Y420A into caspase-2. The expression and purification of the mutants were carried out in the same manner as for wild type caspase-2.
Crystallization and Data Collection
The peptide aldehyde inhibitors were dissolved in dimethyl sulfoxide. Three mg/ml wild type or mutant caspase-2 was allowed to incubate with 1–5 mm aldehyde inhibitors at room temperature for 2 h. Crystals were then grown at room temperature, using the hanging-drop vapor diffusion method with 2 μl of protein or protein-inhibitor complex solution and 2 μl of mother liquor. The well buffer contained 0.1 m HEPES (pH 7.0), 15% PEG 3350, 3 mm DTT. The crystals belonged to space group P212121 and contained two monomers per asymmetric unit. The crystals were harvested in mother liquor with additional 15% PEG 3350 and frozen in liquid nitrogen. Diffraction data sets were collected on beam-line 831 at the Advanced Light Source (ALS) and were processed with the HKL2000 package (15). The crystallographic data are summarized in supplemental Table S1.
Structure Refinement and Model Building
Initial phases were obtained by molecular replacement using the program Molrep in CCP4i with the previously published caspase-2 structure (Protein Data Bank (PDB) ID: 1PYO) as the search model (16). The structure was further refined with Refmac 5 and Phenix and was then fit manually with Coot (17, 18). Both the 2Fo − Fc and the Fo − Fc maps showed the presence of CHO inhibitors in the active site of wild type or mutant caspase-2. Water molecules were added with Coot followed by visual inspection with Coot at the final stage. A total of 5% of the reflections were excluded from refinement and constituted the Rfree set. The final geometry was assessed with PROCHECK (19). Figures were generated with PyMOL (20).
RESULTS AND DISCUSSION
Cloning, Expression and Purification of ΔCARD Caspase-2
Like other caspases, caspase-2 is synthesized as an inactive zymogen. The zymogen sequence includes a long prodomain containing a CARD followed by a large domain, a linker, and a small domain. Caspase-2 undergoes autocatalytic activation to remove the prodomain and linker region to generate a stable dimer consisting of the large subunit (p19) and the small subunit (p12). This p19/p12 dimer self-associates to form the active caspase-2 (variously called a dimer, a tetramer, or a dimer-of-dimers). In this work, we cloned and expressed caspase-2 without the CARD domain in E. coli and purified it by a method similar to that used for other caspases (21). Caspase-2 underwent autoproteolysis at the linker region and was purified as soluble active protein in E. coli Bl21 (DE3) pLysS cells. Assessed by mass spectrometry, the mature caspase-2 contained the p19 large subunit (residues 170–333) and the p12 small subunit (residues 348–452 plus the His6 tag) (supplemental Fig. S1). Caspase-2 expressed in this way has a kcat of 0.60 s−1, and the substrate Ac-VDVAD-AFC has a Km of 25 μm, which is comparable with the activity of the GST-tagged caspase-2 reported previously (22).
Enzymatic Analysis of the Role of P5 in Caspase-2/Substrate Recognition
Previous studies have shown that caspase-2 catalytic efficiency toward pentapeptide substrates is 10–40-fold more efficient than with tetrapeptide substrates and that VDVAD is the optimal sequence for caspase-2 recognition (11–13). To investigate the functional impact of the P5 side chain in caspase-2 substrate recognition, three peptide substrates, Ac-VDVAD-AFC, Ac-ADVAD-AFC, and Ac-DVAD-AFC, were assayed for hydrolysis by caspase-2 (bolded letters for emphasis). As shown in Table 1, truncation of the side chain of the P5 residue (Val → Ala) caused ∼5-fold reduction in catalytic efficiency (kcat/Km). This reduction is manifested by an increased Km and no change in the turnover rate (kcat). Deletion of the P5 residue caused a more dramatic ∼25-fold reduction in catalytic efficiency; the effect appeared in both Km and kcat. For amide bond hydrolysis, formation of the protein-acyl intermediate is rate-limiting, and therefore Km is a reasonable measure of the substrate dissociation constant (23). These data suggest that the side chain of the P5 residue affects the peptide substrate binding to the enzyme, whereas the backbone of the P5 residue significantly affects the enzyme turnover rate.
TABLE 1.
Kinetic data for wild type caspase-2 versus different peptide substrates
|
Km |
kcat |
kcat/Km |
||||
|---|---|---|---|---|---|---|
| μm | Relative | s−1 | Relative | 1/s × μm | Relative | |
| Ac-VDVAD-AFC | 25 | 1.00a | 0.60 | 1.00a | 0.024 | 1.00a |
| Ac-ADVAD-AFC | 150 | 5.80 | 0.81 | 1.40 | 0.0055 | 0.23 |
| Ac-DVAD-AFC | 92 | 3.60 | 0.12 | 0.21 | 0.0013 | 0.057 |
a Relative activity defined by Ac-VDVAD-AFC cleaved by wild type caspase-2. Results are average of n = 3.
To further evaluate the significance of the P5 residue in caspase-2 recognition, inhibition constants for caspase-2 were also measured for the three substrate analogues, Ac-VDVAD-CHO, Ac-ADVAD-CHO, and Ac-DVAD-CHO, which have the same sequences as the fluorogenic substrates (Table 2). Ac-VDVAD-CHO, the canonical sequence, is the strongest inhibitor, with an IC50 of 25 nm. Ac-ADVAD-CHO, which lacks the side chain at the P5 position, is ∼4-fold weaker, and Ac-DVAD-CHO lacking the entire P5 residue is ∼20-fold weaker than Ac-VDVAD-CHO. These data are consistent with kinetic parameters found for the analogous substrates.
TABLE 2.
Inhibition of wild type caspase-2 by peptide aldehydes
| IC50 | Relative activity | |
|---|---|---|
| nm | ||
| Ac-VDVAD-CHO | 25 | 1.00 (defined) |
| Ac-ADVAD-CHO | 110 | 0.23 |
| Ac-DVAD-CHO | 710 | 0.035 |
Structural Investigation into Caspase-2 Pentapeptide Specificity
To investigate the structural basis for the P5 recognition of caspase-2, we determined the crystal structures of apo caspase-2 and caspase-2 in complex with Ac-VDVAD-CHO, Ac-ADVAD-CHO, and Ac-DVAD-CHO (supplemental Table S1). All proteins crystallized in the orthorhombic space group P212121 with two p19/p12 heterodimers per asymmetric unit. Unit cells were of similar size, although apo caspase-2 crystallized in a larger unit cell than inhibited caspase-2. The structures were determined to resolutions of 1.49–2.40 Å with R-factors between 0.19 and 0.23. The terminal residues 170–173 of the p19 subunit and residues 348–353 of the p12 subunit were not visible in any of the structures due to poor electron density.
The overall fold of all the caspase-2 structures including the apo structure was very similar to that of caspase-2·LDESD and to the active conformation for other caspases reported previously (24–26) (Fig. 1). In the casp2·VDVAD and casp2·ADVAD structures, there were two copies of the substrate analog inhibitors, one in each active site in the caspase-2 dimer. Depending on the concentration of Ac-DVAD-CHO used in the co-crystallization condition, the structure of casp2·DVAD contained either one or two copies of DVAD-CHO in the active sites. These variously occupied caspase-2 structures provided a series of snapshots along the enzymatic pathway (discussed below).
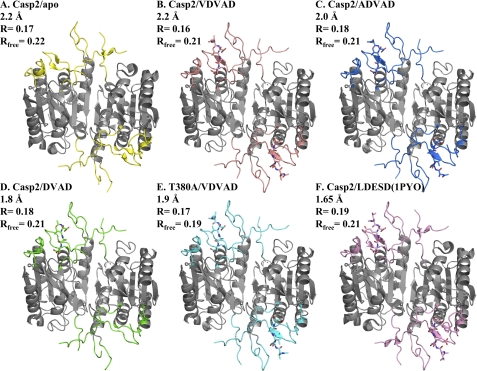
FIGURE 1.
Overall structures of apo and inhibited caspase-2. A–F, apo caspase-2 (PDB ID: 3R7S) (A); caspase-2·VDVAD complex (PDB ID: 3R6G) (B); caspase-2·ADVAD complex (PDB ID: 3R5J) (C); caspase-2·DVAD complex (PDB ID: 3R7B) (D); T380A·VDVAD complex (PDB ID: 3R6L) (D); and previously published structure of caspase-2·LDESD complex (PDB ID: 1PYO) (F). The differences among the structures are located in substrate binding loops, which are colored differently in each structure and are described under “Results” (loop 1 = residues 207–222, loop 2 = residues 318–332, loop 2′ = residues 355–362, loop 3 = residues 374–385, loop 4 = residues 418–429). The rest of the backbone is colored in gray. Yellow, apo casp2·apo; salmon, casp2·VDVAD; marine, casp2·ADVAD; green, casp2·DVAD; cyan, T380A·VDVAD; pink, 1PYO. The same color scheme is used throughout these figures.
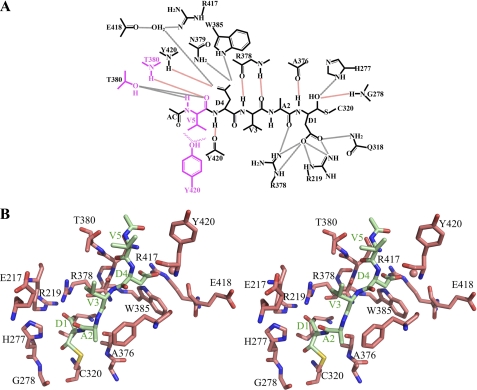
The peptide inhibitors are clearly observed in the electron density map; thus, their positions in the caspase-2 active site are well defined (supplemental Fig. S2). The P1–P4 residues DVAD have the same conformation in all three structures (Figs. 2 and 3 and supplemental Fig. S3). The backbone carbonyl of the P1 Asp is held in position through hydrogen bonds with the side chain of His-277 and backbone nitrogen atom of Gly-278. The P1 Asp side chain is deeply buried in the positively charged S1 pocket formed by the conserved residues Arg-219, Gln-318, and Arg-378, as is similarly found for other caspases (25, 26). The backbone nitrogen of P1 is hydrogen-bonded to the backbone of Ala-376. The carbonyl oxygen of P2 Ala forms a hydrogen bond with the side chain of Arg-378, whereas its side chain interacts with Phe-279 by van der Waal forces. The P3 Val is only involved in main chain hydrogen bonds with Arg-378, and its side chain makes no contacts with the enzyme. The nitrogen atom of P4 Asp forms a hydrogen bond with the carbonyl oxygen of Tyr-420, and its side chain extensively interacts with the enzyme through hydrogen bonds with the backbone of Tyr-420 and the side chains of Trp-385, Asn-379, Arg-417, and Glu-418.
FIGURE 2.
Caspase-2 interactions with peptide inhibitors. A, schematic diagram emphasizing protein/inhibitor contacts for caspase-2·VDVAD-CHO. Backbone hydrogen bonds are shown in black dotted lines, main chain hydrogen bonds are shown in red dotted lines, and hydrophobic interactions are shown in wavy lines. The P5 residue and its interacting residues are shown in magenta. Analogous diagrams for three other caspase-2·inhibitor structures are shown in supplemental Fig. S3. B, stereo image of the substrate-binding groove in the complex of casp2·VDVAD. Caspase-2 is shown in salmon; Ac-VDVAD-CHO is shown in light green.
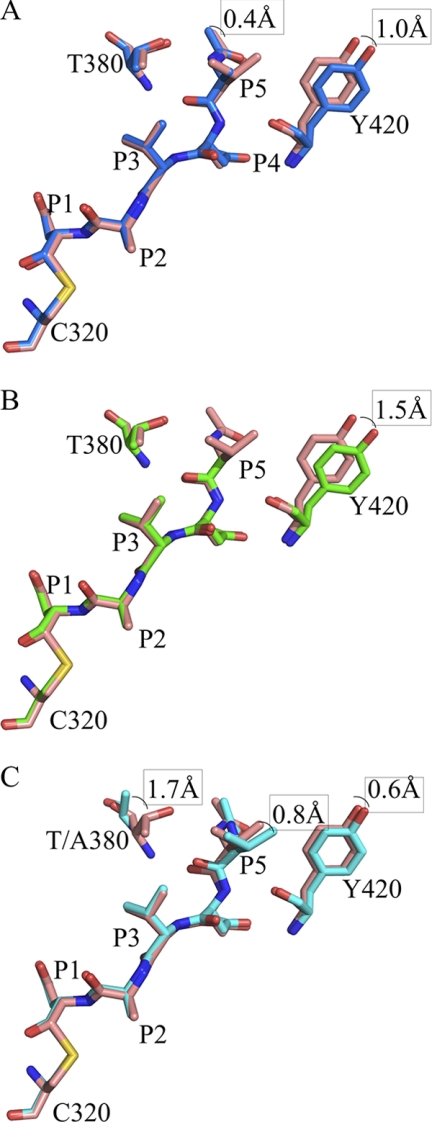
FIGURE 3.
Overlay of the peptide aldehyde and caspase-2 residues described under “Results” for casp2·VDVAD and casp2·ADVAD (A), casp2·VDVAD and casp2·DVAD (3R7B) (B), and casp2·VDVAD and T380A·VDVAD (C).
Not surprisingly, the P5 residue and its interacting residues on caspase-2 are in different conformations in each structure (Fig. 3 and supplemental Fig. S3). In the caspase-2·VDVAD complex, the P5 Val mainly interacts with two residues: Thr-380 and Tyr-420. The inhibitor backbone forms hydrogen bonds with the backbone oxygen atoms from Thr-380 and Tyr-420, as well as the side chain oxygen atom of Thr-380. Additional hydrophobic interactions are observed between the side chains of the P5 Val and Tyr-420 (Fig. 2). The casp2·ADVAD structure is almost identical to the casp2·VDVAD structure, with a root mean square deviation between the two structures of 0.13 Å for 2996 atoms. The most notable change is the side chain of residue Tyr-420, which shifts away from the peptide inhibitor, giving a 1.0 Å shift in the hydroxyl oxygen. Additionally, the P5 Ac-Ala shifts slightly toward Thr-380, reaching a 0.4 Å displacement at the tip of the acetyl group. Taken together, these two shifts cause the distance between the Cα atom of P5 Ala and the Cβ atom of Tyr-420 to be 0.7 Å longer than in the structure of casp2·VDVAD (Fig. 3A). Finally, in the casp2·DVAD structure, Tyr-420 is 1.5 Å farther away from the substrate-binding groove when compared with its position in the casp2·VDVAD structure (measured by the Cβ atoms). In contrast, Thr-380 takes the same position as its equivalent in the pentapeptide structures, presumably because it forms a hydrogen bond with the acetyl group at the N terminus of the peptide (Fig. 3B). These observations support the significance of Thr-380 and Tyr-420 in P5 recognition.
Mutational Studies Confirm the Importance of Thr-380 and Tyr-420 in P5 Recognition
To further probe the importance of Thr-380 and Tyr-420 in mediating substrate specificity, we tested the catalytic activity of T380A and Y420A single mutants as well as the T380A/Y420A double mutant (Table 3). Because the side chain of Thr-380 makes hydrogen bonds with the backbone atoms of P5, we hypothesized that T380A would remove these hydrogen bonds and thus show a catalytic efficiency similar to deletion of the P5 residue. As predicted, the catalytic efficiency of the T380A mutant with Ac-VDVAD-AFC was similar to that of wild type caspase-2 against the tetrapeptide substrate Ac-DVAD-AFC (-fold change in kcat/Km = 0.025 versus 0.057, respectively; Tables 1 and 3). We also expected that the Y420A mutant would lose the hydrophobic interaction between the side chains of Tyr-420 and the P5 Val; thus, the overall effect would be equivalent to the wild type caspase-2 against the Ac-ADVAD-AFC substrate. Consistently, the catalytic efficiency of Y420A with Ac-VDVAD-AFC (Table 3) and the catalytic efficiency of wild type caspase-2 with Ac-ADVAD-AFC (Table 1) are ∼20% of the wild type caspase-2 with Ac-VDVAD-AFC. Finally, the T380A/Y420A double mutant loses both backbone hydrogen bonds and side chain hydrophobic interactions with the P5 residue; therefore it was expected to have much lower activity against the pentapeptide substrate than the wild type enzyme. In fact, the double mutant did not show any activity up to 400 μm. We conclude that the hydrophobic interaction between the side chains of Tyr-420 and P5 Val decreases the binding affinity by 4–5-fold but has no effect on the turnover rate, whereas the hydrogen bonds between the backbone of P5 Val and the side chain of Thr-380 causes reductions in both binding affinity and turnover rate, accounting for ∼40-fold total reduction in catalytic efficiency of caspase-2.
TABLE 3.
Kinetic data for caspase-2 mutants versus Ac-VDVAD-AFC
|
Km |
kcat |
kcat/Km |
||||
|---|---|---|---|---|---|---|
| μm | Relative | s−1 | Relative | 1/s μm | Relative | |
| Wild type | 25 | 1.00a | 0.60 | 1.00a | 0.024 | 1.00a |
| T380A | 220 | 8.60 | 0.130 | 0.22 | 0.00060 | 0.025 |
| Y420A | 84 | 3.30 | 0.520 | 0.87 | 0.0062 | 0.26 |
| T380A/Y420A | No activityb | |||||
a Relative activity defined by Ac-VDVAD-AFC cleaved by wild type caspase-2. Results are average of n = 3.
b Tested up to 400 μm substrate.
The x-ray structure of T380A-caspase-2 with Ac-VDVAD-CHO reveals the basis for the 40-fold loss of activity (Fig. 3C). The overall structure of T380A/VDVAD is very similar to the structure of wild type caspase-2·VDVAD, with a root mean square deviation of 0.14 Å for 3005 atoms (Fig. 1). On the enzyme, the major difference is in loop 3, which contains the T380A mutation. The Cα atom of the residue 380 in the mutant moves 1.7 Å away from the substrate peptide when compared with the wild type (Fig. 3C). On the substrate, the P1–P4 residues align extremely well in the two structures, but the distance between the Cα atoms of P5 Val in the two structures is 0.8 Å. Together, the overall distance between Cα of P5 Val and residue 380 in the enzyme is more than 2 Å longer in the T380A mutant structure than in the wild type. Because P5 Val shifts in the T380A structure and because the Tyr-420-containing loop 4 also shifts in the same direction, the relative distance between P5 Val and Tyr-420 remains the same in the structures of T380A·VDVAD and WT·VDVAD.
In summary, the crystal structures of caspase-2 in complex with different peptide inhibitors are nearly identical except in loops 3 and 4, where Thr-380 and Tyr-420 are located. When these residues are mutated to alanine, the loss of the hydrogen bond between Thr-380 and the P5 backbone causes a 2.3 Å movement in the main chain in residue 380 and a 40-fold loss in catalytic activity, whereas the loss of the hydrophobic interaction between Tyr-420 and the P5 side chain causes a 0.5 Å movement of the side chain of Tyr-420 and a 5-fold loss in activity. These results are consistent with Weber and co-workers (27), who showed that caspase-3 and -6 containing Thr or Ser at the Thr-380-equivalent position and Phe at the position equivalent to Tyr-420 modestly favor pentapeptide over tetrapeptide substrates, whereas caspase-7 and -8 containing Pro at the position equivalent to Thr-380 showed no preference for pentapeptide. These results highlight the importance of a small set of residues in substrate recognition, although the effect of the P5/S5 interaction is much more dramatic for caspase-2 than for other caspases.
Caspase-2 in Action
The crystal structures of caspase-2 in the absence and presence of aldehyde inhibitors DVAD-CHO and VDVAD-CHO provide an interesting set of snapshots along the catalytic pathway. Here, we consider three steps: the mechanism of enzyme activation, substrate binding, and formation of the tetrahedral intermediate after nucleophilic attack.
Substrate Binding Requires Breaking a Salt Bridge
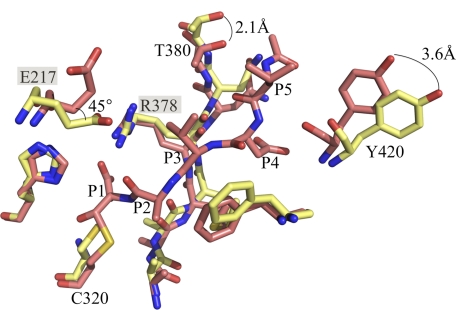
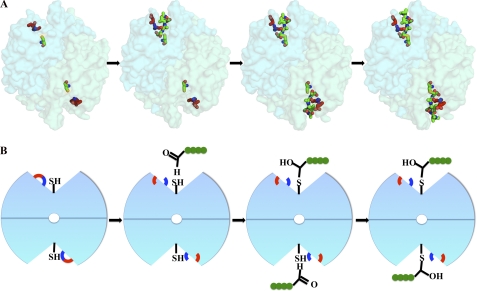
The apo caspase-2 and inhibited caspase-2 structures show the enzyme in very similar conformations; nevertheless, subtle changes are necessary to make the enzyme ready for catalysis. The most significant differences are observed in the four substrate-binding loops. Similar situations have been reported for caspase-3 and -7, where a Tyr side chain in loop 3 occludes a portion of the substrate-binding site in the apo structures. Flipping of this Tyr side chain by ∼90° has been proposed to be an essential step in the activation of caspase-3 and capase-7 (28). This mechanism is not relevant for caspase-2, where the analogous position is an alanine residue; however, caspase-2 may instead require the breaking of a salt bridge during its activation. In the apo caspase-2 structure, Glu-217 in loop 1 forms a salt bridge with the invariant Arg-378 (Fig. 4). In the inhibited caspase-2 structures, the side chain of Glu-217 has rotated 110 degrees around the Cβ-Cγ bond and 45 degrees around the Cγ-Cδ bond. The salt bridge between Glu-217 and Arg-378 is therefore broken in the peptide-bound structures. Arg-378 lies in the S1 pocket and is important for P1 aspartate specificity; thus, breaking the salt bridge must precede substrate binding. The salt bridge is unique to caspase-2 as no other caspase has Glu in the analogous position (Fig. 5).
FIGURE 4.
Comparison between apo caspase-2 (yellow) and caspase-2 bound to VDVAD-CHO (salmon). The salt bridge between Glu-217 and Arg-378 is broken in the casp2·VDVAD complex. The two key residues Thr-380 and Tyr-420 in P5 recognition move 2.1 and 3.6 Å, respectively.
FIGURE 5.
Sequence alignment of human caspases. Caspase-2 is the only human caspase with glutamate at position 217.
Formation of a Half-site Complex
Because the tetrapeptide inhibitor Ac-DVAD-CHO binds weakly to caspase-2, we were able to crystallize the dimeric enzyme bound to either one or two equivalents of Ac-DVAD-CHO by varying the concentration of inhibitor. Crystallization in the presence of 1 mm Ac-DVAD-CHO yields a structure with only one copy of the inhibitor bound to caspase-2. From the electron density, it is clear that the tetrapeptide is fully bound to one site rather than randomly distributed between the two active sites. This half-site binding is not due to crystal packing as the surface contacts are unchanged between singly and doubly inhibited caspase-2 structures. In the occupied active site, the tetrapeptide inhibitor adopts an extended conformation very similar to the P1–P4 residues in the other two complexes (Fig. 3B and supplemental Fig. S3). However, the carbon atom in the aldehyde group is slightly farther away from the thiol sulfur atom of Cys-320 (2.4 Å versus 2.2 Å in the VDVAD structure); additionally, this carbon atom is closer to an sp2 configuration than in the caspase-2·VDVAD structure (Fig. 3B). These two observations suggest that the carbonyl has not fully undergone nucleophilic attack from the thiol group of Cys-320. Thus, this structure is more similar to the enzyme-substrate (ES) complex.
The unoccupied active site is less well defined than the occupied active site, especially in the loop 1, loop 3, and loop 4 regions where several residues are not modeled due to poor electron density. Nonetheless, well defined residues in the empty subunit overlap well with their equivalents in the occupied subunit. The key residues involved in catalysis, including the catalytic dyad and the invariant Arg-378, superpose with each other regardless of the peptide inhibitor binding. Loop 2′, which is critical for active-site formation (29), also overlaps well with the loop 2′ in the occupied subunit. Taken together, these observations indicate that the empty subunit is also in a catalytically active conformation. Thus, this partially occupied structure suggests that both active sites can be formed upon binding of a single substrate.
The effect of half-site reactivity on caspase activation varies among family members. Long prodomain caspases, including caspase-1 and -9, rest in an inactive conformation, and binding to one active site strongly activates the second site and/or stabilizes the dimer interface (30, 31). By contrast, executioner caspase-3, -6, and -7 crystallize in a mostly active conformation and require only minor changes to form the catalytic site. In the case of caspase-2, the Hill coefficient is 1.0, indicating that the turnover of DVAD-AFC or VDVAD-AFC by caspase-2 is not cooperative (supplemental Fig. S4) and that the comparatively small conformational changes required for caspase-2 activation are not rate-limiting. In this way, caspase-2 is more similar to the executioner caspases than to the long prodomain caspases.
Formation of the Tetrahedral Intermediate
By increasing the concentration of Ac-DVAD-CHO to 5 mm, we obtained a second crystal structure of caspase-2·DVAD complex. In this structure, both active sites are occupied by the tetrapeptide aldehyde. Close inspection of the structure reveals that the two active sites are slightly different. In one active site, the C-S bond is long (2.3 Å), as seen in the singly occupied structure. In the other active site, the carbon atom of the aldehyde is clearly covalently linked to the sulfur atom of the catalytic residue Cys-320 via a hemithioacetal bond (2.0 Å; Fig. 6). This suggests that this site of the high concentration complex is more similar to the tetrahedral intermediate (ES≠ complex) prior to the product release. This structure thus represents an intermediate state between the singly occupied tetrapeptide complex structure and doubly occupied VDVAD structure in which both active sites are occupied and in the ES≠ state.
FIGURE 6.
Visualizing of caspase-2 catalysis. A, crystal structures visualized in PyMOL. Glu-217 is shown in red, Arg-378 is shown in blue, catalytic residue Cys-320 is shown in orange, and the backbone peptide inhibitor is shown in green. B, graphics emphasizing the steps in catalysis seen in the corresponding structures: step 1, breaking of the salt bridge between Glu-217 and Arg-378; step 2, binding of substrates sequentially into the two active sites; and step 3, formation of ES≠ complex after nucleophilic attack.
Overall, the structures visualize multiple steps in the catalytic pathway, summarized in Fig. 6. In apo caspase-2, the substrate binding loops are flexible and unstructured, and the protein is held in a non-substrate binding conformation by formation of a salt bridge involving Arg-378. Comparison of apo caspase-2 with the singly occupied caspase-2·DVAD complex suggests that the breaking of the salt bridge and the ordering of catalytic dyad into the active conformation occur upon binding of a single substrate. Subsequent to binding of the substrate in both active sites, the catalytic cysteine nucleophilic attacks the carbonyl carbon on the substrate, forming the ES≠ complex.
Conclusions
The crystal structures of caspase-2 in complex with several peptide inhibitors reveal the molecular basis for the P5 recognition and provide snapshots along the catalytic pathway. Residues Thr-380 and Tyr-420 contain the key elements for binding the P5 residue. When Thr-380 is mutated to alanine, the loss of the hydrogen bond between Thr-380 and the P5 side chain causes a 2.3 Å movement in the main chain in residue 380 and a 40-fold loss in catalytic efficiency. When Tyr-420 is mutated to alanine, the loss of the hydrophobic interaction between Tyr-420 and the P5 side chain causes a 0.5 Å movement of the side chain of Tyr-420 and a 5-fold loss in substrate binding.
Several perhaps discrete catalytic steps are revealed by the structures: 1) activation of caspase-2 involving the breaking of the salt bridge between Glu-217 and Arg-378; 2) formation of a catalytically competent conformation upon binding to a single substrate; and 3) formation of the ES≠ complex after both active sites are occupied by the substrate. These observations will facilitate the design of novel caspase-2-specific probes, which will help to elucidate the true role of caspase-2 in physiological pathways.
Supplementary Material
Acknowledgments
We thank scientists at the CHDI Foundation for scientific insights and helpful discussion, Katherine Augustyn and Peter Lee for cloning, and Nathan Thomsen and Stacie Bulfer for advice on x-ray refinement. We also thank the staff at ALS831 and the Research Collaboratory for Structural Bioinformatics (RCSB) PDB annotation staff for expert technical support.
This work was supported by a grant from the CHDI Foundation.
The atomic coordinates and structure factors (codes 3R7S, 3R6G, 3R5J, 3R7B, 3R7N, and 3R6L) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S4.
- CARD
- caspase recruitment domain
- Ac
- acetyl
- AFC
- 7-amino-4-trifluoromethylcoumarin
- Z
- benzyloxycarbonyl
- FMK
- fluoromethyl ketone
- ES
- enzyme-substrate
- casp2
- caspase-2
- casp3
- caspase-3.
REFERENCES
- 1. Li J., Yuan J. (2008) Oncogene 27, 6194–6206 [DOI] [PubMed] [Google Scholar]
- 2. Lamkanfi M., Declercq W., Kalai M., Saelens X., Vandenabeele P. (2002) Cell Death Differ. 9, 358–361 [DOI] [PubMed] [Google Scholar]
- 3. Crawford E. D., Wells J. A. (2011) Annu. Rev. Biochem. 80, 1055–1087 [DOI] [PubMed] [Google Scholar]
- 4. Boatright K. M., Salvesen G. S. (2003) Curr. Opin. Cell Biol. 15, 725–731 [DOI] [PubMed] [Google Scholar]
- 5. Krumschnabel G., Sohm B., Bock F., Manzl C., Villunger A. (2009) Cell Death Differ. 16, 195–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bergeron L., Perez G. I., Macdonald G., Shi L., Sun Y., Jurisicova A., Varmuza S., Latham K. E., Flaws J. A., Salter J. C., Hara H., Moskowitz M. A., Li E., Greenberg A., Tilly J. L., Yuan J. (1998) Genes Dev. 12, 1304–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bouchier-Hayes L. (2010) J. Cell. Mol. Med. 14, 1212–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shimohama S., Tanino H., Fujimoto S. (1999) Biochem. Biophys. Res. Commun. 256, 381–384 [DOI] [PubMed] [Google Scholar]
- 9. Vakifahmetoglu-Norberg H., Zhivotovsky B. (2010) Trends Cell Biol. 20, 150–159 [DOI] [PubMed] [Google Scholar]
- 10. Kumar S. (2009) Nat. Rev. Cancer 9, 897–903 [DOI] [PubMed] [Google Scholar]
- 11. Talanian R. V., Quinlan C., Trautz S., Hackett M. C., Mankovich J. A., Banach D., Ghayur T., Brady K. D., Wong W. W. (1997) J. Biol. Chem. 272, 9677–9682 [DOI] [PubMed] [Google Scholar]
- 12. Thornberry N. A., Rano T. A., Peterson E. P., Rasper D. M., Timkey T., Garcia-Calvo M., Houtzager V. M., Nordstrom P. A., Roy S., Vaillancourt J. P., Chapman K. T., Nicholson D. W. (1997) J. Biol. Chem. 272, 17907–17911 [DOI] [PubMed] [Google Scholar]
- 13. Benkova B., Lozanov V., Ivanov I. P., Mitev V. (2009) Anal. Biochem. 394, 68–74 [DOI] [PubMed] [Google Scholar]
- 14. Noble J. E., Bailey M. J. (2009) Methods Enzymol. 463, 73–95 [DOI] [PubMed] [Google Scholar]
- 15. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 16. Collaborative Computational Project, Number 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 15299374 [Google Scholar]
- 17. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 20. DeLano W. L. (2010) The PyMOL Molecular Graphics System, version 1.3r1, Schrödinger, LLC, New York [Google Scholar]
- 21. Wolan D. W., Zorn J. A., Gray D. C., Wells J. A. (2009) Science 326, 853–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Baliga B. C., Read S. H., Kumar S. (2004) Cell Death Differ. 11, 1234–1241 [DOI] [PubMed] [Google Scholar]
- 23. Hedstrom L. (2002) Chem. Rev. 102, 4501–4524 [DOI] [PubMed] [Google Scholar]
- 24. Schweizer A., Briand C., Grutter M. G. (2003) J. Biol. Chem. 278, 42441–42447 [DOI] [PubMed] [Google Scholar]
- 25. Shi Y. (2004) Protein Sci. 13, 1979–1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fuentes-Prior P., Salvesen G. S. (2004) Biochem. J. 384, 201–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fang B., Boross P. I., Tozser J., Weber I. T. (2006) J. Mol. Biol. 360, 654–666 [DOI] [PubMed] [Google Scholar]
- 28. Agniswamy J., Fang B., Weber I. T. (2009) Apoptosis 14, 1135–1144 [DOI] [PubMed] [Google Scholar]
- 29. Witkowski W. A., Hardy J. A. (2009) Protein Sci. 18, 1459–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Renatus M., Stennicke H. R., Scott F. L., Liddington R. C., Salvesen G. S. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 14250–14255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Romanowski M. J., Scheer J. M., O'Brien T., McDowell R. S. (2004) Structure 12, 1361–1371 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.