Abstract
Sialic acid-binding immunoglobulin-like lectins (Siglecs) are receptors believed to be important for regulation of cellular activation and inflammation. Several pathogenic microbes bind specific Siglecs via sialic acid-containing structures at the microbial surface, interactions that may result in modulation of host responses. Recently, it was shown that the group B Streptococcus (GBS) binds to human Siglec-5 (hSiglec-5), an inhibitory receptor expressed on macrophages and neutrophils, via the IgA-binding surface β protein, providing the first example of a protein/protein interaction between a pathogenic microbe and a Siglec. Here we show that the hSiglec-5-binding part of β resides in the N-terminal half of the protein, which also harbors the previously determined IgA-binding region. We constructed bacterial mutants expressing variants of the β protein with non-overlapping deletions in the N-terminal half of the protein. Using these mutants and recombinant β fragments, we showed that the hSiglec-5-binding site is located in the most N-terminal part of β (B6N region; amino acids 1–152) and that the hSiglec-5- and IgA-binding domains in β are completely separate. We showed with BIAcoreTM analysis that tandem variants of the hSiglec-5- and IgA-binding domains bind to their respective ligands with high affinity. Finally, we showed that the B6N region, but not the IgA-binding region of β, triggers recruitment of the tyrosine phosphatase SHP-2 to hSiglec-5 in U937 monocytes. Taken together, we have identified and isolated the first microbial non-sialic acid Siglec-binding region that can be used as a tool in studies of the β/hSiglec-5 interaction.
Keywords: Bacteria, Innate Immunity, Ligand-binding Protein, Macrophages, Neutrophil, Tyrosine Protein Phosphatase (Tyrosine Phosphatase), Binding Domain, Streptococcus, Surface Protein
Introduction
The Siglecs3 are a family of mammalian transmembrane receptors that interact with sialic acids and are expressed in the hemopoietic, immune, and nervous systems (1). These lectins contain an N-terminal V-set Ig-like domain mediating sialic acid binding followed by varying numbers of C2-set Ig-like domains (2). There are two subgroups of Siglecs of which the CD33-related Siglecs are primarily expressed on leukocytes and consist so far of Siglecs-3, 5–11, -XII, -14, and -16 in humans. Many human CD33-related Siglecs have one or more immunoreceptor tyrosine-based inhibitory motifs (ITIMs) within their cytoplasmic domain and are proposed to negatively regulate signaling pathways and restrict activation of the cells that express them (1).
Human Siglec-5 (hSiglec-5) is one such CD33-related Siglec and is expressed prominently on human monocytes, macrophages, neutrophils, and dendritic cells (3). Engagement of hSiglec-5 with sialic acid ligands leads to recruitment of the SH1 and SH2 domain-containing tyrosine phosphatases, SHP-1 and SHP-2, to its two intracellular ITIMs (4), initiating an inhibitory intracellular signaling cascade. Interestingly, it has been demonstrated that hSiglec-5 can mediate strong inhibition signals even in the absence of phosphorylation of the ITIMs, indicating more complex signaling properties of this inhibitory leukocyte lectin receptor (5).
A growing body of evidence suggests that several pathogenic microbes, including Neisseria meningitidis, Campylobacter jejuni, group B Streptococcus (GBS), and HIV, have evolved mechanisms to interact with members of the Siglec family (6–9). For some of these interactions, it is believed that the pathogen exploits the inhibitory function of the Siglec receptor, resulting in down-regulation of cellular activation and inflammation. In the great majority of the above cases, the pathogen binds to the Siglec via sialylated surface structures. Recently, however, it was shown that GBS, a major pathogen of human newborns (10, 11), binds to hSiglec-5 via its IgA-binding surface β protein, demonstrating for the first time a functional engagement of a Siglec via a protein/protein interaction (12).
The β protein is a protective surface protein of ∼125 kDa expressed by GBS strains of serotypes Ia, Ib, II, and V (11). This protein has been shown to bind the Fc part of human IgA (13–16) and the human complement inhibitor factor H (FH) (17), suggesting a multifaceted contribution to GBS immune evasion. The IgA-Fc-binding site of β protein has been located to a 73-amino acid residue region (amino acids 153–225) in the N-terminal part of the protein (18) (see Fig. 1A). The β protein binds to the Cα2/Cα3 interdomain in IgA-Fc and blocks the binding of IgA to the human IgA receptor CD89 (FcαRI), which may inhibit IgA effector functions (19). The binding site for FH is completely separate from that of IgA and is located in the C-terminal half of the β protein (17) (see Fig. 1A). FH bound to the β protein at the bacterial surface retains its regulatory activity, indicating that GBS might recruit surface-bound FH to inhibit complement activation (17).
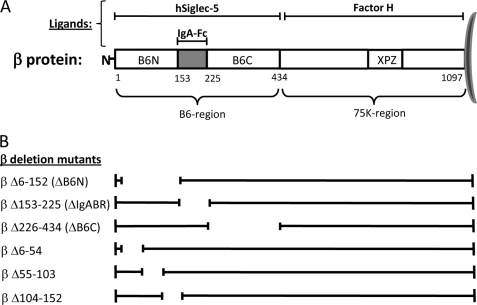
FIGURE 1.
A, schematic representation of protein β demonstrating the binding sites for its ligands hSiglec-5, IgA-Fc, and FH. The protein is divided in two main parts, the B6 and the 75k regions, which correspond to the N- and C-terminal halves of the β protein, respectively. The IgA- and hSiglec-5-binding regions have been shown to be located within the B6 region, whereas FH binds to the 75k region. The XPZ region is a unique sequence in β in which every third amino acid is a proline. The numbering indicates the position of amino acids in the mature β protein. The B6N region comprises amino acids 1–152, the IgA-binding region is located between amino acids 153 and 225, and the B6C region corresponds to amino acids 226–434 in the β protein. B, schematic representation of the β deletion mutants used in this study. The deleted part in each β variant is indicated by a break in the line.
hSiglec-5 is the third ligand identified for the surface β protein (12). Using an isogenic β-negative GBS mutant, it was shown that the β/hSiglec-5 interaction results in recruitment of SHP-2 to hSiglec-5 in human U937 monocytic cells and inhibition of phagocytosis of GBS. Moreover, β protein engagement of hSiglec-5 on primary human neutrophils suppressed their secretion of IL-8, oxidative burst, and formation of neutrophil extracellular traps in response to GBS. Thus, by engaging hSiglec-5 via the surface β protein, GBS may down-regulate the innate immune response from human phagocytes. Comparisons between Siglec-5 from different primates in direct binding experiments to β-expressing GBS indicated that the β-binding site in hSiglec-5 most likely resides in the sialic acid-binding V-set domain. Conversely, the binding site in β for hSiglec-5 was mapped to the N-terminal half of the protein (the B6 region), corresponding to amino acids 1–434 (12). However, it remained unclear whether hSiglec-5 and IgA-Fc have separate or overlapping binding sites in this region.
By using bacterial deletion mutants and recombinant β fragments we show here that the most N-terminal region in β, the so-called B6N region (amino acids 1–152), is necessary and sufficient for binding to hSiglec-5 and that the hSiglec-5-binding region is completely separate from the IgA-binding region (IgABR). Importantly, our study shows that both the hSiglec-5- and IgA-binding domains in β can be studied in isolated form and that the hSiglec-5-binding domain may become a valuable tool for studies of the β/hSiglec-5 interaction.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Cell Lines
The GBS type Ia strain A909 expressing the β protein, its isogenic β-negative mutant Δbac, and the trans-complemented strain Δbac/pLZbac have been described previously (17). A number of clinical GBS isolates analyzed for surface expression of the β protein and binding to hSiglec-5 are presented in supplemental Table S1. GBS was grown in Todd-Hewitt broth (Oxoid, Basingstoke, Hampshire, UK) at 37 °C without shaking or on blood agar. When containing plasmid pLZbac or derivatives thereof, GBS was grown in the presence of spectinomycin (70 μg/ml) to maintain the plasmid. The human monocytic cell line U937 was purchased from ATCC (Teddington, UK).
Proteins and Antibodies
Recombinant hSiglec-5-Fc chimera was purchased from R&D systems (Minneapolis, MN). Purified human IgA was from Cappel Organon-Teknika (Turnhout, Belgium). Fetuin, 3′-sialyllactose, and asialofetuin were from Sigma-Aldrich. Protein β and protein α were purified from streptococcal extracts as described (20, 21). Protein G (Calbiochem), B6N tandem, and IgA-binding tandem constructs were conjugated to CNBr-activated SepharoseTM 4B (GE Healthcare Bio-Sciences AB, Uppsala, Sweden) according to the manufacturer's recommendations. Proteins were radiolabeled with 125I using the chloramine-T method (22). HRP-conjugated goat anti-human IgG was purchased from AbD Serotec (Düsseldorf, Germany). Rabbit anti-human IgA and HRP-conjugated rabbit anti-mouse pAb were from DakoCytomation (Glostrup, Denmark), and the mouse anti-Siglec-5 mAb and HRP-conjugated anti-rabbit IgG were from R&D Systems. Rabbit anti-β, anti-α, and anti-XPZ sera were produced as described (20, 23). Rabbit serum to the GBS serotype Ia polysaccharide capsule was kindly provided by D. L. Kasper (Channing Laboratory, Boston, MA).
Construction of β Protein Mutants with Non-overlapping Deletions in N Terminus
For the construction of the β deletion mutants, plasmid pLZbac, which contains the entire bac sequence encoding the β protein, was used (17). New pLZbac derivatives were created by inverse PCR, generating derivatives of plasmid pLZbac that each lacked a specific part corresponding to regions in the B6 portion of the protein (Fig. 1). The whole pLZbac plasmid was amplified by inverse PCR except for the region in the bac gene to be deleted, and afterward the amplified fragment was religated using an XhoI recognition site introduced through the primers (supplemental Table S2), thus generating novel pLZbac derivatives with different in-frame deletions in the bac gene. In the pLZbac derivative containing a deletion corresponding to the B6N region (pLZΔB6N), we chose to exclude amino acids 6–152 to avoid possible problems with processing and cleavage of the signal sequence. The different pLZbac derivatives were transformed into the isogenic β-negative mutant Δbac (17). In this situation, each bac deletion variant is expressed from the natural bac promoter, and the corresponding β protein variant is transported and anchored to the bacterial surface. To analyze expression of the novel β deletion variants on the surface of the bacteria, we used rabbit antiserum directed against the XPZ region in β, which is located in the C terminus of the protein (see Fig. 1), and is present in all β deletion variants constructed herein (supplemental Fig. S1A). Expression of the type Ia polysaccharide capsule or protein α was analyzed with rabbit anti-Ia-TT and rabbit anti-α serum, respectively (supplemental Fig. S1, B and C).
Construction and Purification of Recombinant β Fragments
The bac gene sequences corresponding to the hSiglec-5-binding B6N region (amino acids 1–152) and IgABR (amino acids 155–225) (see Fig. 1), were amplified from pLZbac by standard PCR using primers listed in supplemental Table S2. The PCR products were then cleaved with BamHI and EcoRI (Fermentas, St. Leon-Rot, Germany) using recognition sequences introduced through the primers and ligated into a BamHI/EcoRI-cleaved plasmid (pGEX-6P-2, GE Healthcare Bio-Sciences AB), thereby fusing the PCR fragments in-frame with the gst gene. The resulting pGEX-6P-2 derivatives, designated pGEX-B6N and pGEX-IgABR, were transformed into Escherichia coli BL21. For expression of the GST fusion proteins, bacteria were grown to an A600 of 0.5 followed by induction with 1 mm isopropyl 1-thio-β-d-galactoside, resulting in overexpression of the GST fusion proteins. Bacteria were lysed with 1.5 mg/ml lysozyme and by repeated freezing and thawing, and after removal of cellular debris by centrifugation, the recombinant proteins were purified on GSTrap columns (GE Healthcare Bio-Sciences AB). Removal of the GST tag was performed according to the manufacturer's instructions, and suspensions containing the purified protein were dialyzed against PBS. To construct tandem variants of the B6N region and IgABR, the corresponding bac sequences were amplified with PCR using primers described in supplemental Table S2. These fragments were then cleaved with BamHI and cloned into plasmids pGEX-B6N and pGEX-IgABR precleaved with BamHI, resulting in novel derivatives of pGEX-B6N and pGEX-IgABR in which two copies of the bac sequence corresponding to the B6N and IgABR, respectively, were fused in tandem and linked with a BamHI cleavage site.
Release of β Protein from GBS with High pH
The β protein was released from different GBS strains with high pH as described (16). Briefly, overnight cultures of bacteria were centrifuged and washed twice with 50 mm Tris-HCl, pH 7.3 and resuspended to 2 × 1010 cfu/ml in 50 mm glycine, pH 11. After incubation for 4 h at 37 °C, the suspensions were spun at 10,000 × g for 15 min at 4 °C, and the supernatants were separated by SDS-PAGE.
SDS-PAGE and Western Blot
The SDS-PAGE and Western blot were performed as described before (22) with the modification that after a 1-h incubation with 5 μg of hSiglec-5-Fc at RT the membrane was washed four times and an HRP-conjugated anti-human IgG diluted 1:10,000 in TBS-T (50 mm Tris-HCl, pH 7.,4, 0.15 m NaCl, 0.25% Tween 20, 0.25% gelatin) was added for another 1 h. The membrane was thoroughly washed and developed using Pierce ECL Western blotting substrates (Thermo Scientific, Waltham, MA) and medical x-ray film (Fuji Photo Film, Düsseldorf, Germany).
To analyze the recombinant proteins, SDS-PAGE and Western blot were performed as described above. The membranes were incubated with hSiglec-5-Fc and HRP-conjugated anti-human IgG as described above or with 5 μg of human IgA for 1 h at RT followed by incubation with rabbit anti-human IgA diluted 1:500 in TBS-T. The membrane was then washed, and HRP-conjugated anti-rabbit pAb diluted 1:1000 in TBS-T was added for a 1-h incubation.
Binding Assays with Whole Bacteria
Overnight cultures of GBS were washed twice in PBSAT (0.12 m NaCl, 0.03 m phosphate, 0.02% NaN3, pH 7.2 supplemented with 0.05% Tween 20), and suspensions of ∼109 cfu/ml were prepared. Bacterial suspensions (180 μl) were mixed with serial dilutions of hSiglec-5-Fc (20 μl) in PBSAT. After a 2-h incubation with shaking at RT, the bacteria were washed twice with PBSAT. Protein G radiolabeled with 125I (∼10,000 cpm/sample in a final volume of 100 μl) was then added, and the mixtures were incubated for 1 h at RT. The bacteria were then washed two times with PBSAT, and the radioactivity in the pellets was determined using a γ-counter. To analyze IgA binding, bacteria were prepared as above, and 180 μl of the different bacterial suspensions was mixed with 20 μl (10,000 cpm/sample) of 125I-labeled IgA. After incubation for 1 h at RT, the samples were washed, and pellets were measured in a γ-counter. Binding is presented as a percentage of total radiolabeled protein G or IgA added.
Binding Assay with Pure Proteins
Purified recombinant B6N (single and tandem) and IgABR (single and tandem) as well as the intact β protein were diluted in 4-fold steps in PBS (highest concentration at 500 nm) and immobilized in the wells of a microtiter plate (Corning Inc. Life Sciences, Lowell, MA) at 4 °C overnight. The wells were washed three times with PBSAT and blocked for 1 h at RT with PBS containing 1% BSA. After three washes with PBSAT, 50 μl of hSiglec-5-Fc at a final concentration of 1 μg/ml was added to each well, and the plate was incubated for 1 h at RT and then washed three times. An HRP-conjugated anti-human IgG diluted 1:10,000 in PBSAT was added and incubated for 1 h at RT. After four additional washings, the plates were developed and measured at 450 nm.
Surface Plasmon Resonance Interaction Analysis (BIAcore)
The B6N (single and tandem) and IgABR (single and tandem) fragments, respectively, as well as intact protein β were diluted in 10 mm sodium acetate (pH 4), and 1500 response units were immobilized via amine coupling to CM5 sensor chip flow chambers, respectively (GE Healthcare). Briefly, proteins were mixed with freshly prepared 100 mm N-hydroxysuccinimide and 400 mm N-ethyl-N′-(dimethylaminopropyl)carbodiimide in equal volumes, and capping of unreacted carboxymethyl sites was achieved by a 1 m ethanolamine, pH 8 injection. A flow chamber subjected to the immobilization protocol but without any addition of protein was used as a control (blank) for each experiment. Human IgA or hSiglec-5 was sequentially diluted in running buffer (10 mm HEPES, 150 mm NaCl, 0.005% Surfactant P20 (BIAcore), pH 7.5) and typically injected over the different surfaces at 100–6 nm and 35 μl/min.
Binding was monitored in a BIAcore 2000 instrument. Between experiments, the surfaces were strictly regenerated with multiple pulses of 2 m NaCl, 1.5 m glycine-HCl, pH 2.0 followed by an extensive wash procedure using running buffer.
After x and y axis normalization of the obtained data, the blank bulk refraction curves from the control flow chamber of each injected concentration were subtracted. Binding curves were displayed, and the association (Ka) and dissociation (Kd) rate constants were determined using BIAevaluation 4.1 software and its equation for 1:1 Langmuir binding. From these values, affinities (KD) were calculated.
Inhibition Assays with Whole Bacteria
An overnight culture of WT strain A909 was washed twice in PBSAT and resuspended to a concentration of ∼109 cfu/ml. Radiolabeled hSiglec-5 or IgA (20 μl of 7.5 × 105 cpm/ml) was preincubated with 20 μl of various concentrations of the B6N or IgABR tandem constructs, respectively, starting at 150 μg/ml, for 30 min at RT. Unlabeled hSiglec-5 and IgA were used as control proteins, and radiolabeled hSiglec-5 and IgA together with 20 μl of PBSAT only were used as positive controls. The protein mixtures were added to 180 μl of bacterial suspension and incubated for 1 h at RT. The bacteria were washed twice in PBSAT, and bound radioactivity was measured in a γ-counter. Inhibition was calculated as a percentage of the decreased binding as compared with the control without inhibitors.
In the inhibition assay with sialic acid-containing proteins, increasing concentrations of fetuin and asialofetuin (0–40 μm) or 3′-sialyllactose and mannose (0–0.5 mm) were preincubated with radiolabeled hSiglec-5 for 30 min at RT. The protein mixtures were incubated with the bacteria, and the assay was performed as described above.
Phagocytosis Assay
Human neutrophils from Sigec-14−/− donors were prepared as described previously (12) using PolyMorphPrep solution (Axis-Shield PoC AS) according to the manufacturer's instructions. GBS was added to neutrophils at a multiplicity of infection of 10. The bacteria and neutrophils were spun together at 500 × g for 5 min to initiate the assay, and 15 min later the total bacterial survival was quantified by serial dilution plating on Todd-Hewitt agar plates.
Analysis of SHP-2 Recruitment to hSiglec-5
U937 monocytes (107) were incubated with Sepharose-conjugated B6N tandem or IgABR tandem constructs for 10 min at 37 °C followed by immediate centrifugation. Cells were resuspended in lysis buffer (20 mm Tris-HCl, pH 8, 137 mm NaCl, 10% glycerol, 1% Nonidet P-40, 2 mm EDTA, protease inhibitor mixture) and rotated end over end for 30 min at 4 °C. The lysates were centrifuged for 20 min at 4 °C at 10,000 × g, and goat anti-Siglec-5 pAb was added to the supernatant and incubated for 2 h at 4 °C. Sepharose-conjugated protein G was added, and the samples were incubated at 4 °C overnight. The Sepharose beads were collected by centrifugation, resuspended in SDS sample buffer, and boiled for 10 min. The released proteins were then separated by SDS-PAGE, and Western blotting was performed as described above using mouse anti-Siglec-5 mAb and HRP-anti-mouse pAb or rabbit anti-SHP-2 and HRP-anti-rabbit antibodies.
Inhibition Assay for Analysis of B6N-binding Site in hSiglec-5
An inhibition assay was developed to analyze the ability of the B6N region to inhibit the binding of Siglec-5 to sialylated ligands. Fetuin was immobilized in the wells of a microtiter plate and incubated at 4 °C overnight. The wells were blocked and washed as described above, and Siglec-5 (1 μg/ml) was preincubated for 30 min in RT with protein β, B6N, or IgABR tandem fragments diluted in 2-fold steps starting at 260 nm. The protein mixtures were added to the plate and incubated for 1 h at RT. After several washings, the HRP-conjugated anti-human IgG was added, incubated, and detected as described above.
RESULTS
The Most N-terminal Part of Protein β Is Necessary for Binding to hSiglec-5
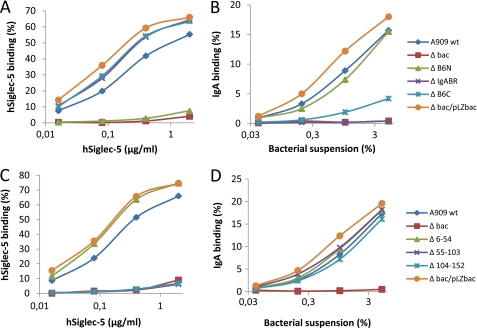
The N-terminal B6 region (Fig. 1A) of the β protein, which contains the IgA-Fc-binding region, was shown previously to be responsible for the interaction with hSiglec-5 (12). To map the binding site for hSiglec-5 in the β protein, different deletion mutants with non-overlapping deletions were constructed, each lacking a specific part of the B6 region (Fig. 1B). The different β-protein deletion mutants were expressed in the Δbac background, and a binding assay with a soluble hSiglec-5-Fc chimera was performed. As shown in Fig. 2A, the β deletion mutant strain lacking the B6N region (ΔB6N) was completely negative for binding to hSiglec-5. In contrast, the mutants lacking the IgA-binding region (ΔIgABR) or B6C region (ΔB6C) bound hSiglec-5 at levels similar to that of the parental strain A909 and the trans-complemented strain (Δbac/pLZbac), both of which express the intact β protein. For comparison, the same β deletion mutant strains were analyzed for binding to human IgA. The mutant lacking the known 73-amino acid IgA-binding region (ΔIgABR) did not show any binding to IgA (Fig. 2B). In contrast, the ΔB6N mutant bound to IgA at the same level as the parental and the trans-complemented GBS strains. The ΔB6C strain showed a decreased binding to IgA as compared with the parental GBS strain, which suggests a conformational change in the IgA-binding region in this deletion mutant. Thus, the most N-terminal region in β, the B6N region, corresponding to amino acids 6–152, is necessary for binding to hSiglec-5.
FIGURE 2.
hSiglec-5-binding domain is located between amino acid residues 55 and 152 in B6N region of β protein. A, the GBS mutant expressing a β protein lacking the B6N region (ΔB6N) does not bind hSiglec-5. B, the GBS mutant expressing a β protein lacking the IgA-binding region (ΔIgA) is negative for binding to human IgA. In addition, the ΔB6C mutant displays a weak binding to human IgA. C, the Δ55–103 and Δ104–152 mutants are negative for binding to hSiglec-5. D, the Δ55–103 and Δ104–152 mutants bind to human IgA. Bacteria were grown overnight and washed in PBSAT. Aliquots of bacterial suspension were incubated for 2 h with increasing concentrations of hSiglec-5 or 125I-labeled IgA. hSiglec-5 was detected with 125I-labeled protein G. Bound radioactivity was measured in a γ-counter. Binding is defined as the percentage of total radiolabeled protein associated with the bacteria. These experiments were performed three times with similar results.
To further pinpoint the hSiglec-5-binding region, additional β mutants with deletions in the B6N region (Fig. 1B) were constructed and analyzed for hSiglec-5 binding. GBS expressing deletion mutants of β lacking amino acid residues 55–103 or 104–152 did not bind to hSiglec-5, whereas the Δ6–54 mutant strain displayed binding to hSiglec-5 similar to that of the parental and trans-complemented strains (Fig. 2C). As expected, all three mutants with a deletion in the B6N region bound to IgA (Fig. 2D). In conclusion, the B6N region of the β protein is responsible for binding to hSiglec-5, and the binding site is most likely situated between amino acid residues 55 and 152.
B6N Region in Protein β Inhibits Phagocytosis by Human Neutrophils
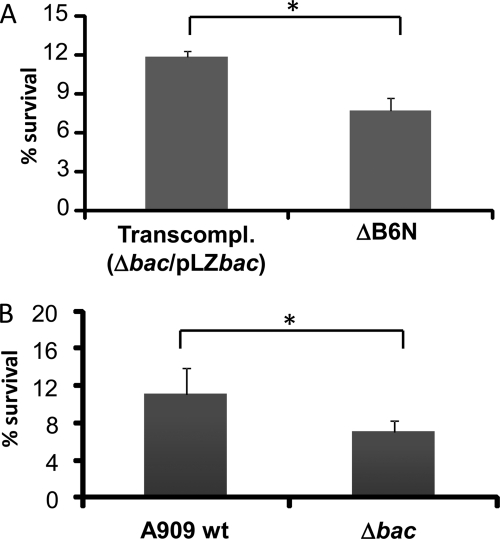
It was shown previously that engagement of hSiglec-5 on human neutrophils by β-expressing GBS inhibits phagocytosis (12). To confirm that this property is correlated to the B6N region, we compared the killing of the trans-complemented strain Δbac/pLZbac, which expresses the intact β protein, and the ΔB6N mutant strain. The reason for this approach is that we wanted to use strains that were grown under the same conditions, i.e. in the presence of spectinomycin (to maintain the plasmid). As shown in Fig. 3A, the ΔB6N mutant strain was killed to a significantly higher extent as compared with the trans-complemented strain. This result shows that the B6N region inhibits phagocytosis of GBS by human neutrophils most likely by interacting with hSiglec-5. As a control, we also included the A909 WT and Δbac mutant GBS strains (Fig. 3B). As expected, this analysis confirmed that β inhibits phagocytosis of GBS by human neutrophils.
FIGURE 3.
B6N region inhibits phagocytosis of GBS by human neutrophils. A, the GBS ΔB6N mutant is phagocytosed to a significantly greater extent compared with the trans-complemented (Transcompl.) strain Δbac/pLZbac, which expresses the intact β protein. B, control experiment showing that the Δbac mutant, which lacks surface β protein, is killed to a greater extent by human neutrophils compared with the WT strain A909. Human neutrophils from Siglec-14−/− individuals were prepared using PolyMorphPrep solution. GBS was added to neutrophils at a multiplicity of infection of 10 and incubated for 15 min. The bacterial survival was determined using serial dilution plating on Todd-Hewitt agar plates. This experiment was performed twice with similar results. *, p ≤ 0.05. Error bars correspond to S.D.
B6N Region in β Is Sufficient for Binding to hSiglec-5
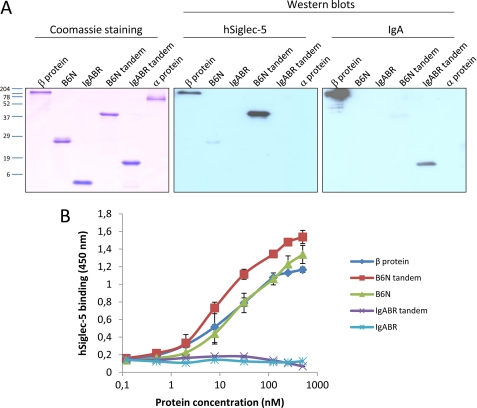
To analyze whether the B6N region is sufficient for binding to hSiglec-5, it was purified recombinantly. Although amino acids 55–152 were found to be necessary for the interaction, we decided to use the whole B6N region (amino acids 1–152) to ensure proper folding of the hSiglec-5-binding region. The IgABR (amino acids 153–225) in β was also purified recombinantly and used for comparison. When the recombinant B6N fragment was analyzed for binding to hSiglec-5 in a Western blot experiment (Fig. 4A), only a weak interaction could be detected. In contrast, when the B6N fragment was coated onto a microtiter plate and incubated with hSiglec-5, it bound to hSiglec-5 in a dose-dependent manner at levels similar to that of the full-length protein β (Fig. 4B). The recombinant IgABR did not bind human IgA when blotted onto a membrane (Fig. 4A), and no binding could be detected in an ELISA (data not shown).
FIGURE 4.
B6N region is sufficient for binding to hSiglec-5. A, Western blot experiment analyzing the ability of the recombinant B6N and IgABR fragments (single copy and tandem, respectively) to bind hSiglec-5. The B6N and IgABR constructs were separated by SDS-PAGE together with full-length protein β (positive control) and protein α (negative control). After blotting and blocking, the membranes were incubated with 5 μg/ml hSiglec-5 followed by HRP-conjugated anti-human IgG or with 5 μg/ml IgA followed by a rabbit anti-human IgA and HRP-conjugated anti-rabbit IgG. B, analysis of the interaction of hSiglec-5 with pure proteins; the recombinant proteins and protein β were coated on microtiter plates at increasing concentrations and incubated with hSiglec-5, which was detected with an HRP-conjugated anti-human IgG. The plates were developed and read at 450 nm. The mean values of three separate experiments are shown, and error bars correspond to S.D.
In an attempt to improve the binding, the recombinant B6N and IgABR proteins were expressed as tandem constructs, i.e. two identical sequences fused consecutively. As demonstrated in Fig. 4A, the B6N tandem protein clearly bound to hSiglec-5 in a Western blot analysis. A similar improvement could be seen for the IgA binding properties of the recombinant IgABR when comparing the single copy protein with the tandem protein in the Western blot experiment (Fig. 4A). The two tandem proteins together with the full-length β protein were also immobilized in microtiter plates and incubated with hSiglec-5. The B6N tandem protein displayed a slightly stronger binding to hSiglec-5 compared with either the single B6N fragment or the intact β protein (Fig. 4B). Neither the single nor tandem IgABR fragments showed binding to hSiglec-5.
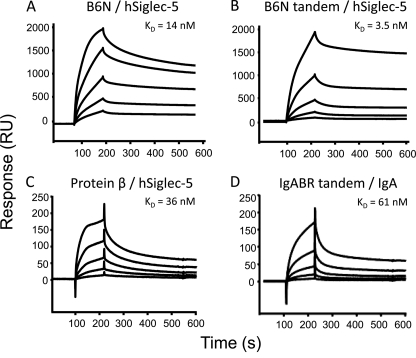
To further analyze the binding properties of the recombinant B6N and IgABR proteins and to establish the affinity constant of the binding, surface plasmon resonance (i.e. BIAcore) analysis was performed. The single copy and tandem proteins, respectively, were immobilized on the surface of CM5 sensor chip flow chambers using amine coupling. The intact β protein was included as a positive control. Subsequently, hSiglec-5 or IgA was injected until saturation was reached. The KD (the affinity) was calculated from the determined Ka and Kd values. The KD for the interaction between B6N and hSiglec-5 was 14 nm (Fig. 5A), whereas the B6N tandem-hSiglec-5 binding had a KD of 3.5 nm (Fig. 5B). BIAcore analysis thus confirmed that the B6N tandem fragment exhibits slightly stronger binding to hSiglec-5 than does the single copy B6N fragment. The KD for the binding between protein β and hSiglec-5 was 36 nm (Fig. 5C). Analysis of the IgABRs showed that only the tandem construct displayed any binding to IgA (Fig. 5D), and the calculated KD was 61 nm. Surprisingly, no binding of IgA to full-length protein β could be detected (data not shown), but this might be due to a formational alteration of the β protein induced upon coupling to the chip. In conclusion, the isolated B6N region is sufficient for binding to hSiglec-5 as both the single copy and tandem constructs bound with high affinity. Moreover, the tandem recombinant IgABR bound human IgA with high affinity.
FIGURE 5.
BIAcore analysis: single and tandem B6N fragments bind hSiglec-5 with high affinity. A, the KD for the interaction between B6N and hSiglec-5 is 14 nm B, the KD for the interaction between B6N tandem and hSiglec-5 is 3.5 nm. C, the KD for the interaction of hSiglec-5 and the intact β protein is 36 nm. D, the KD for the interaction between human IgA and the tandem IgABR is 61 nm. The B6N (single and tandem), IgABR (single and tandem), and intact protein β were diluted in 10 mm sodium acetate, pH 4 and immobilized to sensor chip flow chambers, respectively. The unreacted carboxymethyl sites were blocked with 1 m ethanolamine, pH 8. A flow chamber that was identically activated and blocked was used as a negative control for each experiment. Various concentrations of IgA or hSiglec-5 were injected over the different surfaces at 35 μl/min. Binding was monitored in a BIAcore 2000 instrument. After x and y axis normalization of obtained data, the nonspecific binding from the control flow chamber of each injected concentration was subtracted. Binding curves were displayed, and the association (Ka) and dissociation (Kd) rate constants were determined using BIAevaluation 4.1 software and its equation for 1:1 Langmuir binding. From these values, affinities (KD) were calculated. RU, response units.
Binding Sites of hSiglec-5 and IgA in β Protein Are Separate
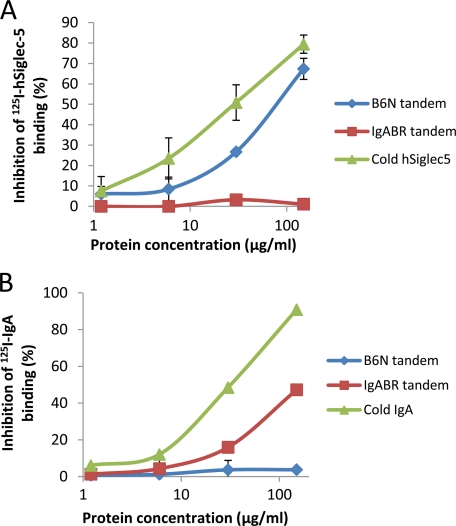
The analyses described above with bacteria expressing deletion mutants of β and recombinant β fragments indicate that hSiglec-5 and IgA have separate binding regions in β. To confirm that the hSiglec-5- and IgA-binding regions in protein β are completely separate and that these two ligands do not sterically prevent the other from binding to β, inhibition experiments were performed in which the abilities of the B6N tandem or the IgABR tandem fragments to inhibit the binding of radiolabeled hSiglec-5 to the β-expressing parental strain A909 were analyzed. A significant and dose-dependent inhibition using the B6N tandem fragment could be detected with a maximum inhibition of 79.5% (Fig. 6A). In contrast, the tandem IgABR did not inhibit the GBS/hSiglec-5 interaction. As expected, the binding of radiolabeled hSiglec-5 could be inhibited with cold hSiglec-5. For comparison, the tandem IgABR, but not the tandem B6N region, inhibited the binding of radiolabeled IgA to WT GBS (Fig. 6B). In conclusion, these results show that the hSiglec-5- and IgA-binding regions in the β protein are completely separate.
FIGURE 6.
hSiglec-5- and IgA-binding regions are distinct and non-overlapping in β protein. A, the B6N tandem fragment inhibits the binding of 125I-hSiglec-5 to A909. B, the IgABR tandem construct inhibits the 125I-IgA binding to A909. An overnight culture of the β-expressing WT strain A909 was washed twice in PBSAT and resuspended to a concentration of ∼109 cfu/ml. Radiolabeled hSiglec-5 or IgA was preincubated with 20 μl of various concentrations of the B6N or IgABR tandem constructs and thereafter added to aliquots of the bacterial suspension and incubated for 1 h at RT. Soluble hSiglec-5 and IgA (cold) were used as positive controls for inhibition of the 125I-hSiglec-5 and 125I-IgA binding, respectively, and radiolabeled hSiglec-5 and IgA together with 20 μl of PBSAT only were used as positive controls. The bacteria were washed twice in PBSAT, and bound radioactivity was measured in a γ-counter. Inhibition was calculated as a percentage of the decreased binding as compared with the control without inhibitors. Mean values of two separate experiments are shown, and the error bars correspond to S.D.
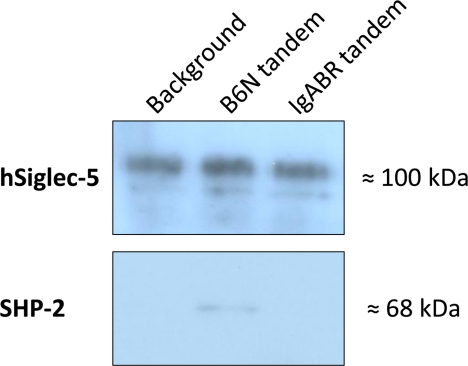
B6N Tandem Fragment Triggers SHP-2 Recruitment to hSiglec-5 in U937 Monocytic Cells
It has been shown previously that β-expressing GBS can trigger the recruitment of SHP-2 to hSiglec-5 (12). To analyze whether the B6N tandem fragment is sufficient to trigger recruitment of SHP-2 to hSiglec-5, an immunoprecipitation experiment was performed. U937 cells were incubated with the B6N or IgABR tandem constructs coupled to Sepharose beads and lysed after which hSiglec-5 was absorbed using a goat anti-hSiglec-5 pAb and protein G-coated Sepharose beads. The absorbed proteins were thereafter separated by SDS-PAGE and blotted onto membranes that subsequently were incubated with anti-hSiglec-5 or anti-SHP-2 antibody. As shown in Fig. 7, SHP-2 was recruited to hSiglec-5 when U937 monocytes were incubated with the B6N tandem. In contrast, no recruitment could be seen after incubation with tandem IgABR. Thus, the interaction between the tandem B6N fragment and hSiglec-5 on U937 cells triggers a key proximal event in hSiglec-5 intracellular signaling.
FIGURE 7.
Engagement of hSiglec-5 on U937 monocytes by B6N tandem fragment results in recruitment of SHP-2. Western blot analysis shows that SHP-2 is coimmunoprecipitated with hSiglec-5 when stimulated with the B6N tandem fragment coupled to Sepharose beads. U937 monocytic cells were incubated with the B6N or IgABR tandem fragments (Sepharose-conjugated) for 10 min. U937 cells alone were used as background. The cells were lysed, and supernatants were incubated with goat anti-hSiglec-5 pAb followed by protein G-conjugated Sepharose beads. The beads were then boiled in SDS sample buffer, and released proteins were separated by SDS-PAGE and blotted to membranes. The membranes were incubated with mouse anti-hSiglec-5 mAb or rabbit anti-SHP-2 followed by HRP-conjugated anti-mouse pAb or anti-rabbit pAb, respectively. A representative experiment of three is shown.
Protein β and Sialic Acid Have Overlapping Binding Sites on Siglec-5
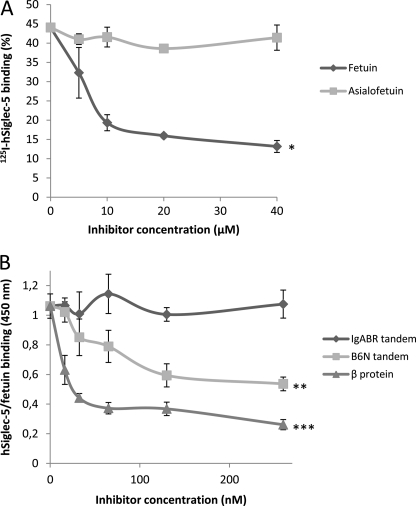
It has been shown previously that the β protein most likely binds to the hSiglec-5 V-set domain, which is the region that contains the binding site for sialic acid (12). To analyze whether the β protein and sialic acid may have overlapping binding sites in hSiglec-5, GBS strain A909 was incubated with 125I-labeled hSiglec-5 together with increasing concentrations of fetuin or asialofetuin. Fetuin is a glycoprotein with terminal sialic acid, whereas asialofetuin is desialylated. Although fetuin strongly inhibited the interaction between 125I-hSiglec-5 and GBS, no inhibition was obtained with asialofetuin (Fig. 8A). Similar inhibition was obtained using 3′-sialyllactose where mannose was included as a negative control (data not shown). Because of problems with radiolabeling of fetuin, we could not perform the reciprocal experiment. Instead, to analyze whether the β protein or the B6N tandem fragment could conversely inhibit the interaction between fetuin and hSiglec-5, an ELISA was performed. Fetuin was immobilized on a microtiter plate; hSiglec-5 with or without preincubation with various concentrations of protein β, B6N, or IgABR tandem constructs was added; and bound hSiglec-5 was detected using an HRP-conjugated anti-human IgG antibody. Both the β protein and the B6N tandem fragment significantly inhibited the interaction between hSiglec-5 and fetuin, whereas preincubation with IgABR tandem did not affect the binding (Fig. 8B). Taken together, these results indicate that the binding sites for sialic acid and protein β in hSiglec-5 are most likely overlapping.
FIGURE 8.
B6N fragment and sialic acid have overlapping binding sites on hSiglec-5. A, addition of sialic acid-containing fetuin inhibits radiolabeled hSiglec-5 binding to A909 WT, whereas asialofetuin does not. Bacteria were grown overnight and washed, and aliquots were incubated with 125I-hSiglec-5 together with increasing concentrations of fetuin or asialofetuin. Bound 125I-labeled protein was then measured in a γ-counter. B, protein β and B6N tandem inhibit the interaction between fetuin and soluble hSiglec-5. hSiglec-5 was preincubated with increasing concentrations of protein β, B6N tandem, or IgABR tandem and thereafter added to a microtiter plate with immobilized fetuin. Bound hSiglec-5 was detected with an HRP-conjugated anti-human IgG. Mean values of three experiments are shown, and error bars indicate S.D. *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001.
hSiglec-5 Binds to β Protein Independently of GBS Serotype
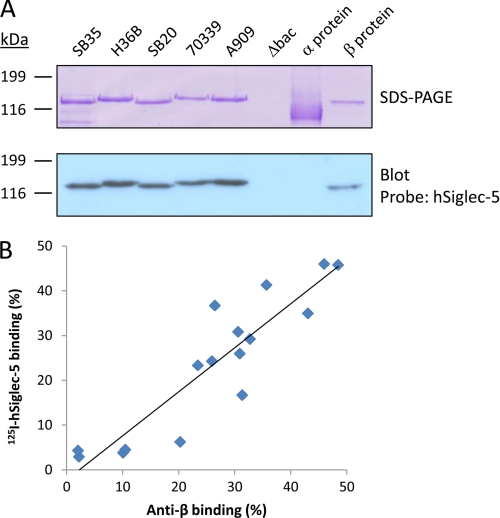
We demonstrated previously that the GBS surface protein β from strain A909 binds to hSiglec-5 in a sialic acid-independent manner (12). To analyze whether this binding occurs for all GBS strains expressing the β protein, we extracted the β protein from five different strains of serotype Ia and Ib, including A909, and analyzed binding to hSiglec-5. As shown in Fig. 9A, all β proteins bound to hSiglec-5, whereas no binding could be detected to the negative control protein α. Binding of 125I-hSiglec-5 could be detected in β-expressing strains of serotypes II, IV, and V, whereas serotype III strains, which are known not to express protein β, were completely negative for binding to hSiglec-5 (Fig. 9B and supplemental Table S1). Most importantly, a correlation between the level of expression of the β protein and hSiglec-5 binding could be demonstrated using a rabbit antiserum against the β protein with a Pearson's correlation coefficient of 0.9 (p = 0.06). To exclude that the polysaccharide capsule affects the ability of GBS to bind hSiglec-5, we compared the binding of hSiglec-5 to isogenic mutants lacking the polysaccharide capsule, the β protein, or both surface structures, respectively (supplemental Fig. S2). This analysis showed very similar binding levels of hSiglec-5 to A909 WT and its isogenic acapsular mutant, excluding that the polysaccharide capsule has a major effect on the ability of GBS to bind hSiglec-5. Taken together, β-expressing GBS strains bind to hSiglec-5 independently of strain serotype, showing that this is a general ability among β-expressing GBS strains.
FIGURE 9.
Protein β binds to hSiglec-5 independently of GBS strain serotype. A, Purified protein β extracted from different Ia and Ib GBS strains bound hSiglec-5, whereas no binding was detected against the control protein α. Bacteria were grown overnight, washed and the β proteins were extracted by elevating the pH in the bacterial suspensions. The β-negative mutant Δbac was used as negative control. After four h incubation, the supernatants were separated by SDS-PAGE and blotted on a nitrocellulose membrane. The membrane was incubated with 5 μg/ml hSiglec-5 followed by a HRP-conjugated anti-human IgG. B, The hSiglec-5 binding is correlated to the level of β-expression. GBS strains of serotypes Ia, Ib, II, III, IV and V were incubated with 125I-labeled hSiglec-5, or rabbit anti-protein β serum followed by 125I-labeled protein G. Bound 125I-labeled protein was then measured in a γ-counter. The mean values of three independently experiments are shown.
DISCUSSION
Sialic acid is the natural ligand for all Siglec receptors, and as there are numerous examples of sialylated glycans in the mammalian glycome, it is thought that these interactions are important for cellular regulation, cell signaling, and endocytosis (1). It is known that many pathogens by various mechanisms incorporate sialic acid into their own surface glycoconjugates (24), such as polysaccharide capsule and LPS. This mechanism represents classical molecular mimicry and is regarded as a virulence mechanism for the pathogen to subvert the innate immune system. Most studies have focused on the role of sialylated microbial surface structures in evasion of complement-mediated phagocytosis (25), but in recent years, it has become evident that microbes may also subvert the innate immune response by interactions with members of the Siglec family. Indeed, GBS binds hSiglec-9 in a sialic acid-dependent manner to suppress neutrophil activation (8). C. jejuni has been shown to bind to Siglec-7, mediating contact to natural killer cells and monocytes, which was suggested to modulate the host inflammatory response (7). However, there are also examples that such interactions may be protective against infections. N. meningitidis interacts with sialoadhesin (Siglec-1) and Siglec-5, resulting in increased pathogen uptake by macrophages (6).
We recently demonstrated the first sialic acid-independent protein/protein interaction with a Siglec receptor (12). We found that the IgA-binding β protein of GBS binds hSiglec-5 expressed on neutrophils and monocytes, an interaction that triggers SHP-2 recruitment to hSiglec-5 and that may lead to evasion of phagocyte responses. The hSiglec-5-binding region in the β-protein was shown to be located in the N-terminal region (amino acids 1–434), which also contains the IgA-Fc-binding site in β. However, it was unclear whether the hSiglec-5- and IgA-binding domains in β represent distinct or overlapping binding sites. In the present study, we identified the hSiglec-5-binding region of the β protein and showed that the hSiglec-5- and IgA-binding sites are completely separate and represent two distinct binding domains in the β protein. Using novel deletion mutants of β expressed in GBS, we showed that the most N-terminal region of β, the so-called B6N region (amino acids 6–152), is necessary for binding to hSiglec-5. We subsequently showed that two β deletion mutants, Δ55–103 and Δ104–152, were negative for binding to hSiglec-5. The most likely explanation for this is that the hSiglec-5-binding region is located between amino acid residues 55 and 152. Because both of these mutants were negative for binding, the hSiglec-5-binding site may be located at the region near the junction between the deletions in these two mutants. Moreover, we could confirm that amino acid residues 153–225 are necessary for binding to human IgA-Fc, which is in good agreement with earlier data indicating this region in IgA binding (18).
To show that the hSiglec-5- and IgA-binding domains are sufficient for binding to their respective ligands, we purified these regions using recombinant methods. The initial analyses indicated that the recombinant B6N region indeed bound to hSiglec-5 but that this interaction seemed to be somewhat weaker when compared with the intact β protein. In particular, binding of hSiglec-5 to B6N in the Western blot experiment was weak, which may be due to alterations of the B6N structure on the blotting membrane. To increase the affinity and to facilitate studies of the B6N region in interactions with hSiglec-5, we decided to construct a variant peptide with two identical copies of B6N fused in tandem. Indeed, by Western blot, ELISA, and BIAcore analyses, we demonstrated that the tandem B6N fragment bound hSiglec-5 with higher affinity compared with the single copy B6N fragment. We could not detect any binding of IgA to the single copy IgABR fragment. Possibly because the IgABR is significantly shorter than the B6N region, it may not be properly folded, thereby affecting interactions with IgA. In contrast, the tandem IgABR exhibited clear binding to human IgA. Together, these data show that the B6N region of β is sufficient for binding to hSiglec-5 and that the hSiglec-5- and IgA-binding domains in β can be studied in isolated form. Importantly, we could not detect any binding of IgA to the B6N fragments, and conversely, no binding of hSiglec-5 was detected to the tandem IgA-binding region. Together with the inhibition data from whole bacteria, we conclusively showed that hSiglec-5 and IgA have separate binding sites in the β protein. Importantly, we demonstrated that the B6N tandem fragment can trigger recruitment of SHP-2 to hSiglec-5 in U937 monocytic cells, indicating that the isolated B6N region could become a valuable tool for studies of various aspects of the hSiglec-5/β protein interaction, including biological and structural studies.
It was shown previously that the β protein binds to the V-set domain of hSiglec-5 (12) that also contains a conserved sialic acid recognition motif (26). However, it is unclear whether the β protein and sialic acid have separate or overlapping binding sites in the V-set domain of hSiglec-5. In this study, we showed that fetuin, but not asialofetuin, inhibits the interaction between hSiglec-5 and B6N. Moreover, the B6N fragment significantly inhibited the interaction between hSigelc-5 and fetuin. The most likely interpretation of these data is that the B6N fragment and sialic acid may have overlapping binding sites in the V-set domain in hSiglec-5. Because of the size of fetuin and B6N, it cannot be excluded that the inhibition seen may be in part due to steric effects. However, because inhibition was also seen using 3′-sialyllactose, the binding sites are most likely overlapping. Future studies using the B6N fragment may reveal important information of the structure of the β·hSiglec-5 complex.
Siglecs have been reported to affect the immune system in different ways over the past years. Most of the human CD33-related Siglecs are regarded as inhibitory receptors as they contain ITIM or ITIM-like motifs, and several examples of biological outcomes after engagements of these Siglecs have been presented. For example, calcium influx of U937 cells was inhibited after engagement of CD33-related Siglecs (27, 28). Avril and co-workers (29) have described that cross-linking of Siglecs-7 and -9 resulted in decreased FcϵRI-mediated serotonin release of erythrocytes. Furthermore, expression of Siglec-5 was demonstrated to down-regulate T-cell receptor signaling (30), and engagement of Siglec-9 was recently shown to inhibit TNF-α production but increase the IL-10 secretion of a macrophage cell line (31). Finally, inactivation of Siglec-F in mice resulted in overproduction and hyper-reactivity of eosinophils, the cell type on which this Siglec is normally found (32).
Recently, a group of activating CD33-related receptors has been established. The antagonizing receptor of hSiglec-5 is Siglec-14, which has an identical sequence in the first two Ig-like domains (33). Immune receptors having similar sequences but counteracting signaling properties are usually called paired receptors. Siglec-14 might function as an activating receptor as it associates with a signaling adapter molecule, DNAX-activating protein 12 (DAP12) (33, 34). Interestingly, some individuals lack expression of Siglec-14, whereas hSiglec-5 is ubiquitously expressed. To be able to specifically study hSiglec-5, we identified Siglec-14-deficient donors and used the U937 monocytic cell line that expresses Siglec-5 and very little or no Siglec-14 (12).
As demonstrated above, the hSiglec-5-binding domain is located in the wall-distal N-terminal part of β, but it is unlikely that the polysaccharide capsule affects this interaction as our analysis showed negligible differences between an isogenic acapsular mutant and an encapsulated parental strain in binding to hSiglec-5 (supplemental Fig. S2). Like many other surface proteins of pathogens, the β protein has several binding sites for different human proteins and thus multiple functions for the bacterial evasion of the immune system. Binding of FH most likely results in inhibition of the complement system (17), whereas the IgA binding might affect IgA-mediated phagocytosis, superoxide generation, and release of enzymes and inflammatory mediators (19). With the demonstration that the β protein also binds to hSiglec-5, a third ligand of this surface protein was identified. Importantly, engagement of hSiglec-5 by β-expressing GBS leads to inhibition of phagocytosis and oxidative burst by phagocytes. Using GBS expressing β-deletion mutants, we show here that the β protein-mediated inhibition of phagocytosis is correlated with the B6N region, providing further evidence that this property is due to the interaction between β and hSiglec-5. We also show a significant correlation between surface expression of the β protein among clinical GBS strains and the ability to bind hSiglec-5, demonstrating that it is a general property of the β protein to bind hSiglec-5. Importantly, binding of hSiglec-5 to the β protein was completely independent of capsular serotype. For one of the strains, BE85/91 (supplemental Table S1), the binding of hSiglec-5 was low, although this strain expresses the β protein at a moderately high level. This result might be due to rare sequence variants in a small number of GBS strains unable to bind hSiglec-5, which has been reported previously for the IgA binding property of β (35). Taken together, the β protein may be involved in important immune evasion mechanisms in β-expressing GBS strains at least in part by interacting with hSiglec-5 on human phagocytes.
Supplementary Material
Acknowledgments
We thank Prof. Gunnar Lindahl and Dr. Margaretha Stålhammar-Carlemalm for the kind gift of the purified α and β proteins and for providing clinical β-expressing GBS isolates. We thank Katie Lorensson for technical help. T. A. thanks Dr. Elisabet Holst for continuous and generous support.
This work was supported in part by the Royal Physiographic Society, the Swedish Government Funding for Clinical Research (ALF), and the Foundations Crafoord, Åke Wibergs, Cornell, Zoégas, Ollie och Elof Ericsson, Österlund, Goljes, and Hierta.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and Tables S1 and S2.
- Siglec
- sialic acid-binding immunoglobulin-like lectin
- GBS
- group B Streptococcus
- IgABR
- IgA-binding region
- FH
- factor H
- ITIM
- immunoreceptor tyrosine-based inhibitory motif
- hSiglec
- human Siglec
- pAb
- polyclonal antibody.
REFERENCES
- 1. Crocker P. R., Paulson J. C., Varki A. (2007) Nat. Rev. Immunol. 7, 255–266 [DOI] [PubMed] [Google Scholar]
- 2. Varki A., Angata T. (2006) Glycobiology 16, 1R–27R [DOI] [PubMed] [Google Scholar]
- 3. Cornish A. L., Freeman S., Forbes G., Ni J., Zhang M., Cepeda M., Gentz R., Augustus M., Carter K. C., Crocker P. R. (1998) Blood 92, 2123–2132 [PubMed] [Google Scholar]
- 4. Ravetch J. V., Lanier L. L. (2000) Science 290, 84–89 [DOI] [PubMed] [Google Scholar]
- 5. Avril T., Freeman S. D., Attrill H., Clarke R. G., Crocker P. R. (2005) J. Biol. Chem. 280, 19843–19851 [DOI] [PubMed] [Google Scholar]
- 6. Jones C., Virji M., Crocker P. R. (2003) Mol. Microbiol. 49, 1213–1225 [DOI] [PubMed] [Google Scholar]
- 7. Avril T., Wagner E. R., Willison H. J., Crocker P. R. (2006) Infect. Immun. 74, 4133–4141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carlin A. F., Uchiyama S., Chang Y. C., Lewis A. L., Nizet V., Varki A. (2009) Blood 113, 3333–3336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rempel H., Calosing C., Sun B., Pulliam L. (2008) PLoS One 3, e1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Edwards M. S., Nizet V., Baker C. J. (2011) in Infectious Diseases of Fetus and Newborn Infant, 7th Ed., pp. 419–469, Elsevier, Philadelphia [Google Scholar]
- 11. Lindahl G., Stålhammar-Carlemalm M., Areschoug T. (2005) Clin. Microbiol. Rev. 18, 102–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Carlin A. F., Chang Y. C., Areschoug T., Lindahl G., Hurtado-Ziola N., King C. C., Varki A., Nizet V. (2009) J. Exp. Med. 206, 1691–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Russell-Jones G. J., Gotschlich E. C., Blake M. S. (1984) J. Exp. Med. 160, 1467–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hedén L. O., Frithz E., Lindahl G. (1991) Eur. J. Immunol. 21, 1481–1490 [DOI] [PubMed] [Google Scholar]
- 15. Jerlström P. G., Chhatwal G. S., Timmis K. N. (1991) Mol. Microbiol. 5, 843–849 [DOI] [PubMed] [Google Scholar]
- 16. Lindahl G., Akerström B., Vaerman J. P., Stenberg L. (1990) Eur. J. Immunol. 20, 2241–2247 [DOI] [PubMed] [Google Scholar]
- 17. Areschoug T., Stålhammar-Carlemalm M., Karlsson I., Lindahl G. (2002) J. Biol. Chem. 277, 12642–12648 [DOI] [PubMed] [Google Scholar]
- 18. Jerlström P. G., Talay S. R., Valentin-Weigand P., Timmis K. N., Chhatwal G. S. (1996) Infect. Immun. 64, 2787–2793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pleass R. J., Areschoug T., Lindahl G., Woof J. M. (2001) J. Biol. Chem. 276, 8197–8204 [DOI] [PubMed] [Google Scholar]
- 20. Stålhammar-Carlemalm M., Stenberg L., Lindahl G. (1993) J. Exp. Med. 177, 1593–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Larsson C., Stålhammar-Carlemalm M., Lindahl G. (1996) Infect. Immun. 64, 3518–3523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stenberg L., O'Toole P., Lindahl G. (1992) Mol. Microbiol. 6, 1185–1194 [DOI] [PubMed] [Google Scholar]
- 23. Areschoug T., Linse S., Stålhammar-Carlemalm M., Hedén L. O., Lindahl G. (2002) J. Bacteriol. 184, 6376–6383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vimr E. R., Kalivoda K. A., Deszo E. L., Steenbergen S. M. (2004) Microbiol. Mol. Biol. Rev. 68, 132–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vimr E., Lichtensteiger C. (2002) Trends Microbiol. 10, 254–257 [DOI] [PubMed] [Google Scholar]
- 26. Zhuravleva M. A., Trandem K., Sun P. D. (2008) J. Mol. Biol. 375, 437–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Paul S. P., Taylor L. S., Stansbury E. K., McVicar D. W. (2000) Blood 96, 483–490 [PubMed] [Google Scholar]
- 28. Ulyanova T., Shah D. D., Thomas M. L. (2001) J. Biol. Chem. 276, 14451–14458 [DOI] [PubMed] [Google Scholar]
- 29. Avril T., Floyd H., Lopez F., Vivier E., Crocker P. R. (2004) J. Immunol. 173, 6841–6849 [DOI] [PubMed] [Google Scholar]
- 30. Nguyen D. H., Hurtado-Ziola N., Gagneux P., Varki A. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 7765–7770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ando M., Tu W., Nishijima K., Iijima S. (2008) Biochem. Biophys. Res. Commun. 369, 878–883 [DOI] [PubMed] [Google Scholar]
- 32. Zhang M., Angata T., Cho J. Y., Miller M., Broide D. H., Varki A. (2007) Blood 109, 4280–4287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Angata T., Hayakawa T., Yamanaka M., Varki A., Nakamura M. (2006) FASEB J. 20, 1964–1973 [DOI] [PubMed] [Google Scholar]
- 34. Lanier L. L., Bakker A. B. (2000) Immunol. Today 21, 611–614 [DOI] [PubMed] [Google Scholar]
- 35. Brady L. J., Boyle M. D. (1989) Infect. Immun. 57, 1573–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.