Background: The disordered TPPP/p25 is a hallmark of synucleinopathies.
Results: Tight binding of TPPP/p25 with β-amyloid results in the formation of massive aggregates both in vitro and in vivo.
Conclusion: The presence of intracellular pathological-like TPPP/p25-β-amyloid aggregates elucidates the partial co-localization of β-amyloid and TPPP/p25 in Lewy body dementia with Alzheimer disease.
Significance: This new type of aggregation may form bridge to conjoin synucleopathies with other neuropathologies.
Keywords: Alzheimer Disease, Amyloid, Neurodegeneration, Parkinson Disease, Synuclein, Tubulin Polymerization Promoting Protein/p25
Abstract
The disordered tubulin polymerization promoting protein (TPPP/p25) was found to be co-enriched in neuronal and glial inclusions with α-synuclein in Parkinson disease and multiple system atrophy, respectively; however, co-occurrence of α-synuclein with β-amyloid (Aβ) in human brain inclusions has been recently reported, suggesting the existence of mixed type pathologies that could result in obstacles in the correct diagnosis and treatment. Here we identified TPPP/p25 as an interacting partner of the soluble Aβ oligomers as major risk factors for Alzheimer disease using ProtoArray human protein microarray. The interactions of oligomeric Aβ with proteins involved in the etiology of neurological disorders were characterized by ELISA, surface plasmon resonance, pelleting experiments, and tubulin polymerization assay. We showed that the Aβ42 tightly bound to TPPP/p25 (Kd = 85 nm) and caused aberrant protein aggregation by inhibiting the physiologically relevant TPPP/p25-derived microtubule assembly. The pair-wise interactions of Aβ42, α-synuclein, and tubulin were found to be relatively weak; however, these three components formed soluble ternary complex exclusively in the absence of TPPP/p25. The aggregation-facilitating activity of TPPP/p25 and its interaction with Aβ was monitored by electron microscopy with purified proteins by pelleting experiments with cell-free extracts as well as by confocal microscopy with CHO cells expressing TPPP/p25 or amyloid. The finding that the interaction of TPPP/p25 with Aβ can produce pathological-like aggregates is tightly coupled with unusual pathology of the Alzheimer disease revealed previously; that is, partial co-localization of Aβ and TPPP/p25 in the case of diffuse Lewy body disease with Alzheimer disease.
Introduction
Alzheimer disease (AD)2 (1) and Parkinson disease (PD) (2), the hallmark proteins of which are Tau/β-amyloid (Aβ) and α-synuclein, respectively, are the most common age-related conformational diseases causing serious socioeconomic problems (3). AD is characterized by two major neuropathological hallmarks, extracellular plaques of Aβ and neurofibrillary tangles consisting of abnormally phosphorylated Tau (4). Aβ is a product of proteolytic processing of amyloid precursor protein (APP) of undetermined function (5). Αβ is a 39–43-amino acid peptide that is the main constituent of amyloid plaque in the brains of AD patients that is a consequence rather than necessarily a cause of cell pathology (6). One of the most common isoforms is Aβ42, which is typically produced by proteolytic cleavage occurring in the trans-Golgi network (7). Accumulation of Aβ42 also occurs intracellularly with cytotoxicity resulting from initial oligomer formation. In the past attention was focused on mature β-sheet-rich amyloid fibrils and recently on the critical role of intraneuronal Aβ aggregates and smaller, soluble Aβ oligomers (8) as risk factors for AD (9, 10).
The β-amyloid hypothesis provided the basis for the therapeutic strategies of AD (11). However, this concept has been in the center of ongoing discussions because the plaque load in AD brains, in contrast to the load of Tau neurofibrillary tangles (12), does not correlate with the disease state. A series of cytosolic and mitochondrial proteins has been identified that bind Aβ aggregates (protofibrils and fibrils) (13).
Recent data have shown that a significant part (even up to 50%) of AD exhibits a third prevalent neuropathology, aggregation of α-synuclein into Lewy bodies (14), whereas Tau pathology was found in PD as well (15). Evidence has been reported for the critical role of the interaction between Aβ and α-synuclein in AD pathology by enhancing mitochondrial failure (16) as well as promoting α-synuclein aggregation with serious toxicity (17).
Tubulin polymerization promoting protein (TPPP/p25) was identified as a disordered protein; its primary target is the microtubular system (18, 19). TPPP/p25 is expressed predominantly in oligodendrocytes of the human brain, where it plays crucial role in their differentiation likely via its role in the rearrangement of the microtubular network during the projection elongation before the onset of myelination (20). However, it was found to be enriched and colocalized with α-synuclein, another disordered protein, in pathological inclusions characteristic for synucleinopathies such as PD and multiple system atrophy (21–23); therefore, it was proposed to be considered as a marker of synucleinopathies. TPPP/p25 promotes the formation of α-synuclein filament, which is probably a crucial pathological process in the cases of certain neurological diseases (23). TPPP/p25 immunolabeled α-synuclein-immunoreactive dystrophic globular neurites at the periphery of β-amyloid plaques in diffuse Lewy body disease with AD (21). Immunopositivity of TPPP/p25 was also documented by immunoelectron microscopy in post mortem brain tissue of AD patients at the pre-tangles but not at the Tau-laden neurofibrillary tangles, and dot-like TPPP/p25 immunoreactivity was also seen in neuronal cytoplasm in areas with abundant Tau pathology in AD (21).
In this paper we identified TPPP/p25 as a potential interacting partner of Aβ42 oligomer mutually influencing their structural and functional properties. Apart from opening new avenues in the research of conformational diseases with mixed type pathology, identification of new protein complexes, ultrastructures, and interfaces could provide a potentially valuable target for pharmacological intervention.
EXPERIMENTAL PROCEDURES
Chemicals
Fmoc amino acids were purchased from IRIS Biotech GmbH (Germany). Other chemicals for peptide synthesis were product of Merck. Peptide synthesis reagent (1,3-diisopropylcarbodiimide, piperidin, 1-hydroxybenzotriazole, diisopropylethylamine, trifluoroacetic acid) and solvents were of reagent grade.
Synthesis of Peptides
Peptide synthesis was carried out on “MULTIPIN NCP” non-cleavable pins (Chiron Technologies) using solid-phase peptide synthesis (Fmoc/t-butyl strategy) according to Geysen et al. (24) with some modifications at the 66-nmol scale. Decapeptides overlapping by 5 amino acid residues were synthesized. The following side chain-protecting groups were used: t-butyl for Asp, Glu, Ser, Thr, and Tyr, t-butoxycarbonyl for Lys and Trp, trityl for Asn, Gln, and His, 2,2,4,6,7-pentamethyldihydrobenzofuran-5-sulfonyl for Arg, and acetamidomethyl for Cys. Coupling was performed with 1,3-diisopropylcarbodiimide/1-hydroxybenzotriazole in N,N-dimethylformamide and monitored with bromphenol blue added to the reaction mixture (25). After the final coupling cycle the Fmoc-protecting groups were removed, and the N terminus of the peptides were acetylated using Ac2O-diisopropylethylamine-N,N-dimethylformamide (5:1:50 (v/v/v)). All side chain protecting groups were cleaved from the peptides with trifluoroacetic acid containing 2.5% ethanedithiol and 2.5% anisole except the acetamidomethyl group of Cys. Peptides were prepared in duplicate, except additional peptides produced only for amino acid analysis as a control for the synthesis.
Αβ42 Preparation
Aβ42 was prepared as described earlier (26). Then 0.2 mg of dry peptide was dissolved in 40 μl of Milli-Q ultrapure water and sonicated for 5 min, and 360 μl of PBS was added to the peptide, which was further sonicated for 5 min. The solution was filtered through a 0.2-μm filter (Millipore) and then was kept at 37 °C for 24 h before use.
TPPP/p25 Purification
Human recombinant TPPP/p25-possessing His tag tail at the N or C terminus was expressed in E. coli BL21 (DE3) cells and isolated on HIS-SelectTM Cartridge (Sigma H8286) as described previously (21). Comparative studies performed with the preparations showed virtually no difference either in the structural or in the interacting features of TPPP/p25 depending on the position of the His tag.
Tubulin Preparation
Tubulin was prepared from bovine brain according to the method of Na and Timasheff (27).
α-Synuclein Purification
Human recombinant α-synuclein was prepared as described previously (28). Protein concentration was determined from the absorbance at 280 nm using an extinction coefficient of 5960 m−1 cm−1.
Purification of Synaptic Plasma Membrane (SPM) Fraction
Highly purified SPM fraction was prepared from the forebrains of rats (Wistar, 200–300 g) as described earlier (29).
Preparation of Extract from Amyloid-expressing CHO7PA2 Cells
Cells were collected at 2000 × g at 4 °C for 15 min and then were diluted into 20 mm Tris buffer, pH 7.0, containing 1 mm EDTA, 1% Triton X-100, and protease inhibitors. The cells were then lysed by sonication with 5 short bursts of 5 s followed by intervals of 30 s for cooling in ice and centrifuged at 16,000 × g at 4 °C for 25 min, and this supernatant was used for a co-precipitation binding assay and affinity chromatography.
Protein Determination
The protein concentration was measured by the Bradford method (30) using the Bio-Rad protein assay kit.
Probing and Data Processing of the ProtoArray Human Protein Microarray
Processing of the ProtoArray Human Protein microarray 4.0 (Invitrogen) was performed according the manufacturer's protocol with small modifications as described in Virok et al. (31). Briefly, 10 μm Aβ oligomer was added on the top of the array and incubated for 1.5 h without shaking at 4 °C. The Aβ binding to the protein array was visualized by a fluorescently labeled monoclonal Aβ antibody (Sigma A3981). Array scanning was carried out using a GenePix Personal 4100A microarray scanner (Molecular Devices, Sunnyvale, CA). The localization of spots on the raw array images was performed by GenePix Pro 7.0 software (Molecular Devices). The generated “gpr” files were further analyzed by the protein-protein interaction module of the Protoarray Prospector Analyzer software (Invitrogen). The significantly Aβ binder protein spots were identified by the Z-factor-based analysis of the Prospector Analyzer software. The Z-factor for a pair of protein spots indicates how far the mean of that spot pair deviates from the mean of the array negative controls comparing the variation associated with that spot pair and negative controls. The negative controls of the same sub-array were chosen as the spot pair for comparison. Two parallel protein array experiments were performed. Proteins were considered significant Aβ binders if each of the four protein spots (1-1 duplicate spots from each array) was found to be significant by the Z factor analysis using the cutoff value of 0.4. The signal intensity of a protein spot was calculated by subtracting the median background value from the median spot value. Protein spot signal intensities were median-normalized so that the median signal intensity of each array became 1. Because every protein had duplicate spots, the average of each duplicate was used as the final signal intensity for a given protein.
The potential cellular compartments that could be influenced by the Aβ binding were investigated by a Gene Ontology (GO) Cellular Content analysis using the DAVID Web-based knowledgebase (32). DAVID analyzes the GO terms relating to the Aβ binding proteins, identifies the GO terms that contain multiple proteins, and calculates a significance value for the observed enrichment compared with all the proteins on the array.
Surface Plasmon Resonance
The direct binding of Aβ to TPPP/p25 was monitored in real time with a BIAcore X instrument (GE Healthcare). The TPPP/p25 was immobilized onto the nickel nitrilotriacetic acid chip through its His tags in 0.01 m Hepes buffer, pH 7.4, containing 0.15 m NaCl and 0.005% P20 detergent. Aβ in PBS buffer was injected onto the immobilized protein surface in various concentrations for 2 min at a flow rate of 10 μl/min at 25 °C. After a 3-min dissociation, the surface was washed with 0.01 m Hepes, pH 7.4, containing 0.15 m NaCl, 50 μm EDTA, and 0.005% P20 detergent. Bound Aβ was removed from the chip with a pulse of 6 m guanidine-HCl solution. All experiments were repeated at least three times.
ELISA
The synthesis of solid-phase peptides on polyethylene pins and immunoscreening with an ELISA type of analysis were carried out similarly to established Pepscan procedures (24). TPPP/p25 peptides coupled to polyethylene pins were tested for Aβ binding by ELISA in 96-well microtiter plates (Greiner Bio-one). Each peptide-carrying pin was immersed in 200 μl of PBS buffer containing 20 mg/ml BSA (blocking buffer) overnight at 4 °C to block nonspecific binding. 4 μm Aβ or 1.5 μm α-synuclein diluted in blocking buffer was added to the wells and incubated for 1 h at room temperature. Each pin was incubated with anti-Aβ (1:7500, Sigma A3981) or anti-α-synuclein (1:5000, Sigma S5566) diluted in blocking buffer for 1 h at room temperature, then with anti-mouse IgG conjugated to horseradish peroxidase (1:5000, dilution in blocking buffer, Sigma) for 1 h at room temperature. Between each incubation step, the wells were washed three times with PBS containing 0.05% Tween 20 for 10 min. The presence of antibodies was detected using o-phenylenediamine in the concentration of 3.7 mm with 0.03% peroxide as substrate solution. The peroxidase-catalyzed reaction was stopped after 10 min with 1 m H2SO4; absorbance was read at 490 nm with a Wallace Victor 2 multiplate reader (PerkinElmer Life Sciences). After completion of the assay, pins were sonicated for 20 min in a water bath with PBS buffer containing 1% SDS and 0.1% 2-mercaptoethanol at 65 °C. The pins were subsequently washed 3 times in hot water (65 °C) and immersed in methanol for 1 min. Pins were allowed to air-dry for a minimum of 20 min and were ready to be used for another assay. Peptides retained their antibody binding capacity for several assays (more than 50). The reaction of a pin-coupled peptide was scored positive (significance level) when the ELISA absorption was significantly higher than the 2-fold average absorption of the peptides.
In the other sets of experiments the microtiter plate was coated with 5 μg/ml (100 μl/well) protein solution (TPPP/p25, tubulin or α-synuclein) in PBS overnight at 4 °C. The wells were blocked with 1 mg/ml BSA in PBS for 1 h at room temperature. Next, the plate was incubated with serial dilutions of an interacting partner (Aβ or other protein) for 1 h at 37 °C in PBS. Where indicated, after the addition of the interacting partner, a further protein was added to the plate in constant concentration (without washing), and the plate was incubated with both partners for 1 h at 37 °C in PBS. Then the plate was sequentially incubated with the corresponding antibody produced against Aβ (1:5000, Sigma A3981) or the appropriate protein (1:5000, tubulin antibody Sigma T6199; α-synuclein antibody Sigma S5566; TPPP/p25 antibody (33) or (21)) and with the secondary IgG-peroxidase conjugate (1:5000, Sigma). Both antibodies were in PBS buffer containing 1 mg/ml BSA and incubated for 1 h at room temperature. Between each incubation steps the wells were washed three times, and o-phenylenediamine was used as the substrate solution as described above.
Turbidity Measurements
The assembly of tubulin (15 μm for paclitaxel-induced, 7 μm for TPPP/p25-induced polymerization) was assessed in polymerization buffer (50 mm MES buffer, pH 6.6, containing 100 mm KCl, 1 mm dithioerythritol, 1 mm MgCl2, and 1 mm ethylene glycol tetraacetic acid) at 37 °C in the absence and presence of Aβ42 (10–15 μm) and/or α-synuclein (5–10 μm). The tubulin polymerization into microtubules was induced by the addition of either 3 μm TPPP/p25 or 20 μm paclitaxel. The optical density was monitored at 350 nm by a Cary 100 spectrophotometer (Varian, Walnut Creek, Australia). At the final state of polymerization, some of the samples were prepared for electron microscopic analysis and analysis by SDS-PAGE.
Pelleting
Aβ peptide was incubated with the proteins for 15 min at 37 °C, or the samples at the quasi end-point of the polymerization were used. After centrifugation at 15,000 × g for 15 min at 37 °C, the pellet and the supernatant fractions were separated. The pellet fraction was washed with PBS buffer and resuspended in PBS buffer. Then the pellet and the supernatant fractions were analyzed by SDS-PAGE, separated on a Tris-Tricine three-layer gel and stained with Coomassie Brilliant Blue R-250 containing 2-mercaptoethanol and dithioerythritol. The S.E. of the determinations was ± 10% (n = 3–5).
Co-precipitation Binding Assay
The extract of amyloid expressing CHO7PA2 cells was incubated overnight at 4 °C with 20 μm human recombinant TPPP/p25. The samples were then centrifuged at 16,000 × g at 4 °C for 15 min. Amyloid expressing CHO7PA2 cells were also transiently transfected with human recombinant TPPP/p25, collected at 2000 × g at 4 °C for 15 min, and then diluted into 20 mm Tris buffer, pH 7.0, containing 1 mm EDTA, 1% Triton X-100, and protease inhibitors. The cells were then lysed by sonication with 5 short bursts of 5 s followed by intervals of 30 s for cooling in ice and were centrifuged at 16,000 × g at 4 °C for 25 min. In both sets of experiments the resulting supernatant and pellet fractions were separated; the pellet fractions were washed with 20 mm Tris buffer, pH 7.0, containing 1 mm EDTA, 1% Triton X-100, and protease inhibitors and resuspended in the same buffer. The samples were analyzed by SDS-PAGE and electrotransferred onto Immobilon-PSQ transfer membranes. The filters were subjected to immunoblotting with an antiserum directed against TPPP/p25 in rat (1:5000 (21)) or with an antibody directed against Aβ42 in mouse (1: 5000, Sigma A8978). Antibody binding was revealed by using anti-rat or anti-mouse IgG coupled with peroxidase, ECL® (enhanced chemiluminescence) Western blotting Detection reagents (Amersham Biosciences), and Kodak X-Omat AR film or 3-amino-9-ethylcarbazole as substrate.
Affinity Chromatography
TPPP/p25 or Aβ42 was immobilized to CNBr-activated Sepharose 4B (Amersham Biosciences) according to the manufacturer's instructions. The TPPP/p25 or Aβ42 bound to Sepharose was packed into column. After each experiment columns were regenerated using 3 cycles of 0.1 m sodium acetate, pH 4.0, buffer containing 0.5 m NaCl and 0.1 m Tris, pH 8.0, buffer containing 0.5 m NaCl. The columns were stored in 20% ethanol, 0.01% NaN3 solution at 4 °C. The affinity columns were equilibrated with 10 mm phosphate buffer, pH 7.2, containing 10 mm NaCl. SPM fraction was loaded to the Aβ42 column, whereas extract of CHO7PA2 cells expressing amyloid was loaded to the TPPP/p25 column. The columns were washed with at least 10 volumes of 10 mm phosphate buffer, pH 7.2, containing 10 mm NaCl. The bound proteins were eluted with 100 mm glycine buffer, pH 3.0, and concentrated using an Amicon ultrafiltration stirred-cell apparatus fitted with a YM-10 or PLAC membrane for the Aβ42 or TPPP/p25 column, respectively. The bound proteins were analyzed by SDS/PAGE, separated on a Tris-Tricine two-layer gel, and stained with Coomassie Brilliant Blue R-250 containing 2-mercaptoethanol and dithioerythritol. A Western blot was carried out as described above for the co-precipitation binding assay.
Electron Microscopy
Microtubule-containing samples were fixed in a mixture of 2% glutaraldehyde, 0.2% tannic acid, and 0.1 m sodium cacodylate, pH 7.4, for 1 h, post-fixed in 0.5% OsO4, and embedded in Durcupan (Fluka, Buchs, Switzerland). Thin sections were contrasted with uranyl acetate and lead citrate and examined in a JEOL CX 100 electron microscope. For negative staining, a drop from the unpelleted samples was applied to Formvar/carbon-coated glow-discharged copper grids for 30 s. The solution was then removed, and the grid was stained with one drop of freshly filtered 1% aqueous uranyl acetate for 30 s. The excess stain was removed by blotting with filter paper.
Cell Cultures
CHO7PA2 cells expressing human APP and processing it to Aβ (kind gift of Dr. Michael Rowan, Dublin) were cultured in DMEM supplemented with 10% FCS, 100 units/ml streptomycin, 100 μg/ml penicillin, 200 μg/ml G418, and 200 μg/ml l-proline (all reagents from Sigma) in a humidified incubator at 37 °C with 5% CO2. The expression of APP/Aβ was induced by withdrawal of the l-proline. The TPPP/p25 stable-expressing clone (CHO10) was selected after subcloning from the CHO-K1 Tet-On cell line transfected with pTRE2hyg-TPPP/p25 (34). Expression of TPPP/p25 in CHO10 cells was induced by doxycycline as described previously (34). Transfection of induced CHO7PA2 cells with human recombinant TPPP/p25 and that of induced CHO10 cells with Aβ42 was carried out with ProteoJuice (Novagen) or Pro-DeliverIn (OZ Bioscience) transfection reagent, respectively, according to the manufacturer's instructions. The cells were grown on 12-mm diameter coverslips for microscopic analysis and on 60-mm dishes for all other experiments.
Immunocytochemistry
For immunofluorescence microscopy analysis, cells were fixed with 2.5% formaldehyde at 37 °C for 10 min. Next, samples were blocked for 30 min in PBS containing 0.1% Triton X-100 and 5% FCS. Samples were stained with a mouse monoclonal Aβ antibody (1:1000, Sigma A8978), a rat polyclonal TPPP/p25 serum (1:300 (21)), and a rabbit LC3 antibody (1:1000, a kind gift of Ron R. Kopito) followed by Alexa-488-, Texas-Red-, and Alexa 633-conjugated anti-mouse, anti-rat, or anti-rabbit antibody, respectively (1:1000, Invitrogen). Samples were washed for 7 min, 3 times with PBS containing 0.1% Triton X-100 between incubations. Nuclei were counterstained with DAPI. For staining cellular membranes, the CHO10 cells were preincubated with 50 μm BODIPY 500/510 dodecanoic acid (BODIPY 3823, Invitrogen) for 1 min before fixation, and LC3 staining was omitted. After processing of samples, the coverslips were mounted with GelMount and sealed with Clarion (reagents from Biomeda).
Microscopy
The pictures of fixed samples were acquired on a Zeiss LSM710 inverted microscope with 63× objective. To minimize the cross-talk between imaged channels, sequential image collection was used. Cells are shown as single confocal section. All images were processed using ZEN software (Zeiss).
RESULTS
Protein Array-based Oligomeric Aβ Interactome Screen
The introduction of protein arrays provided a suitable method for the analysis of Aβ protein interactions on a large scale. We applied two parallel Protoarray 4.0 protein arrays with more than 8100 unique recombinant human proteins immobilized on a matrix for the Aβ interactome analysis. The Aβ binding to the protein array was visualized by a fluorescently labeled monoclonal Aβ antibody, and the fluorescent signal intensities were detected by a regular array scanner. The proteins that bound to the oligomeric Aβ were identified by Z-factor statistical analysis extended with a new type quantitative analysis of the protein array data reported very recently (31) that is based upon the normalization of the signal intensities of the spots by the concentrations. This analysis of the arrays revealed that altogether 2242 proteins displayed significant binding to the oligomeric Aβ (supplemental Table 1). Among the Aβ binding partners, several members of the cellular microtubular network, including tubulin and TPPP/p25, were identified. Our protein chip data were validated by the fact that several previously described Aβ binding proteins such as tubulin β (TUBB) (35), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (36), synuclein (37), CD36 (38), apolipoprotein A-I (APOA1) (39), various ribosomal proteins (31), heat shock proteins (HSP27, HSP60, HSP70) (40), and various histone proteins (41) were identified as Aβ interacting partners. However, it should be added that not all reported interacting partners of Aβ were detected; some of them were not immobilized on the array (ApoE receptors, p75 neurotrophin receptor, nicotinerg receptors) or they did not fulfill all the statistical requirements (integrin β1).
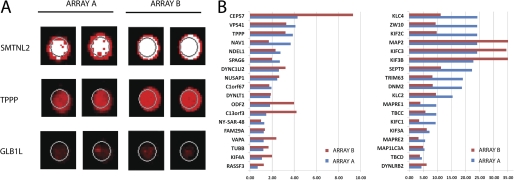
The proteins with related intracellular functions and significant Aβ binding were identified by GO Cellular Content analysis using the DAVID Web-based knowledgebase (32), which quantifies the enrichment of anti-Aβ signal for a given protein compared with that of all proteins on the array. This GO analysis showed that one of the impacted cellular systems displaying distinct affinity to Aβ was the microtubule-related proteins, which includes cytoskeleton, microtubule-associated proteins, microtubule organizing center, and microtubule itself (The GO Cellular Content category “microtubule cytoskeleton” had a p value of 0.000056, “microtubule-associated complex” had a p value of 0.0015, “microtubule organizing center” had a p value of 0.017, and “microtubule” had a p value of 0.03). A key member of this family in which we were specifically interested is the TPPP/p25. The relative Aβ binding intensities of TPPP/p25 were more than 3-fold higher in two different array experiments as compared with the median binding intensity of 1. The spot images of the TPPP/p25 are shown in Fig. 1A together with that of a strong and a weak Aβ-binding protein. The signal intensities of the 37 members of the “microtubule-related proteins” according to the GO Cellular Content category are shown in Fig. 1B. Instead, to extend the ProtoArray experiments by varying the Aβ-oligomer concentration, we carried out extensive studies with isolated proteins relevant to the neurological diseases with mixed type pathology.
FIGURE 1.
Protein array-based interactome analysis of oligomeric Aβ42. A, to explore the interacting protein partners of β-amyloid, 10 μm Aβ42 peptide was hybridized onto the Protoarray 4.0 protein array. Duplicate spot images show a strong Aβ42 binder smoothelin-like protein 2 (SMTNL2), the TPPP/p25, and a weak Aβ42 binder β-galactosidase-1-like protein (GLB1L) on both applied protein arrays. B, the cellular localization of the Aβ42 interacting proteins were characterized using the GO data base. Members of the microtubule GO cellular content category are shown, with their normalized signal intensities on both applied arrays.
Detection and Characterization of the Direct Interactions of Aβ42
Biophysical and biochemical techniques were used to characterize the direct interaction of Aβ42 oligomer with human recombinant TPPP/p25, human recombinant α-synuclein, and tubulin isolated from bovine brain that had been successfully used in our previous studies (26, 42).
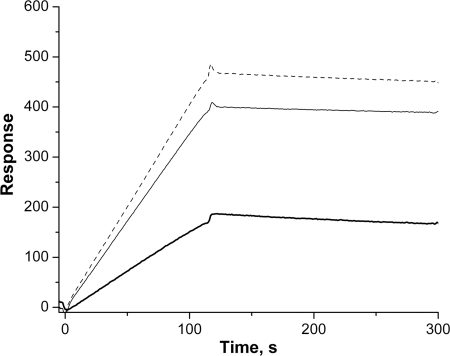
First, the interaction of Aβ42 with TPPP/p25 was tested by surface plasmon resonance, a sensitive method to detect direct complex formation. TPPP/p25 was immobilized onto the surface of a sensorchip, and Aβ42 oligomer solution at different concentrations were injected (binding phase) followed by the injection of buffer alone (dissociation phase). The registered sensorgrams are shown in Fig. 2, which clearly demonstrates the ability of the Aβ42 to associate with the immobilized TPPP/p25. However, as illustrated, the dissociation parts of sensorgrams appeared to be almost horizontal; apparently there is no effective dissociation of amyloid oligomers from the coated TPPP/p25. Thus, the evaluation of the kon and koff values of the interaction were not possible, which is similar to the cases published for other protein systems (19, 43).
FIGURE 2.
TPPP/p25-Aβ42 interaction monitored by surface plasmon resonance. Representative surface plasmon resonance curves are shown. 2.5 μm (bold line), 5 μm (solid line), 7.5 μm (dashed line) Aβ42 was injected onto TPPP/p25 immobilized on the nickel nitrilotriacetic acid chip through its His tag. Three independent experiments were performed. S.E. = ±10%.
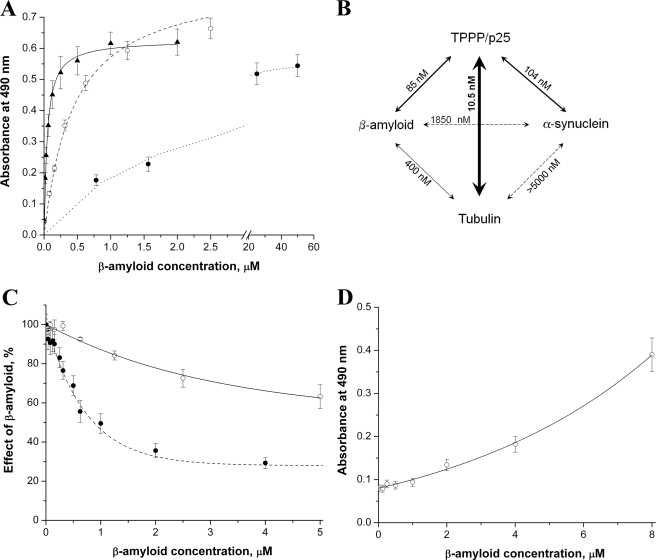
Next, in the ELISA assay, TPPP/p25, α-synuclein, or tubulin were immobilized on the wells of ELISA plates then incubated with Aβ42 oligomer at various concentrations. The binding of the Aβ42 to the immobilized proteins was detected by specific antibody for Aβ42 as described under “Experimental Procedures.” The titration curves are shown in Fig. 3A. The binding constants were evaluated from the computed curves fitted to the experimental points by nonlinear fitting of the hyperbolic saturation curves; accordingly, the apparent dissociation constants (Kd ± S.E.) for interaction of Aβ42 with TPPP/p25, tubulin, or α-synuclein were 0.085 ± 0.016, 0.40 ± 0.03, or 1.85 ± 0.15 μm, respectively (Fig. 3B). These data indicate order of magnitude differences in the binding affinities, and the tightest interaction was found between Aβ42 and TPPP/p25.
FIGURE 3.
The interaction of oligomeric Aβ42 with different proteins followed by ELISA. A, the plate was coated with 0.5 μg/well TPPP/p25 (▴), tubulin (○), or α-synuclein (●), then it was incubated with Aβ42 at different concentrations. B, the apparent dissociation constants (Kd) characteristic for the interactions were evaluated by non-linear fitting of the hyperbolic saturation curves using the Microcal Origin 6.0 software. C, ○, the plate was coated with TPPP/p25 then incubated with Aβ42. After incubation, tubulin was added in constant concentration (100 nm). After further incubation, anti-tubulin was added. ●, the plate was coated with TPPP/p25 and incubated with Aβ42, 100 nm α-synuclein was added, and then anti-α-synuclein was added. The effect of the Aβ42 was calculated as the absorbance at a given Aβ42 concentration divided by the absorbance without Aβ42. D, ○, the plate was coated with α-synuclein, then incubated with Aβ42. After incubation, tubulin was added in constant concentration (100 nm). After further incubation, anti-tubulin was added. The average of three-five independent experiments and the S.E. is shown.
Similar sets of experiments were carried out with TPPP/p25, α-synuclein, and tubulin (no Aβ42) to obtain comparative data for the pair-wise interactions of these proteins; the dissociation constants evaluated from the ELISA experiments are summarized in Fig. 3B. These data show that the affinity of TPPP/p25 to Aβ42 oligomer is weaker than that to tubulin (Kd = 0.0105 ± 0.0012 μm), its physiological interacting partner, but it is in the same order of magnitude as that to α-synuclein (Kd = 0.104 ± 0.011 μm), its pathological interacting partner (23). However, the interactions of α-synuclein with either Aβ42 oligomer or tubulin are weak and can be characterized with a Kd in the micromolar concentration range.
Then the effect of a third partner on bis-protein interactions was investigated by ELISA. In one set of experiments (Fig. 3C) TPPP/p25 was immobilized on the plate, and Aβ42 oligomer was added at various concentrations at constant tubulin or α-synuclein concentration. In the other set, α-synuclein was immobilized, tubulin was constant, and the titration was performed with Aβ42 oligomer as in the other set. In both cases the concentrations of the third partners (the constant ones) were visualized by specific antibodies. If Aβ42 oligomer does not influence the protein-protein interaction, the signal should be constant as a function of Aβ42 oligomer concentration. However, as seen in Fig. 3C, this was not the case; the presence of Aβ42 impeded the association of tubulin as well as that of α-synuclein to the immobilized TPPP/p25. As expected on the basis of the dissociation constants of bis-protein complexes, a higher Aβ42 concentration was required for the displacement of the tubulin than that of α-synuclein from the immobilized TPPP/p25, an indicating alternative binding mechanism (Fig. 3C). Similarly, tubulin impeded the TPPP/p25 interaction with α-synuclein, corresponding to an alternative binding mechanism (data not shown). Rather surprisingly, when TPPP/p25 was omitted from the system, more and more tubulin was associated to the immobilized α-synuclein by increasing the concentration of Aβ42 oligomers (Fig. 3D), which is suggestive for the formation of ternary complex of tubulin-Aβ42-α-synuclein.
Identification of the Binding Domain on TPPP/p25
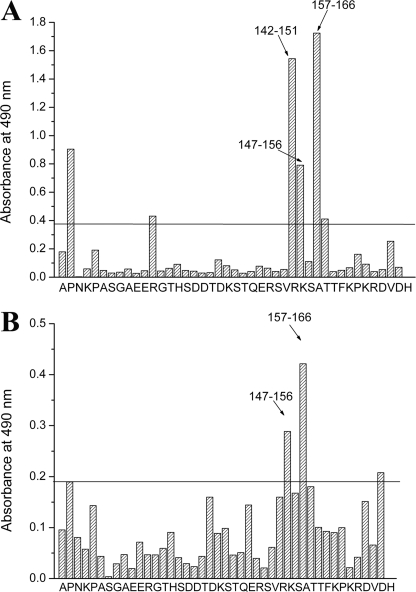
The ELISA-Pepscan method developed by Geysen et al. (24) is suitable to identify the binding motifs involved in heteroassociation, particularly in the case of unstructured proteins. The amino acid sequence of human recombinant TPPP/p25 was used to synthesize a complete set of overlapping decapeptides covalently attached to the surface of pins in a format compatible with standard ELISA. As described under “Experimental Procedures,” pins were incubated with Aβ42 or α-synuclein, and then the reaction was visualized by the addition of Aβ or α-synuclein antibodies, respectively. The reaction of a pin-coupled peptide was scored positive (significance level) when the ELISA absorption was higher than the 2-fold of the average absorption of the peptides. Fig. 4, A and B, show signal intensities along the sequence of TPPP/p25 incubated with Aβ42 or α-synuclein. The 142–166 and 147–166 amino acid sequences are indicated to be the specific binding motifs of TPPP/p25, which are targeted by Aβ42 or α-synuclein.
FIGURE 4.
Mapping of possible binding sites between Aβ42 or α-synuclein and TPPP/p25 by MULTIPIN peptide technology (Pepscan analysis). The amino acid sequence of the TPPP/p25 was used to synthesize a complete set of overlapping decapeptides covalently attached to the surfaces of derivative polyethylene pins in a format compatible with standard ELISA. These overlapping peptides covered the entire sequence of the protein. Pins were coated with 4 μm Aβ42 (A) or 1.5 μm α-synuclein (B), then anti-Aβ or anti-α-synuclein was added, respectively. The absorbances of the peptides (indicated by letters of the first amino acid of the decapeptides) are shown. The reaction of a pin-coupled peptide was scored positive (significance level, indicated by a line) when the ELISA absorption was significantly higher than the 2-fold average absorption of the peptides.
Functional Effect of Aβ42 on TPPP/p25-promoted Tubulin Polymerization-Microtubule Assembly
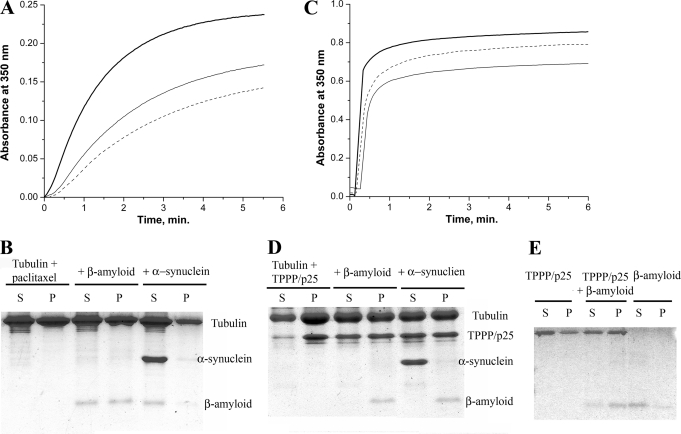
Our large scale protein array experiment (cf. supplemental Table 1) as well as experiments with purified proteins (cf. Fig. 3) provided evidence for the association of Aβ42 with both TPPP/p25 and tubulin. Previously we showed that TPPP/p25 induced assembly and bundling of microtubules as well as tubulin aggregations (18–20, 34, 44). In addition, the association of these proteins with α-synuclein has been reported as well (23). To establish the effect of Aβ42 and its interacting partners on tubulin polymerization, turbidity measurements were performed induced by paclitaxel or TPPP/p25, and the samples at the quasi endpoints were pelleted followed by analysis of the supernatant and pellet fractions.
Typical time courses are shown in Fig. 5, A and C. The initial time course of the TPPP/p25-induced tubulin polymerization does not show lag phase as observed with paclitaxel, indicating that TPPP/p25 induced tubulin aggregation beside microtubule assembly as demonstrated by electron microscopy (19). The addition of Aβ42 oligomer to tubulin at equimolar concentrations resulted in partial inhibition in the polymerization induced by paclitaxel or TPPP/p25 (Fig. 5, A and C), indicating the inhibitory effect of Aβ oligomers on the formation of tubulin assemblies. The pelleting experiments showed that although Aβ42 was partitioned between the supernatant and pellet fractions in the case of paclitaxel-induced tubulin polymerization (Fig. 5B), it was detected exclusively in the pellet (Fig. 5D) when the polymerization was promoted by TPPP/p25. This finding suggests that the self-association of Aβ42 oligomers is promoted by the presence of TPPP/p25; indeed, the interaction of TPPP/p25 with Aβ42 oligomers resulted in aggregation (Fig. 5E). Although the α-synuclein slightly reduced the paclitaxel-induced tubulin polymerization, no protein aggregation could be detected (data not shown).
FIGURE 5.
The effect of the interacting partners on tubulin assembly as determined via a turbidimetric assay followed by pelleting experiments. Tubulin polymerization was induced by paclitaxel (A and B) or by TPPP/p25 (C and D). For the turbidimetric assay (A and C), control (bold line), in the presence of Aβ42 (solid line) or Aβ42 and α-synuclein (dashed line) are shown. For the pelleting experiments (B and D), at the quasi endpoints of the polymerization curves the assay mixtures were centrifuged at 15,000 × g for 15 min at 37 °C, and the pellet (P) and the supernatant (S) fractions were separated followed by SDS-PAGE analysis on Tris-Tricine three-layer gels. Tubulin alone, incubated with Aβ42 or with both Aβ42 and α-synuclein as indicated. The concentration of tubulin was 15 μm (A and B) or 7 μm (C) or 10 μm (D) and the concentration of Aβ42 was 15 μm (A and B) or 10 mm (C) or 25 μm (D), whereas that of the TPPP/p25 was 3 μm (C) or 10 μm (D) and that of the α-synuclein was 10 μm (A, B, and D) or 5 μm (C). E, for the pelleting experiment, 2 μm TPPP/p25 alone, TPPP/p25 incubated with 25 μm Aβ42, and Aβ42 alone as indicated were used. Three-five independent experiments were performed; S.E. for turbidimetry and pelleting were ± 5 and ± 10%, respectively.
To elucidate the ELISA data showing the binding of α-synuclein to tubulin in the presence of Aβ42 oligomer, we measured tubulin polymerization induced by paclitaxel or TPPP/p25 combined with pelleting studies. In the case of paclitaxel-induced polymerization, the mixture of α-synuclein and Aβ42 oligomer further inhibited the polymerization as compared with that without α-synuclein (Fig. 5A), and both proteins appeared in the supernatant (Fig. 5B). In contrast to that, in the presence of TPPP/p25 the tubulin polymerization was slightly inhibited by the mixture of α-synuclein and Aβ42 oligomer (Fig. 5C), and the pellet fraction did not contain α-synuclein, but Aβ42 oligomer was found exclusively in this fraction (Fig. 5D). This finding, therefore, further supports that Aβ42 oligomer and α-synuclein can form a ternary complex with tubulin, a specific soluble ultrastructure that does not occur in the presence of TPPP/p25, displaying an alternative binding mechanism which concerns the ELISA Pepscan data, namely, the α-synuclein and Aβ42 could compete each with other for the common binding motifs on the TPPP/p25, which is maintained in the case of tubulin-bound TPPP/p25.
Ultrastructural Studies; Electron Microscopy
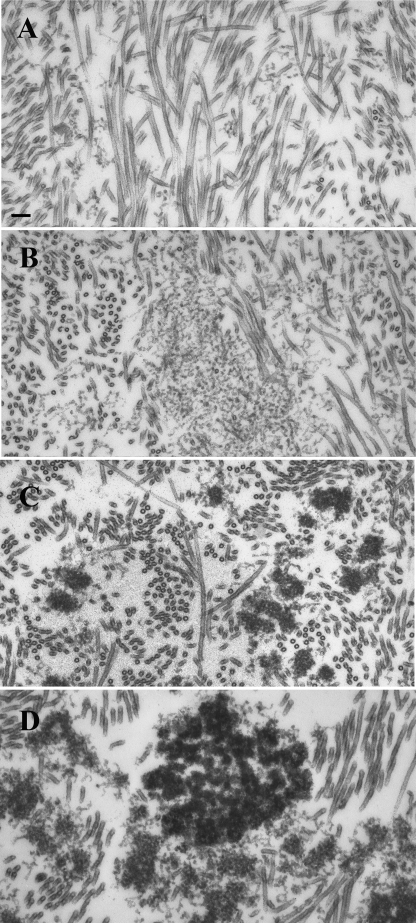
Aberrant (non-physiological) associations of unfolded/misfolded proteins are considered as initiators of pathological protein aggregations leading to formation of inclusions. Our tubulin polymerization and pelleting experiments showed that Aβ42 oligomers affected the microtubule assembly depending on whether the polymerization was induced by paclitaxel or TPPP/p25 (cf. Fig. 5). To visualize the morphologies of the protein assemblies, electron microscopic studies were carried out on sections from resin-embedded pellets. Transmission electron microscopy pictures from samples prepared by the addition of paclitaxel to tubulin solution revealed the presence of large amounts of intact-like microtubules, about 25 nm in diameter, between which small patches of thread-like oligomers were occasionally seen (Fig. 6A). The amount of these threads greatly increased in samples formed in the presence of Αβ42 oligomers without causing visible tubulin aggregation, indicating the effect of Αβ42 on the paclitaxel-induced microtubule assembly (Fig. 6B).
FIGURE 6.
Electron microscopic analysis of the effect of Aβ42 oligomers on paclitaxel- or TPPP/p25-induced tubulin polymerization. Samples were prepared with 20 μm paclitaxel (A and B) or with 1.5 μm TPPP/p25 (C and D) in the presence of 10 μm Aβ42 (B and D) or in its absence (A and C). The tubulin concentration was 10 μm (A and B) or 7 μm (C and D). Scale bar, 100 nm.
Previously we showed (42) that the TPPP/p25-induced tubulin polymerization can produce intact-like microtubules as well as ones effectively bundled by TPPP/p25; however, small tubulin aggregates were also formed as illustrated in Fig. 6C. The addition of Aβ42 oligomers resulted in few but larger aggregates without a significant amount of intact-like microtubules as shown in Fig. 6D. These data suggest that the potency of TPPP/p25 to produce microtubule assembly and bundling coupled with extensive stabilization of the microtubules (42) was impeded due to its interaction with Aβ42.
The Interaction between TPPP/p25 and Aβ at Cell Levels
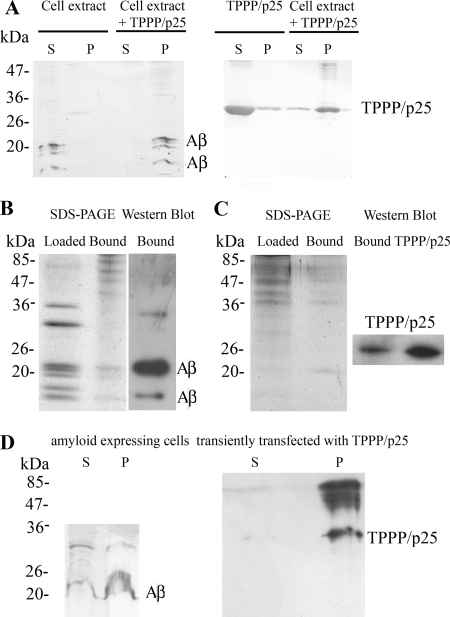
To further investigate the TPPP/p25 and Aβ interaction, three different sets of experiments were carried out. Two kinds of extracts were used for these studies, extracts of amyloid-expressing CHO7PA2 cells and SPM fraction prepared from rat brain, where TPPP/p25 is endogenously expressed. In the first set of experiments, the aggregation of TPPP/p25 and Aβ was studied by a co-precipitation binding assay in amyloid expressing CHO7PA2 cell extract incubated without or with human recombinant TPPP/p25. The partition of the proteins in the supernatant and the pellet fractions was analyzed by Western blot. As shown in Fig. 7A, TPPP/p25 or Aβ alone was found in the supernatant fractions, whereas in the presence of both partners the protein and the different Aβ oligomers/peptides were found in the pellet fraction, indicating their interaction.
FIGURE 7.
Interaction of TPPP/p25 and Aβ at cell level. A, shown is a co-precipitation assay. Extract of amyloid expressing CHO7PA2 cells was incubated without or with 20 μm TPPP/p25 overnight at 4 °C, or 20 μm TPPP/p25 alone was incubated under the same conditions, and after centrifugation, the supernatant (S) and the pellet (P) fractions were subjected to Western blot to test the partition of Aβ and TPPP/p25 as indicated. B, extract of amyloid expressing CHO7PA2 cells loaded to TPPP/p25 affinity column is shown. SDS-PAGE analysis of the cell extract loaded to the column and the bound proteins. After elution, the bound proteins were subjected to Western blot using Aβ antibody. C, extract of the SPM fraction loaded to Aβ affinity column. SDS-PAGE analysis of the SPM extract loaded to the column and the bound proteins. After elution, the bound proteins and 0.9 ng of human recombinant TPPP/p25 were subjected to Western blot using TPPP/p25 antiserum. D, shown is a co-precipitation assay. Amyloid-expressing CHO7PA2 cells were transiently transfected with human recombinant TPPP/p25 and were lysed by sonication. After centrifugation, the supernatant and the pellet fractions were subjected to Western blot to test the partition of Aβ and TPPP/p25 as indicated.
In the second set of experiments the binding of Aβ to TPPP/p25 affinity column was investigated from the same extract used above (Fig. 7B). In the third set the binding of endogenous TPPP/p25 from SPM extract to Aβ was studied by affinity chromatography, where monomeric Aβ42 was immobilized to the column (Fig. 7C). In both cases the bound proteins were analyzed by SDS-PAGE and then subjected to Western blot using Aβ antibody and TPPP/p25 antiserum, respectively. The affinity chromatography experiments corroborated the interaction between TPPP/p25 and Aβ.
In Vivo Non-physiological Aggregation Induced by the Interaction of TPPP/p25 and Amyloid
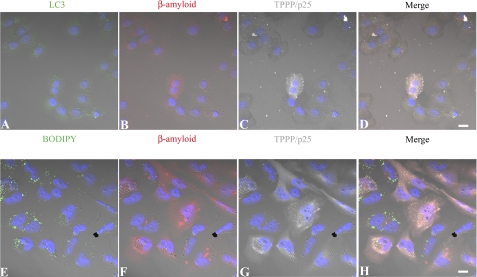
The colocalization of TPPP/p25 and Aβ was visualized by confocal microscopy in amyloid-expressing CHO7PA2 cells transfected with human recombinant TPPP/p25 (Fig. 8, A–D) as well as in CHO10 cells expressing TPPP/p25 and transfected with Aβ42 (Fig. 8, E–H) as described under “Experimental Procedures.” TPPP/p25 and the amyloid were immunostained for TPPP/p25 (gray) and Aβ (red). As shown in Fig. 8, the co-enrichment of TPPP/p25 and amyloid in the aggregates with distinct sizes is visible within the cytoplasm, whereas no protein aggregation can be visualized in the absence of TPPP/p25.
FIGURE 8.
Representative pictures of intracellular Aβ and TPPP/p25 aggregation in CHO7PA2 and CHO10 cells. Amyloid expressing CHO7PA2 cells (A–D) and TPPP/p25 expressing CHO10 cells (E–H) were transiently transfected with human recombinant TPPP/p25 and Aβ42, respectively, and immunostained for TPPP/p25 (C and G, gray), Aβ (B and F, red), and the autophagic marker LC3 (A, green), whereas the general intracellular membrane marker (BODIPY 500/510) was detected by its own signal (E, green). Note the appearance of cytoplasmic aggregates in the case of TPPP/p25 and β-amyloid co-expression on the merged pictures (D and H) that show no co-localization with either vacuole markers. Blue, DAPI. Scale bar, 10 μm.
To study whether the aggregation process was related to vacuolization, the cells were stained with anti-LC3 (Fig. 8, A and D), a specific autophagic marker (kindly provided by Prof. Kopito), or BODIPY (Fig. 8, E and H), a dye-conjugated lipid accumulating in the membranes. None of these markers showed co-labeling with the immunodetected intracellular protein aggregates; therefore, the colocalization of the aggregates did not result from vacuolization but from the mutual interaction of TPPP/p25 and amyloid.
To confirm the intracellular association of the two hallmark proteins, the aggregates formed in amyloid-expressing CHO7PA2 cells transiently transfected with TPPP/p25 were isolated by a pelleting experiment after cell lysis and immunostained for the presence of TPPP/p25 and amyloid (Fig. 7D). The TPPP/p25 was exclusively found in the pellet fraction, whereas the amyloid was found both in the supernatant and the pellet fractions (due to the presence of untransfected cells in the sample). Control data showed that in CHO7PA2 cells not transfected with TPPP/p25, the amyloid was not pelleted at all (Fig. 7A). These data show that their mutual pelleting was detected due to their hetero-association.
DISCUSSION
Cognitive impairment and synaptic dysfunction, which are early changes preceding the accumulation of the hallmark pathological lesions, were found to correlate with the accumulation of intracellular Aβ (45, 46). Recent evidence has also suggested that APP and Aβ accumulate in mitochondrial membrane in AD, and the oligomeric Aβ can induce mitochondrial damage, proteasome dysfunction, calcium dyshomeostasis via structural, and functional alterations (47). In our studies we used a specific amyloid preparation in which the 42-amino acid amyloid peptide occurred in a well established soluble oligomeric form (26), which is considered to be the most toxic form of the Aβ peptides (3). Accumulation of intraneuronal oligomeric Aβ is an early event in the pathogenesis of AD (48).
In this study using protein array we identified 2242 proteins including TPPP/p25 as the interacting partner of the oligomeric Aβ of the 8100 recombinantly expressed human proteins, representing a significant portion of the human proteome (cf. Fig. 1). Several previously described Aβ-binding proteins (35–41) were found among the interacting partners. The recombinant protein expression was performed in an insect cell line; therefore, the eukaryotic posttranslational modifications could be present. Analysis of the Aβ hybridization pattern revealed that more than 2200 human proteins bound to the oligomeric peptide. Although we detected a high number of interactions, it is likely that many of these interactions are not relevant in vivo. The reason could be numerous, e.g. the Aβ and these interacting proteins are not expressed at the same compartment or the binding domains of these proteins are not available for Aβ. The high number of interacting partners in vitro is not surprising because of the structural flexibility of the Aβ peptide, which displays a series of different metastable conformations and interacts with a large number of partner molecules (49).
Ontological analysis of the Aβ binding partners revealed that various members of the microtubular network were its potential interacting partners, which suggests that cumulative impact of Aβ on microtubule function could be significant. TPPP/p25, a modulator of the dynamics and stability of the microtubular network (50), seems to be an interacting partner of Aβ. Although the protein array data suggested that Aβ could bind to TPPP/p25 and other members of the microtubular network, these results should be considered as an output of an initial high-throughput interactome screen, which we validated in this work.
Synergistic interactions among Aβ, Tau, and α-synuclein have been proposed that could mutually promote their accumulations within the inclusions leading to accelerated cognitive dysfunction (51). In fact, deposition of multiple proteins in the brain of demented people is more the rule than exception, which alters the prognosis and therapeutic response. Therefore, TPPP/p25 as a new protein player could be involved in multiple pathological interactions leading to protein aggregations characteristic for a subtype of neurological disorders.
In this work multiple interactions of Aβ42 oligomer as well as that of TPPP/p25 were characterized at molecular and cellular levels. As illustrated in the Fig. 3 scheme, the binding of Aβ42 to TPPP/p25 appears to be the tightest (Kd = 85 nm), whereas its interaction with tubulin and α-synuclein are one and two orders of magnitude weaker, respectively. The binding affinities of other interacting partners to the APP or Aβ peptide, characterized with Kd values also in the nanomolar range, suggest the pathological relevance of these interactions (52–54). Recently, proteomic analysis of hippocampal and cortical tissue from an animal model of AD has been performed where the most important groups of significantly altered proteins included those involved in synaptic plasticity, neurite outgrowth, and microtubule dynamics (55). Moreover, the levels of both tubulin and TPPP/p25 were found to increase both in the cortex and the hippocampus as compared with that of control samples (55), and the increase of TPPP/p25 level was similar to the increase of the α-synuclein level as well as that of the Tau level (55). However, our data offer the first evidence to the direct binding of TPPP/p25 to Aβ42 oligomer, which is stronger than that of the Aβ42 to α-synuclein.
The peptide/proteins used in the present studies are considered as hallmark proteins of neurological diseases; they are disordered or have an extended unfolded region. The studied proteins do not form a ternary complex with TPPP/p25, but they exhibited alternative binding; the formation of binary complexes was detected (cf. Figs. 3C and 5). This is in agreement with the results obtained by ELISA-Pepscan analysis, suggesting (partial) overlap of the binding sequences of TPPP/p25 for α-synuclein and Aβ42 in addition to tubulin as demonstrated previously (42).
A surprising result was obtained when the tubulin, α-synuclein, and Aβ42 system was investigated (cf. Figs. 3D and 5). The association of α-synuclein to tubulin is weak; however, Aβ42 oligomer was able to promote its binding in a concentration-dependent manner indicated by ELISA (cf. Fig. 3D). This soluble ternary complex is likely of functional importance because it causes a more extensive decrease of the tubulin assembly as compared with the decrease without α-synuclein (cf. Fig. 5). The synergistic interaction of these three proteins/peptide, similar to that recently demonstrated in the case of Tau, Aβ42, and α-synuclein leading to more pronounced aggregation coupled with accelerated cognitive dysfunction (51), might be of pathological relevance.
Aβ42 effectively stimulates the oligomerization of α-synuclein (56), and vice versa, the α-synuclein promotes the oligomerization of Aβ42 leading to its in vitro precipitation (57) and formation of hybrid ring-like structures (17). TPPP/p25 can also induce α-synuclein aggregation (23).
Proteomics methods identified TPPP/p25 in various synaptic preparations (58). AD is associated with synapse loss, and emerging evidence links intraneuronal Aβ accumulation to the development of synaptic pathology, which is an early marker for this disease (for review, see Refs. 59–61). Aβ generated from axon-transported APP is released from presynaptic sites and subsequently accumulates close to the nerve terminal. Moreover, it has recently been suggested that monomeric Aβ40 and Aβ42 are the predominant forms required for synaptic plasticity and neuronal survival at physiological circumstances, and Aβ may act as a positive regulator presynaptically and as a negative regulator postsynaptically (61). Previously several synaptosomal proteins were identified to interact with the Aβ peptide including vacuolar proton-pump ATP synthase, glyceraldehyde-3-phosphate dehydrogenase, synapsin I and II, β-tubulin, and 2′,3′-cyclic nucleotide 3′-phosphodiesterase, but for these experiments the fibrillar form of the peptide was used (13). Our affinity chromatographic experiments provided the first evidence that Aβ42 peptide can bind TPPP/25 from the SPM fraction. The identification of synaptosomal molecular partners of Aβ is of great importance both physiologically and pathologically, as there is a bell-shaped relationship between Aβ and synaptic transmission; higher or lower than optimal concentration of Aβ impairs synaptic transmission.
Here we presented in vitro and in vivo evidence for the TPPP/p25-promoted aggregation of Aβ42. Single cell experiments showed the colocalization of TPPP/p25 with amyloid in massive aggregate forms (cf. Fig. 8). We noticed that relatively large particles are formed exclusively in the cells where both TPPP/p25 and Aβ are present, as indicated by their colocalization in CHO cells (cf. Fig. 8), which is the consequence of their mutual interaction within the cytoplasm shown by the isolation of the protein aggregates by pelleting experiment (cf. Fig. 7D).
The formation of protein aggregates with specific ultrastructures might be an early event in AD. In fact, TPPP/p25 was found at the pretangles as well as in neuronal cytoplasm (21), supporting the possibility of its interaction with the intracellular Aβ, which might modify/determine the aggregate formation. In addition, in a previous work we noticed TPPP/p25 immunopositivity with antibody raised against TPPP/p25 peptide for the neurites at the intracellular amyloid plaques in the case of diffuse Lewy body disease with Alzheimer disease (21). Similar immunopositivity for α-synuclein was observed in the case of the same disease. Consequently, the detection of aggregates including amyloid and α-synuclein/TPPP/p25 could be indicative for the development of a new subtype of neurological disorders that forms a functional bridge to conjoin the co-pathologies of synucleopathies and amyloid plaque formation. Further immunohistochemical studies on human brain samples are in progress to identify specific subtypes of dementias.
Acknowledgments
We are grateful to Gergely Róna (Institute of Enzymology, Biological Research Center, Hungarian Academy of Sciences, Budapest, Hungary) for providing the Pro-DeliverIn, BODIPY 500/510 reagents, and the Alexa 633-conjugated secondary antibody.
This work was supported by Hungarian National Scientific Research Fund Grants OTKA T-067963 (to J. Ovádi) and PD 76793 (to J. Oláh), the European Commission (DCI-ALA/19.09.01/10/21526/245–297/ALFA111(2010)29; to J. Ovádi), a János Bolyai Research Scholarship of the Hungarian Academy of Sciences (to J. Oláh), and EC-7 Health Grant 201159 (“Memoload”).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1.
- AD
- Alzheimer disease
- APP
- amyloid precursor protein
- Aβ
- β-amyloid
- GO
- Gene Ontology
- PD
- Parkinson disease
- SPM
- synaptic plasma membrane
- TPPP/p25
- tubulin polymerization promoting protein
- Fmoc
- N-(9-fluorenyl)methoxycarbonyl.
REFERENCES
- 1. Hardy J., Selkoe D. J. (2002) Science 297, 353–356 [DOI] [PubMed] [Google Scholar]
- 2. Spillantini M. G., Crowther R. A., Jakes R., Hasegawa M., Goedert M. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 6469–6473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Irvine G. B., El-Agnaf O. M., Shankar G. M., Walsh D. M. (2008) Mol. Med. 14, 451–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Avila J. (2006) FEBS Lett. 580, 2922–2927 [DOI] [PubMed] [Google Scholar]
- 5. Hiltunen M., van Groen T., Jolkkonen J. (2009) J. Alzheimers Dis. 18, 401–412 [DOI] [PubMed] [Google Scholar]
- 6. Carrell R. W. (2005) Trends Cell Biol. 15, 574–580 [DOI] [PubMed] [Google Scholar]
- 7. Hartmann T., Bieger S. C., Brühl B., Tienari P. J., Ida N., Allsop D., Roberts G. W., Masters C. L., Dotti C. G., Unsicker K., Beyreuther K. (1997) Nat. Med. 3, 1016–1020 [DOI] [PubMed] [Google Scholar]
- 8. Roychaudhuri R., Yang M., Hoshi M. M., Teplow D. B. (2009) J. Biol. Chem. 284, 4749–4753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wirths O., Breyhan H., Cynis H., Schilling S., Demuth H. U., Bayer T. A. (2009) Acta Neuropathol. 118, 487–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shah S. B., Nolan R., Davis E., Stokin G. B., Niesman I., Canto I., Glabe C., Goldstein L. S. (2009) Neurobiol. Dis. 36, 11–25 [DOI] [PubMed] [Google Scholar]
- 11. Hardy J., Allsop D. (1991) Trends Pharmacol. Sci. 12, 383–388 [DOI] [PubMed] [Google Scholar]
- 12. Braak H., Braak E. (1991) Acta Neuropathol. 82, 239–259 [DOI] [PubMed] [Google Scholar]
- 13. Verdier Y., Huszár E., Penke B., Penke Z., Woffendin G., Scigelova M., Fülöp L., Szucs M., Medzihradszky K., Janáky T. (2005) J. Neurochem. 94, 617–628 [DOI] [PubMed] [Google Scholar]
- 14. Crews L., Tsigelny I., Hashimoto M., Masliah E. (2009) Neurotox. Res. 16, 306–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lei P., Ayton S., Finkelstein D. I., Adlard P. A., Masters C. L., Bush A. I. (2010) Int. J. Biochem. Cell Biol. 42, 1775–1778 [DOI] [PubMed] [Google Scholar]
- 16. Kazmierczak A., Strosznajder J. B., Adamczyk A. (2008) Neurochem. Int. 53, 263–269 [DOI] [PubMed] [Google Scholar]
- 17. Tsigelny I. F., Crews L., Desplats P., Shaked G. M., Sharikov Y., Mizuno H., Spencer B., Rockenstein E., Trejo M., Platoshyn O., Yuan J. X., Masliah E. (2008) PLoS One 3, e3135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hlavanda E., Kovács J., Oláh J., Orosz F., Medzihradszky K. F., Ovádi J. (2002) Biochemistry 41, 8657–8664 [DOI] [PubMed] [Google Scholar]
- 19. Tirián L., Hlavanda E., Oláh J., Horváth I., Orosz F., Szabó B., Kovács J., Szabad J., Ovádi J. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 13976–13981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lehotzky A., Lau P., Tokési N., Muja N., Hudson L. D., Ovádi J. (2010) Glia 58, 157–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kovács G. G., László L., Kovács J., Jensen P.H., Lindersson E., Botond G., Molnár T., Perczel A., Hudecz F., Mezo G., Erdei A., Tirián L., Lehotzky A., Gelpi E., Budka H., Ovádi J. (2004) Neurobiol. Dis. 17, 155–162 [DOI] [PubMed] [Google Scholar]
- 22. Orosz F., Kovács G.G, Lehotzky A., Oláh J, Vincze O., Ovádi J. (2004) Biol. Cell 96, 701–711 [DOI] [PubMed] [Google Scholar]
- 23. Lindersson E., Lundvig D., Petersen C., Madsen P., Nyengaard J. R., Højrup P., Moos T., Otzen D., Gai W. P., Blumbergs P. C., Jensen P. H. (2005) J. Biol. Chem. 280, 5703–5715 [DOI] [PubMed] [Google Scholar]
- 24. Geysen H. M., Meloen R. H., Barteling S. J. (1984) Proc. Natl. Acad. Sci. U.S.A. 81, 3998–4002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Krchnák V., Vágner J., Lebl M. (1988) Int. J. Pept. Protein Res. 32, 415–416 [DOI] [PubMed] [Google Scholar]
- 26. Bozso Z., Penke B., Simon D., Laczkó I., Juhász G., Szegedi V., Kasza A., Soós K., Hetényi A., Wéber E., Tóháti H., Csete M., Zarándi M., Fülöp L. (2010) Peptides 31, 248–256 [DOI] [PubMed] [Google Scholar]
- 27. Na G. C., Timasheff S. N. (1986) Biochemistry 25, 6214–6222 [DOI] [PubMed] [Google Scholar]
- 28. Paik S. R., Lee J. H., Kim D. H., Chang C. S., Kim J. (1997) Arch. Biochem. Biophys. 344, 325–334 [DOI] [PubMed] [Google Scholar]
- 29. Fábián G., Bozó B., Szikszay M., Horváth G., Coscia C. J., Szücs M. (2002) J. Pharmacol. Exp. Ther. 302, 774–780 [DOI] [PubMed] [Google Scholar]
- 30. Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 31. Virok D. P., Simon D., Bozsó Z., Rajkó R., Datki Z., Bálint É., Szegedi V., Janáky T., Penke B., Fülöp L. (2011) J. Proteome. Res. 10, 1538–1547 [DOI] [PubMed] [Google Scholar]
- 32. Sherman B. T., Huang da W., Tan Q., Guo Y., Bour S., Liu D., Stephens R., Baseler M. W., Lane H. C., Lempicki R. A. (2007) BMC Bioinformatics 8, 426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Höftberger R., Fink S., Aboul-Enein F., Botond G., Olah J., Berki T., Ovadi J., Lassmann H., Budka H., Kovacs G. G. (2010) Glia 58, 1847–1857 [DOI] [PubMed] [Google Scholar]
- 34. Tokési N., Lehotzky A., Horváth I., Szabó B., Oláh J., Lau P., Ovádi J. (2010) J. Biol. Chem. 285, 17896–17906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Oyama R., Yamamoto H., Titani K. (2000) Biochim. Biophys. Acta 1479, 91–102 [DOI] [PubMed] [Google Scholar]
- 36. Verdier Y., Földi I., Sergeant N., Fülöp L., Penke Z., Janáky T., Szücs M., Penke B. (2008) J. Pept. Sci. 14, 755–762 [DOI] [PubMed] [Google Scholar]
- 37. Henning Jensen P. (2001) Methods Mol. Med. 62, 61–65 [DOI] [PubMed] [Google Scholar]
- 38. Coraci I. S., Husemann J., Berman J. W., Hulette C., Dufour J. H., Campanella G. K., Luster A. D., Silverstein S. C., El-Khoury J. B. (2002) Am. J. Pathol. 160, 101–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Koudinov A. R., Berezov T. T., Kumar A., Koudinova N. V. (1998) Clin. Chim. Acta 270, 75–84 [DOI] [PubMed] [Google Scholar]
- 40. Wilhelmus M. M., de Waal R. M., Verbeek M. M. (2007) Mol. Neurobiol. 35, 203–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Duce J. A., Smith D. P., Blake R. E., Crouch P. J., Li Q. X., Masters C. L., Trounce I. A. (2006) J. Mol. Biol. 361, 493–505 [DOI] [PubMed] [Google Scholar]
- 42. Hlavanda E., Klement E., Kókai E., Kovács J., Vincze O., Tökési N., Orosz F., Medzihradszky K. F., Dombrádi V., Ovádi J. (2007) J. Biol. Chem. 282, 29531–29539 [DOI] [PubMed] [Google Scholar]
- 43. Vincze O., Tökési N., Oláh J., Hlavanda E., Zotter A., Horváth I., Lehotzky A., Tirián L., Medzihradszky K. F., Kovács J., Orosz F., Ovádi J. (2006) Biochemistry 45, 13818–13826 [DOI] [PubMed] [Google Scholar]
- 44. Lehotzky A., Tirián L., Tökési N., Lénárt P., Szabó B., Kovács J., Ovádi J. (2004) J. Cell Sci. 117, 6249–6259 [DOI] [PubMed] [Google Scholar]
- 45. Oddo S., Caccamo A., Shepherd J. D., Murphy M. P., Golde T. E., Kayed R., Metherate R., Mattson M. P., Akbari Y., LaFerla F. M. (2003) Neuron 39, 409–421 [DOI] [PubMed] [Google Scholar]
- 46. Billings L. M., Oddo S., Green K. N., McGaugh J. L., LaFerla F. M. (2005) Neuron 45, 675–688 [DOI] [PubMed] [Google Scholar]
- 47. Reddy P. H. (2009) Exp. Neurol. 218, 286–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. LaFerla F. M., Green K. N., Oddo S. (2007) Nat. Rev. Neurosci. 8, 499–509 [DOI] [PubMed] [Google Scholar]
- 49. Tompa P. (2009) Structure and Function of Intrinsically Disordered Proteins, CRC Press, New York [Google Scholar]
- 50. Ovádi J. (2008) IUBMB Life 60, 637–642 [DOI] [PubMed] [Google Scholar]
- 51. Clinton L. K., Blurton-Jones M., Myczek K., Trojanowski J. Q., LaFerla F. M. (2010) J. Neurosci. 30, 7281–7289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hughes S. R., Khorkova O., Goyal S., Knaeblein J., Heroux J., Riedel N. G., Sahasrabudhe S. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 3275–3280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Golabek A. A., Soto C., Vogel T., Wisniewski T. (1996) J. Biol. Chem. 271, 10602–10606 [DOI] [PubMed] [Google Scholar]
- 54. Ahn H. J., Zamolodchikov D., Cortes-Canteli M., Norris E. H., Glickman J. F., Strickland S. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 21812–21817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Martin B., Brenneman R., Becker K. G., Gucek M., Cole R. N., Maudsley S. (2008) PLoS One 3, e2750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Masliah E., Rockenstein E., Veinbergs I., Sagara Y., Mallory M., Hashimoto M., Mucke L. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 12245–12250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mandal P. K., Pettegrew J. W., Masliah E., Hamilton R. L., Mandal R. (2006) Neurochem. Res. 31, 1153–1162 [DOI] [PubMed] [Google Scholar]
- 58. Orosz F., Lehotzky A., Oláh J., Ovádi J. (2009) in Protein Folding and Misfolding: Neurodegenerative Diseases (Ovádi J., Orosz F. eds) pp. 225–250, Springer-Verlag, New York [Google Scholar]
- 59. Parihar M. S., Brewer G. J. (2010) J. Alzheimers Dis. 22, 741–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gouras G. K., Tampellini D., Takahashi R. H., Capetillo-Zarate E. (2010) Acta Neuropathol. 119, 523–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Palop J. J., Mucke L. (2010) Nat. Neurosci. 13, 812–818 [DOI] [PMC free article] [PubMed] [Google Scholar]