Background: The source of malonyl-CoA required for de novo fatty acid synthesis in mammalian mitochondria is unknown.
Results: Mammalian ACSF3 protein is a mitochondrial malonyl-CoA synthetase.
Conclusion: Free malonate is a precursor for mitochondrial malonyl-CoA.
Significance: Mammalian mitochondria are unusual in utilizing a malonyl-CoA synthetase as a surrogate for the acetyl-CoA carboxylase usually employed in fatty acid synthesis.
Keywords: Biosynthesis, Fatty Acid, Lipogenesis, Mitochondria, Mitochondrial Metabolism, ACSF3, Acyl-CoA Synthetases, Malonate Metabolism, Malonyl-CoA Synthetase
Abstract
The objective of this study was to identify a source of intramitochondrial malonyl-CoA that could be used for de novo fatty acid synthesis in mammalian mitochondria. Because mammalian mitochondria lack an acetyl-CoA carboxylase capable of generating malonyl-CoA inside mitochondria, the possibility that malonate could act as a precursor was investigated. Although malonyl-CoA synthetases have not been identified previously in animals, interrogation of animal protein sequence databases identified candidates that exhibited sequence similarity to known prokaryotic forms. The human candidate protein ACSF3, which has a predicted N-terminal mitochondrial targeting sequence, was cloned, expressed, and characterized as a 65-kDa acyl-CoA synthetase with extremely high specificity for malonate and methylmalonate. An arginine residue implicated in malonate binding by prokaryotic malonyl-CoA synthetases was found to be positionally conserved in animal ACSF3 enzymes and essential for activity. Subcellular fractionation experiments with HEK293T cells confirmed that human ACSF3 is located exclusively in mitochondria, and RNA interference experiments verified that this enzyme is responsible for most, if not all, of the malonyl-CoA synthetase activity in the mitochondria of these cells. In conclusion, unlike fungi, which have an intramitochondrial acetyl-CoA carboxylase, animals require an alternative source of mitochondrial malonyl-CoA; the mitochondrial ACSF3 enzyme is capable of filling this role by utilizing free malonic acid as substrate.
Introduction
Mitochondria synthesize fatty acids de novo using a suite of freestanding enzymes that closely resemble their prokaryotic counterparts. These “type II” fatty acid-synthesizing systems differ from those found in the cytosol of animals, in which the component enzymes are covalently linked in large multifunctional polypeptides, the “type I” systems. The mitochondrial systems are composed of an acyl carrier protein (ACP)2 and malonyltransferase (which together generate the malonyl-ACP substrate used for chain extension by β-ketoacyl synthase) and a trio of β-carbon-processing enzymes (β-ketoacyl reductase, dehydrase, and enoyl reductase) that completely saturate the acyl chain prior to the following round of chain extension. All of these enzymes have been characterized in mammalian mitochondria (1–5), and recent evidence indicates that one of the major functions of the pathway is to generate the octanoyl precursor required for formation of the lipoyl moieties that are essential for post-translational modification of several mitochondrial proteins (6, 7). The source of malonyl-CoA as the substrate for a mitochondrial fatty acid synthase system is unknown (8). Both the α- and β-isoforms of acetyl-CoA carboxylase that have been described in animals generate malonyl-CoA in the cytosol, although the β-isoform is associated with the outer mitochondrial membrane (9–11). The α-form generates malonyl-CoA for utilization by the cytosolic fatty acid synthase, whereas the β-form is believed to provide malonyl-CoA primarily for the regulation of carnitine palmitoyltransferase I, which controls entry of fatty acids into the mitochondria for oxidation. Malonyl-CoA is also produced by the β-oxidation of odd chain length dicarboxylic fatty acids, but this process is thought to be exclusively peroxisomal (12). There are no reports indicating that a malonyl-CoA-translocating system might be present in mitochondrial membranes. An alternative way of generating malonyl-CoA intramitochondrially could be via the mitochondrial propionyl-CoA carboxylase, which has some activity toward acetyl-CoA (13). In an earlier study, we found that mitochondrial extracts from bovine heart were capable of incorporating radiolabeled malonic acid into fatty acids, raising the possibility that malonyl-CoA could be generated by a malonyl-CoA synthetase (7). Malonyl-CoA synthetases have been characterized in prokaryotes (14) but not previously in animals. The goal of this study was to determine whether mammalian mitochondria contain a malonyl-CoA synthetase that could play a role in fatty acid synthesis.
EXPERIMENTAL PROCEDURES
Recombinant ACSF3 Expression and Purification
OmicsLink T7 promoter-driven expression clones encoding ACSF3 (acyl-CoA synthetase family member 3) were obtained as tobacco etch virus protease-cleavable, N-terminally His-tagged (EX-T5643-B01) and GST-tagged (EX-T5643-B03) plasmids from GeneCopoeia (Rockville, MD). Plasmids were chemically transformed into Escherichia coli DH5α and BL21(DE3)pLysS cells according to the GeneCopoeia product protocol. Authenticity of the plasmids was confirmed by DNA sequencing. Several single base substitutions were found in the ACSF3 protein coding region when compared with that of another cDNA in the human sequence database (NM_174917). Only one substitution resulted in an amino acid change, V372M. This replacement, which is located in a region that is not highly conserved, was also reported in another genetic variant (AAH28399). Overnight culture of transformed BL21(DE3)pLysS clones in Terrific broth medium containing 0.4% glucose, 50 μg/ml carbenicillin, and 34 μg/ml chloramphenicol was used to inoculate 120 ml of Terrific broth medium containing 0.2% glycerol and carbenicillin at 37 °C. When an absorbance unit of 1 (600 nm) was reached, the entire culture was transferred to a fermentor filled with 1.9 liters of Terrific broth medium with 0.2% glycerol and enriched with 2 g/liter (NH4)2HPO4. Cells were grown at 37 °C again to an absorbance unit of 1, cooled to 20 °C, induced with 1 mm isopropyl β-d-thiogalactopyranoside, and left overnight. Pelleted cells were diluted in 3 volumes of 50 mm NaCl, 50 mm Tris-HCl (pH 8), 5 mm EDTA, 5 μg/ml leupeptin, 1 mm E-64, 5 μg/ml α1-antitrypsin, and 1 mm PMSF and lysed in a French press; the extract was centrifuged at 17,000 × g for 20 min; and the supernatant flash-frozen in liquid nitrogen at −80 °C. The GST-ACSF3 fusion protein was purified from the lysate using glutathione-Sepharose 4B beads (GE Healthcare) essentially according to the manufacturer's batch protocol except that 1% Triton X-100 was added to the lysate, TBS was used in place of PBS, and the fusion protein was eluted with 10 mm reduced glutathione in 50 mm Tris-HCl (pH 8). The fusion protein was concentrated by centrifugal ultrafiltration; cleaved by treatment with AcTEVTM protease (Invitrogen) for 2 days at 4 °C; and dialyzed against 0.1 m MOPS (pH 7.1), 1 mm EDTA, and 10% glycerol. The products were separated using glutathione-Sepharose 4B beads. The ACSF3 protein was recovered in the unbound fraction.
Mutagenesis
Two recombinant ACSF3 mutants were made from the GST-ACSF3 plasmid using a QuikChange site-directed mutagenesis kit, and primers were designed using the Agilent QuikChange primer Design program: R354A (CG → GC), 5′-acaccctgctggaggcgtatggcatgaccg-3′ and 5′-cggtcatgccatacgcctccagcagggtgt-3′; and R354L (G → T), 5′-caccctgctggagctgtatggcatgaccg-3′ and 5′-cggtcatgccatacagctccagcagggtg-3′. The R354A and R354L plasmids were transformed into E. coli XL1-Gold and BL21(DE3)pLysS competent cells, and authenticity of the mutants was confirmed by DNA sequencing. The mutant proteins were expressed and purified essentially as described for the wild-type protein.
Assay of Malonyl-CoA Synthetase Activity
Enzyme activity was assayed routinely by incubating samples at 37 °C in a 50-μl reaction mixture that included 100 mm MOPS (pH 7.1), 2 mm tris(2-carboxyethyl)phosphine, 3 mm MgCl2, 200 μm CoA, 2 mm ATP, and 0.1 mm [2-14C]malonate. Control reactions lacked ATP. In some assays, CoA was replaced by 10 μm human mitochondrial holo-ACP, prepared as described previously (5), and reactions were continued for either 2 h or overnight. In specificity studies, various di- and monocarboxylic substrates at 100 μm (or at 20 μm for a very long chain) were used in place of malonate. Purified ACSF3 protein (1 or 3 ng/μl) was used in kinetic or specificity assays, respectively. Reactions were stopped after 30 min (kinetic assays) or 2 h (specificity assays) either by the addition of cold 10% TCA for substrates with fewer than eight carbon atoms or by flash-freezing for longer substrates. Samples were centrifuged at 12,000 × g for 10 min at 4 °C, and the supernatants were analyzed by HPLC on SpheriSorb C18 column (300 Å, 5 μm, 4.6 × 250 mm; Waters) at 20 °C. Products were identified by comparison of their elution times with those of authentic standards and quantitated either by radioactivity monitoring or, for nonradioactive substrates, from the absorbance at 258 nm attributable to the CoA thioester. In some cases, the appearance of an absorbance peak corresponding to AMP was also used to detect activity. Stock solution of octanoic and lauric acids were prepared in methanol. The final concentration of methanol in the assay was 2%. Stock solutions of palmitic, oleic, and lignoceric acids were prepared in buffered α-cyclodextrin (15). α-Cyclodextrin at concentrations up to 2 mm did not affect ACSF3 activity with malonate.
Three different HPLC solvent systems were utilized for product identification and quantitation. For [2-14C]malonic acid, [1-14C]acetic acid, [1-14C]butyric acid, and succinic acid, the column was developed at 0.8 ml/min using a two-step gradient: 50 mm sodium phosphate (pH 5.6) to 50% 50 mm sodium phosphate (pH 5.6) and 20% methanol over 10 min and then to 100% 50 mm sodium phosphate (pH 5.6) and 20% methanol over 20 min and held for a further 10 min (gradient system 1). For methylmalonic, oxalic, octanoic, lauric, palmitic, lignoceric, and oleic acids, the column was developed at 1 ml/min with 50 mm sodium phosphate (pH 5.6) for 10 min, followed by a three-step gradient to 40% 50 mm sodium phosphate (pH 5.6) and 10% acetonitrile over 10 min, to 100% 50 mm sodium phosphate (pH 5.6) and 10% acetonitrile over 2 min, and to 90% acetonitrile over 18 min and then continued at 90% acetonitrile for 5 min (gradient system 2). Reaction products with lignoceric and oleic acids were also analyzed on an Inertsil phenyl column (150 Å, 5 μm, 4.6 × 150 mm; Varian) developed in 95% 50 mm potassium phosphate and 5% acetonitrile at a flow rate of 1.5 ml/min for 5 min, followed by three-step gradient: 95% 50 mm potassium phosphate and 5% acetonitrile to 70% 50 mm potassium phosphate and 30% acetonitrile over 3 min, then 70% 50 mm potassium phosphate and 30% acetonitrile to 31% 50 mm potassium phosphate and 69% acetonitrile over 13 min, and finally 31% 50 mm potassium phosphate and 69% acetonitrile to 20% 50 mm potassium phosphate and 80% acetonitrile over 2 min and held for a further 2 min (gradient system 3). Values for Km and kcat were calculated using EnzymeKinetics (Trinity Software).
Electrophoresis and Western Analysis
SDS-PAGE was performed on 10 or 9% gels that were subsequently stained with Pro-Blue Staining Kit (Owl Separation Systems/Fisher Scientific) or blotted onto a PVDF membrane and analyzed as described previously (7). Samples used for quantification were transferred overnight in the cold in 4 mm 3-(cyclohexylamino) propane sulfonic acid (pH 11) in a Bio-Rad Mini-PROTEAN® II transfer unit with the voltage set at 30 V. All secondary antibodies used in Western analyses were tagged with horseradish peroxidase, which was detected with a chemiluminescent substrate, Pierce ECL or SuperSignal West Femto (Thermo Scientific). The primary polyclonal antibody against human ACSF3 protein (LOC197322) was obtained from Abnova. Some mitochondrial preparations were precipitated with ice-cold 20% TCA in acetone before solubilization in the sample buffer. Images were scanned as transparencies and quantified with NIH ImageJ (1.45i). Each sample was analyzed by SDS-PAGE at least twice, and only images that contained limited numbers of black pixels were used for quantification.
HEK293T Cell Culture and siRNA Knockdown
HEK293T cell culture, siRNA knockdown, and purification of mitochondria were performed as described previously (6) with minor changes: knockdown experiments were scaled up using T-25 flasks, and 10 mm EDTA in Dulbecco's phosphate-buffered saline was used to detach cells. These mitochondrial preparations were used to assay for malonyl-CoA synthetase activity and for overall activity of the fatty acid biosynthetic pathway as described below.
Preparation of Crude Microsomal and Mitochondrial Preparations from HEK293T Cells
Cells were homogenized and centrifuged at 650 × g for 10 min to remove debris. Mitochondria were sedimented at 10,500 × g for 30 min, the pellet was washed with homogenization buffer, and mitochondria were finally sedimented again at 10,500 × g for 15 min. Both supernatants were combined and centrifuged at 100,000 × g for 1 h to sediment the microsomal fraction. The microsomal pellet was washed with homogenization buffer and resedimented. Final microsomal and mitochondrial pellets were resuspended in 100 mm MOPS (pH 7.1), 1 mm DTT, 1 mm EDTA, and 10% glycerol; flash-frozen, and stored at −80 °C.
Subcellular Fractionation of HEK293T Cell Lysates and Density Gradient Purification of Mitochondria
Debris-free cell homogenate derived from 15 × 106 cells was centrifuged at 100,000 × g for 1 h. The pellet was washed and resuspended in 0.3 ml of homogenization buffer and then applied at 4 °C to the top of a five-layer 10–30% discontinuous iodixanol gradient (OptiPrep) in a total gradient volume of 4.75 ml. Samples were centrifuged for 3 h at 100,000 × g in an SW 50.1 rotor (Beckman) at 4 °C, 0.1 ml was taken from the top of the gradient, and the remainder was collected in 0.35-ml fractions. A 0.225-ml portion of each fraction was diluted with 0.225 ml of homogenization buffer, and organelles were pelleted at 100,000 × g for 1 h. The resulting pellets were suspended in 50 μl of solubilization buffer (1% Triton X-100, 50 mm imidazole-HCl (pH 7.4), 2 mm aminohexanoic acid, 1 mm EDTA, 1 mm DTT, and protease inhibitors). After 20 min on ice, extracts were centrifuged at 12,000 × g for 10 min, and supernatants were used for activity assays and Western analysis. The remaining portions of the gradient fractions were treated with methanol and chloroform (16), and the precipitates were dissolved in sample buffer and analyzed by Western blotting.
Assay of Overall Activity of the Mitochondrial Pathway for Fatty Acid Biosynthesis Using Malonate as a Substrate
Assays and product analysis were carried out as described previously (7) with some modifications. The mitochondrial extract was replaced with whole mitochondria (derived from either HEK293T cells or mouse tissues; 1.2 mg/ml mitochondrial protein), and reaction mixtures contained additionally 0.2% Triton X-100 and 2.5 μm rotenone. Reactions were stopped after 5 h by the addition of 0.35 ml of ice-cold TCA to 10% and 75 μg of BSA and washed twice with 0.4 ml of ice-cold 5% TCA and five times with 0.3 ml of ice-cold 2.4 m ammonium sulfate. This procedure ensured complete washing of free radioactivity, leaving only covalently bound radioactive products (data not shown).
Other Methods
Bovine and mouse mitochondria were isolated as described (7) and purified by differential centrifugation (6) or on a five-layer discontinuous iodixanol gradient as described above. Protein concentrations were determined using the bicinchoninic acid reagent (Pierce) in the presence of 0.16% SDS. Citrate synthase was assayed essentially as described (17). [2-14C]Malonate, free of contamination by [14C]acetate, was obtained from Moravek (Brea, CA).
RESULTS
Identification of a Candidate Malonyl-CoA Synthetase
Interrogation of the human protein sequence database using malonyl-CoA synthetase sequences from Rhizobium trifolii and Bradyrhizobium japonicum as probes identified a number of putative acyl-CoA synthetases with significant sequence similarity. Among the highest scoring proteins were ACSM2 (acyl-CoA synthetase member 2), a butyryl-CoA synthetase, and ACSF3, a previously uncharacterized protein, each with ∼60% sequence similarity and 28% identity (supplemental Fig. 1). ACSF3 and ACSM2 are acronyms for acyl-CoA synthetases (ACS) according to the classification scheme introduced by Watkins et al. (18). “M2” indicates member 2 of the group active toward medium chain substrates, and “F3” indicates member 3 of the generic family having uncertain specificity.
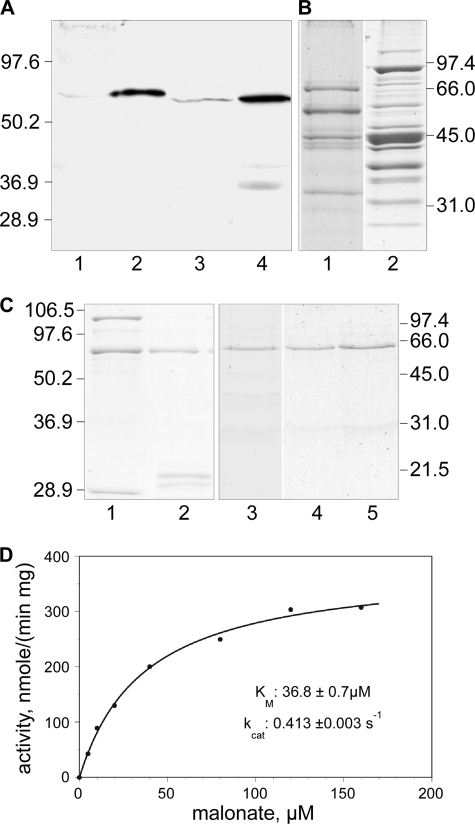
Using the available crystal structure of ACSM2 with a butyryl moiety bound at the active site (Protein Data Bank code 3EQ6) (19), we identified all amino acids within 6 Å of C4 of the butyryl moiety. By aligning the other acyl-CoA synthetases, positionally equivalent residues were identified. Only one of these proteins, ACSF3, included an arginine residue in the putative active site region that also aligned with an arginine in the malonyl-CoA synthetases from Rhizobium leguminosarum and B. japonicum. We reasoned that this arginine might facilitate the binding of a malonyl moiety through formation of a salt bridge. This arginine residue is positioned within a sequence motif (ERYGMTE) that is conserved in the prokaryotic malonyl-CoA synthetases and in ACSF3 proteins but not in other homologous acyl-CoA synthetases. The predicted molecular mass of the ACSF3 protein is 64,131 Da, and, compared with the malonyl-CoA synthetases of R. trifolii and B. japonicum, the hypothetical human protein includes an N-terminal extension that is predominantly basic in character and predicted by iPSORT (20), MitoProt II (21), and Target 1.1 (22, 23) to have the properties of a mitochondrial targeting sequence (supplemental Fig. 1). Highly homologous ACSF3 and ACSM2 sequences were also found in the bovine, rat, and mouse databases. On this basis, we concluded that ACSF3 was the best candidate for the role of a mitochondrial malonyl-CoA synthetase. However, there was one piece of published information that argued against this conclusion: ACSF3 had previously been tentatively identified as an acyl-CoA synthetase that purportedly exhibits a preference for very long chain fatty acids (18). With this reservation in mind, we initiated attempts to purify a malonyl-CoA synthetase directly from bovine and mouse heart mitochondria but were thwarted by the low abundance of the enzyme and poor yields, particularly in low ionic strength media. For example, although we were able to obtain substantial enrichment of the ACSF3 proteins from both sources, as judged by Western analysis (Fig. 1A), general staining of these preparations subjected to SDS-PAGE indicated that the ACSF3 proteins remained minor components (Fig. 1B). Indeed, although we were able to identify all of the major mitochondrial proteins in the partially purified bovine preparation by in situ tryptic digestion and mass spectrometric analysis, none of them corresponded to bovine ACSF3 (data not shown). Nevertheless, we were able to establish that these partially purified preparations were capable of synthesizing malonyl-CoA from malonate, ATP, Mg2+, and CoA and contained a protein of ∼60 kDa that was recognized by antibodies against ACSF3 (Fig. 1A). We therefore elected to attempt cloning and expression of this protein for detailed biochemical analyses.
FIGURE 1.
Purification and properties of ACSF3. A, Western analysis of bovine and mouse mitochondrial extracts probed with antibody against ACSF3. Lanes 1 and 3, 60 μg of mitochondrial protein from bovine and mouse heart extracts, respectively; lanes 2 and 4, 10 μg of protein from partially purified bovine and mouse preparations, respectively. B, general protein staining of partially purified bovine (lane 1) and mouse (lane 2) ACSF3 preparations. C, General protein staining of recombinant ACSF3 protein preparations. Lane 1, purified GST-ACSF3; lane 2, GST-ACSF3 after treatment with tobacco etch virus protease; lanes 3–5, final purified preparations of wild-type ACSF3 and mutants R354A and R354L, respectively. D, malonyl-CoA synthetase activity of the ACSF3 preparation. Assays were performed at 37 °C for 30 min, and reaction components were separated by HPLC.
Expression of the Human ACSF3 Protein
Two constructs encoding the protein were expressed in E. coli, one as a GST fusion protein and the other with an N-terminal hexahistidine tag. Only the GST fusion construct generated detectable amounts of soluble protein. The GST fusion protein was purified; the N-terminal GST domain was removed by proteolysis; and the free synthetase was isolated as a homogeneous protein of 65 kDa, close to the value of 64,940 Da predicted for ACSF3 protein containing an additional 8 residues remaining from the engineered tobacco etch virus protease site (Fig. 1C).
Properties of the Human ACSF3 Protein
The purified enzyme converted malonate to malonyl-CoA with high efficiency (kcat = 0.413 ± 0.003 s−1) and exhibited a Km for malonate of 36.8 ± 0.7 μm (Fig. 1D). Specificity studies (Table 1) revealed that methylmalonate was activated at 70% of the rate observed for malonate, and weak activity was observed with acetate (0.04% of the rate observed with malonate). No detectable activity was found with other dicarboxylic acids (oxalate or succinate) or with the fatty acids (butyrate, octanoate, laurate, palmitate, oleate, or lignocerate). Activity was absolutely dependent on the presence of ATP, Mg2+, and CoA. Human mitochondrial holo-ACP could not replace CoA. These data support the identification of the enzyme as a malonyl-CoA synthetase.
TABLE 1.
Substrate specificity of recombinant ACSF3
Reactions were run for 2 h at 37 °C, and products were analyzed by HPLC as described under “Experimental Procedures.” The 100% value for malonate corresponds to 177 nmol min−1 · mg−1. Representative chromatograms are included in supplemental Fig. 2.
| Substrate | Concentration | ACSF3 activity |
|---|---|---|
| μm | % | |
| Malonate | 100 | 100.0 ± 3.0 |
| Methylmalonate | 100 | 70.4 ± 1.2 |
| Dicarboxylic fatty acids (C2 and C4) | 100 | No activity |
| Acetate | 100 | 0.04 ± 0.00 |
| Saturated fatty acids (C4, C8, and C12) | 100 | No activity |
| Saturated fatty acids (C16 and C24) | 20 | No activity |
| Oleic acid | 20 | No activity |
Mutagenesis of the Human Malonyl-CoA Synthetase
Arg-354 is located within the conserved motif ERYGMTE and is predicted to be positioned at the active site. Engineered Ala-354 and Leu-354 mutants were found to be completely devoid of malonyl-CoA synthetase activity, although the Ala-354 mutant retained the same very low activity toward acetate that we observed with the wild-type enzyme. This evidence supports the possibility that Arg-354 facilitates substrate binding in the human malonyl-CoA synthetase by interacting with the 3-carboxylate of malonate.
Subcellular Localization of the Human Malonyl-CoA Synthetase
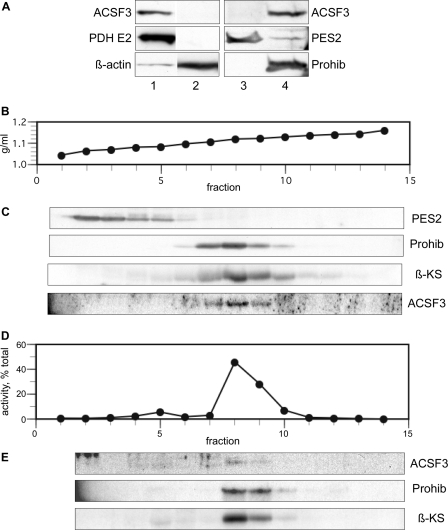
Fractionation of extracts of (human) HEK293T cells by differential centrifugation or on an iodixanol density gradient (monitored by Western blotting with various antibodies and activity assay) revealed that the malonyl-CoA synthetase sedimented together with the mitochondrial markers pyruvate dehydrogenase, mitochondrial β-ketoacyl synthase, and prohibitin (Fig. 2). No malonyl-CoA synthetase was detected in the post-mitochondrial supernatant and the microsomal fraction.
FIGURE 2.
Subcellular localization of ACSF3 in HEK293T cells. A, Western analysis of the mitochondrial fraction from 1.2 × 106 HEK293T cells (lane 1), the post-mitochondrial supernatant from 0.7 × 106 cells (lane 2), the microsomal fraction from 5.8 × 106 cells (lane 3), and the mitochondrial fraction from 5.8 × 106 cells (lane 4). Blots were probed with antibodies against ACSF3, the pyruvate dehydrogenase E2 subunit (PDH E2; a mitochondrial marker), prostaglandin E synthase 2 (PES2; a microsomal marker), β-actin (a cytosolic marker), and prohibitin (Prohib; a mitochondrial marker). B–E, subcellular fractionation of HEK293T cell extracts by centrifugation on an iodixanol density gradient. B, iodixanol density. C, Western analysis of gradient fractions after precipitation with methanol/chloroform. Pellets analyzed for ACSF3 were additionally washed with ice-cold acetone. β-KS, β-ketoacyl synthase (a mitochondrial marker). D, malonyl-CoA synthetase activity. Fractions were diluted and recentrifuged at 100,000 × g for 1 h, the pellet was extracted with 1% Triton X-100, and extracts were assayed as described under “Experimental Procedures.” E, Western analysis of 1% Triton X-100 extracts.
Effect of ACSF3 Knockdown in Cell Culture
HEK293T cells were exposed to siRNAs targeting either ACSF3 or mitochondrial ACP or to vehicle alone and cultured for 48 h. Cell proliferation was decreased by 30–36% in cultures exposed to either ACSF3 or ACP siRNAs for 48 h. As we had found previously (6), cells treated with ACP siRNA exhibited defective lipoylation of mitochondrial proteins. However, cells treated with ACSF3 siRNA exhibited normal protein lipoylation profiles (data not shown). Isolated mitochondrial preparations were analyzed by Western analysis, enzyme activity assays, and de novo fatty acid synthesis assays (Fig. 3). In cells treated with ACSF3 siRNA, the level of ACSF3 protein was reduced to 16% (Fig. 3, A and B), and the level of malonyl-CoA synthetase activity was reduced to 37% (Fig. 3C) compared with cells treated with vehicle alone. In contrast, neither the amount nor the enzyme activity of ACSF3 protein was reduced in cells treated with ACP siRNA (Fig. 3, A and B). The levels of β-ketoacyl synthase and prohibitin were relatively unaffected by exposure to either siRNA (Fig. 3, A and B). These results demonstrated that the ACSF3 protein accounts for most, if not all, of the malonyl-CoA synthetase activity in mammalian mitochondria.
FIGURE 3.
Knockdown of malonyl-CoA synthetase in HEK293T cells by RNA interference. A, Western analysis of mitochondrial extracts from HEK293T cells treated with vehicle only (lane 1), 20 nm ACP siRNA (lane 2), or 75 nm ACSF3 siRNA (lane 3). Loading was normalized on the basis of citrate synthase activity (14.5 units/lane). B, digitalized Western blot data. β-KS, β-ketoacyl synthase. C, malonyl-CoA synthetase activity in mitochondrial extracts. The 100% value for malonyl-CoA synthetase activity corresponds to 2.9 pmol/min, normalized for 1 unit of citrate synthase activity. D, analysis of ACP-linked thioesters formed from [2-14C]malonate metabolism by mitochondrial preparations derived from siRNA-treated HEK293T cells. The metabolites detected included malonyl-ACP and its decarboxylation product, acetyl-ACP, and the two major products of fatty acid synthesis, octanoyl-ACP and hexanoyl-ACP. The 100% value for the [14C]acetyl-ACP plus [14C]malonyl-ACP (14C Acet & Mal-ACP) pool size was 6.4 μm; acetyl-ACP accounted for 35 ± 7% of the total pool. The 100% value for [14C]hexanoyl-ACP was 0.51 μm, and that for [14C]octanoyl-ACP was 0.62 μm, both expressed in terms of malonyl moieties. The data represent means of at least two experiments, with probability values of Student's t test, when <0.1, shown.
To evaluate the effect of ACSF3 knockdown on mitochondrial function, we assessed the ability of mitochondria to utilize [2-14C]malonate for fatty acid synthesis. The presence in these mitochondrial preparations of substantial thioesterase activity against malonyl-CoA precluded estimation of the amount of this metabolite formed (7). However, we were able to reliably assay the amount of the relatively stable ACP-linked products formed from malonate via malonyl-CoA. They included malonyl-ACP, acetyl-ACP, and the acyl-ACPs produced using the malonyl extender moieties. Acetyl-ACP is formed from malonyl-ACP in an uncoupled decarboxylation reaction catalyzed by the mitochondrial β-ketoacyl synthase (4, 7). Formation of acetyl-ACP is blocked by cerulenin (7), an inhibitor of β-ketoacyl synthase. There is no other known pathway for the formation of acetyl-ACP because the mitochondrial malonyltransferase shuttles exclusively malonyl moieties between CoA and ACP thiols (5, 24). Thus, the total amount of radiolabeled malonyl-ACP and acetyl-ACP is representative of the malonyl-ACP moieties derived from [2-14C]malonate that were not utilized for fatty acid synthesis. The combined amount of these radiolabeled malonyl-ACP and acetyl-ACP thioesters formed was reduced by 33% in cells treated with ACSF3 siRNA (Fig. 3D). The major fatty acyl-ACP products were hexanoyl-ACP and octanoyl-ACP, and the amounts formed were reduced by 50 and 17%, respectively, in cells treated with ACSF3 siRNA (Fig. 3D). These results confirmed that ACSF3 is involved in the formation of the malonyl-ACP moieties required as substrates for the mitochondrial de novo fatty acid synthesis pathway.
Expression of ACSF3 in Various Mouse Tissues
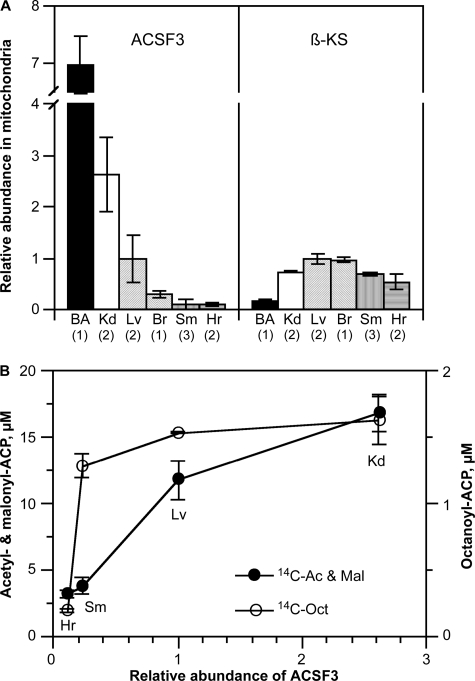
ACSF3 protein was detectable in all of the mouse tissues examined; the highest levels were found in brown adipose tissue, kidney, and liver (Fig. 4A). Mitochondrial β-ketoacyl synthase was also broadly expressed, but the lowest level was found in brown adipose tissue (Fig. 4A). It is unclear why the tissue expression profiles differ for these two enzymes. The molecular mass of the mature mitochondrial ACSF3 protein is 6.2 ± 0.2 kDa shorter than that anticipated for full-length mouse ACSF3, presumably because of cleavage of the mitochondrial targeting sequence. A similar difference between full-length and mature mitochondrial ACSF3 proteins was found in human HEK293T cells. When the ability of mitochondria from these different tissues to utilize [2-14C]malonate was compared, the formation of malonyl-ACP and acetyl-ACP was found to be dependent on the tissue level of ACSF3, again consistent with the involvement of this enzyme in the delivery of malonyl moieties for fatty acid synthesis (Fig. 4B). However, the formation of the octanoyl-ACP product did not correlate with tissue ACSF3 content, indicating that other component(s) of the pathway likely determine the overall rate of product formation. Indeed, the tissue content of β-ketoacyl synthase correlated well with the ability to synthesize octanoyl-ACP but, as expected, did not correlate with the ability to synthesize malonyl-ACP and acetyl-ACP from malonate (data not shown).
FIGURE 4.
Relative abundance of ACSF3 and β-ketoacyl synthase in mitochondria isolated from mouse tissues. A, proteins were detected by Western blotting, and the data were digitized. Numbers in parentheses indicate the number of different mitochondrial preparations analyzed. BA, brown adipose tissue; Kd, kidney; Lv, liver; Br, brain; Sm, skeletal muscle; Hr, heart. B, relationship between products formed from malonate in mitochondrial preparations from different tissues and the relative abundance of ACSF3. Mitochondria used in these experiments were purified on a discontinuous iodixanol gradient as described under “Experimental Procedures.” 14C Ac & Mal, [14C]acetyl-ACP plus [14C]malonyl-ACP; 14C-Oct, [14C]octanoyl-ACP.
DISCUSSION
More than 30 years ago, rat brain mitochondria were found to have a “malonate-activating” activity, but work in several laboratories concluded that the conversion of malonate to malonyl-CoA proceeded indirectly by transfer of the CoA moiety from acetoacetyl-CoA and succinyl-CoA (25, 26). Subsequently, bona fide malonyl-CoA synthetases have been identified in several organisms (Pseudomonas fluorescens (27), B. japonicum (28), R. trifolii (29), and pea leaves (30)) but not in animals. This study has provided compelling evidence indicating that malonyl-CoA synthetases are also found in the mitochondria of animals and are encoded by the nuclear ACSF3 gene. (i) The mammalian malonyl-CoA synthetases exhibit substantial sequence similarity to their prokaryotic and plant counterparts and, in particular, contain an arginine residue essential for activity within a sequence motif (ERYGMTE) that is conserved uniquely in known prokaryotic malonyl synthetases. (ii) Sequence analysis and subcellular fractionation studies indicate that animal ACSF3 enzymes possess N-terminal extensions that target the proteins to mitochondria. (iii) Knockdown of human ACSF3 by RNA interference eliminates most of the malonyl-CoA synthetase activity in HEK293T cell mitochondria and reduces the amount of malonyl-ACP formed from free malonate.
The earlier identification of ACSF3 as a very long chain acyl-CoA synthetase (18) was based on indirect evidence. A full-length cDNA encoding human ACSF3 was cloned into a mammalian expression vector and expressed in COS-1 cells. When the cells were subsequently lysed and the whole extract was assayed for acyl-CoA synthetase activity, the specific activities were increased by 77% with C24:0, 22% with C8:0, and 7.5% with C16:0 compared with extracts from control cells. Because the expressed ACSF3 was not isolated for assay, it is likely that the observed changes in activity resulted from an indirect effect of protein expression. On the basis of the new evidence presented in this study, we suggest that the notation for this enzyme can now be refined to ACSMal to reflect its high specificity for malonate.
In an earlier study, we found that siRNA-mediated knockdown of mitochondrial ACP in HEK293T cells dramatically compromised the protein lipoylation pathway by limiting the availability of octanoyl moieties used as the precursor for lipoyl formation (6). However, in this study, knockdown of ACSF3 did not alter the lipoylation status of any of the mitochondrial proteins. We suspect that the absence of an effect on protein lipoylation indicates that the level of ACSF3 enzyme present in HEK293T cell mitochondria is far from rate-limiting. Indeed, experiments with mitochondria from skeletal muscle indicated that malonate was as good a substrate for octanoyl-ACP formation as malonyl-CoA (106.1 ± 1.1 and 105.3 ± 2.7 pmol of malonyl unit h−1·mg−1 of mitochondrial protein, respectively). Thus, even when ACSF3 activity was lowered to 37% of normal levels by siRNA treatment, sufficient activity remained to provide adequate amounts of octanoyl-ACP required to service the protein lipoylation pathway. We have made similar observations using siRNAs directed against other components of the mitochondrial fatty acid biosynthetic pathway in HEK293T cells. Knockdown efficiencies of ∼75% for malonyltransferase and enoyl reductase were insufficient to compromise the lipoylation status of any of the mitochondrial proteins.3
The metabolism of malonate, the ACSF3 substrate, has not been studied extensively in animals. However, malonate is known to be present in substantial concentrations, and levels as high as 200 nmol/g have been measured in normal brain (31). Higher levels (up to 50 mm achieved by direct injection) are toxic in vitro due to inhibition of succinate dehydrogenase and depletion of ATP (32, 33). Thus, ACSF3, with a Km for malonate of only 37 μm, is capable of operating at Vmax within the likely physiological concentration of this metabolite, far below levels that would inhibit succinate dehydrogenase.
The origin of mitochondrial malonate is uncertain, but it appears likely that it can be recruited from both exogenous and endogenous sources. Free malonate is found in plants, especially legumes, and standard rodent chow contains abundant malonate. Malonate is readily taken up by mitochondria via a transporter that mediates the exchange of phosphate and dicarboxylates (malate, succinate, oxalacetate, malonate) against phosphate and other dicarboxylates (34, 35). It has also been suggested that, in animals, malonate may be produced from oxalacetate in mitochondria, possibly driven by H2O2 generated from the respiratory chain (36) by a reaction first described in plants (37).
Since our study was completed, the crystal structure (Protein Data Bank code 3NYR) was published for a previously uncharacterized malonyl-CoA synthetase from Streptomyces coelicolor (38). This enzyme is active toward malonate, methylmalonate, ethylmalonate, methoxymalonate, and hydroxymalonate and exhibits 37% sequence identity to the human ACSF3 enzyme. The crystal structure of the enzyme with malonyl-CoA bound at the active center revealed that the arginine residue within the conserved motif ERYGMTE forms a salt bridge with the 3-carboxylate of the malonyl moiety, validating our conclusion, based on sequence analysis and mutagenesis, that, in the human ACSF3 enzyme, Arg-354 plays a critical role in substrate recognition. A serine residue also implicated in malonate binding by the S. coelicolor enzyme is also positionally conserved in mammalian ACSF3 enzymes but not in other acyl-CoA synthetases (supplemental Fig. 1).
In conclusion, the entire suite of enzymes required for the de novo biosynthesis of fatty acids has now been identified in the mitochondria of plants, fungi, and animals. Although the mitochondrial enzymes involved in the pathway share many similarities in eukaryotes, different enzymes are employed to generate the malonyl moieties required as chain extenders. Fungal mitochondria appear to be the only type possessing an acetyl-CoA carboxylase capable of performing this task (39). Plants and animals are able to use a malonyl-CoA synthetase as a surrogate enzyme for production of malonyl-CoA (Ref. 30 and this study).
Supplementary Material
Acknowledgment
We thank Ayesha Moghul for skillful technical assistance.
Note Added in Proof
An advance online publication has reported that a plant homolog of the mammalian ACSF3s is a malonyl-CoA synthetase that is essential for growth and development of Arabidopsis (Chen, H., Kim, H. U., and Browse, J. (2011) The Plant Cell, doi:10.1105/tpc.111.086140). The authors also cloned and expressed the human ACSF3 protein and found it to have malonyl-CoA synthetase activity.
This work was supported in part by National Institutes of Health Grant GM069717.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2 and an additional reference.
D. Feng and S. Smith, unpublished data.
- ACP
- acyl carrier protein.
REFERENCES
- 1. Autio K. J., Kastaniotis A. J., Pospiech H., Miinalainen I. J., Schonauer M. S., Dieckmann C. L., Hiltunen J. K. (2008) FASEB J. 22, 569–578 [DOI] [PubMed] [Google Scholar]
- 2. Chen Z., Kastaniotis A. J., Miinalainen I. J., Rajaram V., Wierenga R. K., Hiltunen J. K. (2009) FASEB J. 23, 3682–3691 [DOI] [PubMed] [Google Scholar]
- 3. Miinalainen I. J., Chen Z. J., Torkko J. M., Pirilä P. L., Sormunen R. T., Bergmann U., Qin Y. M., Hiltunen J. K. (2003) J. Biol. Chem. 278, 20154–20161 [DOI] [PubMed] [Google Scholar]
- 4. Zhang L., Joshi A. K., Hofmann J., Schweizer E., Smith S. (2005) J. Biol. Chem. 280, 12422–12429 [DOI] [PubMed] [Google Scholar]
- 5. Zhang L., Joshi A. K., Smith S. (2003) J. Biol. Chem. 278, 40067–40074 [DOI] [PubMed] [Google Scholar]
- 6. Feng D., Witkowski A., Smith S. (2009) J. Biol. Chem. 284, 11436–11445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Witkowski A., Joshi A. K., Smith S. (2007) J. Biol. Chem. 282, 14178–14185 [DOI] [PubMed] [Google Scholar]
- 8. Hiltunen J. K., Chen Z., Haapalainen A. M., Wierenga R. K., Kastaniotis A. J. (2010) Prog. Lipid Res. 49, 27–45 [DOI] [PubMed] [Google Scholar]
- 9. López-Casillas F., Bai D. H., Luo X. C., Kong I. S., Hermodson M. A., Kim K. H. (1988) Proc. Natl. Acad. Sci. U.S.A. 85, 5784–5788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Widmer J., Fassihi K. S., Schlichter S. C., Wheeler K. S., Crute B. E., King N., Nutile-McMenemy N., Noll W. W., Daniel S., Ha J., Kim K. H., Witters L. A. (1996) Biochem. J. 316, 915–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Abu-Elheiga L., Jayakumar A., Baldini A., Chirala S. S., Wakil S. J. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 4011–4015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Suzuki H., Yamada J., Watanabe T., Suga T. (1989) Biochim. Biophys. Acta 990, 25–30 [DOI] [PubMed] [Google Scholar]
- 13. Jang S. H., Cheesbrough T. M., Kolattukudy P. E. (1989) J. Biol. Chem. 264, 3500–3505 [PubMed] [Google Scholar]
- 14. Jung J. W., An J. H., Na K. B., Kim Y. S., Lee W. (2000) Protein Sci. 9, 1294–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Singh I., Kishimoto Y. (1983) J. Lipid Res. 24, 662–665 [PubMed] [Google Scholar]
- 16. Wessel D., Flügge U. I. (1984) Anal. Biochem. 138, 141–143 [DOI] [PubMed] [Google Scholar]
- 17. Srere P. A. (1969) Methods Enzymol. 13, 3–11 [DOI] [PubMed] [Google Scholar]
- 18. Watkins P. A., Maiguel D., Jia Z., Pevsner J. (2007) J. Lipid Res. 48, 2736–2750 [DOI] [PubMed] [Google Scholar]
- 19. Kochan G., Pilka E. S., von Delft F., Oppermann U., Yue W. W. (2009) J. Mol. Biol. 388, 997–1008 [DOI] [PubMed] [Google Scholar]
- 20. Bannai H., Tamada Y., Maruyama O., Nakai K., Miyano S. (2002) Bioinformatics 18, 298–305 [DOI] [PubMed] [Google Scholar]
- 21. Claros M. G., Vincens P. (1996) Eur. J. Biochem. 241, 779–786 [DOI] [PubMed] [Google Scholar]
- 22. Emanuelsson O., Nielsen H., Brunak S., von Heijne G. (2000) J. Mol. Biol. 300, 1005–1016 [DOI] [PubMed] [Google Scholar]
- 23. Nielsen H., Engelbrecht J., Brunak S., von Heijne G. (1997) Protein Eng. 10, 1–6 [DOI] [PubMed] [Google Scholar]
- 24. Bunkoczi G., Misquitta S., Wu X., Lee W. H., Rojkova A., Kochan G., Kavanagh K. L., Oppermann U., Smith S. (2009) Chem. Biol. 16, 667–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Menon G. K., Stern J. R. (1960) J. Biol. Chem. 235, 3393–3398 [PubMed] [Google Scholar]
- 26. Koeppen A. H., Mitzen E. J., Ammoumi A. A. (1974) Biochemistry 13, 3589–3595 [DOI] [PubMed] [Google Scholar]
- 27. Kim Y. S., Bang S. K. (1985) J. Biol. Chem. 260, 5098–5104 [PubMed] [Google Scholar]
- 28. Kim Y. S., Chae H. Z. (1991) Biochem. J. 273, 511–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. An J. H., Kim Y. S. (1998) Eur. J. Biochem. 257, 395–402 [DOI] [PubMed] [Google Scholar]
- 30. Gueguen V., Macherel D., Jaquinod M., Douce R., Bourguignon J. (2000) J. Biol. Chem. 275, 5016–5025 [DOI] [PubMed] [Google Scholar]
- 31. Mitzen E. J., Koeppen A. H. (1984) J. Neurochem. 43, 499–506 [DOI] [PubMed] [Google Scholar]
- 32. Stokes A. H., Bernard L. P., Nicklas W. J., Zeevalk G. D. (2001) J. Neurosci. Res. 64, 43–52 [DOI] [PubMed] [Google Scholar]
- 33. Fernandez-Gomez F. J., Galindo M. F., Gómez-Lázaro M., Yuste V. J., Comella J. X., Aguirre N., Jordán J. (2005) Br. J. Pharmacol. 144, 528–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Saint-Macary M., Foucher B. (1980) Biochem. Biophys. Res. Commun. 96, 457–462 [DOI] [PubMed] [Google Scholar]
- 35. Schoolwerth A. C., LaNoue K. F. (1985) Annu. Rev. Physiol. 47, 143–171 [DOI] [PubMed] [Google Scholar]
- 36. Wojtovich A. P., Brookes P. S. (2008) Biochim. Biophys. Acta 1777, 882–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pattee H. E., Shannon L. M. (1965) Bot. Gaz. 126, 179–181 [Google Scholar]
- 38. Hughes A. J., Keatinge-Clay A. (2011) Chem. Biol. 18, 165–176 [DOI] [PubMed] [Google Scholar]
- 39. Hoja U., Marthol S., Hofmann J., Stegner S., Schulz R., Meier S., Greiner E., Schweizer E. (2004) J. Biol. Chem. 279, 21779–21786 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.