Background: PKD inhibits actin-driven directed cell migration.
Results: PKD regulates cofilin activity through LIMK and PAK4.
Conclusion: PKD increases the net amount of inactive cofilin in cells.
Significance: The regulation of cofilin activity at multiple levels explains the inhibitory effects of PKD on directed cell migration.
Keywords: Actin, Cell Migration, Cell Motility, Serine/Threonine Protein Kinase, Signal Transduction, Cofilin, LIMK, PAK4, PKD, Slingshot
Abstract
Dynamic reorganization of the actin cytoskeleton at the leading edge is required for directed cell migration. Cofilin, a small actin-binding protein with F-actin severing activities, is a key enzyme initiating such actin remodeling processes. Cofilin activity is tightly regulated by phosphorylation and dephosphorylation events that are mediated by LIM kinase (LIMK) and the phosphatase slingshot (SSH), respectively. Protein kinase D (PKD) is a serine/threonine kinase that inhibits actin-driven directed cell migration by phosphorylation and inactivation of SSH. Here, we show that PKD can also regulate LIMK through direct phosphorylation and activation of its upstream kinase p21-activated kinase 4 (PAK4). Therefore, active PKD increases the net amount of phosphorylated inactive cofilin in cells through both pathways. The regulation of cofilin activity at multiple levels may explain the inhibitory effects of PKD on barbed end formation as well as on directed cell migration.
Introduction
Directed migration of cells requires rapid actin reorganization to mediate membrane protrusion and lamellipodium extension (1, 2). Actin turnover is concerted by a multitude of proteins including cofilin, a small actin-binding protein with actin severing activities (3, 4). Cofilin severs existing F-actin filaments at the leading edge of the lamellipodium and generates new barbed ends, which allow filament elongation and dendritic branching through the Arp2/3 complex. The net effect of signaling cascades regulating cofilin activity determines cell migration (5, 6). Consequently, cofilin activity is tightly regulated at several levels involving its binding to phosphatidylinositol 4,5-phosphate or cortactin as well as covalent modification through phosphorylation (for review, see Refs. 7, 8).
Phosphorylation at serine residue 3 (Ser-3) abolishes cofilin binding to actin and therefore its actin severing activities (9), which subsequently results in decreased cell motility (7). Phosphorylation of cofilin at Ser-3 is mediated by LIM kinases (LIMKs2; Lin-11/Isl-1/Mec-3 kinases), of which two enzymes (LIMK1, LIMK2) are ubiquitously expressed (10, 11). LIMKs are activated by phosphorylation of Thr-508/Thr-505 (LIMK1/2) within the activation loop of their kinase domain. This can be mediated by the group II p21-activated kinase 4 (PAK4) (12). Other cofilin kinases, testicular protein kinases, have been identified, but their activation occurs independently of PAK proteins (13). The dephosphorylation of cofilin at Ser-3 reactivates its actin severing activity. This is mediated through phosphatases of the slingshot (SSH) family (14, 15), which also can be regulated in a PAK4-dependent manner (12, 16). Moreover, the phosphatase chronophin (pyridoxyl-5-phosphate phosphatase) has also been shown to dephosphorylate cofilin (17).
Like PAK family kinases, which act as downstream effectors of Rac and Cdc42 (18), members of the protein kinase D (PKD) family of serine/threonine kinases are also activated by small RhoGTPases (19). The cellular mechanisms of how PKDs impact cell motility are currently not well defined. When expressed as constitutively active alleles, PKD1 and PKD2 inhibit directed cell migration in many tumor cell lines. PKD1 interacts with actin (20) and is regulated by RhoA and kinases that are involved in regulating directed cell migration and adhesion (21). PKD1 targets, whose phosphorylation may promote its inhibitory effects on the motile phenotype, include EVL-1 (22), RIN1 (23), E-cadherin (24), β-catenin (25), Hsp27 (26), and cortactin (27). However, in many cases the functional consequences of their phosphorylation by PKD1 are still ill defined. One mechanism of how PKD1 modulates cell motility is that it inhibits the formation of F-actin free barbed ends which has been linked to phosphorylation of SSH1L, resulting in a decrease in the pool of active cofilin (28). Such inhibition of SSH1L is mediated by the phosphorylation-dependent recruitment of 14-3-3 proteins (14, 29). However, the inhibition of SSH alone does not explain the dramatic inhibitory effects of active PKD on barbed end formation and actin incorporation observed in previous work (29).
Here, we describe PAK4, a kinase that has been shown to activate LIMK and increase cofilin phosphorylation levels, as a substrate for all three PKD isoforms. We show that PKD phosphorylates PAK4 in its activation loop in vitro and in vivo. This translates to increased activity of PAK4 and its downstream target LIMK as well as to increased levels of phospho-cofilin. We show that the regulation of this pathway by PKD, in addition to inhibiting SSH, decreases the net amount of unphosphorylated and active cofilin. PKD-mediated inhibition of cofilin through both of its regulatory pathways may explain its dramatic effects on actin incorporation at the leading edge and on directed cell motility.
EXPERIMENTAL PROCEDURES
Cell Lines, Antibodies, and Reagents
NMuMG cells (obtained from American Type Culture Collection (ATCC)) were maintained in DMEM with 10 μg/ml insulin and 10% FBS. HeLa cells (obtained from ATCC) were maintained in DMEM with 10% FBS. HuMEC cells (obtained from Invitrogen) were maintained in HMEC Culture System from Invitrogen. Anti-GST and anti-14-3-3 antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-HA, anti-FLAG (M2), and anti-actin were from Sigma-Aldrich. Anti-PKD1 was from Abgent (San Diego, CA). Anti-PKD2 antibody was from Millipore (Billerica, MA). Anti-PKD3 and anti-SSH1L were from Bethyl Laboratories (Montgomery, TX). Anti-pMOTIF (PKD substrate antibody), anti-LIMK, anti-pT508-LIMK, anti-PAK4, anti-pS474-PAK4, anti-cofilin, and anti-pS3-cofilin were from Cell Signaling Technology (Danvers, MA). The anti-pS978-SSH1L antibody was described previously (14). Secondary HRP-linked antibodies were from Roche Applied Science. Secondary antibodies (Alexa Fluor 568 F(ab′)2 fragment of goat-anti-mouse IgG or Alexa Fluor 546 F(ab′)2 fragment of goat anti-rabbit) were from Invitrogen. Mirus HeLa-Monster (Madison, WI) was used for transient transfection of HeLa, GenJet Reagent II (SignaGen Laboratories, Ijamsville, MD) for NMuMG, and Lipofectamine 2000 (Invitrogen) for HuMEC. CID755673 was from TOCRIS Bioscience (Ellisville, MO).
DNA Constructs
Expression plasmids for constitutively active FLAG-tagged PKD2 (PKD2.CA, PKD2.S706E/S710E mutation) and GST-tagged PKD3 (PKD3.CA, PKD3.S731E/S735E mutation) were generated by site-directed mutagenesis using previously described plasmids as templates (30, 31). The expression plasmid for HA-tagged constitutively active PKD1 (PKD1.CA, PKD1.S738E/S742E mutation) was described before (32). The expression plasmid for FLAG-tagged cofilin in pcDNA4/TO was described previously (28). FLAG-tagged human PAK4 was amplified from a HeLa cDNA library using 5′-GGCGGATCCATGGACTATAAGGACGATGATGACAAATTTGGGAAGAGGAAGAAGCGGGTGGAG-3′ and 5′-GGCCTCGAGTCATCTGGTGCGGTTCTGGCGCAT-3′ as primers and cloned into pcDNA4/TO via BamHI and XhoI. FLAG-tagged human LIMK1 (used in supplemental Fig. 1) was amplified from a HeLa cDNA library using 5′-CGCAAGCTTATGGACTATAAGGACGATGATGACAAAAGGTGGACGCTACTTTGTTGCACC-3′ and 5′-CGCGATATCTCAGTCGGGGACCTCAGGGTGGGC-3′ as primers and cloned into pcDNA4/TO via HindIII and EcoRV. GST-PAK4 was generated by cloning full-length human PAK4 into pGEX4-T1 via BamHI and XhoI. Site-directed mutagenesis was carried out using the QuikChange kit (Stratagene, La Jolla, CA) and 5′-GGCAAGCTGGTGGCCGTCATGAAGATGGACCTGCGCAAG-3′ and 5′-CTTGCGCAGGTCCATCTTCATGACGGCCACCAGCTTGCC-3′ as primers to generate a kinase-dead PAK4 (PAK4.K350M).
In Vitro Kinase Assays
Kinase assays with GST fusion proteins were carried out by adding 250 ng of active, purified PKD1 (Millipore), PKD2, or PKD3 (both from Enzo Life Sciences, Plymouth Meeting, PA) to 2 μg of purified GST or GST fusion protein in a volume of 40 μl of kinase buffer (50 mm Tris, pH 7.4, 10 mm MgCl2, and 2 mm DTT) supplemented with 100 μm ATP. The kinase reaction (30 min, room temperature) was stopped by adding 2× Laemmli buffer.
Immunoblotting, Immunoprecipitation, and PAGE
Cells were washed twice with ice-cold PBS (140 mm NaCl, 2.7 mm KCl, 8 mm Na2HPO4, 1.5 mm KH2PO4, pH 7.2) and lysed with lysis buffer A (50 mm Tris-HCl, pH 7.4, 1% Triton X-100, 150 mm NaCl, 5 mm EDTA, pH 7.4) plus Protease Inhibitor Mixture (PIC; Sigma-Aldrich). Lysates were vortexed and incubated on ice for 30 min. Following centrifugation (13,000 rpm, 15 min, 4 °C) the protein concentration of lysates was determined. Proteins of interest were immunoprecipitated by a 1-h incubation with a specific antibody (2 μg) followed by a 30-min incubation with protein G-Sepharose (Amersham Biosciences). Immune-complexes were washed three times with TBS (50 mm Tris-HCl, pH 7.4, 150 mm NaCl) and resolved in 20 μl of TBS and 2× Laemmli buffer (33). Samples were subjected to SDS-PAGE. Following PAGE, proteins were transferred to nitrocellulose membranes and visualized by immunostaining.
Analysis of Free Actin Filament Barbed Ends
Cells were transfected and reseeded on 8-well ibiTreat μ-Slides (Ibidi; Integrated BioDiagnostics, Martinsried, Germany). Cells were serum-starved (16 h). The medium was removed, cells were stimulated with EGF (50 ng/ml, 1 min), washed with prewarmed PBS, permeabilized, and labeled with 0.4 μm actin from rabbit muscle conjugated to Alexa Fluor 568 in permeabilization buffer (20 mm HEPES, 138 mm KCl, 4 mm MgCl2, 3 mm EGTA, 0.2 mg/ml saponin, 1% BSA) plus 1 mm ATP for 15 s at 37 °C. The cells were fixed with 4% paraformaldehyde in modified Krebs buffer (145 mm NaCl, 5 mm KCl, 1.2 mm CaCl2, 1.3 mm MgCl2, 1.2 mm NaH2PO2, 10 mm glucose, 20 mm HEPES, pH 7.4) at room temperature for 10 min. Samples were examined using a IX81 DSU Spinning Disc Confocal microscope from Olympus.
Immunofluorescence
Cells were transfected as indicated in 8-well ibiTreat μ-Slides (Ibidi). The next day cells were washed twice with PBS. Following fixation with 4% paraformaldehyde (15 min, 37 °C), cells were washed three times in PBS and then permeabilized with 0.1% Triton X-100 in PBS for 2 min at room temperature. Samples were blocked with 3% BSA and 0.05% Tween 20 in PBS (blocking solution) for 30 min at room temperature and then incubated for 2 h at room temperature with primary antibodies (anti-FLAG 1:5000 for PAK4, or with an anti-phospho-S474-PAK4 antibody 1:2000) diluted in blocking solution. Cells were washed five times with PBS and incubated with secondary antibodies (Alexa Fluor 568 F(ab′)2 fragment of goat anti-mouse IgG or Alexa Fluor 546 F(ab′)2 fragment of goat anti-rabbit; both Invitrogen), diluted (1:2000) in blocking solution for 2 h at room temperature. F-actin was stained together with secondary antibodies by incubating with phalloidin (Alexa Fluor 633-phalloidin, shown as cyan pseudo color staining) in blocking solution. After extensive washes in PBS, cells were mounted in Ibidi mounting medium (Ibidi). Samples were examined using an IX81 DSU Spinning Disc Confocal microscope from Olympus with a 40x objective.
Impedance-based Real-time Cell Migration Analysis
Cells were transfected as indicated and after 24 h were seeded on Transwell CIM-plate 16 (Roche Applied Bioscience). After 2 h of attachment, cell migration toward NIH-3T3 conditioned medium was continuously monitored in real-time using the xCELLigence RTCA DP instrument (Roche Applied Science). Error bars (gray) in Fig. 1C represent four experiments.
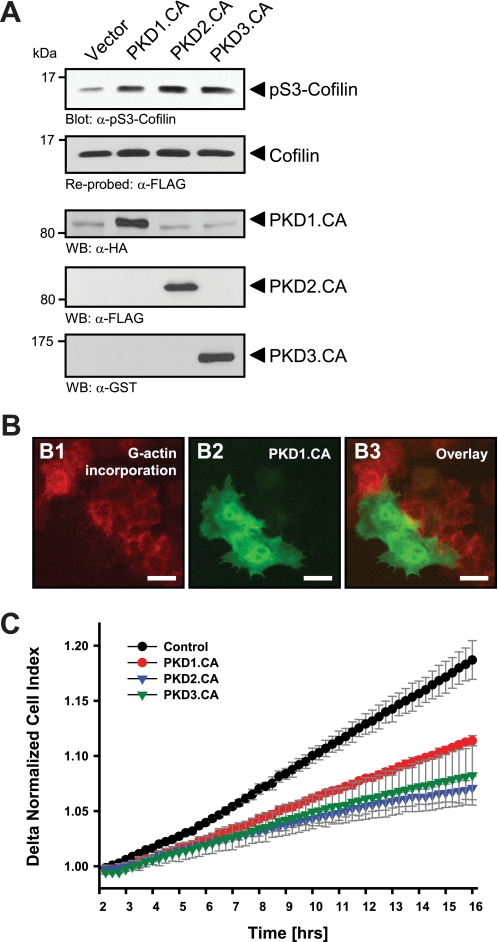
FIGURE 1.
Active PKD isoforms effectively inhibit cofilin activity and directed cell migration. A, HeLa cells (0.65 × 106 cells, 6-cm dish) were co-transfected with cofilin and control vector, constitutively active PKD1, PKD2, or PKD3 as indicated. Lysates were analyzed for cofilin phosphorylation at Ser-3 or total cofilin using α-pS3-coflin or α-FLAG antibodies, respectively. Expression of active PKD alleles was controlled using antibodies directed against their tags (α-HA for PKD1.CA, α-FLAG for PKD2.CA, and α-GST for PKD3.CA). B, free barbed end-induced actin incorporation at the leading edge is blocked by active PKD. HeLa cells (5 × 104 cells, 8-well ibiTreat μ-slide) were transfected with control vector or constitutively active GFP-tagged PKD1 (PKD1.CA). To analyze cofilin-mediated actin incorporation F-actin free barbed ends were performed as described under “Experimental Procedures.” Scale bars are 20 μm. C, all active forms of PKD reduce directed cell migration. HeLa cells (5 × 105 cells, 6-cm dish) were transfected with control vector, constitutively active PKD1 (PKD1.CA), PKD2 (PKD2.CA), or PKD3 (PKD3.CA). After 2 h of attachment, cell migration toward NIH-3T3 conditioned medium over 14 h was monitored continuously in real-time using a Transwell CIM-plate 16 and the xCELLigence RTCA DP instrument. Error bars (gray) represent four experiments.
RESULTS
Active PKD Isoforms Inhibit Cofilin Activity and Directed Cell Migration
The generation of F-actin free barbed ends through ADF/cofilin correlates with the ability of cells to migrate (3). The ectopic expression of active alleles of all three PKD isoforms, PKD1, PKD2, and PKD3, significantly increased phosphorylation of cofilin at Ser-3, suggesting its inactivation through PKD-regulated signaling mechanisms (Fig. 1A). Increased phospho-cofilin levels correlated with a complete block of F-actin free barbed end formation and actin incorporation in cells where active PKD was expressed (Fig. 1B), suggesting that PKD effectively inhibits cofilin activity.
Next, we tested whether this translates to an inhibition of cofilin-mediated cell migration. Previously we have shown that in HeLa and MTLn3 cells the expression of constitutively active PKD1 significantly decreased cell migration in Boyden chamber/Transwell chemotaxis assays (28). Utilizing impedance-based Transwell assays (xCELLigence RTCA DP system, Transwell CIM-plates 16 from Roche Applied Science), we performed time lapse studies of cell migration toward a chemotactic stimulus over a time period of 14 h and found that active alleles of all three PKD isoforms decreased directed cell migration (Fig. 1C).
Active PKD Isoforms Phosphorylate and Inactivate SSH1L
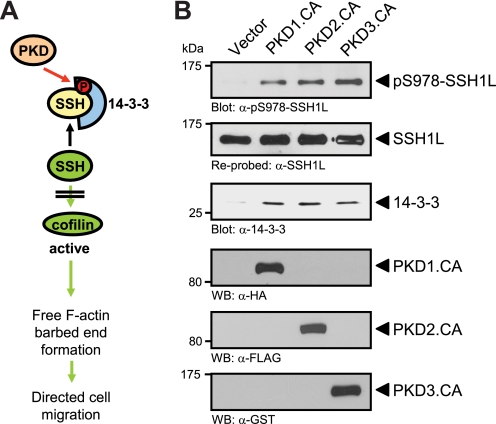
The inactivating phosphorylation of cofilin can be maintained through activation of the PAK/LIMK pathway or through inhibition of SSH phosphatases (12, 15, 16). PKD effects on cofilin-mediated F-actin barbed end formation are mediated in part through regulation of SSH1L (28). We and others have shown that PKD1 and PKD2 directly phosphorylate SSH1L at its amino acid residue Ser-978 and that this generates a binding site for 14-3-3 proteins (Fig. 2A) (28, 34). Further, the binding of 14-3-3 proteins mediates inactivation of SSH1L and loss of interaction with F-actin (14). Here, we show that active PKD3 also mediates SSH1L phosphorylation at Ser-978 and binding to 14-3-3, further confirming the involvement of all three PKD isoforms in negatively regulating this phosphatase (Fig. 2B).
FIGURE 2.
Active PKD isoforms phosphorylate and inactivate SSH1L. A, schematic shows how PKD regulates SSH1L and cofilin activity (from Ref. 28). B, HeLa cells (0.5 × 106, 6-cm dish) were transfected with tagged, constitutively active alleles of PKD1, PKD2, or PKD3, as indicated. SSH1L was immunoprecipitated and analyzed for PKD-mediated phosphorylations at Ser-978 using a phospho-specific antibody. Samples were then stripped and reprobed for total SSH1L. Samples were also analyzed for co-immunoprecipitated 14-3-3 protein using a pan-14-3-3 antibody. For input control, Western blot analysis was performed on lysates to determine expression of active PKD1 (α-HA staining), PKD2 (α-FLAG staining), and PKD3 (α-GST staining).
The net effect of this signaling event is an increase in phospho-cofilin levels, due to decreased dephosphorylation by SSH1L. However, the dramatic effects of active PKD on cofilin-mediated F-actin severing could not be fully rescued with a SSH1L mutant that lacks PKD phosphorylation sites (28), and we hypothesized that PKD may also impact cofilin activity through other pathways. Therefore, we analyzed whether active PKDs also regulate the PAK4/LIMK pathway to exert its dramatic effects on inhibiting actin incorporation at the leading edge.
Active PKD and Active PAK4 Both Localize at the Leading Edge
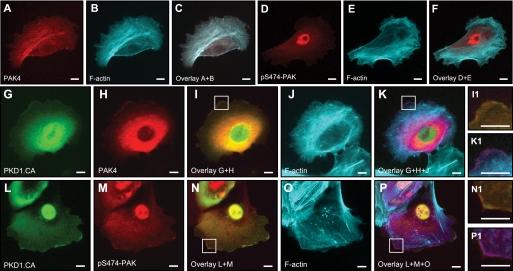
The group II p21-activated kinase PAK4 has been described to regulate cofilin activity negatively through LIMK (7, 16). PAK4 is co-localized with F-actin in HuMEC cells (Fig. 3, A–C), but shows only little basal activity at these F-actin structures, as measured by analysis of its phosphorylation status at Ser-474 (Fig. 3, D and E). When co-expressed with active PKD1 (PKD1.CA), PAK4 localizes to F-actin filaments at the leading edge and the cytosol (Fig. 3, G–K). Ectopically expressed active PKD1 also co-localized with PAK4 and F-actin at the leading edge (Fig. 3, I1 and K1). Similar co-localization with PKD1.CA and actin was observed when active PAK4 was determined with an anti-pS474-PAK4 antibody (Fig. 3, L–P). The co-localization of active PAK4 and active PKD1 suggested that PKD may contribute to PAK4 activity at the leading edge of migrating cells. Therefore, we tested whether PAK4 can be a PKD substrate.
FIGURE 3.
Active PKD and active PAK4 localize to the leading edge. HuMEC cells (1 × 104 cells, 8-well ibiTreat μ-slide) were co-transfected with FLAG-tagged PAK4 and either empty vector or GFP-tagged constitutively active PKD1 (PKD1.CA) as indicated, and samples were subjected to immunofluorescence analysis. Samples were indirectly labeled with α-FLAG for PAK4 (A and H) or with α-Ser(P)-474-PAK4/5/6 antibody (D and M) and secondary Alexa Fluor 568 antibody. F-actin was stained with phalloidin (Alexa Fluor 633, shown as cyan pseudo color in B, E, J, and O). C and F, overlay of actin with PAK4 or pS474-PAK4/5/6 staining. I and N, overlay of GFP-PKD1.CA with PAK4 or pS474-PAK staining. Insets are magnified ×3.2 in I1 and N1. K and P, overlays of G+H+J and L+M+O. Insets are magnified ×3.2 in K1 and P1. Scale bars indicate 10 μm.
PKD Phosphorylates PAK4 in Its Activation Loop in Vitro
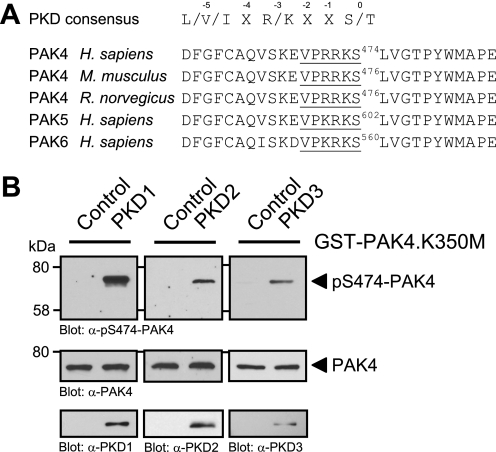
The minimal consensus motif (Fig. 4A) for PKD substrates requires a leucine, isoleucine, or valine at the −5, and an arginine or lysine at the −3 amino acid positions relative to the targeted serine or threonine residue at position 0 (35). On analysis of the PAK4 amino acid sequence for potential PKD1 sites we found that all members of the group II PAK family, PAK4–PAK6, contain a PKD phosphorylation motif in their kinase domains which is conserved in PAK4 through species (Fig. 4A). Moreover, the serine potentially targeted by PKD (i.e. Ser-474 in human PAK4) represents a critical residue in the activation loop of the kinase. It was shown previously that phosphorylation of this residue is required for PAK4 activity (16).
FIGURE 4.
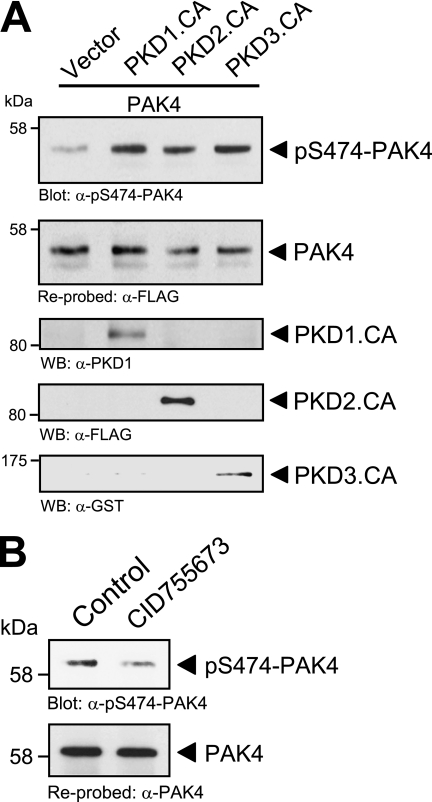
PKD phosphorylates PAK4 in its activation loop in vitro. A, analysis of group II p21-activated kinases (PAK4–PAK6) for PKD phosphorylation sites showed that a serine residue, important for PAK activation, lies within an ideal PKD consensus motif. B, purified GST fusion protein of full-length kinase-dead PAK4 (GST-PAK4.K350M) was subjected to in vitro kinase assays with purified PKD1, PKD2, and PKD3. To analyze substrate phosphorylation, Western blots of resolved proteins were probed with α-pS474-PAK4. Control blots show equal substrate loading (α-PAK4) and input of PKD1 (α-PKD1), PKD2 (α-PKD2), or PKD3 (α-PKD3).
Therefore, we determined whether PKD can phosphorylate PAK4 at this activation loop serine. To test this we performed in vitro kinase assays with purified PKD isoforms PKD1, PKD2, and PKD3 and a bacterially expressed GST fusion protein of kinase-dead PAK4 (PAK4.K350M mutant) to exclude autophosphorylation events (Fig. 4B). Phosphorylation of PAK4 was determined using a phosphospecific pS474-PAK4 antibody. We found that all three PKD isoforms, PKD1, PKD2, and PKD3, phosphorylate PAK4 at Ser-474 in vitro. This suggests that PKD contributes to PAK4 activity by direct phosphorylation of its activation loop.
PKD Regulates PAK4 Activity in Cells
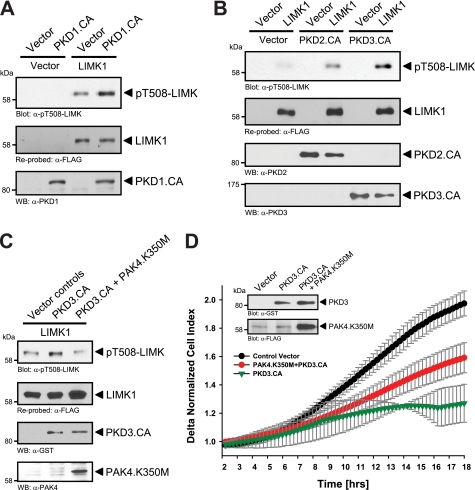
To test whether phosphorylation and activation of PAK4 through PKD enzymes occur in cells we used normal murine mammary gland (NMuMG) cells because these cells have low basal levels of PAK4 activity. Ectopic expression of active forms of PKD1, PKD2, and PKD3 induced PAK4 phosphorylation at Ser-474, suggesting that all three PKD enzymes can increase PAK4 activity in vivo (Fig. 5A). Moreover, treatment of cells with the PKD inhibitor CID755673 decreased basal levels of PAK4 phosphorylation at Ser-474 (Fig. 5B). Next, we tested whether PKD-mediated activation of PAK4 translates to increased activity of LIMK. LIMK was previously described as a physiological target for PAK4, and PAK4 can directly phosphorylate the activation loop threonine of this kinase (16). As predicted, the ectopic expression of active PKD1 increased LIMK activity. This was evaluated by analyzing LIMK phosphorylation at its activation loop threonine at amino acid position 508 (Fig. 6A). Similar results were obtained with active alleles of PKD2 and PKD3 (Fig. 6B). We also tested the possibility that PKD1 may phosphorylate and activate LIMK directly. Therefore, we used the PKD substrate motif antibody (pMOTIF). This antibody was previously generated using a degenerated peptide library and detects PKD-mediated phosphorylations (26). Using this antibody, we found that LIMK is not a direct PKD substrate (supplemental Fig. 1). Our data suggest that PKD-mediated activation of PAK4 indeed confers to increased activity of its substrate LIMK. Moreover, expression of kinase-dead PAK4 (PAK4.K350M) effectively blocked PKD3-mediated increase in LIMK activity, as judged by its phosphorylation at Thr-508, showing that LIMK is indeed downstream of PAK4 in this pathway (Fig. 6C). Moreover, combination of constitutively active PKD3 with a kinase-dead PAK4 allele in impedance-based Transwell assays showed that expression of a kinase-dead PAK4 partially rescues cells from PKD3-mediated inhibition of directed cell migration (Fig. 1C). However, we did not observe a full rescue, which can be explained because PKD also has additional targets at the leading edge of cells including SSH (28), EVL-1 (22), Hsp27 (26), or cortactin (27) which all may contribute to directed cell migration.
FIGURE 5.
PKD regulates PAK4 activity in vivo. A, NMuMG cells (3 × 105, 6-cm dish) were co-transfected with FLAG-tagged PAK4 and either vector control or constitutively active PKD1, PKD2, or PKD3, as indicated. PAK4 was immunoprecipitated (α-FLAG) and analyzed for phosphorylation at Ser-474 (α-pS474-PAK4). Blots were stripped and analyzed for total PAK4 (α-FLAG). Control blots were performed on lysates to determine expression of active PKD1 (α-PKD1 staining), PKD2 (α-FLAG staining), and PKD3 (α-GST staining). B, HeLa cells (0.5 × 106, 6-cm dish) were treated with the PKD inhibitor CID755673 (20 μm) for 1 h. Endogenous PAK4 was immunoprecipitated (α-PAK4), and samples were analyzed for phosphorylation at Ser-474 (α-pS474-PAK4). Blots were stripped and analyzed for total PAK4 (α-PAK4).
FIGURE 6.
PKD regulates activity of the PAK4 substrate LIMK1. A, NMuMG cells (5 × 105, 6-cm dish) were co-transfected with FLAG-tagged LIMK or active PKD1 or respective controls. LIMK was immunoprecipitated (α-FLAG) and analyzed for phosphorylation (α-pT508-LIMK). Blots were reprobed for LIMK expression (α-FLAG), and control blots were probed for PKD1. B, NMuMG cells (5 × 105, 6-cm dish) were co-transfected with FLAG-tagged LIMK, active PKD2, active PKD3, or respective controls. LIMK was immunoprecipitated (α-FLAG) and analyzed for activating phosphorylation at Thr-508 (α-pT508-LIMK). Blots were reprobed for LIMK expression (α-FLAG), control blots were probed for PKD2 and PKD3. C, NMuMG cells (5 × 105, 6-cm dish) were co-transfected with vector controls, FLAG-tagged LIMK, GST-tagged constitutively active PKD3 (PKD3.CA), and dominant negative PAK4 (PAK4.K350M) as indicated. LIMK was immunoprecipitated (α-FLAG) and analyzed for activating phosphorylation at Thr-508 (α-pT508-LIMK). Control Western blotting was performed for PAK4 (α-PAK4) and PKD3 (α-GST) expression. D, HeLa cells (5 × 105 cells, 6-cm dish) were transfected with control vector, constitutively active PKD3 (PKD3.CA), and kinase-dead PAK4 (PAK4.K350M) as indicated. After 2 h of attachment, cell migration toward NIH-3T3 conditioned medium over 16 h was continuously monitored in real-time using Transwell CIM-plate 16 and the xCELLigence RTCA DP instrument. Error bars (gray) represent four experiments. Control Western blotting was performed for PKD3.CA (α-GST) and PAK4.K350M (α-FLAG) expression.
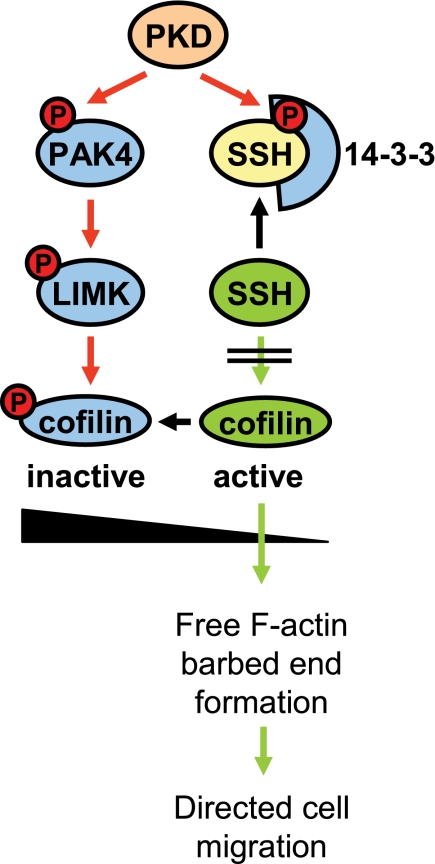
Taken together, our data suggest that all three PKD isoforms regulate cofilin activity through inhibition of slingshot and increase of LIMK activity, with the net effect of increasing the inactive phospho-cofilin pool, which confers to decreased directed cell migration (Fig. 7).
FIGURE 7.
Proposed model in which PKD regulates cofilin activity via both PAK4/LIMK and SSH pathways. PKD enzymes regulate cofilin activity by phosphorylation of SSH1L at Ser-978. This generates a binding motif for 14-3-3 proteins which sequester SSH1L in the cytosol. Here, we put forward that PKD isoenzymes also regulate cofilin activity via a second pathway where PKD phosphorylates and activates PAK4. This mediates activation of the PAK4 downstream target LIMK, resulting in further increased levels of inactive phospho-cofilin. This confers to the observed complete block of cofilin-induced barbed end formation and actin incorporation at the leading edge and may explain the effects of active PKD on inhibiting directed cell migration.
DISCUSSION
Dependent on their subcellular localization, PKD isoforms regulate a variety of cellular functions (for review see Ref. 36), including membrane receptor signaling (19), transport processes at the Golgi (31, 37), protection from oxidative stress at the mitochondria (38), and transcriptional regulation in the nucleus (39). However, the roles of PKD enzymes in the regulation of cell motility are not well defined. PKD1 is highly active at cell-cell contacts in normal epithelial tissue, where it decreases cell motility (24). Furthermore, PKD1 expression levels are down-regulated in breast, gastric, and prostate cancer, and a reverse correlation between PKD1 expression and invasive behavior was demonstrated for these cancers (24, 29, 40). Recent studies have suggested an involvement of PKD enzymes in processes that regulate actin reorganization and actin cytoskeleton-driven directed cell migration (20, 24, 41).
The rapid actin remodeling events at the progressing leading edge of migrating cells require activity of cofilin (5), an actin-severing enzyme that generates F-actin free barbed ends (3). Cofilin also is highly expressed in multiple cancers including pancreatic cancer and invasive breast cancer, but moderately in less invasive cells (5, 42). In cells cofilin activity is regulated at multiple levels. Cofilin is inactivated by phosphorylation at Ser-3 by LIMK (3, 7), and this event is reversed by the cofilin phosphatase slingshot (SSH1L) (14). We and others have shown previously that SSH1L is negatively regulated by PKD1 (28) and PKD2 (34) and that this results in sequestration of slingshot in the cytosol, mediating a net increase in cellular phospho-cofilin. Here, we expand this knowledge by showing that PKD3 also induces similar negative regulatory signaling, namely to phosphorylate slingshot at its Ser-978 residue, leading to binding of 14-3-3 proteins (Fig. 2). However, the significant effects on directed cell migration, as well as complete block of cofilin-induced F-actin free barbed end formation and actin incorporation at the leading edge in presence of active PKD (Fig. 1B) suggested that PKD may have additional targets in pathways that negatively regulate cofilin activity.
To test this, we first analyzed LIMK for PKD-mediated phosphorylations, but found no evidence that LIMK could be a direct target for PKD (supplemental Fig. 1). We then analyzed upstream activators for LIMK as potential PKD targets. LIMK activation was shown through the p21-activated kinases PAK1 and PAK4 (16, 18) and the RhoA downstream kinase ROCK (Rho-associated coiled-coil-containing kinase) (43). PKD, in some signaling pathways, can act downstream of ROCK (21), but analysis of its amino acid sequence did not provide any indication of a PKD phosphorylation motif (data not shown). PAK1 can be localized to the leading edge of motile cells and is known to regulate cell morphology and cytoskeletal reorganization (44); however, an in-depth analysis of its amino acid sequence indicated that this kinase also lacks PKD phosphorylation motifs. In contrast, group II p21-activated kinases PAK4, PAK5, and PAK6 have an ideal PKD substrate motif in their activation loop with the serine residue critical for PAK activation (Ser-474 in human PAK4) as a potential PKD site (Fig. 4A). From this group, PAK4 was formerly implicated in cytoskeletal reorganization and filopodia formation (45), whereas PAK5 principally seems to be localized at the mitochondria and PAK6 in the nucleus (46). Additionally, PAK4 may have a role in Ras-mediated transformation and was found to be overexpressed in 78% of a variety of human cancer cell lines (47).
Active PKD1 co-localizes with PAK4 and F-actin at the leading edge (Fig. 3K) and a similar co-localization could be observed when samples were probed with an antibody recognizing active PAK4 (Fig. 3P). However, staining with the pS474-PAK4 antibody showed additional nuclear staining (Fig. 3M), although total PAK4 does not show nuclear staining (i.e. Fig. 3A). Nuclear staining can be explained because the amino acids surrounding the activation loop serine in all three group II PAKs are identical (see Fig. 4A), and the pS474-PAK4 antibody also recognizes active PAK5 and PAK6. All group II PAKs are expressed in HuMEC cells, but PAK4 is the major expressed isoform (supplemental Fig. 2A). Of these, PAK6 is localized in the nucleus (supplemental Fig. 2B). This suggests that the nuclear staining observed when samples are stained with the pS474-PAK4 antibody may represent nuclear active PAK6.
In vitro kinase assays using purified proteins showed that all three PKD isoforms can directly phosphorylate PAK4 in its activation loop Ser-474 (Fig. 4B). This signaling also occurs in cells (Fig. 5) and leads to PAK4 activity as measured by analysis of the activity of its downstream target LIMK (Fig. 6, A and B). We also showed that the PKD-mediated increase in LIMK is PAK4-dependent (Fig. 6C). Therefore, with this PKD-PAK4-LIMK signaling pathway we define a second mechanism by which PKD induces inactivation of cofilin (Fig. 7).
Similar to what we have shown for PKD (Fig. 2B and Ref. 28), previously it was shown that PAK4 can negatively regulate SSH1L by phosphorylation events that mediate binding to 14-3-3 proteins (12). Therefore, it is possible that PKD1-activated PAK4 also regulates SSH1L activity. However, so far we were not able to show that PAK4 contributes to PKD1-mediated phosphorylation of SSH1L at Ser-978 nor to PKD-mediated binding of 14-3-3 (data not shown). Therefore, it is possible that PAK4 phosphorylates a second site in SSH1L that contributes to 14-3-3 binding.
The ability of PKD to decrease cofilin activity at multiple levels, including the inhibition of SSH1L and activation of the PAK4-LIMK pathway, could explain the dramatic effects on F-actin free barbed end formation and directed cell migration observed when PKD is constitutively active. Going forward, this knowledge may be of benefit for the development of therapeutic strategies to reactivate or hyperactivate PKD enzymes in cancer with the overall goal to block tumor expansion or metastasis.
Supplementary Material
Acknowledgments
We thank our colleagues in the Storz laboratory for helpful discussions, Dr. T. Seufferlein for PKD2, and Dr. V. Malhotra for PKD3 expression plasmids.
This work was supported, in whole or in part, by National Institutes of Health Grants GM086438 and CA140182 (to P. S.). This work was also supported by Bankhead-Coley Grant 10BG11 from the Florida Department of Health (to P. S.) and by Ministry of Education, Culture, Science, Sports, and Technology of Japan Grants 21370086 and 22121501 (to K. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- LIMK
- Lin-11/Isl-1/Mec-3 kinase
- CA
- constitutively active
- PAK4
- p21-activated kinase 4
- SSH
- slingshot.
REFERENCES
- 1. Bamburg J. R., McGough A., Ono S. (1999) Trends Cell Biol. 9, 364–370 [DOI] [PubMed] [Google Scholar]
- 2. Pollard T. D., Borisy G. G. (2003) Cell 112, 453–465 [DOI] [PubMed] [Google Scholar]
- 3. Mouneimne G., DesMarais V., Sidani M., Scemes E., Wang W., Song X., Eddy R., Condeelis J. (2006) Curr. Biol. 16, 2193–2205 [DOI] [PubMed] [Google Scholar]
- 4. Yamaguchi H., Condeelis J. (2007) Biochim. Biophys. Acta 1773, 642–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang W., Eddy R., Condeelis J. (2007) Nat. Rev. Cancer 7, 429–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang W., Mouneimne G., Sidani M., Wyckoff J., Chen X., Makris A., Goswami S., Bresnick A. R., Condeelis J. S. (2006) J. Cell Biol. 173, 395–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Oser M., Condeelis J. (2009) J. Cell. Biochem. 108, 1252–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Huang T. Y., DerMardirossian C., Bokoch G. M. (2006) Curr. Opin. Cell Biol. 18, 26–31 [DOI] [PubMed] [Google Scholar]
- 9. Agnew B. J., Minamide L. S., Bamburg J. R. (1995) J. Biol. Chem. 270, 17582–17587 [DOI] [PubMed] [Google Scholar]
- 10. Ikebe C., Ohashi K., Fujimori T., Bernard O., Noda T., Robertson E. J., Mizuno K. (1997) Genomics 46, 504–508 [DOI] [PubMed] [Google Scholar]
- 11. Mizuno K., Okano I., Ohashi K., Nunoue K., Kuma K., Miyata T., Nakamura T. (1994) Oncogene 9, 1605–1612 [PubMed] [Google Scholar]
- 12. Soosairajah J., Maiti S., Wiggan O., Sarmiere P., Moussi N., Sarcevic B., Sampath R., Bamburg J. R., Bernard O. (2005) EMBO J. 24, 473–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Toshima J., Toshima J. Y., Amano T., Yang N., Narumiya S., Mizuno K. (2001) Mol. Biol. Cell 12, 1131–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nagata-Ohashi K., Ohta Y., Goto K., Chiba S., Mori R., Nishita M., Ohashi K., Kousaka K., Iwamatsu A., Niwa R., Uemura T., Mizuno K. (2004) J. Cell Biol. 165, 465–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Niwa R., Nagata-Ohashi K., Takeichi M., Mizuno K., Uemura T. (2002) Cell 108, 233–246 [DOI] [PubMed] [Google Scholar]
- 16. Dan C., Kelly A., Bernard O., Minden A. (2001) J. Biol. Chem. 276, 32115–32121 [DOI] [PubMed] [Google Scholar]
- 17. Gohla A., Birkenfeld J., Bokoch G. M. (2005) Nat. Cell Biol. 7, 21–29 [DOI] [PubMed] [Google Scholar]
- 18. Edwards D. C., Sanders L. C., Bokoch G. M., Gill G. N. (1999) Nat. Cell Biol. 1, 253–259 [DOI] [PubMed] [Google Scholar]
- 19. Rozengurt E., Rey O., Waldron R. T. (2005) J. Biol. Chem. 280, 13205–13208 [DOI] [PubMed] [Google Scholar]
- 20. Eiseler T., Schmid M. A., Topbas F., Pfizenmaier K., Hausser A. (2007) FEBS Lett. 581, 4279–4287 [DOI] [PubMed] [Google Scholar]
- 21. Cowell C. F., Yan I. K., Eiseler T., Leightner A. C., Döppler H., Storz P. (2009) J. Cell. Biochem. 106, 714–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Janssens K., De Kimpe L., Balsamo M., Vandoninck S., Vandenheede J. R., Gertler F., Van Lint J. (2009) Cell. Signal. 21, 282–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ziegler S., Eiseler T., Scholz R. P., Beck A., Link G., Hausser A. (2011) Mol. Biol. Cell 22, 570–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jaggi M., Rao P. S., Smith D. J., Wheelock M. J., Johnson K. R., Hemstreet G. P., Balaji K. C. (2005) Cancer Res. 65, 483–492 [PubMed] [Google Scholar]
- 25. Du C., Jaggi M., Zhang C., Balaji K. C. (2009) Cancer Res. 69, 1117–1124 [DOI] [PubMed] [Google Scholar]
- 26. Döppler H., Storz P., Li J., Comb M. J., Toker A. (2005) J. Biol. Chem. 280, 15013–15019 [DOI] [PubMed] [Google Scholar]
- 27. Eiseler T., Hausser A., De Kimpe L., Van Lint J., Pfizenmaier K. (2010) J. Biol. Chem. 285, 18672–18683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Eiseler T., Döppler H., Yan I. K., Kitatani K., Mizuno K., Storz P. (2009) Nat. Cell Biol. 11, 545–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Eiseler T., Döppler H., Yan I. K., Goodison S., Storz P. (2009) Breast Cancer Res. 11, R13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sturany S., Van Lint J., Muller F., Wilda M., Hameister H., Hocker M., Brey A., Gern U., Vandenheede J., Gress T., Adler G., Seufferlein T. (2001) J. Biol. Chem. 276, 3310–3318 [DOI] [PubMed] [Google Scholar]
- 31. Yeaman C., Ayala M. I., Wright J. R., Bard F., Bossard C., Ang A., Maeda Y., Seufferlein T., Mellman I., Nelson W. J., Malhotra V. (2004) Nat. Cell Biol. 6, 106–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Storz P., Döppler H., Johannes F. J., Toker A. (2003) J. Biol. Chem. 278, 17969–17976 [DOI] [PubMed] [Google Scholar]
- 33. Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 34. Peterburs P., Heering J., Link G., Pfizenmaier K., Olayioye M. A., Hausser A. (2009) Cancer Res. 69, 5634–5638 [DOI] [PubMed] [Google Scholar]
- 35. Hutti J. E., Jarrell E. T., Chang J. D., Abbott D. W., Storz P., Toker A., Cantley L. C., Turk B. E. (2004) Nat. Methods 1, 27–29 [DOI] [PubMed] [Google Scholar]
- 36. Wang Q. J. (2006) Trends Pharmacol. Sci. 27, 317–323 [DOI] [PubMed] [Google Scholar]
- 37. Hausser A., Storz P., Märtens S., Link G., Toker A., Pfizenmaier K. (2005) Nat. Cell Biol. 7, 880–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Storz P., Döppler H., Toker A. (2005) Mol. Cell. Biol. 25, 8520–8530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. von Blume J., Knippschild U., Dequiedt F., Giamas G., Beck A., Auer A., Van Lint J., Adler G., Seufferlein T. (2007) EMBO J. 26, 4619–4633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kim M., Jang H. R., Kim J. H., Noh S. M., Song K. S., Cho J. S., Jeong H. Y., Norman J. C., Caswell P. T., Kang G. H., Kim S. Y., Yoo H. S., Kim Y. S. (2008) Carcinogenesis 29, 629–637 [DOI] [PubMed] [Google Scholar]
- 41. Riol-Blanco L., Iglesias T., Sánchez-Sánchez N., de la Rosa G., Sánchez-Ruiloba L., Cabrera-Poch N., Torres A., Longo I., García-Bordas J., Longo N., Tejedor A., Sánchez-Mateos P., Rodríguez-Fernández J. L. (2004) Eur. J. Immunol. 34, 108–118 [DOI] [PubMed] [Google Scholar]
- 42. Sinha P., Hütter G., Köttgen E., Dietel M., Schadendorf D., Lage H. (1999) Electrophoresis 20, 2952–2960 [DOI] [PubMed] [Google Scholar]
- 43. Ohashi K., Nagata K., Maekawa M., Ishizaki T., Narumiya S., Mizuno K. (2000) J. Biol. Chem. 275, 3577–3582 [DOI] [PubMed] [Google Scholar]
- 44. Sells M. A., Boyd J. T., Chernoff J. (1999) J. Cell Biol. 145, 837–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Abo A., Qu J., Cammarano M. S., Dan C., Fritsch A., Baud V., Belisle B., Minden A. (1998) EMBO J. 17, 6527–6540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Eswaran J., Soundararajan M., Knapp S. (2009) Cancer Metastasis Rev. 28, 209–217 [DOI] [PubMed] [Google Scholar]
- 47. Callow M. G., Clairvoyant F., Zhu S., Schryver B., Whyte D. B., Bischoff J. R., Jallal B., Smeal T. (2002) J. Biol. Chem. 277, 550–558 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.