Background: Antizyme is a regulator of cell proliferation, inhibiting this process when overexpressed.
Results: Antizyme overexpression does not attenuate cell proliferation and viability in cells whose polyamine supply is secured.
Conclusion: Antizyme affects cell proliferation and viability only by modulating polyamine metabolism.
Significance: This result emphasizes the functional relationship of antizyme to cellular polyamine metabolism.
Keywords: Cyclin D1, Decarboxylase, p73, Polyamines, Proliferation, Aurora-A, Antizyme, Ornithine Decarboxylase (ODC)
Abstract
Antizymes are key regulators of cellular polyamine metabolism that negatively regulate cell proliferation and are therefore regarded as tumor suppressors. Although the regulation of antizyme (Az) synthesis by polyamines and the ability of Az to regulate cellular polyamine levels suggest the centrality of polyamine metabolism to its antiproliferative function, recent studies have suggested that antizymes might also regulate cell proliferation by targeting to degradation proteins that do not belong to the cellular polyamine metabolic pathway. Using a co-degradation assay, we show here that, although they efficiently stimulated the degradation of ornithine decarboxylase (ODC), Az1 and Az2 did not affect or had a negligible effect on the degradation of cyclin D1, Aurora-A, and a p73 variant lacking the N-terminal transactivation domain whose degradation was reported recently to be stimulated by Az1. Furthermore, we demonstrate that, although Az1 and Az2 could not be constitutively expressed in transfected cells, they could be stably expressed in cells that express trypanosome ODC, a form of ODC that does not bind Az and therefore maintains a constant level of cellular polyamines. Taken together, our results clearly demonstrate that Az1 and Az2 affect cell proliferation and viability solely by modulating cellular polyamine metabolism.
Introduction
The polyamines regulate fundamental cellular processes, most profoundly those supporting cell proliferation (1, 2). For maximal effectiveness, polyamines must be kept within a narrow optimal range. Deviation from the optimal level results, at the lower end, in inhibition of cell proliferation and, at the upper end, in cytotoxicity and cell death (3, 4). It is therefore not surprising that optimal levels of cellular polyamines are maintained by tight regulation at several control levels. The central player of the main regulatory circuit is a small polyamine-induced protein termed antizyme (Az)3 (5–7). Polyamines promote Az expression by stimulating programmed +1 ribosomal frameshifting that combines two different open reading frames to produce a full-length functional protein (8, 9). Az, whose affinity for ornithine decarboxylase (ODC) subunits is greater than the affinity these subunits display toward each other, binds to transient ODC monomers, preventing their reassociation to form active ODC. It then targets them to ubiquitin-independent degradation by the 26 S proteasome (10). In addition, Az inhibits uptake and stimulates excretion of polyamines via a yet unresolved mechanism (11). Az inhibits cell proliferation and displays antitumor activity, and therefore, it is regarded as a tumor suppressor (12–15). Mammalian cells express three characterized members of the Az family of proteins (6, 16). While one of them, Az3, is testis-specific and observed only in haploid germinal cells (17, 18), the other two, the prototypical Az1 and Az2, are ubiquitously expressed, with Az1 being expressed at much higher levels (5, 6, 19).
The tight regulation of Az synthesis by polyamines, together with the ability of Az to regulate cellular polyamine levels, suggests that Az might exert its antiproliferative effect exclusively by regulating cellular polyamine metabolism. However, several recent studies put forward an alternative possibility, suggesting that Az might also inhibit cell proliferation by targeting to ubiquitin-independent degradation growth-regulating proteins that do not belong to the cellular polyamine metabolic pathway. These include Smad1, a key transducer of the bone morphogenetic proteins (20–22); the cell cycle regulators cyclin D1 and Aurora-A (23, 24); and the anti-apoptotic N-terminally truncated form of p73 (ΔNp73) (25).
In this study, we tested these two alternative possibilities. First, we compared the ability of Az to target these proteins to degradation relative to its ability to stimulate ODC degradation. Second, we monitored the effect of Az in cells displaying constant levels of polyamines due to the expression of trypanosome ODC, an ODC variant that is refractory to the destabilizing effect of Az (26, 27). We show here that, although greatly stimulating ODC degradation, Az1 and Az2 did not stimulate or exerted a negligible effect on cyclin D1, Aurora-A, and ΔNp73 degradation under the same experimental conditions. Moreover, we show that both antizymes could be efficiently expressed in trypanosome ODC-expressing cells without exerting an antiproliferative effect or affecting the viability of these cells. Our results therefore demonstrate that polyamine metabolism represents the only cellular target for the antiproliferative effect of Az.
EXPERIMENTAL PROCEDURES
Cell Culture Conditions and Transfections
NIH3T3 mouse fibroblast and HEK-293T cell lines were grown in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% (v/v) fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin (Biological Industries). NIH3T3 cells were transfected using JetPEI (Source BioScience Autogen) following the manufacturer's instructions. HEK-293 cells were transfected using the calcium phosphate method (28). NIH3T3 cells expressing the tetracycline-responsive repressor were grown and stimulated for the expression of Az as described (29).
Cloning and Construction of Plasmids
Az, trypanosome ODC, cyclin D1, Aurora-A, ΔNp73, and the Tet repressor were cloned into different variants of the bicistronic pEFIRES vectors (30), which differ in their selectable markers, conferring resistance to puromycin, neomycin, and hygromycin B, respectively. Thus, we were able to achieve stable expression of three of these proteins in the same transfected cells. For inducible expression, Az cDNA was cloned downstream of a Tet-responsive promoter in a pcDNA3-based plasmid. A nucleotide was deleted from Az to permit expression of a full-length protein without requiring frameshifting. Mutations, deletions, and tags were introduced using the overlap extension method (31, 32).
Western Blot Analysis
Cells were lysed in buffer containing 50 mm Tris-HCl (pH 8), 150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mm dithiothreitol, and a mixture of protease inhibitors (Sigma). Cellular extracts containing equal amounts of protein were heated for 5 min in sample buffer and fractionated by SDS-PAGE. The proteins were then electroblotted onto nitrocellulose membrane, and specific proteins were identified by incubation with the indicated antibodies, followed by horseradish peroxidase-conjugated anti-IgG antibodies. Signals were developed using EZ-ECL (Biological Industries), and the membranes were exposed to x-ray film.
In Vitro Degradation Assay
ODC, cyclin D1, Aurora-A, and Az were translated in vitro using TNT reaction mixture (Promega) in the presence of [35S]methionine. The synthesized proteins were resolved by electrophoresis, and the molarity of the synthesized proteins was normalized by dividing the radioactivity in the relevant band by the number of methionine residues in each of these proteins. Equal molar amounts of the tested proteins and of Az were then incubated in a degradation reaction containing 40 mm Tris-HCl (pH 7.5), 5 mm MgCl2, 2 mm dithiothreitol, 0.5 mm ATP, 10 mm phosphocreatine, 1.6 mg/ml creatine phosphokinase, and 6 μl of reticulocyte lysate (Promega) at 37 °C for the indicated times. The proteins were then resolved by electrophoresis, and the radioactivity present in individual bands was determined using a Fuji BAS-2500 phosphoimager.
In Vivo Degradation Assay
The degradation rate was determined by the addition of cycloheximide (20 μg/ml) to the growth medium. Cells were harvested at the indicated times, cellular extracts were prepared, and the amount of the tested proteins was determined by Western blot analysis. The degradation rate was quantified using ImageJ.
ODC Activity Assay
200 μg of protein from cellular extracts or portions of reticulocyte lysate were brought to 100 μl with ODC assay buffer (25 mm Tris-HCl (pH 7.5), 2.5 mm DTT, 0.1 mm EDTA, 0.2 mm pyridoxal phosphate, and 33 mm l-ornithine) containing 0.5 μCi of l-[14C]ornithine. The reaction was incubated at 37 °C for 2 h in a 96-well plate. The liberated [14C]CO2 was trapped in a covering 3-mm paper soaked with saturated barium hydroxide solution. The paper was washed with acetone and dried, and the results were quantified using the Fuji BAS-2500 phosphoimager.
Polyamine Analysis
Cells grown in 10-cm dishes were harvested, sedimented, and resuspended in 100 μl of PBS. The cells were lysed in 3% perchloric acid, and precipitated material was removed by centrifugation for 5 min at 13,000 rpm. The supernatant was collected for polyamine analysis, whereas the pellet was used for normalization by DNA quantification. (DNA was quantified by resuspension of the pellet in 400 μl of 4% diphenylamine (Sigma) in acetic acid, 400 μl of 10% perchloric acid, and 20 μl of 1:500 acetaldehyde (Sigma), followed by incubation for 16 h at 30 °C and absorbance determination at 595 and 700 nm.) For polyamine analysis, 100 μl of the perchloric acid supernatant were mixed with 200 μl of 6 mg/ml dansyl chloride (in acetone). After the addition of 10 mg of sodium carbonate, the mixture was incubated for 16 h in the dark. To neutralize residual dansyl chloride, 50 μl of 100 mg/ml l-proline solution were added for 1 h at room temperature. Dansylated derivatives were extracted into 250 μl of toluene. Portions of 50–100 μl were spotted on Silica Gel 60 F254 TLC plates (Merck). The dansylated derivatives were then resolved by thin layer chromatography using ethyl acetate/cyclohexane (1:1.5) as a solvent and visualized by UV illumination. Dansylated derivatives of known polyamines served as markers.
Determination of Growth Rate
Cells were plated in 12-well plates and grown in medium supplemented with 10% FBS. At the indicated times following induction of Az expression, the cells were trypsinized and counted using a bright-line counting chamber (Hausser Scientific, Horsham, PA).
RESULTS
Az Differentially Stimulates ODC Degradation
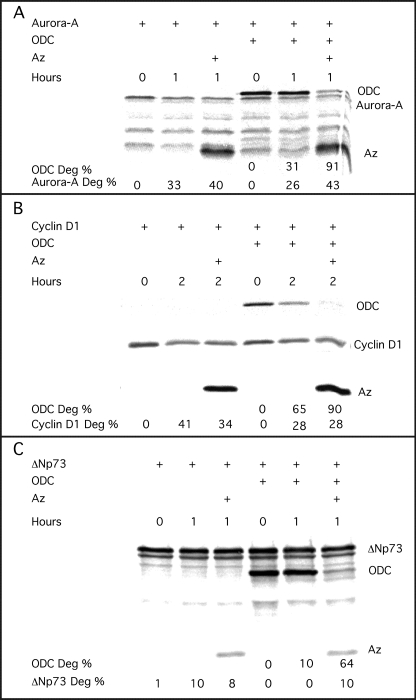
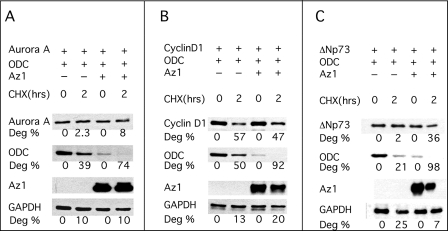
Az expression and function are tightly associated with cellular polyamine metabolism. However, it was suggested recently that Az might exert its antiproliferative effect not only through targeting ODC to ubiquitin-independent degradation and to inhibiting polyamine uptake but also by stimulating degradation of the growth-regulating proteins cyclin D1, Aurora-A, and ΔNp73, which do not belong to polyamine metabolism (23–25). However, in these studies, the ability of Az to stimulate the degradation of these proteins was not compared with its ability to stimulate ODC degradation. We therefore set out to compare the ability of Az1, the prototype and most studied member of this protein family, to stimulate their degradation with its ability to stimulate ODC degradation. We tested its effect both in an in vitro reticulocyte lysate-based degradation mixture and in transfected cells. For the in vitro degradation reaction, Az1 and the tested proteins were translated in reticulocyte lysate in the presence of [35S]methionine. Next, we performed three degradation reactions containing cyclin D1 and ODC with or without Az1, Aurora-A and ODC with or without Az1, and ΔNp73 and ODC with or without Az1. In all three reactions, Az1 greatly stimulated the degradation of ODC but failed to stimulate or had a negligible effect on the degradation of cyclin D1, Aurora-A, and ΔNp73 (Fig. 1, A–C). To test for the effect of Az1 in cells, constructs encoding Az1, each of the tested proteins, and ODC were transiently cotransfected into HEK-293T cells, and cycloheximide was added to the growth medium 24 h post-transfection. Cellular extracts were prepared at the indicated times, and the degradation rates of the tested proteins were determined by Western blot analysis. The results showed that, although dramatically stimulating ODC degradation, Az1 did not affect the degradation rate of cotransfected cyclin D1, Aurora-A, or ΔNp73 (Fig. 2, A–C).
FIGURE 1.
Az1 stimulates ODC but not cyclin D1, Aurora-A, and ΔNp73 degradation in vitro in a reticulocyte lysate-based degradation reaction. Az1, ODC, Aurora-A, cyclin D1, and ΔNp73 were synthesized in vitro in reticulocyte lysate. As described under “Experimental Procedures,” we performed three degradation (Deg) reactions containing Az, ODC, and Aurora-A (A), cyclin D1 (B), and ΔNp73 (C). A reaction lacking Az was performed as a control. Following the indicated times of incubation at 37 °C, the material was resolved by SDS-PAGE, and the protein bands were visualized and quantified using the Fuji BAS-2500 phosphoimager. Each of the above experiments was repeated twice, yielding similar results.
FIGURE 2.
Az1 stimulates ODC but not cyclin D1, Aurora-A, and ΔNp73 degradation in vivo in transfected NIH3T3 cells. Constructs encoding Az1 and ODC were transfected into HEK-293T cells together with a construct encoding Aurora-A (A), cyclin D1 (B), or ΔNp73 (C). Cycloheximide (CHX) was added to the growth medium 24 h post-transfection, the cells were harvested at the indicated times, and the levels of the individual tested proteins were determined by Western blot analysis using anti-FLAG antibodies. Anti-actin antibodies were used to normalize the amount of protein loaded in each lane. The experiments presented were repeated three times, yielding practically identical results. Deg, degradation.
Cells Expressing Trypanosome ODC Tolerate Constitutive Az1 Expression
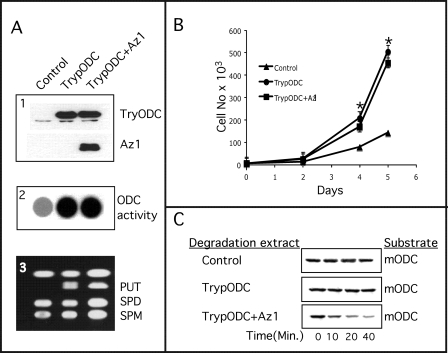
Although the above results suggest that Az1 displays clear specificity for components of cellular polyamine metabolism, we cannot exclude the possibility that Az affects cell proliferation by affecting other cellular proteins. We therefore wanted to test in a general unbiased way whether Az1 affects cell proliferation solely through manipulating cellular polyamine metabolism or whether it does so also by affecting other growth-regulating proteins that do not belong to the polyamine metabolic pathway. In this respect, it is important to note that we failed to stably express Az in transfected cells. We therefore established an NIH3T3-derived cell line that stably expresses trypanosome ODC. This form of ODC does not bind Az and therefore is not targeted to degradation in the presence of Az (26, 27). Because the polyamine level in these cells is expected to remain steady, we inferred that, if Az regulates growth solely through manipulating cellular polyamines, it should not exert any antiproliferative effect in these cells. As expected, stable expression of Az1 in the trypanosome ODC-expressing cells (Fig. 3A, panel 1) did not reduce the elevated ODC activity and polyamine levels attributed to the expression of trypanosome ODC (Fig. 3A, panels 2 and 3). Importantly, Az expression did not reduce the proliferation rate of these cells. The rate was practically identical to that of cells expressing only trypanosome ODC, which is higher than that of cells carrying empty vectors (Fig. 3B). That the expressed Az1 was active was demonstrated by the ability of cellular extracts of the Az1-transfected cells to stimulate degradation of mouse ODC in an in vitro degradation reaction (Fig. 3C).
FIGURE 3.
Expression of trypanosome ODC enables constitutive Az1 expression. A, NIH3T3 mouse fibroblasts were stably transfected with constructs encoding trypanosome ODC (TrypODC) or trypanosome ODC and Az1 or with the compatible empty vectors (Control; panel 1). ODC activity (panel 2) and cellular polyamines (panel 3) were determined as described under “Experimental Procedures.” PUT, putrescine; SPD, spermidine; SPM, spermine. B, the growth rates of the three cell lines were determined by daily counting of cells as described under “Experimental Procedures.” *, significant difference from control cells using Student's t test (p < 0.02). C, the functionality of transfected Az1 was determined by mixing cellular extract prepared from the three cell lines in an in vitro degradation reaction using in vitro synthesized [35S]methionine-labeled mouse ODC (mODC) as a substrate.
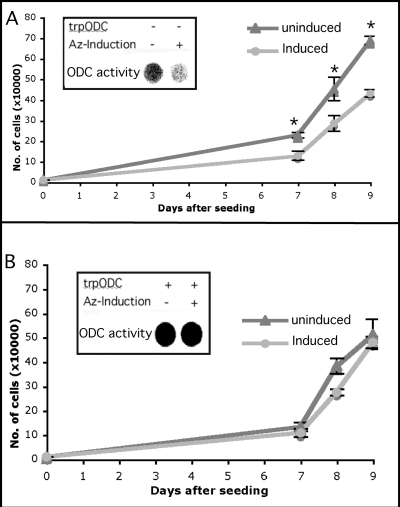
Inducible Az1 Expression Inhibits Proliferation of NIH3T3 Cells but Not That of Trypanosome ODC-expressing Cells
Although unlikely, it can be argued that, during selection of stable transformants, in addition to the adaptation of polyamine metabolism, Az expression may also provoke adaptation through other cellular components. We therefore set out to test whether trypanosome ODC expression is sufficient to protect cells from the consequences of inducible expression of Az. For this purpose, Az1 was cloned downstream of a Tet-inducible promoter, and the resulting construct was used to stably transform NIH3T3 cells already stably transformed with a trypanosome ODC-encoding construct or with an empty vector (control cells). As shown in Fig. 4, although induction of Az expression inhibited ODC activity and growth of control cells (Fig. 4A), ODC activity and growth of the trypanosome ODC-expressing cells remained unaffected (Fig. 4B). This further supports the notion that Az1 affects cell proliferation only by manipulating cellular polyamine metabolism.
FIGURE 4.
Trypanosome ODC protects against the antiproliferative effect of induced Az. A construct encoding Az1 cDNA cloned downstream of a Tet-responsive promoter was transfected into NIH3T3 cells previously transformed with a plasmid encoding a Tet-controlled transactivator and a trypanosome ODC (trpODC)-encoding construct (B) or the compatible empty vector (A). Successful induction of Az was demonstrated by inhibition of ODC activity in cells transformed with the empty vector (insets). The growth rates of the various cells was determined by cell counting at days 7, 8, and 9 following seeding and induction. *, significant difference from control cells using Student's t test (p < 0.02).
Az2 Affects Cell Proliferation Solely by Regulating Polyamine Metabolism
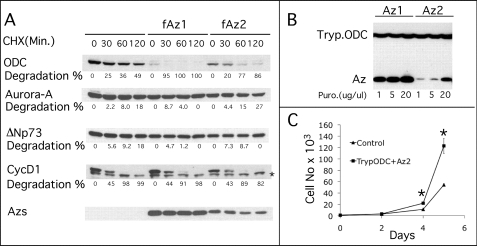
As mentioned above, Az1, the prototype of the Az family of proteins, is not the only form of Az expressed in mammalian cells. The other two forms are Az3, which is testis-specific, and Az2, which is ubiquitously expressed like Az1 but to significantly lower levels (33). Because Az2 is evolutionarily conserved to an even higher degree than Az1 (19), its minority coexistence with Az1 in the same cells is of interest and may suggest that it has a different cellular role. In support of this possibility is a recent demonstration that Az2 is predominantly nuclear, whereas Az1 is cytoplasmic (34). We therefore set out to determine whether Az2 might regulate cell proliferation by affecting proteins that do not belong to the polyamine metabolic pathway. The ability of Az2 to stimulate protein degradation was tested only in cells, as it was demonstrated previously that Az2 does not stimulate ODC degradation in an in vitro degradation reaction (29, 35). Simultaneous cotransfection of Az with all four tested proteins demonstrated that, like Az1, Az2 did not stimulate the degradation of cyclin D1, Aurora-A, and ΔNp73 but efficiently stimulated ODC degradation (Fig. 5A). Furthermore, we demonstrated that, as with Az1, stable expression of trypanosome ODC permitted the selection of cells that constitutively express Az2 without affecting cell viability (Fig. 5B). As with Az1, Az2 did not affect growth rate when expressed in trypanosome ODC-expressing cells (Fig. 5C). Interestingly however, although expressed from an identical construct as Az1, Az2 was expressed to significantly lower levels compared with Az1, and amplification of its expression unit was required to achieve levels that were equivalent to those of Az1 observed in the initial transformants (Fig. 5B). These results demonstrated that, like Az1, Az2 could be constitutively expressed in cells provided that the supply of polyamines was preserved.
FIGURE 5.
Az2 stimulates ODC but not cyclin D1, Aurora-A, and ΔNp73 degradation and can be stably expressed in trypanosome ODC-expressing cells. A, HEK-293T cells were cotransfected with constructs encoding FLAG-tagged (f) Az2, ODC, Aurora-A, cyclin D1, and ΔNp73. Cycloheximide (CHX) was added to the growth medium 24 h post-transfection, cellular extracts were prepared at the indicated times, and the amounts of the individual tested proteins were determined by Western blot analysis using anti-FLAG antibodies. The data presented are representative of three repetitions. The asterisk denotes a nonspecific band. B, HEK-293T cells stably expressing trypanosome ODC (Tryp.ODC) were transfected with Az1 or Az2-expressing constructs. Focuses were initially selected with 1 mg/ml puromycin (Puro), followed by amplification of the expression unit by increasing puromycin to 5, and 20 mg/ml. Cellular extracts were prepared, and the expression levels of trypanosome ODC, Az1, and Az2 were determined by Western blot analysis. C, the growth rate of cells expressing trypanosome ODC and Az2 was compared with that of control cells transfected with two empty vectors by daily counting of cells as described under “Experimental Procedures.” *, significant difference from control cells using Student's t test (p < 0.02).
DISCUSSION
We have provided here compelling evidence that Az1 and Az2 affect mammalian cell proliferation and viability solely through affecting cellular polyamine metabolism. Az is a small polyamine-induced protein that inhibits mammalian cell proliferation and is therefore regarded as a tumor suppressor (12–15). Az binds to transient ODC monomeric subunits, preventing their association to form active dimers and targeting them to ubiquitin-independent degradation by the 26 S proteasome (7, 10). In addition to stimulating ODC degradation, Az regulates polyamine transport across the plasma membrane through a yet undefined mechanism (11, 36, 37). Because the efficiency of Az synthesis is regulated by intracellular polyamine concentrations (8, 9), whereas, at the same time, Az regulates cellular polyamine levels, it can be assumed that Az affects cell proliferation by regulating cellular polyamine metabolism. On the other hand, it can be argued that it is rather unlikely that such a system was evolutionarily selected for degrading a single cellular protein (ODC) and for affecting a single metabolic pathway. Indeed, recent studies raised the possibility that Az might affect cell proliferation and viability through stimulating the degradation of additional proteins such as Smad1, cyclin D1, Aurora-A, and ΔNp73, which can regulate growth independently of cellular polyamine metabolism (20–25). However, these studies did not compare the efficiency of their degradation with the efficiency with which Az stimulates ODC degradation. This is particularly important, as Az acts catalytically in stimulating ODC degradation, being effective even when present in inferior molar amounts (38). We have shown here that the two major forms of Az, Az1 and Az2, efficiently stimulated ODC degradation but failed to stimulate or negligibly affected the degradation of cyclin D1, Aurora-A, and ΔNp73. A reflection of the efficient stimulation of ODC degradation by Az was significantly reduced basal levels of ODC in cotransfected cells (Figs. 2 and 5). No reduction in the level of the other three tested proteins was observed.
The observation that both antizymes did not stimulate cyclin D1, Aurora-A, and ΔNp73 degradation does not rule out the possibility that Az negatively regulates cell proliferation by affecting the expression or functionality of other growth-regulating proteins. We therefore sought to determine in a general way whether Az1 and Az2 regulate cell growth solely by affecting cellular polyamine metabolism or whether they do so also by affecting other growth-regulating proteins. For this purpose, we established NIH3T3 and HEK-293 cells stably expressing trypanosome ODC, which is refractory to the deleterious effects of Az because it does not bind Az. Because the expression of trypanosome ODC provides a steady supply of polyamines, in contrast to wild-type cells, whose polyamines are depleted by forcefully expressed Az, we inferred that these cells will survive stable Az expression if components of polyamine metabolism are the only targets of Az but will die or their proliferation will be severely retarded if Az has targets outside the polyamine metabolic pathway that are crucial for cell proliferation and viability. Our results showed that, although it is impossible to express Az stably in wild-type cells, both Az1 and Az2 did not affect the growth rate or viability of the trypanosome ODC-expressing cells.
On the basis of the data we have presented here, we conclude that both Az1 and Az2 affect the growth and viability of mammalian cells solely by targeting components of cellular polyamine metabolism. We cannot rule out, however, the possibility that Az might regulate the degradation of (or influence in a different way) other cellular proteins that do not affect cell proliferation and viability.
Acknowledgment
We thank Karen Rae Bone for helpful comments.
This work was supported by grants from the Israel Academy of Science and Humanities.
- Az
- antizyme
- ODC
- ornithine decarboxylase.
REFERENCES
- 1. Igarashi K., Kashiwagi K. (2010) Int. J. Biochem. Cell Biol. 42, 39–51 [DOI] [PubMed] [Google Scholar]
- 2. Pegg A. E. (2009) IUBMB Life 61, 880–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Meyskens F. L., Jr., Gerner E. W. (1999) Clin. Cancer Res. 5, 945–951 [PubMed] [Google Scholar]
- 4. Tobias K. E., Kahana C. (1995) Cell Growth Differ. 6, 1279–1285 [PubMed] [Google Scholar]
- 5. Kahana C. (2009) Essays Biochem. 46, 47–61 [DOI] [PubMed] [Google Scholar]
- 6. Kahana C. (2009) Cell. Mol. Life Sci. 66, 2479–2488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Murakami Y., Matsufuji S., Hayashi S., Tanahashi N., Tanaka K. (2000) Biochem. Biophys. Res. Commun. 267, 1–6 [DOI] [PubMed] [Google Scholar]
- 8. Matsufuji S., Matsufuji T., Miyazaki Y., Murakami Y., Atkins J. F., Gesteland R. F., Hayashi S. (1995) Cell 80, 51–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rom E., Kahana C. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 3959–3963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Murakami Y., Matsufuji S., Kameji T., Hayashi S., Igarashi K., Tamura T., Tanaka K., Ichihara A. (1992) Nature 360, 597–599 [DOI] [PubMed] [Google Scholar]
- 11. Mitchell J. L., Judd G. G., Bareyal-Leyser A., Ling S. Y. (1994) Biochem. J. 299, 19–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fong L. Y., Feith D. J., Pegg A. E. (2003) Cancer Res. 63, 3945–3954 [PubMed] [Google Scholar]
- 13. Iwata S., Sato Y., Asada M., Takagi M., Tsujimoto A., Inaba T., Yamada T., Sakamoto S., Yata J., Shimogori T., Igarashi K., Mizutani S. (1999) Oncogene 18, 165–172 [DOI] [PubMed] [Google Scholar]
- 14. Koike C., Chao D. T., Zetter B. R. (1999) Cancer Res. 59, 6109–6112 [PubMed] [Google Scholar]
- 15. Tsuji T., Usui S., Aida T., Tachikawa T., Hu G. F., Sasaki A., Matsumura T., Todd R., Wong D. T. (2001) Oncogene 20, 24–33 [DOI] [PubMed] [Google Scholar]
- 16. Ivanov I. P., Atkins J. F. (2007) Nucleic Acids Res. 35, 1842–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ivanov I. P., Rohrwasser A., Terreros D. A., Gesteland R. F., Atkins J. F. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 4808–4813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tosaka Y., Tanaka H., Yano Y., Masai K., Nozaki M., Yomogida K., Otani S., Nojima H., Nishimune Y. (2000) Genes Cells 5, 265–276 [DOI] [PubMed] [Google Scholar]
- 19. Ivanov I. P., Gesteland R. F., Atkins J. F. (2000) Nucleic Acids Res. 28, 3185–3196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang T. (2003) Front. Biosci. 8, d1109–d1127 [DOI] [PubMed] [Google Scholar]
- 21. Gruendler C., Lin Y., Farley J., Wang T. (2001) J. Biol. Chem. 276, 46533–46543 [DOI] [PubMed] [Google Scholar]
- 22. Lin Y., Martin J., Gruendler C., Farley J., Meng X., Li B. Y., Lechleider R., Huff C., Kim R. H., Grasser W. A., Paralkar V., Wang T. (2002) BMC Cell Biol. 3, 15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lim S. K., Gopalan G. (2007) Oncogene 26, 6593–6603 [DOI] [PubMed] [Google Scholar]
- 24. Newman R. M., Mobascher A., Mangold U., Koike C., Diah S., Schmidt M., Finley D., Zetter B. R. (2004) J. Biol. Chem. 279, 41504–41511 [DOI] [PubMed] [Google Scholar]
- 25. Dulloo I., Gopalan G., Melino G., Sabapathy K. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 4902–4907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ghoda L., Phillips M. A., Bass K. E., Wang C. C., Coffino P. (1990) J. Biol. Chem. 265, 11823–11826 [PubMed] [Google Scholar]
- 27. Li X., Coffino P. (1992) Mol. Cell. Biol. 12, 3556–3562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Graham F. L., van der Eb A. J. (1973) Virology 54, 536–539 [DOI] [PubMed] [Google Scholar]
- 29. Snapir Z., Keren-Paz A., Bercovich Z., Kahana C. (2009) Biochem. J. 419, 99–103 [DOI] [PubMed] [Google Scholar]
- 30. Hobbs S., Jitrapakdee S., Wallace J. C. (1998) Biochem. Biophys. Res. Commun. 252, 368–372 [DOI] [PubMed] [Google Scholar]
- 31. Higuchi R., Krummel B., Saiki R. K. (1988) Nucleic Acids Res. 16, 7351–7367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. (1989) Gene 77, 51–59 [DOI] [PubMed] [Google Scholar]
- 33. Ivanov I. P., Gesteland R. F., Atkins J. F. (1998) Genomics 52, 119–129 [DOI] [PubMed] [Google Scholar]
- 34. Murai N., Shimizu A., Murakami Y., Matsufuji S. (2009) J. Cell. Biochem. 108, 1012–1021 [DOI] [PubMed] [Google Scholar]
- 35. Chen H., MacDonald A., Coffino P. (2002) J. Biol. Chem. 277, 45957–45961 [DOI] [PubMed] [Google Scholar]
- 36. He Y., Suzuki T., Kashiwagi K., Igarashi K. (1994) Biochem. Biophys. Res. Commun. 203, 608–614 [DOI] [PubMed] [Google Scholar]
- 37. Suzuki T., He Y., Kashiwagi K., Murakami Y., Hayashi S., Igarashi K. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 8930–8934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mamroud-Kidron E., Omer-Itsicovich M., Bercovich Z., Tobias K. E., Rom E., Kahana C. (1994) Eur. J. Biochem. 226, 547–554 [DOI] [PubMed] [Google Scholar]