Abstract
Pathogenic bacteria require iron to replicate inside mammalian hosts. Recent studies indicate that heme acquisition in Gram-positive bacteria is mediated by proteins containing one or more near-iron transporter (NEAT) domains. Bacillus anthracis is a spore-forming, Gram-positive pathogen and the causative agent of anthrax disease. The rapid, extensive, and efficient replication of B. anthracis in host tissues makes this pathogen an excellent model organism for the study of bacterial heme acquisition. B. anthracis secretes two NEAT hemophores, IsdX1 and IsdX2. IsdX1 contains a single NEAT domain, whereas IsdX2 has five, a novel property among hemophores. To understand the functional significance of harboring multiple, non-identical NEAT domains, we purified each individual NEAT domain of IsdX2 as a GST fusion and analyzed the specific function of each domain as it relates to heme acquisition and transport. NEAT domains 1, 3, 4, and 5 all bind heme, with domain 5 having the highest affinity. All NEATs associate with hemoglobin, but only NEAT1 and -5 can extract heme from hemoglobin, seemingly by a specific and active process. NEAT1, -3, and -4 transfer heme to IsdC, a cell wall-anchored anthrax NEAT protein. These results indicate that IsdX2 has all the features required to acquire heme from the host and transport heme to the bacterial cell wall. Additionally, these results suggest that IsdX2 may accelerate iron import rates by acting as a “heme sponge” that enhances B. anthracis replication in iron-starved environments.
Keywords: Bacteria, Heme, Hemoglobin, Iron, Porphyrin, Bacillus anthracis, Hemophore, IsdX2, Near-iron Transporter
Introduction
Pathogenic bacteria must acquire iron from their mammalian hosts to survive and replicate during infection (1). However, the host sequesters iron within heme, which is further bound tightly to hemoproteins, such as hemoglobin (Hb) (2, 3). In response, bacteria have evolved protein-based systems that scavenge heme from host proteins and import this heme-iron, thereby alleviating iron deficiency and promoting bacterial replication (4–6).
Recent studies suggest Gram-positive bacteria utilize a conserved protein module, a near-iron transporter (NEAT)3 domain, to acquire heme from host hemoproteins (7–9). Proteins harboring this domain may bind heme, Hb, or both and initiate NEAT-NEAT heme transfer to receptor proteins localized on the cell envelope (10–14). Bacterial strains lacking genes encoding NEATs grow poorly on heme or Hb as an iron source and generally are less virulent than their wild-type counterparts (15–17). There is also evidence that recombinant NEAT proteins can serve as effective vaccines (18–21). However, the structural and molecular mechanisms of NEAT function remain to be determined.
Bacillus anthracis is a Gram-positive, spore-forming bacterium that is the causative agent of anthrax disease and a weapon of bioterrorism (22, 23). Infection begins when spores enter a host and are phagocytosed by resident macrophages (24, 25). Germination at the site of infection or in regional lymph nodes leads to the escape of rapidly replicating vegetative cells into hematogenous tissues, resulting in high bacterial cell numbers (26–29). The multifaceted ability of this pathogen to replicate efficiently in several host tissues, including blood, makes it an ideal model system for the study of iron uptake processes.
Along these lines, B. anthracis secretes two NEAT-containing hemophores (iron-regulated surface determinants 1 and 2 (IsdX1 and IsdX2)) that promote the growth of this pathogen on Hb as a sole iron source (14, 16, 30). IsdX1 harbors a single NEAT domain that can bind and transfer heme to cell wall-anchored IsdC (16, 30). The second hemophore, IsdX2, contains five NEAT domains (30). These findings raise interesting questions as to why a bacterial hemophore would harbor five potentially functionally redundant NEAT domains. In this report, we describe the properties of each recombinant IsdX2 NEAT domain and find that this hemophore is capable of performing all of the functions needed for heme acquisition, including the ability to bind heme and Hb, extract heme from Hb, and transfer heme to a downstream receptor. These properties are novel for a bacterial hemophore.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Reagents, and Cloning
Escherichia coli strains (DH5α or XL1-Blue) were grown in Luria broth (LB) supplemented with 50 μg/ml ampicillin (Fisher). B. anthracis strain Sterne 34F2 was grown in LB, and chromosomal DNA was extracted using the Wizard genomic DNA purification kit (Promega). The primer pairs, neatn-BamHI and neatn-EcoRI (supplemental Table S1), allowed each NEAT domain of isdX2 to be PCR-amplified individually off the chromosome. Forward primers each encoded a BamHI restriction site. The reverse primer for each NEAT had an artificial stop codon (UAA) inserted before the EcoRI restriction site. The resulting PCR product was digested using BamHI and EcoRI restriction enzymes (New England Biolabs). The insert was ligated and cloned between the BamHI/EcoRI sites of the vector pGEX2TK to create a protein fusion to glutathione S-transferase (GST): pGEX2TK-gst-neatn. DNA was then transformed into chemically competent DH5α and XL1-Blue. pGEX2TK-gst-isdC in XL1-Blue was previously cloned by Maresso et al. (15) and used for IsdC expression.
Protein Purification
E. coli XL1-Blue strains harboring gst-neatn or gst-isdC were grown in LB supplemented with 50 μg/ml ampicillin. Each protein was expressed using 1.5 mm isopropyl β-d-thiogalactopyranoside (Sigma) induction for 2 h at 37 °C or overnight at 30 °C. The 2-h induction leads to approximately a 2–3-fold reduction in the amount of heme co-purifying with each NEAT domain when compared with overnight induction. Cells were centrifuged (6,000 × g) and resuspended in phosphate-buffered saline (PBS; 137 mm NaCl, 2.7 mm KCl, 10 mm sodium phosphate dibasic, 2 mm potassium phosphate monobasic, pH 7.4) or 50 mm Tris-HCl, pH 7.0. Bacteria were lysed using a French press and centrifuged at 14,000 × g, and supernatants were applied to glutathione-Sepharose resin (Amersham Biosciences). After two 30-ml washes in buffer, each protein was eluted off the column after incubation with 50 units of thrombin (Calbiochem) to isolate NEATn or 25 mm reduced glutathione (Calbiochem) to isolate GST-NEATn/-IsdC. Thrombin was removed from protein preparations using aminobenzamidine resin (Sigma). For the heme-binding assays in Fig. 3, heme was removed by adjusting the pH to ∼3, followed by heme removal using methyl ethyl ketone (31). However, this method leads to substantial protein precipitation and could not be used to make apoprotein for the heme scavenging assays shown in Fig. 4. For these assays, the NEAT proteins were purified using the 2-h induction described above. The concentration of recombinant proteins was determined using the bicinchoninic acid method (Pierce) or the Bradford assay (Bio-Rad) (32). The molar extinction coefficients for each recombinant NEAT were calculated from the amino acid sequence according to the method of Pace et al. (33) (NEAT1 (11,460 m−1 cm−1), NEAT2 (4,470 m−1 cm−1), NEAT3 (12,950 m−1 cm−1), NEAT4 (12,950 m−1 cm−1), and NEAT5 (12,950 m−1 cm−1)).
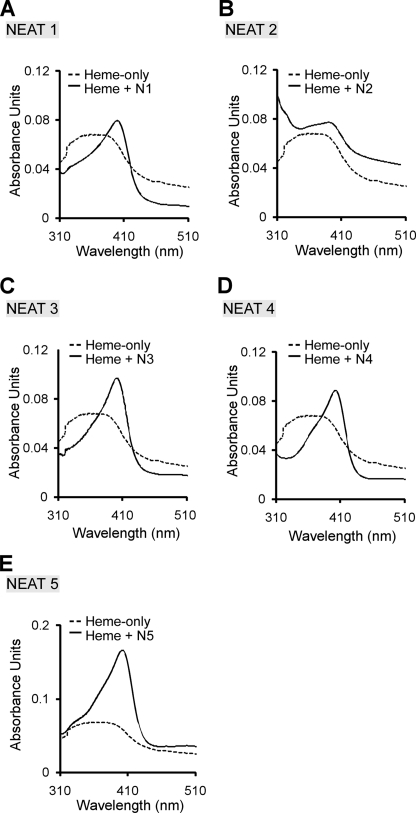
FIGURE 3.
Heme binding by the IsdX2 NEAT domains. A–E, each apo-NEAT (4.5 μm) was incubated with 2.5 μm heme for 15 min (solid lines). Spectroscopic scans of NEAT1 (A), NEAT2 (B), NEAT3 (C), NEAT4 (D), and NEAT5 (E) are shown and compared with the heme-only control reactions (2.5 μm; dotted lines).
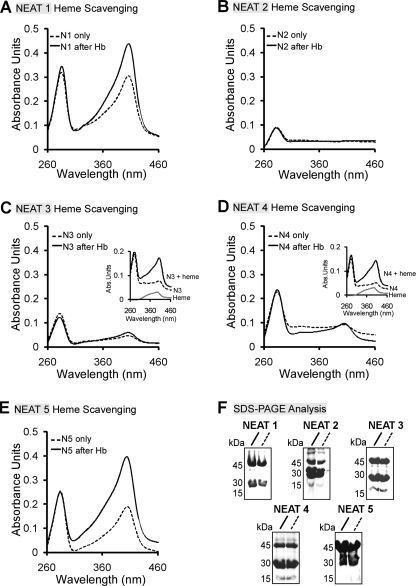
FIGURE 4.
Heme scavenging from Hb by the IsdX2 NEAT domains. A–E, NEAT domain 1 (12 μm; A), 2 (16.5 μm; B), 3 (15 μm; C), 4 (13.5 μm; D), or 5 (15 μm; E) was coupled to glutathione-Sepharose, and the complexes were incubated with holo-Hb (2.5 μm monomer). NEAT proteins complexed to the resin were next separated from Hb, the resin was washed, GST-NEATs were eluted, and each eluate (solid lines) was analyzed by comparing its Soret intensity with an equivalent amount of GST-NEAT protein incubated with Tris-HCl buffer instead of Hb (dotted lines; compare solid with dotted lines at 400 nm). Quantitation of the molar concentration of heme in each reaction after elution yielded the following values: NEAT1 + buffer/+Hb = 0.7 μm/2.5 μm; NEAT3 + buffer/+Hb = 0.16 μm/0.28 μm; NEAT4 + buffer/+Hb = 0.17 μm/0.22 μm; and NEAT5 + buffer/+Hb = 0.83/2.6 μm. Insets C and D, the ability of NEAT3 and -4 to bind heme was tested by incubating the NEAT-only elutions from the heme scavenging experiments with 2.5 μm heme. F, Coomassie. Eluates (10 μl) from each GST-NEAT elution were analyzed by SDS-PAGE. GST-NEATn migrates at ∼42 kDa. The bands at ∼26 and 14 kDa for each GST-NEATn elution represent free GST and NEAT, respectively, due to some proteolysis during the purification and/or experiment. Each result is a single representation of three independent experiments.
Measurement of Heme Binding
For a qualitative measure of heme binding, E. coli cells expressing GST-NEATn were grown in 1.5 liters of LB for 3 h, followed by overnight induction at 30 °C. Each protein was then purified as described above and scanned from 250 to 650 nm. For a more quantitative measure of heme binding, each apo-NEATn (4.5 μm) was incubated in Tris-HCl, pH 7, with 2.5 μm hemin (oxidized heme) for 15 min at 25 °C, and the UV-visible spectrum was measured (250–650 nm) using a DU800 spectrophotometer (Beckman-Coulter, London, UK). The absorbance at ∼400 nm (Soret band) was recorded for each sample as well as a hemin-only control.
Heme Scavenging from Hemoglobin
GST-NEATn (NEAT1, 12 μm; NEAT2, 16.5 μm; NEAT3, 15 μm; NEAT4, 13.5 μm; NEAT5, 15 μm) were expressed for 2 h, purified from E. coli as stated above, immobilized on 2 ml of glutathione-Sepharose resin, and washed with 30 ml of Tris-HCl (50 mm; pH 7.0). The resin-NEAT complex was incubated with 1 ml (2.5 μm monomer) of bovine Hb (Sigma) at 25 °C. After 30 min, the supernatant (Hb) was removed, and the resin-NEAT was washed with 25 ml of buffer. Next, each GST-tagged NEATn was eluted with 0.8 ml of 25 mm glutathione, and its relative heme content was determined by Soret spectroscopy and compared with an equivalent reaction in which GST-NEATn was incubated with buffer (50 mm Tris-HCl, pH 7.0) instead of Hb. Additionally, all elutions were subjected to the pyridine hemochrome assay to determine molar concentrations of heme, as described previously (34). SDS-PAGE was performed on each sample to verify that any differences in the Soret absorbance were not due to differences in NEAT protein amounts or carryover of Hb into NEAT elutions. To test whether NEAT3 and -4 were still capable of binding heme, the elutions that were incubated with the buffer control were mixed with heme (2.5 μm), and the absorbance was measured from 250 to 650 nm.
Association with Hemoglobin
A BIAcore 3000 biosensor (Amersham Biosciences) was used to measure the interaction of each NEAT with Hb. Briefly, holo- or apo-Hb (in 50 mm Tris-HCl, pH 7.0) was covalently coupled to a CM5 sensor chip at 25 °C to a density of 3600 (holo-Hb) and 8000 (apo-Hb) response units (RUs) using amine chemistry as described previously (35, 36). NEAT1 (1–20 μm), NEAT2 (1–15 μm), NEAT3 (1–17 μm), NEAT4 (1–14 μm), or NEAT5 (1–12 μm) in HBS-N buffer (0.01 m HEPES, 0.15 m NaCl, pH 7.4) was injected at 20 μl/min for 300 s at 25 °C. Data were obtained for each reaction using “double referencing,” where parallel injections of analyte are flowed over a control surface and then over immobilized Hb. The kinetics and affinity constants were calculated using BIAevaluation 4.1 software (Amersham Biosciences), and the data fit as described (37). All fits for this kinetic analysis had a χ2 value of <2. S.D. values for each NEAT protein were calculated from three different concentrations of NEAT injected from three independent experiments (n = 9).
Heme Transfer from IsdX2 NEAT Domains to IsdC
GST-IsdC was purified using affinity chromatography as described above, eluted with glutathione, and dialyzed in two applications of 50 mm Tris-HCl, pH 7.0. NEAT1, -3, -4, and -5 were purified from E. coli as described previously, loaded with heme while on the resin by running 20 ml of 3 μm heme over the column, followed by one 25-ml wash with Tris-HCl, and then cleaved with thrombin. Examination of the Soret absorbance for each preparation indicated that each NEAT was equivalently heme-loaded (data not shown). GST-IsdC (7 μm) was immobilized on 250 μl of glutathione-Sepharose beads, and resin was washed three times in 1 ml of Tris-HCl buffer and incubated for 1 h at room temperature with NEAT1 (12 μm), NEAT3 (14 μm), NEAT4 (6 μm), or NEAT5 (7.5 μm). Resin was sedimented by centrifugation, and supernatants containing NEATn were removed. After washing, eluates containing GST-IsdC were collected by eluting with 125 μl of 25 mm glutathione. Each sample was next analyzed from 250 to 650 nm and compared with the spectrum for a GST-IsdC-only control. Heme occupancy measurements were calculated for each GST-IsdC reaction by dividing the Soret band maxima absorbance unit (∼400 nm) for each IsdC elution by the 280 nm absorbance unit for the same sample. Because the extinction coefficients for GST-IsdC would be identical for all eluates, this ratio is a valid measure of the relative heme content in a particular sample. All elutions were also examined by SDS-PAGE.
Heme Dissociation
Holo-NEAT1, -3, -4, and -5 were generated as described for the IsdC transfer studies. NEAT preparations (NEAT1 at 1.1 μm, NEAT3 at 0.93 μm, NEAT4 at 1.5 μm, and NEAT5 at 0.87 μm) were incubated with 47 μm apomyoglobin (Mb) in PBS, pH 7.4, at room temperature for 20 h (30, 38). A Cary spectrophotometer (Agilent) or HP spectrophotometer (Aligent) was used to measure spectra from 250–700 nm as a function of time. Changes in absorbance at 382 nm (control) minus that at 408 nm (Mb-specific) were plotted against time to generate heme dissociation curves.
RESULTS
In Silico Analysis of IsdX2 and Protein Purification
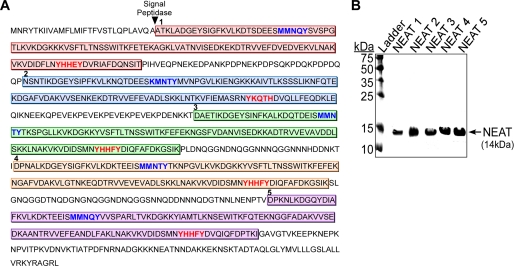
Most Gram-positive pathogenic bacteria harbor genes encoding for NEAT domain proteins, and several of these proteins contain more than one NEAT (16, 17, 39–44). This finding raises interesting questions concerning the functional role of multi-NEAT proteins in heme uptake. A bioinformatics analysis of IsdX2 from B. anthracis identified five non-identical NEAT domains, each ∼125 amino acids in length, within 885 total residues (Fig. 1A). A comparison of each NEAT indicates that four of the five NEATs contain a YXXXY sequence within the proposed heme-binding pocket (8, 16, 45–47) (Fig. 1A, red text, and supplemental Fig. S1). The most obvious difference is the substitution of a histidine for the second tyrosine in IsdX2 NEAT domain 2 (Fig. 1A, red text), suggesting that this domain may function differently than the other four domains, an idea that we have confirmed experimentally in succeeding sections. Other than this change, sequence data alone are insufficient in predicting the function of these five NEAT domains.
FIGURE 1.
In silico analysis and purification of IsdX2 NEAT domains. A, the full 885-amino acid sequence of IsdX2 is shown. The lip region (blue text) and the YXXXY signature sequence within the β8-sheet (red text) are indicated for each color-coded NEAT domain. The arrowhead indicates the predicted site of cleavage by signal peptidase. B, each NEAT protein was expressed in E. coli and purified by GST affinity chromatography. Approximately 5 μg of each thrombin-cleaved preparation was applied to SDS-PAGE and analyzed by Coomassie stain. All NEAT domains were obtained in a pure form at the correct predicted size (14 kDa).
To begin to understand the functional role of IsdX2, we took a reductionist approach and analyzed each individual NEAT domain separately. This strategy simplifies the assignment of a function to a single domain, avoids the use of full-length IsdX2 (which is difficult to purify because of its susceptibility to proteolysis), and is consistent with existing literature that indicates individual NEAT domains form functional units (41, 42, 46–50). B. anthracis genomic DNA encoding each NEAT was amplified by PCR and cloned into pGEX2TK to create a fusion to GST (15, 16, 30, 41, 42, 45, 46, 49, 51–53). Recombinant GST-NEATn protein was expressed in E. coli and purified by affinity chromatography, and each NEAT domain was released from GST by thrombin proteolysis. SDS-PAGE analysis of each preparation (Fig. 1B) indicated a single polypeptide of ∼14 kDa (theoretical mass of NEAT1, 14 kDa; NEAT2, 14.2 kDa; NEAT3, 14.1 kDa; NEAT4, 14.1 kDa; NEAT5, 14.1 kDa), indicating that each NEAT domain migrated at the expected theoretical mass.
Heme Binding by the IsdX2 NEAT Domains
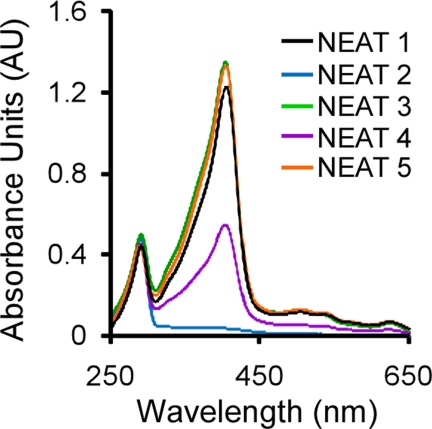
To analyze the heme binding function of each NEAT, we first sought to determine if recombinant NEATs expressed for 16 h could bind endogenous heme produced by E. coli. Each NEAT protein was purified as described under “Experimental Procedures” and assayed for bound heme by measuring the Soret absorbance at ∼400 nm, a well documented spectroscopic assay used to detect the presence of iron-porphyrin (12, 16, 41, 43, 50–52, 54, 55). As indicated by the spectral scans from 250 to 650 nm shown in Fig. 2, NEAT domains 1, 3, 4, and 5 demonstrated intense Soret signals, suggesting co-purification with iron-porphyrin. In contrast, purified NEAT2 (Fig. 2, blue) shows no absorbance at 400 nm, suggesting that it is unable to bind heme.
FIGURE 2.
Analysis of the purified IsdX2 NEAT domains. Each recombinant NEAT domain was purified as described under “Experimental Procedures.” The resulting recombinant proteins (∼15 μm) were analyzed for the presence of a Soret band at 400 nm. Specific Soret wavelengths were as follows: 405 nm for NEAT1 (black); 404 nm for NEAT3 (green) and NEAT5 (orange); 403 nm for NEAT4 (purple). NEAT2 does not absorb light at the Soret wavelength, suggesting that it cannot bind heme (blue).
To confirm the heme-binding activity of these NEATs, heme was removed from each preparation, and apoproteins were incubated with hemin, followed by measurement of the Soret spectra (Fig. 3, A–E, compare solid with dotted lines). When 2.5 μm heme is incubated with NEAT domains 1, 3, 4, or 5, the Soret band becomes red-shifted and narrows (compared with the heme-only control reaction), a property that is characteristic of proteins that specifically bind heme (8, 15). These data show that NEAT domains 1, 3, 4, and 5 of IsdX2 bind heme, whereas NEAT domain 2 binds heme very poorly if at all.
Heme Scavenging by IsdX2 NEAT Domains
To determine if any of the NEAT domains of IsdX2 can acquire heme from hemoglobin, each GST-tagged NEAT domain was immobilized on glutathione-Sepharose resin, the entire complex was incubated with or without bovine holo-Hb for 30 min, and eluted GST-NEATn fractions were analyzed for heme by Soret spectroscopy and quantitated by the pyridine hemochrome assay. For NEAT domains 1 and 5, increases in the Soret peak intensities were observed after incubation with Hb (Fig. 4, A and E; compare solid with dotted lines at 400 nm). Indeed, calculation of the molar heme amount (values reported in the legend to Fig. 4) for each sample suggested that NEAT1 and -5 had more heme relative to the amount of heme originally present on each NEAT (3.6- and 3.1-fold increase, respectively). In contrast, no or little change in Soret absorbance was observed for NEAT domains 3 and 4 (Fig. 4, C and D), and little change was observed in the heme content before and after the experiment (1.7- and 1.3-fold, respectively). Upon incubation of NEAT2 with Hb, no Soret absorbance was observed (Fig. 4B), most likely because this NEAT cannot bind heme (Figs. 2 (blue) and 3B). These differences cannot be accounted for by different amounts of GST-NEAT in each preparation (Fig. 4F, SDS-PAGE of each elution with GST-NEAT at 45 kDa, GST at 26 kDa, or NEAT at 14 kDa).
To rule out the possibility that the reason for the lack of heme scavenging by NEAT3 and -4 was due to non-functioning protein preparations, we performed an additional experiment: the elutions for NEAT3 and -4 were subjected to a separate heme binding analysis. When free heme (2.5 μm) was added to these samples, an increase and red shift of the Soret band was observed when compared with a heme-only control reaction (Fig. 4, C and D, inset, spectral scans). These data suggest that the NEATs (NEAT3 and -4) in these samples can readily bind heme, suggesting that their inability to scavenge heme is not due to any residual bound heme carried over from the purification. Instead, these results suggest that there are functional differences between the NEAT domains in IsdX2, with NEAT1 and -5 having the capacity to extract heme from Hb but NEAT2, -3, and -4 lacking this ability.
The Association of the IsdX2 NEAT Domains with Hemoglobin
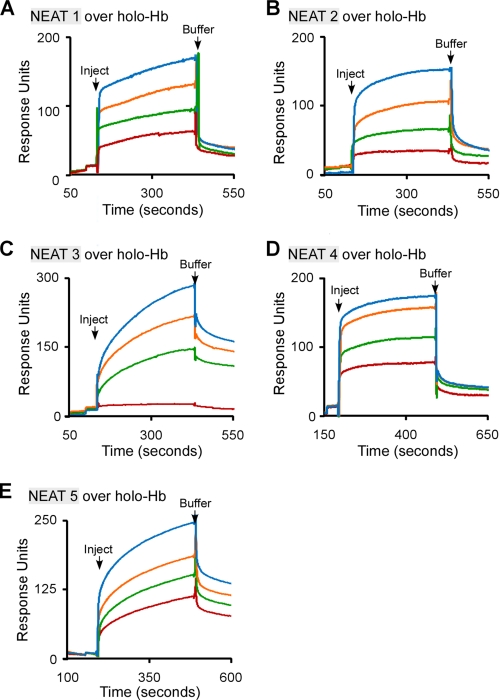
Although the study of bacterial hemophores has increased in recent years, very little is known about the mechanisms by which heme is taken from Hb (56, 57). For example, the most well studied hemophore, HasA, from the Gram-negative pathogen Serratia marcescens, seems to acquire heme from Hb passively (i.e. without a physical interaction) (58–60). To understand how the NEATs of IsdX2 acquire heme from Hb, we tested each NEAT domain to see if it associates with holo-Hb, using surface plasmon resonance spectroscopy. Holo-Hb was covalently coupled to a carboxymethyl chip using amine chemistry. Increasing amounts of each recombinant NEAT were then infused over holo-Hb, and RUs were measured as reported previously (37).
As indicated in Fig. 5, A–E, a dose-dependent increase in RUs was seen for all recombinant NEAT proteins infused, suggesting a physical interaction with holo-Hb. NEAT1 and -5, the IsdX2 domains that scavenge heme from Hb (Fig. 4, A and E), associated with holo-Hb with apparent affinities (reported in the legend to Fig. 5) that were ∼75- and 4-fold greater than the non-scavenging NEAT2, -3, and -4. Little or no response (association) was observed when the experiments were repeated using the highest concentration of each NEAT infused over apo-Hb, indicating that these interactions are dependent on Hb being in the heme-bound state (supplemental Fig. S2). Close examination of the response curves in Fig. 5 suggests that the interaction with holo-Hb may be biphasic, which may suggest that there are two binding sites on Hb (one with high affinity and one with low affinity). Given the complex nature of mammalian hemoglobin (the protein exists as a dimer of α and β subunits, both of whose confirmation and affinity for heme are different (61)), the biphasic nature may suggest that there is preference for one subunit. A test of this hypothesis will require additional studies. Regardless, the complete association and dissociation phases are used to calculate the KD, meaning the reported values represent a “composite” affinity of both phases. In all cases, the χ2 values (how well the models fit the actual data) were low (<2) and rigorous (see “Experimental Procedures”), indicating the fits correctly model the experimental sensorgrams.
FIGURE 5.
The association of the IsdX2 NEAT domains with holo-Hb. A–E, recombinant NEAT1 (1–20 μm; A), NEAT2 (1–15 μm; B), NEAT3 (1–17 μm; C), NEAT4 (1–14 μm; D), or NEAT5 (1–12 μm; E) was injected at a constant flow rate of 20 μl/min over 3600 RU of immobilized holo-Hb. Association and dissociation phases were monitored for 300 s by observing changes in the RUs with time. The dissociation constants (KD) were calculated assuming a 1:1 binding model (see “Experimental Procedures”) and represent the mean and S.D. values of at least three different concentrations from three independent determinations. KD was as follows: NEAT1 = 4.1 ± 1.4 × 10−8 m; NEAT2 = 4.4 ± 1.7 × 10−6 m; NEAT3 = 4.6 ± 0.8 × 10−6 m; NEAT4 = 1.8 ± 0.1 × 10−6 m; and NEAT5 = 7.5 ± 3 × 10−7. All sensorgrams analyzed had a χ2 value of <2. The S.D. values for all values were calculated from the average of two concentrations for each NEAT/IsdX2 sensorgram, from three independent experiments with multiple preparations of the same protein (n = 9).
Heme Transfer from IsdX2 to IsdC
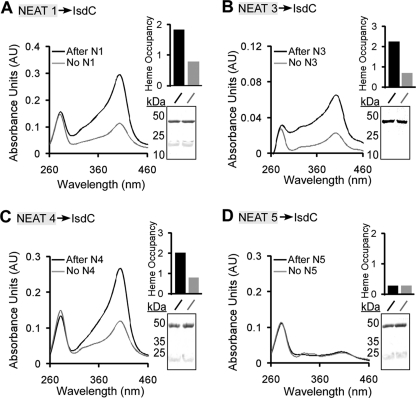
To determine if IsdX2 can transfer its heme to the B. anthracis cell wall-anchored protein IsdC, recombinant GST-IsdC was immobilized on glutathione-Sepharose resin, the entire complex was incubated with or without holo-NEATn, and eluted GST-IsdC was analyzed for bound heme by absorption spectroscopy. As indicated in Fig. 6, the Soret band for GST-IsdC increased after incubation with NEAT domains 1, 3, and 4 (Fig. 6, A–C). Quantitation of the data by calculating the ratio of the Soret absorbance (A400 nm) to the protein absorbance (A280 nm) (a valid method when the extinction coefficients are identical for GST-IsdC eluates in this analysis) indicates that the increase in Soret is due to heme transfer and not differences in protein (Fig. 6, A–D, insets). However, no significant increase in Soret absorbance was observed when IsdC was incubated with NEAT domain 5 (Fig. 6D), despite this preparation being heme-loaded (supplemental Fig. S3). Additionally, the increase in the Soret absorbance for IsdC incubated with NEAT1, -3, or -4 could not be accounted for by different amounts of analyzed GST-IsdC or contamination with free NEAT (Fig. 6, A–D, insets showing an SDS-PAGE analysis of each IsdC elution with GST-IsdC at 45 kDa and cleaved IsdC at 22 kDa). Collectively, the data suggest that NEAT domains 1, 3, and 4 transfer heme to IsdC.
FIGURE 6.
Heme transfer from the IsdX2 domains to IsdC. A–D, holo-NEAT domain 1 (12 μm; A), 3 (14 μm; B), 4 (6 μm; C), or 5 (7.5 μm; D) was incubated with GST-IsdC (7 μm) that was coupled to glutathione-Sepharose resin, and complexes were washed, GST-IsdC was eluted, and eluate was analyzed by Soret spectroscopy (Fig. 6, A–D, compare black with gray lines at 400 nm). The relative heme occupancy represents the ratio of the absorbance at 400 nm to that at 280 nm (black bars, IsdC after incubation with a NEAT domain; gray bars, IsdC after incubation with a buffer control). Insets A–D, the reactions (10 μl) from each GST-IsdC elution were analyzed by SDS-PAGE. GST-IsdC migrates at ∼45 kDa, and holo-NEATs migrate at 14 kDa. The bands at 22 kDa represent free IsdC due to some proteolysis during the experiment. Each result is a single representation of three independent experiments.
Heme Dissociation from the IsdX2 NEAT Domains
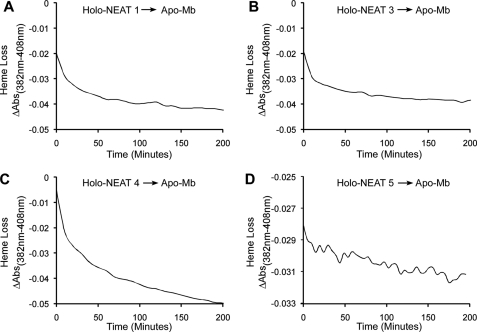
Plotting absorbance changes after titration of apoprotein with free hemin is complicated by hemin dimerization and the micromolar amounts of apoprotein and hemin needed for the measurement. Thus, affinity calculations made from such data are almost always an underestimate of the true affinity. However, estimates of heme affinity can be made by measuring the rate of heme dissociation from heme-binding proteins (38). To determine the rates of heme loss from each heme-binding IsdX2 NEAT domain, holo-NEAT1, -3, -4, and -5 were incubated with the heme-scavenging reagent apo-Mb, which contains H64Y and V68F mutations to amplify spectral changes and maintain an exceptionally low KD for heme (38). This high affinity apo-Mb promotes unidirectional flow from the heme donor to the scavenging reagent, and the observed time courses provide a measure of the rate and extent of heme dissociation from each NEAT (30).
As indicated in Fig. 7, A–C, NEAT1 and -3 lost 100% of the heme initially loaded on the protein within 200 min, and NEAT4 lost 100% of the heme within 300 min. However, only 35% of the heme was lost from NEAT5 (Fig. 7D) after 20 h of incubation with apo-Mb. Table 1 indicates the relative dissociation rates calculated for each NEAT domain, with two phases detected for each. The cause of the very slow phases could be slow hemin loss from higher affinity conformations. These data suggest that NEAT5 has the highest affinity for heme (∼10−13 m, the KD for apo-Mb (38, 62)), whereas NEAT1, -3, and -4 are probably less than this value. Thus, the lack of heme transfer from NEAT5 to IsdC is probably due to the higher affinity of NEAT5, suggesting that transfer to IsdC by IsdX2 is affinity-mediated and determined by the off rates of thermal heme dissociation from each NEAT domain.
FIGURE 7.
Dissociation of heme from IsdX2 NEAT domains. A–D, holo-NEAT1 (1.17 μm; A), holo-NEAT3 (0.93 μm; B), holo-NEAT4 (1.5 μm; C), or holo-NEAT5 (0.87 μm; D) was mixed with 47 μm apo-Mb for 20 h. Spectral changes at 408 nm, relative to a control wavelength at 382 nm, were measured using a conventional spectrophotometer. The rate constants derived from these experiments are reported in Table 1.
TABLE 1.
Rates of heme dissociation from IsdX2 NEAT domains
Conventional spectroscopy was used to measure heme dissociation from IsdX2 NEAT domains, as described under “Experimental Procedures.” Each NEAT had two phases of dissociation kinetics, an initial rapid phase followed by a slower rate of heme loss. All kd values were calculated as described previously (30). NA, not applicable.
| Protein | kd, phase 1 | kd, phase 2 | Heme transfer, t = 20 h |
|---|---|---|---|
| min−1 | min−1 | ||
| NEAT1 | 0.07 | 0.005 | Complete |
| NEAT3 | 0.07 | 0.005 | Complete |
| NEAT4 | 0.04 | 0.003 | Complete |
| NEAT5 | NA | 0.0005 | 35% transferred |
DISCUSSION
Examination of the function of each NEAT domain of IsdX2 indicates the following: (i) NEAT1, -3, -4, and -5 bind heme, with NEAT5 having the highest affinity; (ii) NEAT1 and -5 readily extract heme from Hb; (iii) all of the NEATs appear to associate with Hb, but the two heme-scavenging NEATs (NEAT1 and -5) interact with the highest apparent affinity; and (iv) NEAT1, -3, and -4 transfer heme to a downstream NEAT protein, IsdC (summarized in Table 2). These data, in combination with our previous report demonstrating that IsdX1 can transfer heme to full-length IsdX2 (30), suggest that IsdX2 is a multifunctional secreted protein that harbors all of the heme acquisition activities often associated with multiple protein systems, as is observed for the cell wall-anchored proteins of Staphylococcus aureus (10, 14). In essence, IsdX2 may act as a “heme sponge,” effectively soaking up heme from multiple sources and increasing the localized concentration of heme around the bacillus for eventual delivery to the cell surface.
TABLE 2.
A summary of the biological function of each IsdX2 NEAT domain
NA, not applicable.
| Protein | Lip region | β-Hairpin (YXXXY) | Heme binding | Heme scavenging | Heme transfer to IsdC |
|---|---|---|---|---|---|
| NEAT1 | MMNQY | YHHEY | Yes | Yes | Yes |
| NEAT2 | KMNTY | YKQTH | No | No | NA |
| NEAT3 | MMNTY | YHHFY | Yes | No | Yes |
| NEAT4 | MMNTY | YHHFY | Yes | No | Yes |
| NEAT5 | MMNQY | YHHFY | Yes | Yes | No |
A growing body of evidence indicates that NEAT domains are major mediators of heme uptake in Gram-positive pathogenic bacteria. In some cases, genetic deletion of one or more NEAT proteins reduces virulence, presumably by diminishing the ability of the bacterium to acquire iron and replicate during infection (13, 15–17). More recent studies highlight the potential utilization of NEAT proteins in vaccine assembly. For example, the vaccination of mice with antibodies raised against two recombinant NEAT proteins, IsdA and IsdB from S. aureus, protected mice against abscess formation and a lethal challenge of Staphylococci (19). This suggests that individual NEAT domains can serve as immunogens for induction of neutralizing antibodies, and protection is probably due to inhibition of NEAT function by the antibody binding to the heme binding structure. Interestingly, IsdX2 was one of the top five most robust inducers of antibody production in a serum screen of potential anthrax immunogens (63). Such studies highlight the importance of understanding the mechanism of action of NEAT proteins for both vaccine and anti-infective development.
Along these lines, the crystal structure of the NEAT protein IsdC revealed two regions that may mediate the properties of heme binding, extraction, and transfer (46–49). The first is a β-hairpin structure that often contains a YXXXY heme-binding signature on the eighth β-sheet. The second region, the flexible 310-helical lip, extends over the distal side of the heme and may act as a “lock” that facilitates heme scavenging and transfer. It is likely that attempts to improve recombinant NEAT vaccines or develop small molecule inhibitors of NEAT activity will revolve around an understanding of the mechanistic properties of these two regions.
An alignment of each IsdX2 NEAT domain with NEAT domains of B. anthracis and S. aureus revealed a correlation with amino acid sequence and NEAT function (supplemental Fig. S1). First, with respect to the β-hairpin region, the first tyrosine of the YXXXY signature sequence is thought to be the fifth axial ligand and coordinates the heme iron, a result confirmed in the crystal structures of IsdC and IsdA and supported by mutagenesis data (42, 46, 48). The second tyrosine probably stabilizes this linkage by hydrogen bonding to the first. Interestingly, IsdX2 NEAT domain 2, which has a histidine in place of the second tyrosine (supplemental Fig. S1, arrowhead indicates the position of this residue), does not bind heme (Figs. 2 (blue) and 3B), consistent with the proposed role of the second Tyr side chain in stabilization of the proximal iron ligand (first Tyr). A similar observation has been made for the first NEAT domain of IsdB and the first two NEAT domains of IsdH, both of which also lack the second tyrosine and instead act as hemoprotein receptors (8, 12, 17, 40, 42, 50, 64). However, unlike these NEAT domains, NEAT domain 2 of IsdX2 binds Hb poorly, and its rather rapid rate of Hb dissociation would seemingly preclude it from serving as a “receptor” for host Hb (Fig. 5B). However, we cannot rule out the possibility that NEAT2 may serve a structural role, perhaps positioning the other NEATs to efficiently interact with Hb for heme extraction or heme transfer to IsdC. Future studies will be geared toward testing these hypotheses.
There are also differences in the lip region that may correlate with the heme scavenging function of each IsdX2 NEAT domain (supplemental Fig. S1, underlined residues). For example, NEAT1 and -5, which both actively scavenge heme from Hb, contain the amino acid sequence MMNQY, whereas the heme-binding NEATs that do not scavenge heme from Hb (NEAT3 and -4) contain an MMNTY. Whether or not the ability to extract heme from Hb depends on a glutamine in the fourth position remains to be determined; however, it should be noted that IsdX1, which can also scavenge heme from Hb, harbors an arginine at this position, suggesting that heme scavenging functions may rely on specific side chains and are probably dependent on more than one residue. These results also suggest that the mechanism of heme binding may be separate from that of heme scavenging from Hb.
One of the interesting questions that arises from these studies is why a single bacterial hemophore would evolve to contain multiple, functionally distinct domains. One possibility is to accelerate heme acquisition and import for the pathogen. A single protein that can receive and transfer heme to the cell surface is expected to enhance bacterial replication by catalytically promoting the import of iron. Although this study focused on the properties of each individual NEAT domain, it is possible that the NEATs may act together in the context of the full-length protein.
The data for heme transfer to IsdC indicates that there are important inherent differences between the IsdX2 NEAT domains with respect to heme uptake by the bacterium. NEAT5 is inefficient at donating heme to IsdC, in contrast to NEAT1, -3, and -4. This is probably due to the low heme dissociation rates (higher affinity) observed for NEAT5, suggesting that IsdX2-mediated heme transfer to IsdC may be a passive event that is dependent on the thermal rate of heme dissociation from the NEAT domains instead of being governed by protein-protein interactions. This passive transfer of heme from IsdX2 to IsdC is in contrast to that seen for transfer from IsdX1 and BslK, both of which have slower rates of heme loss and actively transfer heme to IsdC via direct protein-protein interactions (30, 51).
Although IsdX2 clearly transfers heme to IsdC, we cannot rule out the possibility that other B. anthracis NEAT proteins or as yet unidentified proteins can receive heme from NEAT5 or the full-length IsdX2 protein. Such candidate proteins include the recently identified BslK, an interesting NEAT protein with three S-layer homology domains, or BAS0520, a NEAT-containing protein that is up-regulated in low iron environments and important for virulence in an inhalational model of anthrax disease (51, 65). How the seemingly multifunctional role of IsdX2 in heme import relates to the potentially redundant role of these other cell-associated NEAT proteins remains to be determined, and studies are currently ongoing to answer these questions.
This study allows us to expand the current model of heme acquisition in B. anthracis. Upon infection and exposure to Hb, extracellular IsdX2, through NEAT1 and -5, associates with holo-Hb, leading to active heme capture. Additionally, any unoccupied NEATs (NEAT3 and -4) can bind free heme released in the vicinity, allowing four NEAT domains to essentially “soak up” large amounts of heme. Spontaneous heme dissociation from IsdX2 NEAT domains 1, 3, and 4 allows heme transfer to cell surface-anchored IsdC, presumably by a passive mechanism. IsdC can then rapidly transfer this heme to the membrane ABC-like transporter IsdEFD, which subsequently pumps heme into the cytosol for degradation by the heme monooxygenase IsdG, thereby liberating the iron. By scavenging heme from Hb, accepting heme from IsdX1, and possessing a high rate of heme dissociation, IsdX2 increases the useable heme concentration around the cell surface and may accelerate the rate of heme-iron uptake, which in turn would correlate with increases in growth and replication of B. anthracis under conditions of iron starvation.
Supplementary Material
Acknowledgments
We thank Miriam Balderas and Chris Nobles for reading the manuscript and providing suggestions.
This work was supported, in whole or in part, by National Institutes of Health Grants AI069697 (to A. W. M.), GM035649 and HL047020 (to J. S. O.), and GM84348 (to M. F.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S4.
- NEAT
- near-iron transporter
- Mb
- myoglobin
- RU
- response unit.
REFERENCES
- 1. Crosa J. H., Mey A. R., Payne S. M. (2004) Iron Transport in Bacteria, American Society of Microbiology Press, Washington, D. C [Google Scholar]
- 2. Heinemann I. U., Jahn M., Jahn D. (2008) Arch. Biochem. Biophys. 474, 238–251 [DOI] [PubMed] [Google Scholar]
- 3. De Domenico I., McVey Ward D., Kaplan J. (2008) Nat. Rev. Mol. Cell Biol. 9, 72–81 [DOI] [PubMed] [Google Scholar]
- 4. Wandersman C., Delepelaire P. (2004) Annu. Rev. Microbiol. 58, 611–647 [DOI] [PubMed] [Google Scholar]
- 5. Cescau S., Cwerman H., Létoffé S., Delepelaire P., Wandersman C., Biville F. (2007) Biometals 20, 603–613 [DOI] [PubMed] [Google Scholar]
- 6. Faraldo-Gómez J. D., Sansom M. S. (2003) Nat. Rev. Mol. Cell Biol. 4, 105–116 [DOI] [PubMed] [Google Scholar]
- 7. Andrade M. A., Ciccarelli F. D., Perez-Iratxeta C., Bork P. (2002) Genome Biol. 3, RESEARCH0047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mazmanian S. K., Skaar E. P., Gaspar A. H., Humayun M., Gornicki P., Jelenska J., Joachmiak A., Missiakas D. M., Schneewind O. (2003) Science 299, 906–909 [DOI] [PubMed] [Google Scholar]
- 9. Daou N., Buisson C., Gohar M., Vidic J., Bierne H., Kallassy M., Lereclus D., Nielsen-LeRoux C. (2009) PLoS Pathog. 5, e1000675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Grigg J. C., Ukpabi G., Gaudin C. F., Murphy M. E. (2010) J. Inorg. Biochem. 104, 341–348 [DOI] [PubMed] [Google Scholar]
- 11. Zhu H., Xie G., Liu M., Olson J. S., Fabian M., Dooley D. M., Lei B. (2008) J. Biol. Chem. 283, 18450–18460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Muryoi N., Tiedemann M. T., Pluym M., Cheung J., Heinrichs D. E., Stillman M. J. (2008) J. Biol. Chem. 283, 28125–28136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gat O., Zaide G., Inbar I., Grosfeld H., Chitlaru T., Levy H., Shafferman A. (2008) Mol. Microbiol. 70, 983–999 [DOI] [PubMed] [Google Scholar]
- 14. Honsa E. S., Maresso A. W. (2011) Biometals 3, 533–545 [DOI] [PubMed] [Google Scholar]
- 15. Maresso A. W., Chapa T. J., Schneewind O. (2006) J. Bacteriol. 188, 8145–8152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Maresso A. W., Garufi G., Schneewind O. (2008) PLoS Pathog. 4, e1000132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Torres V. J., Pishchany G., Humayun M., Schneewind O., Skaar E. P. (2006) J. Bacteriol. 188, 8421–8429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stranger-Jones Y. K., Bae T., Schneewind O. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 16942–16947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim H. K., DeDent A., Cheng A. G., McAdow M., Bagnoli F., Missiakas D. M., Schneewind O. (2010) Vaccine 28, 6382–6392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kuklin N. A., Clark D. J., Secore S., Cook J., Cope L. D., McNeely T., Noble L., Brown M. J., Zorman J. K., Wang X. M., Pancari G., Fan H., Isett K., Burgess B., Bryan J., Brownlow M., George H., Meinz M., Liddell M. E., Kelly R., Schultz L., Montgomery D., Onishi J., Losada M., Martin M., Ebert T., Tan C. Y., Schofield T. L., Nagy E., Meineke A., Joyce J. G., Kurtz M. B., Caulfield M. J., Jansen K. U., McClements W., Anderson A. S. (2006) Infect. Immun. 74, 2215–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ebert T., Smith S., Pancari G., Clark D., Hampton R., Secore S., Towne V., Fan H., Wang X. M., Wu X., Ernst R., Harvey B. R., Finnefrock A. C., Wang F., Tan C., Durr E., Cope L., Anderson A., An Z., McNeely T. (2010) Hum. Antibodies 19, 113–128 [DOI] [PubMed] [Google Scholar]
- 22. Inglesby T. V., O'Toole T., Henderson D. A., Bartlett J. G., Ascher M. S., Eitzen E., Friedlander A. M., Gerberding J., Hauer J., Hughes J., McDade J., Osterholm M. T., Parker G., Perl T. M., Russell P. K., Tonat K. (2002) JAMA 287, 2236–2252 [DOI] [PubMed] [Google Scholar]
- 23. Mock M., Fouet A. (2001) Annu. Rev. Microbiol. 55, 647–671 [DOI] [PubMed] [Google Scholar]
- 24. Ross J. M. (1955) Br. J. Exp. Pathol. 36, 336–339 [PMC free article] [PubMed] [Google Scholar]
- 25. Abramova A. A., Grinberg L. M. (1993) Arkh. Patol. 55, 18–23 [PubMed] [Google Scholar]
- 26. Klichko V. I., Miller J., Wu A., Popov S. G., Alibek K. (2003) Biochem. Biophys. Res. Commun. 303, 855–862 [DOI] [PubMed] [Google Scholar]
- 27. Dixon T. C., Fadl A. A., Koehler T. M., Swanson J. A., Hanna P. C. (2000) Cell Microbiol. 2, 453–463 [DOI] [PubMed] [Google Scholar]
- 28. Shannon J. G., Ross C. L., Koehler T. M., Rest R. F. (2003) Infect. Immun. 71, 3183–3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Guidi-Rontani C., Weber-Levy M., Labruyère E., Mock M. (1999) Mol. Microbiol. 31, 9–17 [DOI] [PubMed] [Google Scholar]
- 30. Fabian M., Solomaha E., Olson J. S., Maresso A. W. (2009) J. Biol. Chem. 284, 32138–32146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ascoli F., Fanelli M. R., Antonini E. (1981) Methods Enzymol. 76, 72–87 [DOI] [PubMed] [Google Scholar]
- 32. Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 33. Pace C. N., Vajdos F., Fee L., Grimsley G., Gray T. (1995) Protein Sci. 4, 2411–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Berry E. A., Trumpower B. L. (1987) Anal. Biochem. 161, 1–15 [DOI] [PubMed] [Google Scholar]
- 35. Howell S., Kenmore M., Kirkland M., Badley R. A. (1998) J. Mol. Recognit. 11, 200–203 [DOI] [PubMed] [Google Scholar]
- 36. Murphy M., Jason-Moller L., Bruno J. (2006) Curr. Protoc. Protein Sci. Chapter 19, Unit 19.14 [DOI] [PubMed] [Google Scholar]
- 37. Dementieva I. S., Tereshko V., McCrossan Z. A., Solomaha E., Araki D., Xu C., Grigorieff N., Goldstein S. A. (2009) J. Mol. Biol. 387, 175–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hargrove M. S., Singleton E. W., Quillin M. L., Ortiz L. A., Phillips G. N., Jr., Olson J. S., Mathews A. J. (1994) J. Biol. Chem. 269, 4207–4214 [DOI] [PubMed] [Google Scholar]
- 39. Meehan M., Burke F. M., Macken S., Owen P. (2010) Microbiology 156, 1824–1835 [DOI] [PubMed] [Google Scholar]
- 40. Dryla A., Hoffmann B., Gelbmann D., Giefing C., Hanner M., Meinke A., Anderson A. S., Koppensteiner W., Konrat R., von Gabain A., Nagy E. (2007) J. Bacteriol. 189, 254–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pilpa R. M., Fadeev E. A., Villareal V. A., Wong M. L., Phillips M., Clubb R. T. (2006) J. Mol. Biol. 360, 435–447 [DOI] [PubMed] [Google Scholar]
- 42. Pilpa R. M., Robson S. A., Villareal V. A., Wong M. L., Phillips M., Clubb R. T. (2009) J. Biol. Chem. 284, 1166–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ouattara M., Cunha E. B., Li X., Huang Y. S., Dixon D., Eichenbaum Z. (2010) Mol. Microbiol. 78, 739–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhu H., Liu M., Lei B. (2008) BMC Microbiol. 8, 15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Grigg J. C., Vermeiren C. L., Heinrichs D. E., Murphy M. E. (2007) Mol. Microbiol. 63, 139–149 [DOI] [PubMed] [Google Scholar]
- 46. Pluym M., Muryoi N., Heinrichs D. E., Stillman M. J. (2008) J. Inorg Biochem. 102, 480–488 [DOI] [PubMed] [Google Scholar]
- 47. Gaudin C. F., Grigg J. C., Arrieta A. L., Murphy M. E. (2011) Biochemistry 50, 5443–5452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sharp K. H., Schneider S., Cockayne A., Paoli M. (2007) J. Biol. Chem. 282, 10625–10631 [DOI] [PubMed] [Google Scholar]
- 49. Villareal V. A., Pilpa R. M., Robson S. A., Fadeev E. A., Clubb R. T. (2008) J. Biol. Chem. 283, 31591–31600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Watanabe M., Tanaka Y., Suenaga A., Kuroda M., Yao M., Watanabe N., Arisaka F., Ohta T., Tanaka I., Tsumoto K. (2008) J. Biol. Chem. 283, 28649–28659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tarlovsky Y., Fabian M., Solomaha E., Honsa E., Olson J. S., Maresso A. W. (2010) J. Bacteriol. 192, 3503–3511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Vermeiren C. L., Pluym M., Mack J., Heinrichs D. E., Stillman M. J. (2006) Biochemistry 45, 12867–12875 [DOI] [PubMed] [Google Scholar]
- 53. Grigg J. C., Vermeiren C. L., Heinrichs D. E., Murphy M. E. (2007) J. Biol. Chem. 282, 28815–28822 [DOI] [PubMed] [Google Scholar]
- 54. Hillier J., Hoffman J. F. (1953) J. Cell. Physiol. 42, 203–247 [DOI] [PubMed] [Google Scholar]
- 55. Liu M., Tanaka W. N., Zhu H., Xie G., Dooley D. M., Lei B. (2008) J. Biol. Chem. 283, 6668–6676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gao J. L., Nguyen K. A., Hunter N. (2010) J. Biol. Chem. 285, 40028–40038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yukl E. T., Jepkorir G., Alontaga A. Y., Pautsch L., Rodriguez J. C., Rivera M., Moenne-Loccoz P. (2010) Biochemistry 49, 6646–6654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Izadi-Pruneyre N., Huché F., Lukat-Rodgers G. S., Lecroisey A., Gilli R., Rodgers K. R., Wandersman C., Delepelaire P. (2006) J. Biol. Chem. 281, 25541–25550 [DOI] [PubMed] [Google Scholar]
- 59. Létoffé S., Nato F., Goldberg M. E., Wandersman C. (1999) Mol. Microbiol. 33, 546–555 [DOI] [PubMed] [Google Scholar]
- 60. Létoffé S., Ghigo J. M., Wandersman C. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 9876–9880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hargrove M. S., Whitaker T., Olson J. S., Vali R. J., Mathews A. J. (1997) J. Biol. Chem. 272, 17385–17389 [DOI] [PubMed] [Google Scholar]
- 62. Culbertson D. S., Olson J. S. (2010) Biochemistry 49, 6052–6063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chitlaru T., Gat O., Grosfeld H., Inbar I., Gozlan Y., Shafferman A. (2007) Infect. Immun. 75, 2841–2852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Dryla A., Gelbmann D., von Gabain A., Nagy E. (2003) Mol. Microbiol. 49, 37–53 [DOI] [PubMed] [Google Scholar]
- 65. Carlson P. E., Jr., Carr K. A., Janes B. K., Anderson E. C., Hanna P. C. (2009) PLoS One 4, e6988 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.