Abstract
Rod and cone photoreceptor neurons utilize discrete PDE6 enzymes that are crucial for phototransduction. Rod PDE6 is composed of heterodimeric catalytic subunits (αβ), while the catalytic core of cone PDE6 (α′) is a homodimer. It is not known if variations between PDE6 subunits preclude rod PDE6 catalytic subunits from coupling to the cone phototransduction pathway. To study this issue, we generated a cone-dominated mouse model lacking cone PDE6 (Nrl−/− cpfl1). In this animal model, using several independent experimental approaches, we demonstrated the expression of rod PDE6 (αβ) and the absence of cone PDE6 (α′) catalytic subunits. The rod PDE6 enzyme expressed in cone cells is active and contributes to the hydrolysis of cGMP in response to light. In addition, rod PDE6 expressed in cone cells couples to the light signaling pathway to produce S-cone responses. However, S-cone responses and light-dependent cGMP hydrolysis were eliminated when the β-subunit of rod PDE6 was removed (Nrl−/− cpfl1 rd). We conclude that either rod or cone PDE6 can effectively couple to the cone phototransduction pathway to mediate visual signaling. Interestingly, we also found that functional PDE6 is required for trafficking of M-opsin to cone outer segments.
Keywords: Cyclic GMP (cGMP), Phosphodiesterases, Protein Assembly, Protein Targeting, Signal Transduction
Introduction
Vertebrate visual perception is mediated by two types of photoreceptor cells, rods, and cones (1–3). Rods mediate vision in dim light and respond to a single photon. In contrast, cones mediate vision in bright light, are less sensitive than rods to light, respond faster, and adapt to light stimuli over several orders of magnitude (1). Although the transduction mechanism used by both rods and cones to detect and respond to light are similar, the protein components that mediate the visual signaling are distinct (3). The differences between rod and cone responses are likely due to levels and variations in the protein transduction machinery (2, 3). Phototransduction is initiated when photoisomerized rhodopsin or cone opsin activates transducin, which in turn activates light-activated rod phosphodiesterase-6 (PDE6),2 the effector of the cascade (2, 3). PDE6 hydrolyzes cGMP resulting in closure of cation channels and subsequent hyperpolarization of photoreceptor cell membranes (4). No differences in light-dependent signaling between rods and cones are observed at the level of opsin activation (5). In contrast, cell-specific transducin subunits contributes to altered signaling properties between rods and cones (6). However, this finding is contradicted by another study showing that rod and cone transducin are able to substitute for each other efficiently (7).
The last step in the phototransduction cascade, the activation of PDE6 by transducin subunits is different between rods and cones. Apart from transducin, rods and cones contain distinct PDE6 subunits (4). Rod PDE6 exists as a heteromer with two catalytic subunits (αβ) and two inhibitory (γ) subunits. Rod PDE6 is the only member of a vast family of PDE proteins that functions as a catalytic heterodimer. Cone PDE6 is composed of two identical catalytic subunits (α′) with distinct inhibitory subunits (γ′) (8–10). Furthermore, the catalytic subunits of rod PDE6 are differentially lipid modified. The α-subunit is farnesylated whereas the β- subunit is geranylgeranylated (11, 12). The catalytic subunits of cone PDE6 on the other hand are likely geranylgeranylated (8). Within the PDE6 structure, the catalytic domain is most conserved and is enzymatically equivalent among PDE6 subunits (13, 14). The largest differences among PDE6 subunits reside in the regulatory, non-catalytic cGMP binding GAF domains (15, 16). The affinity for cGMP toward non-catalytic GAF domains varies between rod and cone PDE6 with rod PDE6 exhibiting higher affinity toward cGMP (17). The variation in cGMP binding affinity is thought to affect the transducin-dependent activation of PDE6 and subsequent removal of inhibition by γ- subunits of PDE6 (13, 14). However, it is not known if the changes in properties of GAF domains between rod and cone PDE6 dictate their interaction with cell-specific transducin and inhibitory subunits (PDE6 γ) and affect the ability of rod PDE6 to function in cone photoreceptor cells.
In this study, we generated mice with an all cone retina that has no cone PDE6 (Nrl−/− cpfl1) and we used them as a model to study achromatopsia caused by defects in PDE6α′ subunit (18). Our analysis of this model revealed that cone cells in this animal express catalytic subunits of rod PDE6 (αβ). This made it possible for us to use this model to investigate how rod PDE6 couples to the cone phototransduction pathway and to decipher the importance of PDE6 in cones.
EXPERIMENTAL PROCEDURES
Animals
Nrl mutant mice (obtained from Dr. Anand Swaroop) were crossed with cpfl1 mutant mice to generate heterozygous animals. These heterozygotes were then bred to create homozygous Nrl−/− cpfl1 mutant mice. These mice were further crossed with rd1 mutant mice to create Nrl−/− cpfl1 rd/+ mutant animals. After several rounds of breeding rd heterozygotes, Nrl−/− cpfl1 rd mice were identified by genotyping using the primers listed (supplemental Table S1). The conditions used for the polymerase chain reaction (PCR) amplification of Nrl, rd1, and cpfl1 mutant alleles were 95 °C for 1 min followed by 30 cycles of (95 °C, 30 s; 56 °C, 30 s; 72 °C, 30 s). Animals were maintained in complete darkness, or cyclic light conditions, and physiological experiments were performed under dim red illumination using a Kodak number 1 Safelight filter (transmittance > 560 nm). Animals were handled and maintained according to the guidelines established by Institutional Animal Care and Use Committee of the West Virginia University.
RT-PCR
Isolated retinas from enucleated mouse eyes were flash-frozen on dry ice in TRIzol reagent (Invitrogen). Total RNA was isolated as per manufacturer's instructions. Oligo (dT)-primed reverse transcription reactions were performed with 2.5 μg of total RNA by using SuperScript III (Invitrogen) to obtain cDNA, which was then used as a template in PCR. The conditions used for PCR were 95 °C for 2 min followed by 95 °C for 30 s, 58 °C for 45 s, and 72 °C for 45 s, for 30 cycles. The primers used in this analysis are listed in the supplemental Table S2. All experiments were repeated three times.
Immunohistochemistry
Mouse eyes were enucleated, punctured with a fine needle in the dorsal region of the eye and incubated for 10 min in 4% paraformaldehyde (Electron Microscopy Sciences) in phosphate-buffered saline (1× PBS; 137 mm NaCl, 2.7 mm KCl, 4.3 mm Na2HPO4, and 1.47 mm KH2PO4) at room temperature. To make eyecups, eyes were removed from the fixative, cornea and lens were dissected away and dorsal region were marked by longitudinal cut. Eyecups were further fixed for 2–3 h at room temperature, then cryoprotected in PBS containing 20% sucrose overnight at 4 °C followed by incubation in 1:1 ratio of PBS containing 20% sucrose and OCT (Tissue-Tek) for 2 h at 4 °C. The eyecups were embedded in OCT and stored at −80 °C. Retinal sections (16 μm thick) were cut using cryostat (Leica CM1850) and mounted on Superfrost Plus slides (Fisher Scientific). For immunocytochemistry, sections were washed (three times for 10 min) in 1× PBST (1× PBS with 0.1% Triton X-100) and incubated for 1 h with blocking buffer (2% goat serum (Invitrogen), 0.1% Triton X-100, and 0.05% sodium azide in 1× PBS). Primary antibodies were incubated overnight at 4 °C. Sections were washed with 1× PBST (three times for 10 min) and incubated with secondary antibody (Alexa Fluor-488, or Alexa Fluor-568, LI-COR Biosciences) for 1 h at room temperature. After three washes with 1× PBST, sections were mounted with Fluoromount-G (Southern Biotech) and coverslipped. Imaging of stained retinal sections were performed at the WVU Microscope Imaging Facility with a Zeiss LSM 510 laser scanning confocal microscope using excitation wavelengths of 488, 543, and 633 nm. Primary antibodies used in this study were: Gα transducin (GαT1) polyclonal (Santa Cruz Biotechnology), cone transducin (GαT2) polyclonal (Santa Cruz Biotechnology), blue-, and red/green-cone opsin polyclonal (Chemicon International), GC-E/F (David Garbers), rod PDE6αβγ polyclonal (MOE, cytosignal), rod PDE6α, PDE6β, and PDEγ subunit specific antibodies (ABR), cone PDEγ′ (Vadim Arshavsky) and cone PDE6α' polyclonal antibody (3184P) (19). All primary antibodies were used at 1:1,000 dilution and secondary antibodies at 1:2,000 dilution, unless noted otherwise. Rhodamine-conjugated peanut agglutinin (PNA, Vector Laboratories) and TO-PRO-3 nuclear stain (Invitrogen) was used at 1:500 dilution for 1 h during secondary antibody incubation.
Immunoblot Analysis
Dissected retina (2) were homogenized by sonication (Microson Ultrasonic cell disruptor, 5 pulses 10 s at power setting 6) in 150 μl of 1× urea-SDS buffer (6 m urea, 125 mm Tris-HCl, pH 6.8, 4% SDS, 0.2% bromphenol blue, 10 mm dithiothreitol) in a 1.5-ml microcentrifuge tube on ice. After homogenization, protein concentration was measured with NanoDrop spectrophotometer (ND-1000, Thermo Scientific). Equal concentrations (150 μg) of total protein samples were separated by SDS-PAGE gel electrophoresis, transferred to Immobilon-FL membrane (Millipore), and probed with indicated antibodies. The primary antibodies as listed in the previous section were used at 1:2000 dilution. The secondary antibodies, odyssey goat anti-rabbit Alexa 680 and odyssey goat anti-mouse Alexa 680 (LI-COR Biosciences) were used at 1:50,000. Primary antibodies were diluted in 1:1 ratio of blocking buffer (Rockland) and 1× PBST (1× PBS/0.1% Tween-20). Secondary antibodies were diluted with 1× PBST. Membranes were scanned using an Odyssey Infrared Imaging System (LI-COR Biosciences).
Electroretinogram (ERG)
Following anesthesia with 5% isofluorane with 2.5% oxygen, pupils were dilated with phenylephrine HCl (1%) and tropicamide (1%). During ERG recording, mice were supported on an adjustable stage with a built-in heating device. The ERGs were differentially recorded from a pair of silver ring that directly contacted with eye through the artificial methylcellulose. A needle electrode placed subcutaneously on the forehead served as reference electrode. Mice were placed into a Ganzfeld chamber and light flashes were delivered at varying intensities. Dark-adapted ERGs were performed after mice were kept in the dark overnight. Light-adapted ERGs were performed after mice were exposed to constant light (30 cd · m−2) for 10 min, and flashes were administered in the presence of this constant background lighting. ERGs were performed with a UTAS-E4000 Visual Electrodiagnostic Test System using EMWIN 8.1.1 software (LKC Technologies). For recording M- or S-opsin activated responses, the maximum light intensities of monochromatic stimuli for the green (530 nm) and UV (360 nm) at 0.7 log cd s/m2 were used.
ERG responses were fitted using Michaelis-Menten equation (also referred as Naka-Rushton equation), A = (Amax × I)/(I + Ih) where A is the amplitude of the a- or b-wave, Amax is the asymptotic maximum amplitude, a measure of cone responsiveness to light and I is the light intensity or flash strength. Ih is the light intensity needed to elicit half-maximal ERG response, provides a measure of cone photoreceptor sensitivity. Plot of b-wave amplitude arising mostly from bipolar cells is a reflection of photoreceptor activity. Curve fitting was performed with GraphPad Prizm 4 software.
Immunoprecipitation (IP)
Eight frozen retinas were homogenized in 800 μl of IP buffer (10 mm Tris-HCl, pH 7.5, 100 mm KCl, 20 mm NaCl, 1 mm MgCl2) containing protease and phosphatase inhibitors and 10 mm iodoacetamide using a pellet pestle (VWR) in a 1.5 ml Eppendorf tube on ice (5 s × 4). After homogenization, Triton X-100 was added to a final concentration of 1%. Homogenized retinal extracts were pre-cleared by addition of 10 μl of immunopure immobilized protein A plus beads (Fisher) by incubating at 4 °C for 1 h. Supernatants were collected by centrifuging at 10,000 × g (Eppendorf 5424) for 5 min at 4 °C. 400 μl each of supernatant was used for IP with ROS-I antibody and control IgG. We used 1.5 μg of purified ROS-I monoclonal antibody for each pull-down experiment. After IP, proteins were separated by 4–20% SDS-polyacrylamide gel (Bio-Rad) and immunoblotting was performed as described earlier with catalytic subunit specific rod or cone PDE6 antibodies.
cGMP Measurement
Dissected retina from enucleated mouse eyes were homogenized in 0.1 m HCl (20, 21). We estimated protein concentration in retinal homogenate using nanodrop spectrophotometer. The acidic supernatant after boiling and centrifugation of retinal homogenates (1 mg/ml protein) was used to measure cGMP levels with direct cGMP assay kit enzyme-linked immunoassay (ImmunoDesign). Each reaction was performed in duplicate and results are an average of three independent experiments. To minimize the individual variations between mice, retinas were pooled from different mice for each time point assayed.
RESULTS
Creation of Cone-dominated Animal Model (Nrl−/− cpfl1) Lacking Cone PDE6
To study the role of cone PDE6 in a cone-rich retina, we utilized a mouse model lacking the Nrl transcription factor. The photoreceptor layer of Nrl−/− retina are populated by cone photoreceptor cells (22). We generated an all-cone mouse model lacking cone PDE6 by crossing Nrl−/− with cpfl1 mice. Cpfl1 mutant mice contain a spontaneous mutation with an insertion in intron 4 and single nucleotide deletion in exon 7 of Pde6c gene (18). These changes in cone PDE6α′ lead to cone photoreceptor dysfunction and progressive degeneration of cones in cpfl1 mice. The majority of photoreceptor cells in the cpfl1 retina, rods, are spared in this mouse model and are functional (Fig. 1A) (18). The heterozygous mice obtained from this cross were further bred to generate Nrl−/−cpfl1 and Nrl−/−cpfl1/+ heterozygous mice. We used Nrl−/− or Nrl−/−cpfl1/+ mice as controls throughout this work. No differences in photoreceptor viability or light-dependent electrical responses were observed between Nrl−/− or Nrl−/− cpfl1/+ mice (data not shown). The genotypes of the animals used in this study were determined as described in the methods and verified by sequencing (supplemental Fig. S1 and data not shown). In addition, absence of cone PDE6 in Nrl−/− cpfl1 mice was confirmed by Western blotting (Fig. 3B), immunoprecipitation (Fig. 4A) and mass spectrometry (supplemental Fig. S4).
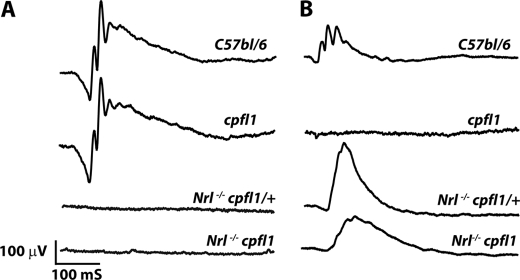
FIGURE 1.
Light-dependent ERG. A, scotopic ERG measuring rod function in Nrl−/− cpfl1/+ and Nrl−/− cpfl1 mice at P30 (n = 3). As controls, we measured responses from C57Bl/6 and cpfl1 mice. The light intensity used to measure scotopic ERG was −0.8 log cd s/m2. B, photopic ERG measuring cone function in Nrl−/− cpfl1/+ and Nrl−/− cpfl1 mice at P30 (n = 3). C57bl/6 and cpfl1 mice serve as positive and negative controls, respectively. A typical response from each animal is shown. Photopic ERGs were measured at 0.4 log cd s/m2 xenon white flash with steady background light.
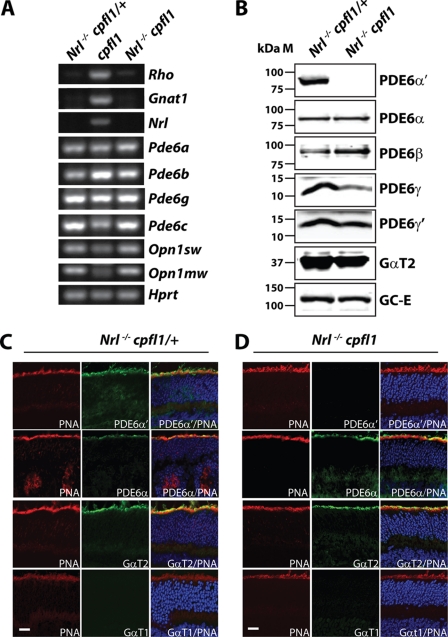
FIGURE 3.
Expression of rod PDE6 in cone photoreceptor cells. RT-PCR analysis using retinal RNA extracted from in Nrl−/− cpfl1/+ and Nrl−/− cpfl1 mice at P12 (A). Expression of rod specific genes in the middle panel from retinal tissue lacking cone PDE6 (cpfl1) serves as positive control. Hprt, a housekeeping gene serves as loading control (A). Immunoblot analysis with indicated antibodies investigating the expression levels of proteins in retinal extracts from P30 Nrl−/− cpfl1/+ and Nrl−/− cpfl1 mice. Equal amounts (150 μg) of total proteins were loaded in each lane (B). Immunolocalization of rod and cone specific PDE6 and transducin in frozen retinal sections (P30) from Nrl−/− cpfl1/+ (C) and Nrl−/− cpfl1 mice (D). TO-PRO-3 stained nuclei are shown in blue, and peanut agglutinin (PNA)-stained cones are depicted in red. Cone PDE6α′, rod PDE6α, cone α-transducin (GαT2), and rod α-transducin (GαT1) staining are shown in green. (Scale bar: 10 μm.)
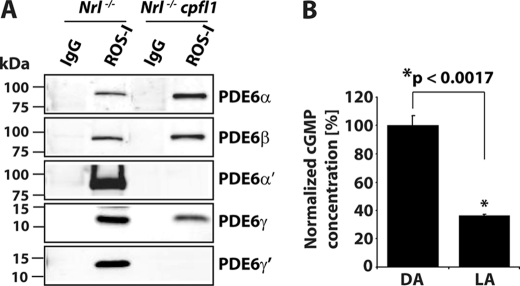
FIGURE 4.
Rod PDE6 expressed in Nrl−/−cpfl1 mice is functionally active. A, immunoprecipitation (IP) of assembled rod PDE6 αβγ subunits from retinal extracts of Nrl−/− and Nrl−/− cpfl1 mice at P30 using ROS-I monoclonal antibody. After ROS-I IP, immunoblots were probed with rod or cone-specific PDE6 antibodies as indicated. Control IP with nonspecific mouse IgG is shown. B, amount of total cGMP, measured in dark (DA) and light-adapted (LA) retina from Nrl−/− cpfl1 mice. The data are presented as mean ± S.E. n = 3, *, p < 0.0017 compared with dark-adapted mice. Light-adapted retinas were obtained after mice (P30) were exposed to constant white light (71 cd/m2) in the ERG Ganzfeld for 15 min. Dark-adapted retinas were obtained from mice after overnight adaptation.
Cone Photoreceptor Cells in Nrl−/− cpfl1 Mice Respond to Light
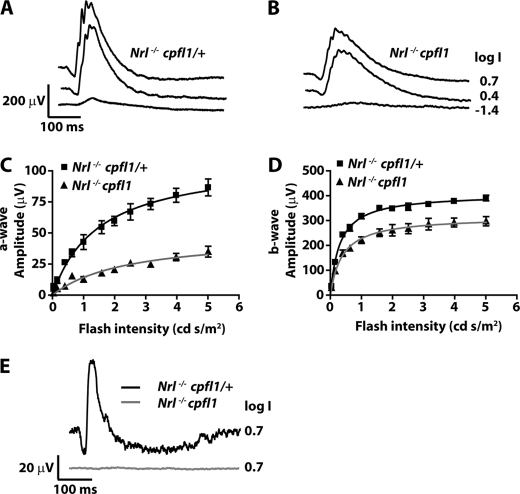
To assess retinal function in the all-cone mouse model lacking cone PDE6, we performed ERGs. ERG measurements under dark and light-adapted conditions reflect rod and cone photoreceptor cells activities, respectively. The a-wave of ERG originates from photoreceptor activity, while the majority of b-wave is comprised of activity from downstream bipolar neurons. In agreement with findings that photoreceptor cells in Nrl−/− mice are cone-like, Nrl−/− cpfl1/+ mice lacked rod electroretinogram (ERG) response (Fig. 1A) but exhibited a robust cone ERG response (Fig. 1B) (22, 23). As expected, Nrl−/− cpfl1 mice lacked rod responses (Fig. 1A). Surprisingly, light-adapted ERG response indicative of functional cone cells was observed in these mice (Fig. 1B). To determine the origin of the cone response in Nrl−/− cpfl1 mice, we measured ERGs using light of wavelengths attributed to activation of either S- or M-opsin (22–24). Responses arising from S-opsin were isolated using light of monochromatic stimuli at 360 nm. Nrl−/− cpfl1 and littermate control Nrl−/− cpfl1/+ mice showed S-cone-mediated response (Fig. 2, A and B). The amplitudes of S-cone ERG a- and b-waves obtained over varying flash light intensities were fitted using the Michaelis-Menten equation (Fig. 2, C and D). The parameters obtained from curve-fitting are indicated in Table 1. Both a- and b-wave responses were reduced in Nrl−/− cpfl1 mice. The maximum amplitude was reduced by 55% (a-wave) and 23% (b-wave) (Table 1). In addition, a slight reduction in the sensitivity of the a-wave responses was also observed (Table 1 and supplemental Fig. S2).
FIGURE 2.
Cone-isolated ERGs. Light adapted ERGs from Nrl−/− cpfl1/+ (A) and Nrl−/− cpfl1 mice (B) at P30 to increasing light intensities of short wavelength monochromatic stimuli (360 nm) ranging from −3.6 to 0.7 log cd s/m2. Selected traces at the indicated light intensities are shown. Responses obtained from ERG recording were plotted against light intensities (C and D). Curves were fitted using Michaelis-Menten function as described under “Experimental Procedures.” Photoreceptor response (a-wave) and downstream bipolar response (b-wave) are depicted in panels C and D, respectively. Responses depicted are an average ± S.E. response from both eyes of three mice. M-cone ERG in Nrl−/− cpfl1/+ and Nrl−/− cpfl1 mice measured in response to monochromatic stimuli at 530 nm (E). Responses depicted are an average ± S.E. response from both eyes of three mice.
TABLE 1.
The fitting parameters (sensitivity and maximum amplitude) of the amplitude-intensity plots in Fig. 2, C and D
The data shown are from at least three animals of each genotype shown as mean ± S.E.
| Ih | Amax | |
|---|---|---|
| cd.s/m2 | μV | |
| a-wave | ||
| Nrl−/−cpfl1/+ | 1.5 ± 0.25 | 109.3 ± 7.15 |
| Nrl−/−cpfl1 | 2.4 ± 0.62 | 48.52 ± 5.89 |
| b-wave | ||
| Nrl−/−cpfl1/+ | 0.31 ± 0.02 | 407.6 ± 5.68 |
| Nrl−/−cpfl1 | 0.39 ± 0.05 | 315.7 ± 9.79 |
The responses derived from the M-opsin driven phototransduction cascade were recorded using monochromatic light stimuli at 530 nm (22–24). In contrast to controls, Nrl−/− cpfl1 mice did not exhibit significant M-cone responses (Fig. 2E). Altogether, our results suggested that the majority of cone response in Nrl−/− cpfl1 mice arise from S-opsin-mediated signaling.
Cone Photoreceptor Cells in Nrl-deficient Mice Express Rod PDE6 Subunits
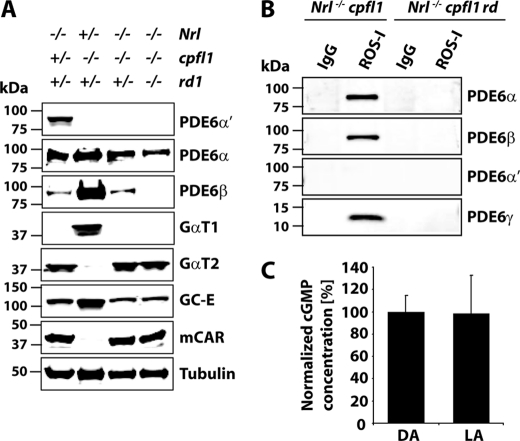
To investigate if observed cone responses in Nrl−/− cpfl1 mice are due to expression of rod PDE6 subunits, we checked the message levels of rod PDE6 subunits. Our RT-PCR results using retinal cDNA prepared from Nrl−/− cpfl1/+ and Nrl−/− cpfl1 mice showed expression of rod Pde6a, Pde6b, and Pde6g genes (Fig. 3A). In contrast, expression of rod-specific genes, Gnat1 (rod transducin) and Rho (rod opsin) were not observed. We also confirmed the lack of Nrl expression in Nrl−/− cpfl1/+ and Nrl−/− cpfl1 mice (Fig. 3A).
To verify the expression of rod PDE6 protein in retina obtained from Nrl−/− cpfl1/+ and Nrl−/− cpfl1 mice, we performed immunoblotting of retinal extracts using catalytic subunit-specific PDE6 antibodies. The specificity of the antibodies was tested using extracts from tissue culture cells transfected with plasmids expressing individual mouse rod or cone PDE6 subunits. Antibodies against PDE6-α and β subunits specifically recognized the respective PDE6 subunits from the retinas. Importantly, under our experimental conditions, rod subunit specific antibodies did not recognize cone PDE6 (supplemental Fig. S3). Rod PDE6 catalytic subunits (α and β) were present and the cone PDE6 catalytic subunit was absent in Nrl−/− cpfl1 mice (Fig. 3B). Surprisingly, both rod and cone PDE6 inhibitory subunits (γ and γ′) were present in both Nrl−/− cpfl1/+ and Nrl−/− cpfl1 mice (Fig. 3B). Similar levels of cone photoreceptor markers, cone transducin (GαT2) and guanylate cyclase (GC-E) were found in both animal models (Fig. 3B). In contrast, rod-specific proteins, such as transducin (GαT1) and guanylate cyclase (GC-F) were undetectable (supplemental Fig. S3B).
In agreement with our immunoblotting results, cone PDE6 was absent in retinal sections from Nrl−/− cpfl1 mice (Fig. 3D, top row). In control retinal sections from Nrl−/− cpfl1/+ mice, we observed expression of cone PDE6 in outer segments of photoreceptor cells (Fig. 3C, top row). Rod PDE6 catalytic subunits, cone transducin (GαT2) and guanylate cyclase-E (GC-E) were expressed in photoreceptor outer segments of both animal models (Fig. 3, C and D and supplemental Fig. S5). However, rod transducin was undetectable (Fig. 3, C and D, last row).
Rod PDE6 Expressed in Nrl−/− cpfl1 Mice Is Assembled and Functional
To verify the assembly status of rod PDE6 subunits present in NRL-deficient cones, we used ROS-I antibody to immunoprecipitate PDE6. ROS-I is a monoclonal antibody that recognizes assembled and functional PDE6 subunits (25, 26). Immunoblotting using PDE6 specific antibodies demonstrated that ROS-I recognized rod and cone PDE6 subunits in retinal extracts from Nrl−/− mice (Fig. 4A). In Nrl−/− cpfl1 mice, rod PDE6 catalytic subunits along with rod PDE6γ were detected (Fig. 4A). Although, cone PDE6γ′ was present in retinal extracts from Nrl−/− cpfl1 mice, we did not detect PDE6γ′ in ROS-I pull-downs (Fig. 4A). We independently confirmed the presence of assembled rod PDE6 catalytic subunits in the retinal extracts from Nrl−/− cpfl1 mice by mass spectrometry. ROS-I immunoprecipitates were separated on a PAGE gel and the bands corresponding to the size of rod and cone PDE6 catalytic subunits were subjected to mass spectrometric analysis. MALDI followed by MS/MS analysis identified several peptides unique for rod PDE6α and β subunits (supplemental Fig. S4). To examine if rod PDE6 present in Nrl−/− cpfl1 mice was functional, we measured light-dependent changes in cGMP levels in the retina. Retinal cGMP levels were reduced by 70% in response to light (Fig. 4B). Collectively, these results demonstrated the expression of functional rod PDE6 in Nrl−/− cpfl1 mice.
Light Response in Nrl−/− cpfl1 Mice Is Mediated through Rod PDE6 Subunits
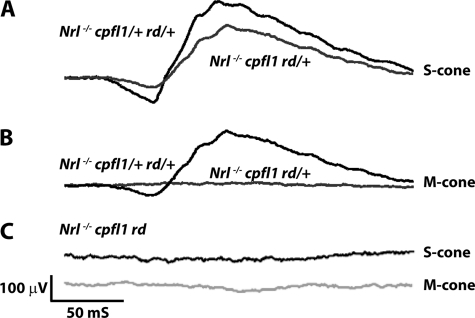
To demonstrate the ability of rod PDE6 to participate in the cone phototransduction pathway, we genetically eliminated the β-subunit of rod PDE6 in Nrl−/− cpfl1 mice. We crossed Nrl−/− cpfl1 mice with rd1 animals to generate Nrl−/− cpfl1 rd mice (27, 28). In agreement with our earlier results, littermate controls (Nrl−/− cpfl1 rd/+) exhibited S-cone response (Fig. 5A). The removal of rod PDE6β subunit in Nrl−/− cpfl1 rd mice completely abrogated the S-cone response (Fig. 5C). Both cpfl1 mutant mouse models lack M-cone response in comparison to Nrl−/− cpfl1/+ rd/+ littermate control (Fig. 5, B and C).
FIGURE 5.
S-cone ERG is eliminated in Nrl−/− mice with defective rod and cone PDE6. Light-adapted S-cone responses in controls, Nrl−/− cpfl1/+ rd/+ (A), Nrl−/− cpfl1 rd/+ (A) and in mice lacking rod PDE6β subunit, Nrl−/− cpfl1 rd mice (C). M-cone responses in Nrl−/− cpfl1 rd/+ (B) and Nrl−/− cpfl1 rd (C) and in control, Nrl−/− cpfl1/+ rd/+ mice (B). All recordings were performed using littermate controls at P30 with light intensity of 0.7 log cd s/m2.
Immunoblot analysis of retinal extracts from Nrl−/− cpfl1 rd mice revealed that rod PDE6β was undetectable (Fig. 6A). ROS-I did not recognize PDE6 subunits in retinal extracts from Nrl−/− cpfl1 rd mice (Fig. 6B). In addition, the unassembled rod PDE6 in Nrl−/− cpfl1 rd mice were not functional, as cGMP levels unaltered in response to light (Fig. 6C). Notably, unlike in the rd1 mouse model, the stability of PDE6α was unaffected (Fig. 6A) (29). The levels of cone transducin, cone arrestin, and guanylate cyclase (GC-E), which served as controls, remain unchanged (Fig. 6A).
FIGURE 6.
Rod PDE6 functions as an obligate heteromer. A, equal amounts (150 μg) of retinal extracts from littermate controls at P30 were used for immunoblot analysis with indicated antibodies. B, assessing the assembly of PDE6 by ROS-I. IP using mouse IgGs served as controls. Retina from P30 mice were used for these experiments. C, cGMP levels measured in the dark- (DA) and light-adapted (LA) retinas from Nrl−/− cpfl1 rd mice at P30. Light- and dark-adapted retinas were obtained as described earlier.
Transport of M-opsin Is Impaired in the Absence of Functional PDE6
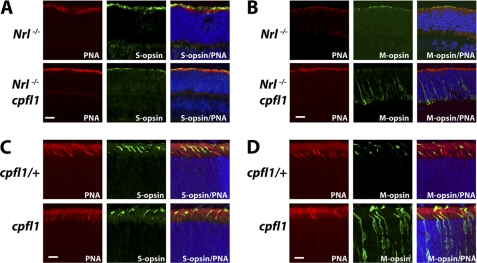
In adult mice, M-opsin is severely reduced in retinas from Nrl−/− cpfl1 mice (supplemental Fig. S5D). In agreement with this finding, M-cone responses were absent in Nrl−/− cpfl1 mice (Fig. 2E). To identify the reason behind this loss of M-opsin, we examined the retinal sections for presence of cone opsin at earlier stages of retinal development (P12), when there were no signs of cell death (supplemental Fig. S6). M-opsin was present in the photoreceptor outer segments of control Nrl−/− mice. On the other hand, M-opsin in Nrl−/− cpfl1 mice were mis-localized to the inner segments and synaptic region of photoreceptor cells (Fig. 7B). In contrast, S-opsin was localized to outer segments in photoreceptor cells from Nrl−/− and Nrl−/− cpfl1 mice (Fig. 7A).
FIGURE 7.
Functional PDE6 is crucial for localization of M-opsin to outer segments. Frozen retinal sections were probed with anti S-opsin (green) and peanut agglutinin (red). S-opsin is present in outer segments irrespective of the functional status of PDE6 (A, C). M -opsin (green) is present in outer segments in Nrl−/− and cpfl1/+ mice (B, D: upper panel) but is mislocalized to synaptic and nuclear layer of retina from Nrl−/− cpfl1 and cpfl1 mice (B, D: lower panel). All retinal sections were from P12 mice. (Scale bar: 10 μm.)
To investigate if the need for functional PDE6 in transport of M-opsin is universal and not a unique characteristic of retina lacking Nrl, we examined the localization of opsin in retina from cpfl1 mice at P12 (18). As described earlier, cpfl1 mice contain normal complement of functioning rods but lack both S- and M-opsin-mediated photoresponse (Fig. 1). In these mice, rod PDE6 catalytic subunits (αβ) are not expressed in cones (Data not shown). We observed similar results as Nrl−/− cpfl1 mice with M-opsin mislocalized to inner segments, nuclei and synaptic regions (Fig. 7D). S-opsin was localized normally to photoreceptor outer segments and did not depend on the presence of PDE6 catalytic subunits (Fig. 7C). S-opsin localization was also unaffected in Nrl−/− cpfl1 rd mice that lacked S-cone response (supplemental Fig. S7).
DISCUSSION
This work establishes the ability of rod PDE6 to functionally substitute for cone PDE6 to mediate visual signaling in vivo. Light-dependent PDE6 activation in cones lacking cone PDE6 was abrogated when rod PDE6-β subunit was removed in Nrl−/− cpfl1 rd mice. In this animal model, despite the presence of an α-subunit, PDE6 was not functional. This finding implies that irrespective of cell type, rod PDE6 functions as an obligatory heteromer in vivo. Additionally, our results reveal the need for functional PDE6 in M-opsin trafficking, but not S-opsin.
We demonstrated the presence of rod PDE6 in cone photoreceptor cells lacking NRL using multiple lines of evidence as listed below; 1) RT-PCR with subunit specific primers showing the expression of pde6a and pde6b message, 2) Western blotting with subunit specific antibodies establishing the presence of rod PDE6 protein subunits and absence of cone PDE6 catalytic subunit, 3) mass spectrometry confirming the presence of rod PDE6 subunits with 100% confidence. In contrast, we did not detect cone PDE6 subunits and 4) Removal of PDE6β subunit using a genetic approach from Nrl−/− cpfl1 mice eliminated light-dependent cGMP hydrolysis and visual response. Our results also concur with previous findings demonstrating the expression of rod PDE6 subunits in retina from adult Nrl−/− mice by RT-PCR (30, 31).
Retina lacking NRL transcription factor are enriched with cones expressing S-opsin due to conversion of rods to S-cones en masse (22). Incidentally, in Nrl−/− cpfl1 mice, majority of light responses were from activated S-opsin. Native M-cones present in the NRL deficient retina similar to those present in cpfl1 mice do not express PDE6 and therefore lack light responses.
Quantitative Western blotting using GST-rod or -cone PDE6 catalytic subunit as standards show that rod PDE6 present in Nrl−/− cpfl1 is 10 times lower than cone PDE6 catalytic subunits in Nrl−/− retinas (data not shown). Maximal cGMP PDE activity was reduced by 23-fold in retina from Nrl−/− cpfl1 mice (data not shown). Altogether, these results suggest that rod PDE6 is expressed at low levels in converted rods present in mice lacking NRL. The leaky expression of rod PDE6 subunits is likely due to partial requirement for NRL protein in transcription of rod PDE6 message.
Interestingly, we observed robust expression of both rod and cone PDE6 inhibitory subunits in cones lacking NRL. These results show that transcription of PDE6 inhibitory subunits is independent of NRL protein. However, in retina from Nrl−/− cpfl1 mice, we observe only assembled rod PDE6 subunits. These results suggest that although cone PDE6γ′ is present, they do not associate with rod catalytic subunits. Alternatively, ROS-I may not recognize hybrid complex of rod catalytic subunits with cone inhibitory subunit.
While the expression of cone opsin and cone transducin were observed, rod opsin and rod transducin subunits were not expressed in retina lacking NRL. These results imply that in Nrl−/− cpfl1 mice, the S-opsin response from converted rods is due to activation of rod PDE6 by cone transducin. Although, we observed consistent S-opsin response, the a- and b-wave amplitudes are reduced in Nrl−/− cpfl1 mice. The reduction in ERG response is likely due to reduced overall levels of PDE6 activity in these animals. On the other hand, the reduction in ERG could be a reflection of changes in cone dimensions and numbers. Further ultrastructural studies are needed to address these possibilities. In addition, the sensitivity of ERG response (a-wave) to light was reduced in Nrl−/− cpfl1 animals. However, the decrease in sensitivity when rod PDE6 is coupled to cone transducin was modest (Table 1 and supplemental Fig. S2). This result suggests that the coupling efficiency of cone transducin to either rod or cone PDE6 subunits to hyperpolarize photoreceptor membranes in response to light is similar in vivo. Our results should be interpreted with caution as multiple changes may occur in cones due to lack of NRL and cone PDE6 catalytic subunit. Despite these limitations, given the difficulties in expressing functional PDE6, animal models characterized in this study showed that rod PDE6 can effectively couple to the cone visual signaling pathway.
An interesting finding from our study is the need for functional cone PDE6 in localization of M-opsin. This defect was selective as localization of S-opsin was not affected. In cpfl1 mice lacking cone ERG response, S-opsin was localized to outer segments. No rod or cone PDE6 subunits were present in cone cells from cpfl1 animals (data not shown). However, lack of PDE6 selectively affected the localization of M-opsin. Mis-localization of M-opsin was not a secondary defect caused by cell death as we observed this trafficking defect at early stages of retinal development when there was little or no cell death (supplemental Fig. S6). Mis-localization of M-opsin was strain independent and was observed in retina from cpfl1 mice with functional rod cells, and in cone enriched Nrl−/− cpfl1 and in Nrl−/− cpfl1 rd mice. Another possibility for observation of M-opsin trafficking defect could be altered translocation of M-opsin containing cone nuclei (32). Further studies are needed to pinpoint the exact reason(s) behind the need for functional PDE6 in M-opsin localization.
Our study also shows that, irrespective of cell type, rod PDE6 needs to form heteromers in vivo to be functional. The reason behind hetero-dimerization of catalytic subunits is not known. Deletion of one catalytic subunit of rod PDE6 altered the stability of cognate partner (29, 33). However, in our all-cone mouse model lacking cone PDE6 and rod PDE6β, the rod PDE6α subunit was stable but was not active. Therefore, our studies rule out the possibility that hetero-dimerization of rod PDE6 is solely required to maintain the stability of PDE6 subunits. Alternatively, hetero-dimerization could be a mechanism to control the amount of functional PDE6 enzyme present in photoreceptor cells.
Supplementary Material
Acknowledgments
We thank Drs. Wolfgang Baehr (mouse cone arrestin), David Garber (GC-E and F), Vadim Arshavsky (cone PDE6 γ), and Ted Wensel (ROS1 and PDE6 γ) for their generous gift of antibodies. The Nrl mutant mice were obtained from Dr. Anand Swaroop. We are indebted to Drs. Maxim Sokolov, James Hurley, Neal Peachey, and Vernon Odom for suggestions and the members of the Dr. Ramamurthy laboratory for their support throughout this study.
This work was supported, in whole or in part, by National Institutes of Health Grant R01EY017035 (to V. R.), WV Lions Eye Bank, Lions Club International Foundation, and Research to Prevent Blindness (RPB) challenge grant to West Virginia University.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7 and Tables S1 and S2.
- PDE6
- cGMP phosphodiesterase type 6
- Cpfl1
- cone photoreceptor function loss 1
- NRL
- neural retina leucine zipper transcription factor
- RD1
- retinal degeneration 1
- GAF domains
- mammalian cGMP binding PDEs Anabaena adenylyl cyclases
- Hprt
- hypoxanthine guanine phosphoribosyl transferase
- Opn1sw
- short-wavelength sensitive opsin
- Opn1mw
- middle-wavelength sensitive opsin
- KO
- knockout.
REFERENCES
- 1. Pugh E. N., Jr., Cobbs W. H. (1986) Vision Res. 26, 1613–1643 [DOI] [PubMed] [Google Scholar]
- 2. Fu Y., Yau K. W. (2007) Pflugers Arch 454, 805–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Burns M. E., Arshavsky V. Y. (2005) Neuron 48, 387–401 [DOI] [PubMed] [Google Scholar]
- 4. Zhang X., Cote R. H. (2005) Front Biosci. 10, 1191–1204 [DOI] [PubMed] [Google Scholar]
- 5. Kefalov V., Fu Y., Marsh-Armstrong N., Yau K. W. (2003) Nature 425, 526–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen C. K., Woodruff M. L., Chen F. S., Shim H., Cilluffo M. C., Fain G. L. (2010) J. Physiol. 588, 3231–3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Deng W. T., Sakurai K., Liu J., Dinculescu A., Li J., Pang J., Min S. H., Chiodo V. A., Boye S. L., Chang B., Kefalov V. J., Hauswirth W. W. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 17681–17686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li T. S., Volpp K., Applebury M. L. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 293–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gillespie P. G., Beavo J. A. (1988) J. Biol. Chem. 263, 8133–8141 [PubMed] [Google Scholar]
- 10. Hamilton S. E., Hurley J. B. (1990) J. Biol. Chem. 265, 11259–11264 [PubMed] [Google Scholar]
- 11. Anant J. S., Ong O. C., Xie H. Y., Clarke S., O'Brien P. J., Fung B. K. (1992) J. Biol. Chem. 267, 687–690 [PubMed] [Google Scholar]
- 12. Wensel T. G. (2008) Vision Res. 48, 2052–2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mou H., Cote R. H. (2001) J. Biol. Chem. 276, 27527–27534 [DOI] [PubMed] [Google Scholar]
- 14. Muradov H., Boyd K. K., Artemyev N. O. (2010) J. Biol. Chem. 285, 39828–39834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Muradov H., Boyd K. K., Artemyev N. O. (2004) Vision Res. 44, 2437–2444 [DOI] [PubMed] [Google Scholar]
- 16. Zhang X. J., Cahill K. B., Elfenbein A., Arshavsky V. Y., Cote R. H. (2008) J. Biol. Chem. 283, 29699–29705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gillespie P. G., Beavo J. A. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 4311–4315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chang B., Grau T., Dangel S., Hurd R., Jurklies B., Sener E. C., Andreasson S., Dollfus H., Baumann B., Bolz S., Artemyev N., Kohl S., Heckenlively J., Wissinger B. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 19581–19586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kirschman L. T., Kolandaivelu S., Frederick J. M., Dang L., Goldberg A. F., Baehr W., Ramamurthy V. (2010) Hum. Mol. Genet. 19, 1076–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Farber D. B., Lolley R. N. (1982) Methods Enzymol. 81, 551–556 [DOI] [PubMed] [Google Scholar]
- 21. Ramamurthy V., Niemi G. A., Reh T. A., Hurley J. B. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 13897–13902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mears A. J., Kondo M., Swain P. K., Takada Y., Bush R. A., Saunders T. L., Sieving P. A., Swaroop A. (2001) Nat. Genet. 29, 447–452 [DOI] [PubMed] [Google Scholar]
- 23. Daniele L. L., Lillo C., Lyubarsky A. L., Nikonov S. S., Philp N., Mears A. J., Swaroop A., Williams D. S., Pugh E. N., Jr. (2005) Invest. Ophthalmol Vis. Sci. 46, 2156–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nikonov S. S., Daniele L. L., Zhu X., Craft C. M., Swaroop A., Pugh E. N., Jr. (2005) J. Gen. Physiol. 125, 287–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hurwitz R. L., Bunt-Milam A. H., Beavo J. A. (1984) J. Biol. Chem. 259, 8612–8618 [PubMed] [Google Scholar]
- 26. Kolandaivelu S., Huang J., Hurley J. B., Ramamurthy V. (2009) J. Biol. Chem. 284, 30853–30861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bowes C., Li T., Danciger M., Baxter L. C., Applebury M. L., Farber D. B. (1990) Nature 347, 677–680 [DOI] [PubMed] [Google Scholar]
- 28. Pittler S. J., Baehr W. (1991) Prog Clin. Biol. Res. 362, 33–66 [PubMed] [Google Scholar]
- 29. Farber D. B. (1995) Invest. Ophthalmol Vis. Sci. 36, 263–275 [PubMed] [Google Scholar]
- 30. Pittler S. J., Zhang Y., Chen S., Mears A. J., Zack D. J., Ren Z., Swain P. K., Yao S., Swaroop A., White J. B. (2004) J. Biol. Chem. 279, 19800–19807 [DOI] [PubMed] [Google Scholar]
- 31. Farjo R., Skaggs J. S., Nagel B. A., Quiambao A. B., Nash Z. A., Fliesler S. J., Naash M. I. (2006) J. Cell Biol. 173, 59–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Trifunović D., Dengler K., Michalakis S., Zrenner E., Wissinger B., Paquet-Durand F. (2010) J. Comp. Neurol 518, 3604–3617 [DOI] [PubMed] [Google Scholar]
- 33. Sakamoto K., McCluskey M., Wensel T. G., Naggert J. K., Nishina P. M. (2009) Hum. Mol. Genet. 18, 178–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.