Abstract
Rev1 and DNA polymerase ζ (Polζ) are involved in the tolerance of DNA damage by translesion synthesis (TLS). The proliferating cell nuclear antigen (PCNA), the auxiliary factor of nuclear DNA polymerases, plays an important role in regulating the access of TLS polymerases to the primer terminus. Both Rev1 and Polζ lack the conserved hydrophobic motif that is used by many proteins for the interaction with PCNA at its interdomain connector loop. We have previously reported that the interaction of yeast Polζ with PCNA occurs at an unusual site near the monomer-monomer interface of the trimeric PCNA. Using GST pull-down assays, PCNA-coupled affinity beads pull-down and gel filtration chromatography, we show that the same region is required for the physical interaction of PCNA with the polymerase-associated domain (PAD) of Rev1. The interaction is disrupted by the pol30-113 mutation that results in a double amino acid substitution at the monomer-monomer interface of PCNA. Genetic analysis of the epistatic relationship of the pol30-113 mutation with an array of DNA repair and damage tolerance mutations indicated that PCNA-113 is specifically defective in the Rev1/Polζ-dependent TLS pathway. Taken together, the data suggest that Polζ and Rev1 are unique among PCNA-interacting proteins in using the novel binding site near the intermolecular interface of PCNA. The new mode of Rev1-PCNA binding described here suggests a mechanism by which Rev1 adopts a catalytically inactive configuration at the replication fork.
Keywords: DNA Damage, DNA Polymerase, Mutagenesis Mechanisms, Yeast, Yeast Genetics, PCNA, Rev1, Translesion DNA Synthesis
Introduction
Cellular DNA is continuously attacked by endogenous and exogenous agents that damage the bases and the DNA backbone. Damage that is not repaired prior to the S phase of cell cycle can block the replication machinery, because most lesions cannot be accommodated in the highly selective active site of replicative DNA polymerases (1, 2). Replication stalling activates several damage tolerance mechanisms that help bypass the damage in either an accurate or mutagenic manner. Translesion DNA synthesis (TLS)3 is an important damage tolerance pathway, in which specialized DNA polymerases are recruited to synthesize DNA through template lesions (3). Structural studies showed that TLS polymerases have a more open active site that allows them to accommodate a variety of DNA lesions and catalyze polymerization on damaged templates (4). The accuracy of TLS can vary depending on the particular lesion and the DNA polymerases involved. It is, however, an inherently mutagenic process, because the damage can alter the coding properties of the bases, and because TLS polymerases generally have low fidelity (2).
In human cells, TLS polymerases include Y family enzymes Polη, Polι, Polκ, and REV1, and the B family enzyme Polζ. Although the Y family polymerases share little amino acid sequence similarity with the classical DNA polymerases, they have a similar overall “right-hand” architecture with the “palm,” “thumb,” and “fingers” domains. The thumb and fingers domains of the Y family enzymes, however, are short and do not make as many contacts with the DNA as they do in the other polymerases. Instead, the Y-family polymerases possess an additional DNA-binding domain called the “little finger,” “polymerase-associated domain (PAD),” or “wrist” (4). Whereas the eukaryotic REV1 protein shares these structural features, its role in TLS is unique. First, Rev1 is a deoxycytidyl transferase that is very specific for incorporating a C opposite G, abasic sites and damaged bases (5–11). This is in contrast to all other known DNA polymerases that utilize all four nucleotides for DNA synthesis. Second, although the catalytic activity of Rev1 is apparently used during the bypass of some lesions in vivo, particularly abasic sites and 1,N(6)-ethenoadenine, the essential role of Rev1 in most types of TLS is structural (3, 12). Yeast and mammalian REV1 interact with multiple DNA polymerases, including all other Y family enzymes, Polζ, and an accessory subunit of the replicative DNA polymerase δ, Pol32 (3, 13). This led to a model wherein REV1 plays a central organizing role in TLS by providing a docking site for other TLS polymerases and facilitating polymerase exchange.
In addition to Rev1, DNA polymerase processivity factor proliferating cell nuclear antigen (PCNA) plays an important role in regulating the activity of eukaryotic TLS polymerases. In vitro, PCNA stimulates TLS by Polη, Polι, Polκ, and Polζ (14–17). In yeast and human cells, exposure to DNA damaging agents induces monoubiquitylation of PCNA at Lys164 by Rad6-Rad18 complex (18–20). This modification is required for Polη and Polζ-dependent TLS, and for TLS-mediated mutagenesis (18, 20, 21). The ubiquitylation of PCNA enhances its association with the Y-family polymerases Polη, Polι, and Rev1 due to the presence of ubiquitin-binding domains in these enzymes (22–26). The ubiquitylated PCNA was also shown to be more efficient in stimulating the TLS activity of Polη and Rev1 in vitro than the unmodified PCNA (27).
Several studies addressed a question of how the two central TLS regulators, REV1 and PCNA, interact. Although it is clear that the ubiquitin-binding motifs of Rev1 mediate binding to the ubiquitin moiety of the modified PCNA (23, 26), controversial data exist in regard to which part of REV1 binds to PCNA itself. Most PCNA partners carry a conserved PCNA-binding motif QXX(L/I)XXFF (PIP box) that mediates their interaction with the interdomain connector loop or the C terminus of PCNA (28). REV1, however, lacks this motif. In one study, the PCNA-binding site in the human REV1 was mapped to a ∼125 amino acid region that lies downstream of the catalytic core and encompasses the ubiquitin-binding motifs (29). Studies with mouse Rev1 implicated its N-terminal part containing the BRCT domain in the interaction with PCNA (30). Stimulation of the activity of yeast Rev1 by PCNA required ∼200 C-terminal amino acids of Rev1, a segment that overlaps with the proposed PCNA-binding site in the human REV1 (26). The latter study also suggested that regions other than the C terminus contribute to PCNA binding, although binding at these other sites is insufficient for the stimulation of Rev1 activity. Which PCNA regions mediate the interaction with REV1 remained unknown.
We previously showed that a mutation resulting in a double amino acid substitution at the monomer-monomer interface of PCNA trimer (pol30-113) abolished UV-induced mutagenesis, indicating a defect in Polζ/Rev1-dependent TLS (31). The defect was not due to the inability of PCNA-113 to undergo monoubiquitylation, since the protein was readily ubiquitylated both in vitro in a reconstituted Rad6/Rad18-dependent reaction and in vivo in response to DNA damage. PCNA-113, however, was completely defective in stimulating the TLS activity of Polζ in vitro, suggesting that the region near the subunit interface of PCNA is critical for the interaction with Polζ (31). This binding at a novel binding site on PCNA is consistent with the fact that, like Rev1, Polζ lacks the consensus PCNA-binding motif. Although the defective functional interaction with Polζ would be sufficient to cause UV-immutability in the pol30-113 strains, there remains a possibility that PCNA-113 has altered interactions with other factors required for mutagenesis as well.
The present study was inspired by a previously reported crystal structure of a complex formed by the Escherichia coli β-clamp and the “little finger” domain of E. coli Y-family DNA polymerase Pol IV (32). This structure revealed significant contacts between the “little finger” and the region near the intermolecular interface of the β-clamp. The “little finger” domain of Pol IV is structurally and functionally analogous to the polymerase-associated domain (PAD) present in eukaryotic Y-family DNA polymerases, including Rev1. At the same time, Rev1, along with Polζ, is required for DNA damage-induced mutagenesis. In this study, we tested a hypothesis that the region of yeast PCNA marked by the pol30–113 mutation is involved in the interaction with the PAD of Rev1. We also further investigated the role of this novel protein binding site on PCNA in the control of DNA damage tolerance in vivo.
EXPERIMENTAL PROCEDURES
Plasmids
Yeast expression vector pRS425-GALGST (14) was kindly provided by Peter Burgers (Washington University School of Medicine, St. Louis). Fragments of Saccharomyces cerevisiae REV1 encoding for amino acids 297–746 and 621–746 were amplified by PCR and cloned in frame with glutathione S-transferase (GST) into pRS425-GALGST. The pGEX-5X-1-Pol32 plasmid4containing the open reading frame of S. cerevisiae POL32 cloned in-frame with GST into the E. coli expression vector pGEX-5X-1 was used for the yeast Pol32 overproduction. A deletion of the region encoding for the nine C-terminal amino acids of Pol32 (PCNA-interacting motif) was created in pGEX-5X-1-Pol32 by site-directed mutagenesis by using a QuickChange site-directed mutagenesis kit from Stratagene. The plasmid pBL228 and its derivative containing the pol30-113 allele (31) were used for the overproduction of yeast PCNA and PCNA-113, respectively, in E. coli. The pol30-79 mutation, resulting in alanine substitutions for Leu-126 and Ile-128 in the interdomain connector loop of PCNA (33), was created in pBL228 by site-directed mutagenesis.
Strains
Saccharomyces cerevisiae strain BJ2168 (MATa ura3-52 trp1-289 leu2-3,112 prb1-1122 prc1-407 pep4-3; Ref. 34) was used for overproduction and purification of GST-tagged Rev1 fragments. Strains used in the UV sensitivity assays are described in Table 1. The rad14, rad52, rad18, rev3, and rev1 mutants were isogenic to E134; the rad5, mms2, and ubc13 mutants were isogenic to BY4742; and the rad30 mutant was isogenic to 1A-PSD105. The chromosomal wild-type POL30 gene of the rad14, rad18, rev3, rev1, rad5, mms2, ubc13, and rad30 mutants and the corresponding wild-type strains was replaced by the pol30-113 allele as described previously (31). To construct double rad52Δ pol30-113 and rev3Δ pol30-113 mutants, the RAD52 and REV3 genes of the pol30-113 mutant of E134 (PS2001) were disrupted with a selectable kanMX cassette (35). E. coli strain BL21 (DE3) was used for overproduction and purification of GST-tagged Pol32 and untagged PCNA.
TABLE 1.
S. cerevisiae strains used in the UV sensitivity assay
| Strain | DNA damage tolerance defect | Genotype | Source |
|---|---|---|---|
| E134 | - | MATα ade5-1 lys2::InsEA14 trp1-289 his7-2 leu2-3,112 ura3-52 | Ref. 51 |
| PS2001 | pol30-113 | MATα ade5-1 lys2::InsEA14 trp1-289 his7-2 leu2-3,112 ura3-52 pol30-113 | This study |
| PS750 | rad14 | MATα ade5-1 lys2::InsEA14 trp1-289 his7-2 leu2-3,112 ura3-52 rad14Δ::hygB | Ref. 36 |
| PS2009 | rad14 pol30-113 | MATα ade5-1 lys2::InsEA14 trp1-289 his7-2 leu2-3,112 ura3-52 rad14Δ::hygB pol30-113 | This study |
| PS2025 | rad52 | MATα ade5-1 lys2::InsEA14 trp1-289 his7-2 leu2-3,112 ura3-52 rad52Δ::kanMX | This study |
| PS2027 | rad52 pol30-113 | MATα ade5-1 lys2::InsEA14 trp1-289 his7-2 leu2-3,112 ura3-52 rad52Δ::kanMX pol30-113 | This study |
| DD31 | rad18 | MATα ade5-1 lys2::InsEA14 trp1-289 his7-2 leu2-3,112 ura3-52 rad18Δ::kanMX | D. L. Daee and P. V. Shcherbakova, unpublished |
| PS2007 | rad18 pol30-113 | MATα ade5-1 lys2::InsEA14 trp1-289 his7-2 leu2-3,112 ura3-52 rad18Δ::kanMX pol30-113 | This study |
| PS446 | rev3 | MATα ade5-1 lys2::InsEA14 trp1-289 his7-2 leu2-3,112 ura3-52 rev3Δ::LEU2 | Ref. 31 |
| PS2005 | rev3 pol30-113 | MATα ade5-1 lys2::InsEA14 trp1-289 his7-2 leu2-3,112 ura3-52 rev3Δ::LEU2 pol30-113 | This study |
| PS487 | rev1 | MATα ade5-1 lys2::InsEA14 trp1-289 his7-2 leu2-3,112 ura3-52 rev1Δ::kanMX | M. R. Northam and P. V. Shcherbakova, unpublished |
| PS2003 | rev1 pol30-113 | MATα ade5-1 lys2::InsEA14 trp1-289 his7-2 leu2-3,112 ura3-52 rev1Δ::kanMX pol30-113 | This study |
| BY4742 | - | MATα his3Δ leu2Δ lys2Δ ura3Δ | Research Genetics |
| PS2011 | pol30-113 | MATα his3Δ leu2Δ lys2Δ ura3Δ pol30-113 | This study |
| BY4742 rad5Δ | rad5 | MATα his3Δ leu2Δ lys2Δ ura3Δ rad5Δ::kanMX | Research Genetics |
| PS2013 | rad5 pol30-113 | MATα his3Δ leu2Δ lys2Δ ura3Δ rad5Δ::kanMX pol30-113 | This study |
| BY4742 mms2Δ | mms2 | MATα his3Δ leu2Δ lys2Δ ura3Δ mms2Δ::kanMX | Research Genetics |
| PS2015 | mms2 pol30-113 | MATα his3Δ leu2Δ lys2Δ ura3Δ mms2Δ::kanMX pol30-113 | This study |
| BY4742 ubc13Δ | ubc13 | MATα his3Δ leu2Δ lys2Δ ura3Δ ubc13Δ::kanMX | Research Genetics |
| PS2017 | ubc13 pol30-113 | MATα his3Δ leu2Δ lys2Δ ura3Δ ubc13Δ::kanMX pol30-113 | This study |
| 1A-PSD105 | - | MATα ura3-52 ade2-101 leu2-3.112 | P. V. Shcherbakova, unpublished |
| PS2019 | pol30-113 | MATα ura3-52 ade2-101 leu2-3.112 pol30-113 | This study |
| PS546 | rad30 | MATα ura3-52 ade2-101 leu2-3.112 rad30Δ::kanMX | A. Lupu and P. V. Shcherbakova, unpublished |
| PS2023 | rad30 pol30-113 | MATα ura3-52 ade2-101 leu2-3.112 rad30Δ::kanMX pol30-113 | This study |
Purification of Rev1 Fragments
The yeast BJ2168 strain carrying pRS425-GALGST-Rev1(297–746) or pRS425-GALGST-Rev1(621–746) was grown at 30 °C in 3% glycerol/2% ethanol/0.1% glucose medium selective for the plasmids, and expression was then induced by incubating in yeast extract/peptone/0.1% glucose/2% galactose medium for 16–20 h. The cells (∼70 g) were harvested and washed with 500 ml of ice-cold water. The GST-tagged Rev1 proteins were purified on a glutathione-Sepharose 4B FF column by using a protocol described previously (5). To obtain untagged proteins, the GST-fused proteins bound to glutathione-Sepharose beads were treated overnight at 4 °C with PreScission protease (Amersham Biosciences).
PCNA Purification
The E. coli BL21 (DE3) containing pBL228, pBL228-pol30–113 or pBL228-pol30–79 was grown in LB medium containing ampicillin (50 mg/liter) at 37 °C to an A600 of 0.6 and induced with 1 mm IPTG for 3 h. The cells were harvested, resuspended in buffer containing 50 mm Tris, pH 7.5, 100 mm NaCl, and 10% sucrose, and frozen at −80 °C. All subsequent operations were carried out at 4 °C. After thawing, lysozyme was added to a concentration of 0.2 mg/ml, and cells were incubated for 30 min on a rocker. Cells were then sonicated to complete lysis, and cell extract was cleared by centrifugation at 20,000 × g for 30 min. Solid ammonium sulfate (0.325 g/ml) was stirred in the lysate, and the precipitate was spun down for 30 min at 10,000 × g. Supernatant was dialyzed against 25 mm Tris pH 7.4, 10% (v/v) glycerol, 1 mm EDTA, 5 mm DTT (Buffer 1) containing 0.1 m NaCl and applied onto HiTrap (1-ml) column equilibrated with Buffer 1 containing 0.1 m NaCl. PCNA was eluted with a 20-ml linear gradient from 0.1 to 1 m NaCl in Buffer 1. The fractions containing PCNA were pooled and dialyzed overnight against 25 mm phosphate buffer, pH 7.0, containing 0.01% Nonidet P-40, 10% glycerol and 5 mm DTT (Buffer 2). The dialyzed extract was loaded onto MonoS HR 5/5 (1-ml) column equilibrated with Buffer 2 and eluted with 5 ml of 500 mm phosphate buffer, pH 7.0, containing 0.01% Nonidet P-40, 10% glycerol and 5 mm DTT (Buffer 3). The flow through fractions containing PCNA were pooled and loaded onto Hydroxyapatite (Bio-scale CHT2–1, 2-ml) column equilibrated with Buffer 2. PCNA was eluted with a 40-ml linear gradient of 25–500 mm phosphate buffer (Buffer 2 to Buffer 3).
Purification of GST-tagged Pol32 Protein
The E. coli BL21 (DE3) containing pGEX-5X-1-Pol32 was grown in LB medium containing ampicillin (50 mg/L) at 37 °C to an A600 of 0.6–0.8. The temperature was then shifted to 30 °C, and IPTG was added to a final concentration of 2 mm. Cells were harvested after 5 h. Cell lysis and the nucleic acid precipitation were performed as described previously (37). To the cleared lysate, ammonium sulfate was added with stirring to a concentration of 0.28 g/ml. After incubation at 4 °C for 4 h, the lysate was spun at 20,000 × g for 1 h. The pellet was dissolved in 100 mm sodium phosphate buffer (pH 7.5) containing 0.15 m NaCl and 2 mm DTT and dialyzed against the same buffer overnight. The dialyzed extract was loaded onto 1-ml GSTrap FF column equilibrated with the same buffer, and GST-Pol32 was eluted with 20 mm glutathione (pH 7.5).
GST Pull-down Assays
GST fusion proteins (20 μg) coupled to glutathione-Sepharose beads were incubated with PCNA (20 μg) in 30 μl of buffer I (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 5 mm dithiothreitol, 0.01% Nonidet P-40, 10% glycerol) at 4 °C for 4 h on a rocker. The beads were spun down, and the unbound protein was collected. The beads were thoroughly washed three times with 10 volumes of buffer I. The bound proteins were eluted with 20 μl of sodium dodecyl sulfate (SDS) loading buffer. Proteins in various fractions were separated by electrophoresis in 4–12% SDS-polyacrylamide gel and detected by silver or Coomassie staining.
PCNA-coupled Affinity Beads Pull-down
Wild-type PCNA and PCNA-113 were coupled to Affi-Gel 15 beads (Bio-Rad) according to the manufacturer instructions. To achieve stable coupling, ∼10 mg of purified PCNA or PCNA-113 was dialyzed against coupling buffer (0.1 m MOPS pH 7.5, 80 mm CaCl2) at 4 °C for 12 h with three subsequent buffer changes. Affi-Gel 15 matrix pre-washed with cold water was mixed with PCNA in a final volume of 1 ml overnight. Coupling to the beads was monitored by the decrease in protein concentration in the supernatant. 20 μg of Rev1 fragments were incubated with 20 μl of either wild-type or mutant PCNA beads for 1 h at 4 °C in 20 mm Tris-HCl (pH 7.4), 200 mm NaCl, 1 mm DTT, 0.1 mm EDTA, 0.01% Nonidet P-40, and 10% glycerol. The unbound protein was removed by centrifugation, and beads were washed four times with the same buffer containing 1 m NaCl. The beads were resuspended in an equal volume of 2× loading buffer and incubated at 37 °C for 15 min. The eluted proteins were separated by SDS-PAGE and detected by silver staining.
Gel Filtration Analysis of PCNA Interaction with the PAD of Rev1
5 μg of Rev1(621–746) alone, 5 μg of PCNA alone, or a mixture of the two proteins were incubated in 500 μl of buffer B containing 20 mm Tris-HCl (pH 7.5), 100 mm NaCl, 1 mm EDTA, 1 mm dithiothreitol, 0.01% Nonidet P-40, and 10% glycerol for 60 min at 4 °C followed by incubation for 10 min at 25 °C. The proteins were then subjected to gel filtration on a Superdex 200 PC 3.2/30 column (Amersham Biosciences) equilibrated with buffer B at a flow rate of 0.25 ml/min at 4 °C. Fractions were collected, and the proteins were separated by electrophoresis in a 4–12% sodium dodecyl sulfate-polyacrylamide gel and detected by silver staining.
RESULTS
The PAD of Rev1 Interacts with PCNA at the Site Marked by the pol30-113 Mutation
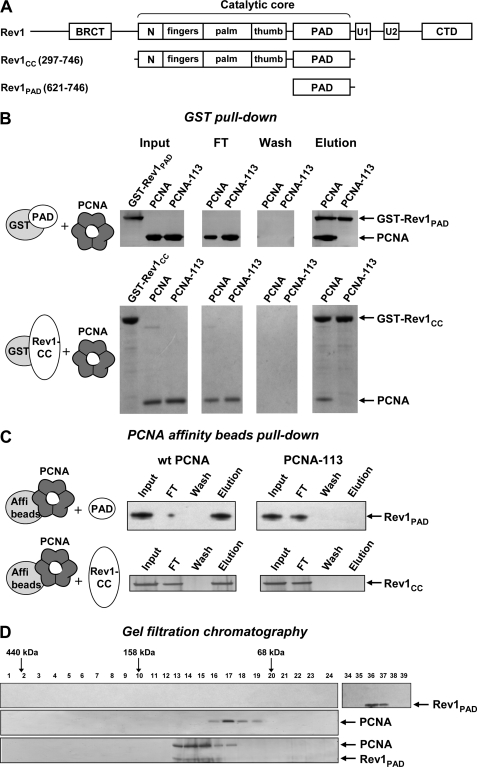
The first set of experiments was designed to determine whether the PAD of Rev1 binds to PCNA, and, if so, whether the interaction involves the region near the intermolecular interface of the PCNA trimer. To this end, we overproduced in yeast and purified two GST-tagged Rev1 fragments, one containing just the PAD (amino acids 621–746) and another containing the full catalytic core (CC) of Rev1 (amino acids 297–746) that includes the PAD (Fig. 1A). We also overproduced in E. coli and purified untagged yeast PCNA and its variant with a double amino acid substitution (E113G, L151S) at the subunit interface (PCNA-113). We first studied the interaction of Rev1 fragments with PCNA variants using the GST pull-down assay (Fig. 1B). Consistent with our hypothesis, we found that both GST-Rev1-PAD and GST-Rev1-CC efficiently pulled down PCNA but not PCNA-113. No nonspecific binding of PCNA or PCNA-113 to GST was observed (supplemental Fig. S1). We previously reported that PCNA-113 has a minor defect in trimer stability that is negligible in vivo, but becomes pronounced at low protein concentrations in vitro (31). Native PAGE analysis showed that PCNA-113 is predominantly a trimer at the concentration used in the GST pull-down experiment (data not shown). This indicated that the Rev1 interaction defect in PCNA-113 is not due to the inability to form the trimeric ring, but is likely caused by the local structural changes brought about by the two amino acid substitutions.
FIGURE 1.
The PAD of Rev1 interacts with PCNA at the site marked by the pol30-113 mutation. A, structure of S. cerevisiae Rev1 and its fragments used in this study. N, N-digit domain (50). U1 and U2, ubiquitin-binding motifs (23). CTD, C-terminal domain (40). B, purified GST-tagged Rev1 fragments were incubated with PCNA and glutathione-Sepharose beads and washed extensively. The bound proteins were eluted with sodium dodecyl sulfate (SDS) sample buffer, separated by electrophoresis in 4–12% SDS-polyacrylamide gel and detected by silver (top panel) or Coomassie (bottom panel) staining. FT, flow-through fraction. C, PCNA or PCNA-113 were coupled to Affi-gel 15 beads (Bio-Rad) and incubated with purified untagged Rev1 fragments. After extensive washing, the bound Rev1 fragments were eluted with SDS sample buffer, separated by electrophoresis in 4–12% SDS-polyacrylamide gel and detected by silver staining. D, Rev1-PAD (top), wild-type PCNA (middle), or the mixture of PCNA and Rev1-PAD preincubated for 1 h at 4 °C (bottom) was analyzed by gel filtration on a Superdex 200 PC 3.2/30 column (Amersham Biosciences). Fraction numbers are shown above the gel image. Ferritin (440 kDa), aldolase (158 kDa), and albumin (68 kDa) eluted at the positions indicated by the arrows. Cytochrome c (12 kDa) eluted in fractions 43–44 (not shown).
To provide additional evidence for the specific interaction of Rev1-PAD with PCNA, we purified untagged Rev1 fragments and studied their binding to PCNA covalently coupled to Affi-Gel 15 beads (Fig. 1C). The PCNA-coupled beads efficiently pulled down both Rev1-PAD and Rev1-CC. This interaction is specific, because we observed no binding of Rev1 fragments to BSA-coupled Affi-Gel 15 beads used as a control (supplemental Fig. S2). Neither Rev1 fragment was able to bind to PCNA-113-coupled beads, confirming that the Rev1-PCNA interaction requires the region near the intermolecular interface of PCNA.
Further, we used gel filtration to examine whether untagged Rev1-PAD can form a stable complex with PCNA in vitro. As shown in Fig. 1D (middle panel), PCNA alone elutes from the gel filtration column at a position corresponding to the molecular weight of the homotrimer. However, when PCNA was preincubated with Rev1-PAD, the vast majority of PCNA co-elutes with Rev1-PAD in earlier fractions with an apparent molecular mass of 130 kDa (Fig. 1D, bottom panel). When Rev1-PAD alone was subjected to gel filtration, it eluted much later (fractions 36–37 according to the numbering used in Fig. 1D), in agreement with its molecular mass of 14 kDa (upper panel). The gel filtration analysis, thus, indicated that PCNA and the PAD of Rev1 interact, consistent with the results of GST pull-down and PCNA affinity beads pull-down assays.
The pol30-113 Mutation Does Not Affect the Interaction of PCNA with the Pol32 Subunit of DNA Polymerase δ
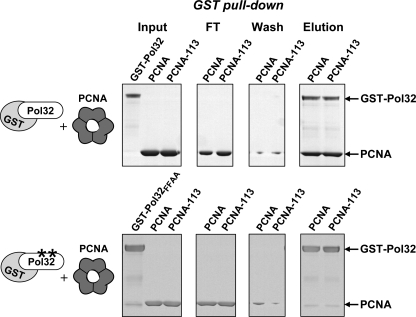
Most proteins that interact with PCNA possess the PIP box that mediates their binding to the interdomain connector loop or the carboxyl terminus of PCNA (28). The studies described in Fig. 1 suggested that Rev1 uses a different binding site near the subunit interface of PCNA, because the interaction between the PAD of Rev1 and PCNA was disrupted by amino acid substitutions in this region. Similarly, we previously observed that the pol30-113 mutation disrupts the functional interaction of PCNA with Polζ (31). At the same time, PCNA-113 was proficient in stimulating DNA synthesis by Polδ and Polη that are thought to interact with PCNA through the PIP box (31). The GST pull-down assays (Fig. 2, top) showed that PCNA-113 is also fully capable of interacting with a subunit of Polδ (Pol32) that carries the PIP box. Consistent with the previous studies (37), the interaction of Pol32 with both wild-type PCNA and PCNA-113 was disrupted by a deletion of nine C-terminal amino acids of Pol32 containing the PCNA-binding motif (Fig. 2, bottom). This demonstrated that PCNA-113, while being defective in the interaction with Rev1 and Polζ, retains the ability to interact with proteins that bind to regions other than the subunit interface.
FIGURE 2.
Interaction of wild-type PCNA and PCNA-113 with Pol32. Purified GST-tagged Pol32 (top) and its derivative with a double amino acid substitution in the PCNA interaction motif (bottom) were incubated with wild-type PCNA or PCNA-113 and glutathione-Sepharose beads and analyzed as in Fig. 1B.
Interaction between the Catalytic Core of Rev1 and PCNA Does Not Involve the Interdomain Connector Loop of PCNA
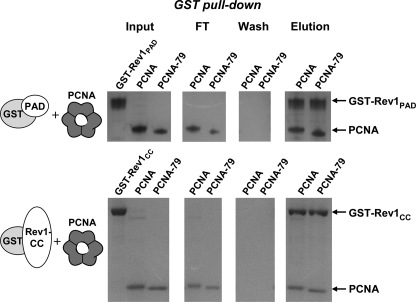
Experiments shown in Figs. 1 and 2 suggested that the mode of interaction of the catalytic core of Rev1 with PCNA is different from that used by PIP box-containing proteins. To further support this conclusion, we utilized a previously characterized PCNA mutant (PCNA-79) with a double amino acid substitution in the interdomain connector loop of PCNA. The PCNA-79 has been shown to be defective in the physical and functional interaction with Polδ (33). We overproduced in E. coli and purified PCNA-79 and studied its interaction with Rev1-PAD and Rev1-CC in the GST pull-down assays. As expected, PCNA-79 was fully capable of interacting with the Rev1 fragments (Fig. 3). Consistent with the earlier report (33), we observed no interaction of PCNA-79 with GST-Pol32 (data not shown).
FIGURE 3.
The protein binding site at the interdomain connector loop of PCNA is dispensable for the interaction with the catalytic core of Rev1. Purified GST-tagged Rev1 fragments were incubated with wild-type PCNA or PCNA-79 and glutathione-Sepharose beads and analyzed as in Fig. 1B.
Yeast pol30-113 Mutant Is Specifically Defective in the Rev1/Polζ-dependent TLS
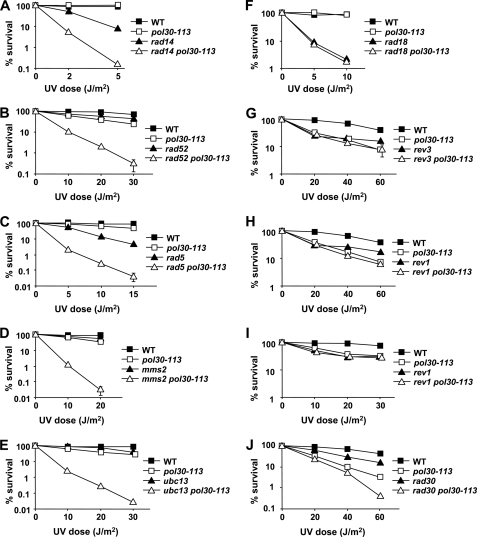
The biochemical data (Ref. 31 and Figs. 1 and 2) suggested that the pol30-113 mutation disrupts the binding of PCNA to Polζ and Rev1, but not to Polδ or Polη. In agreement with this, the yeast pol30-113 strains were completely deficient in UV-induced mutagenesis that depends on Polζ and Rev1, but showed no defect in DNA replication that requires the interaction of PCNA with Polδ (31). PCNA, however, is involved in multiple other processes including several DNA repair pathways, homologous recombination and the Rad5/Mms2/Ubc13-dependent damage tolerance pathway. To determine which of these processes are affected by the pol30-113 mutation, we determined epistatic relationships of pol30-113 with other DNA repair or damage tolerance mutations in regard to UV sensitivity.
The combinations of the pol30-113 mutation with deletions of several genes led to a synergistic increase in UV sensitivity. These included the RAD14 and RAD52 genes controlling nucleotide excision repair and homologous recombination, respectively (Fig. 4, A and B), as well as RAD5, MMS2, and UBC13 genes controlling the error-free branch of the Rad18-dependent DNA damage tolerance pathway (Fig. 4, C–E). This indicated that, in each of these five cases, the pol30-113 mutation and the other DNA repair or damage tolerance mutation disrupted different, non-overlapping pathways that provide resistance to UV-induced damage. Thus, nucleotide excision repair, homologous recombination and the Rad5-dependent damage tolerance are functional in the pol30-113 mutants.
FIGURE 4.
Effect of the pol30-113 mutation on survival of DNA repair and DNA damage tolerance mutants after UV irradiation. Yeast cells from appropriately diluted logarithmic cultures were plated onto YPD medium, immediately irradiated with 254 nm UV light as indicated and incubated for 4 days at 30 °C. Survival was determined by dividing the number of colonies on UV-irradiated plates by the number of colonies on untreated plates. Data are averages for three to six independent experiments. Standard errors are shown where the size of the error bar exceeds the size of the plot symbol.
In contrast, when the pol30-113 mutation was introduced into the rad18Δ strain, no further increase in UV sensitivity was seen above that conferred by the rad18Δ mutation alone (Fig. 4F). The RAD18 gene encodes the ubiquitin ligase that is responsible for the monoubiquitylation of PCNA in response to DNA damage (18). Thus, the Rad18 protein is required for all DNA damage tolerance pathways regulated by the ubiquitylation of PCNA: the Rev1/Polζ-dependent mutagenic TLS, Polη-dependent non-mutagenic TLS and the Rad5/Mms2/Ubc13-dependent error-free damage tolerance. Because the rad18Δ mutation is epistatic to pol30-113 (Fig. 4F), we conclude that the PCNA defect does not impair any damage response pathways other than those controlled by the RAD18. The pol30-113 mutant, however, is much more resistant to UV irradiation than the rad18Δ (Fig. 4F), consistent with the observation that the Rad5/Mms2/Ubc13-dependent branch of the RAD18 pathway is intact in the PCNA mutant (Fig. 4, C–E).
To further investigate which of the Rad18-dependent TLS subpathways is affected by the pol30-113 mutation in vivo, we combined it with the mutations inactivating Polζ (rev3Δ), Rev1 (rev1Δ), or Polη (rad30Δ). The UV sensitivity of the double rev3Δ pol30-113 and rev1Δ pol30-113 mutants was indistinguishable from that of the single pol30-113 mutant (Fig. 4, G and H). This indicated that the pol30-113 mutation and the inactivation of Polζ and Rev1 disrupt the same damage tolerance pathway, and, therefore, the protein binding site near the monomer-monomer interface of PCNA is essential for the Rev1/Polζ-dependent TLS. Interestingly, while the UV sensitivity of the single pol30-113 mutant was identical to that of the single rev3Δ or rev1Δ mutants at a lower dose of UV (20 J/m2), the rev3Δ and rev1Δ mutants showed a significantly higher survival at higher UV doses (Fig. 4, G and H). A more detailed comparison of the survival of rev1Δ, pol30-113 and the double rev1Δ pol30-113 mutants at lower UV doses showed that the sensitivities of all three strains were indistinguishable at doses up to 30 J/m2 (Fig. 4I). In contrast, the double rad30Δ pol30-113 mutant was significantly more sensitive to UV than either rad30Δ or pol30-113 single mutants (Fig. 4J), indicating that the pol30-113 mutation does not disrupt Polη-dependent TLS.
DISCUSSION
In this work, we report three novel observations relevant to the interaction of the key regulators of TLS, Rev1 and PCNA. First, we showed that the PAD of yeast Rev1 interacts with PCNA in vitro. Second, we showed that this interaction requires the non-traditional protein binding site near the monomer-monomer interface of the trimeric PCNA. Third, the epistatic analysis demonstrated that yeast mutants lacking this protein binding site are specifically defective in the Rev1/Polζ-dependent TLS and not in other DNA repair or damage tolerance pathways. In our earlier study, we observed that the binding site near the intermolecular interface of PCNA is also essential for the functional interaction of Polζ with PCNA (31). Taken together, these studies suggest that the non-traditional binding site is uniquely used by the proteins in the Rev1/Polζ pathway, while most PCNA partners bind to other sites, such as the interdomain connector loop or the C terminus of PCNA. The presence of the additional binding site increases the number of proteins that could potentially be simultaneously bound to PCNA, and, thus, expands the opportunities for the regulation of various processes that PCNA is involved in.
Several conflicting reports on the nature of Rev1-PCNA interaction have been published previously. Guo et al. (30) reported that the mouse Rev1 interacts with PCNA via its N-terminal part containing the BRCT domain. The region required for the functional interaction of the yeast Rev1 with PCNA was found to be confined to ∼200 C-terminal amino acids of Rev1 (26). A contribution of regions other than the C terminus to the physical interaction with PCNA has also been suggested (26). Earlier, the PCNA binding by the human REV1 was shown to involve a ∼125 amino acid region downstream of the catalytic core, and therefore, of the PAD (29). The interaction of PCNA with this portion of the human REV1, however, was only observed in a mammalian two-hybrid system, and could thus be indirect. Finally, another study found no physical or functional interaction between yeast Rev1 and PCNA (38). The demonstration of the ability of the PAD of Rev1 to bind to PCNA in the present study further illustrates the complexity of the Rev1-PCNA interaction. The apparent discrepancies could, in part, reflect species-specific differences. However, as discussed below, the interaction of the PAD/“little finger” domains with the site at monomer-monomer interface of processivity clamps is apparently conserved even between prokaryotic and eukaryotic species. It is likely, therefore, that this interaction could occur in the mammalian cells as well. It is possible that Rev1 possesses multiple PCNA-binding sites that could be used individually or in combination to mediate various modes of interaction. Such versatile PCNA interaction ability could allow the Rev1 protein to perform its complex functions in the regulation of TLS.
It remains to be established what amino acid sequence motifs mediate protein binding to the intermolecular interface area on PCNA. A recent study described an alternative PCNA-binding motif (K/R)(F/Y/W)(L/I/V/A)(L/I/V/A)(K/R) conserved in many proteins that lack the classical QXX(L/I)XXFF motif (39). This motif is present in Polζ (39), but is absent in the PAD of Rev1. Interestingly, yeast Rev1 contains an exact match to this motif, KYLIK, in its C-terminal part (amino acids 913–917). The motif overlaps with a previously identified stretch of amino acids that is conserved in REV1 proteins from various species (40). The PCNA-binding motif as defined by Gilliam and coauthors, however, is only present in S. cerevisiae and closely related yeasts. Alanine substitutions for the first three residues in this motif lead to increased UV sensitivity and a defect in UV-induced mutagenesis (40). It would be interesting to determine whether the KYLIK sequence could contribute to PCNA binding by the yeast Rev1.
In addition to PCNA-113, two other PCNA variants with amino acid changes at the subunit interface have been studied. A screen for mutants defective in UV-induced mutagenesis identified a G178S substitution in PCNA located within 5 Å of Leu-151 mutated in pol30-113 (41). In contrast to the pol30-113 mutation, the G178S substitution disrupted all the Rad6/Rad18-dependent DNA damage tolerance mechanisms, including TLS by Polζ/Rev1, Polη and the error-free recombination-dependent mechanism. It was, therefore, suggested that the G178S mutation prevented ubiquitylation of Lys-164 in PCNA (41), although this has not been directly tested. In another study, a crystal structure was reported for a PCNA variant carrying one of the two amino acid substitutions present in pol30-113, E113G (42). The structural change in the intermolecular interface region brought about by the E113G substitution was found to be similar to the one in the G178S variant (43). Both G178S and E113G changes were found to impair the ability of PCNA to stimulate Polη (43). Despite the similar position in the PCNA structure, the pol30-113 mutation has no significant effect on PCNA ubiquitylation (31) or the Rad5-dependent branch of the Rad18 pathway (Fig. 4, C–E and G–I). We observed only a very mild (∼30%) reduction in the stimulation of Polη in vitro (31) and no defect in Polη-dependent TLS in pol30-113 mutants in vivo (Fig. 4, G–J). This suggests that the structural changes introduced by the pol30-113 mutation (double E113G,L151S substitution) are different from those in PCNA-E113G and PCNA-G178S. In addition, the absence of Polη stimulation by PCNA-E113G could reflect the fact that this PCNA variant is expected to have a severe defect in the trimer stability under the assay conditions used in that study (a low PCNA concentration and no macromolecular crowding agent; Refs. 42, 43). Future studies of PCNA-113 structure could help characterize the changes in the protein that alter its interactions with TLS polymerases.
The PAD/“little finger” domain is unique to Y family DNA polymerases and plays an important role in DNA binding by these enzymes (44). The PAD domains of different Y family DNA polymerases have no amino acid sequence similarity but share the same overall architecture. Binding of the PAD of Rev1 and the little finger of E. coli Pol IV near the intermolecular interface of the corresponding processivity clamps (Fig. 1 and Ref. 32) suggests that this mode of clamp binding may be conserved within the Y family. However, with the exception of REV1, all other eukaryotic Y-family DNA polymerases, Polη, Polι, and Polκ, carry the PIP box that mediates their association with PCNA (45–47). In addition, the Y-family polymerases associate with ubiquitylated PCNA through their ubiquitin-binding motifs (22–26). The presence of a structurally conserved PAD in all Y family polymerases suggests a possibility that, like Rev1, the other family members might bind to the site near the subunit interface of PCNA through the PAD. This interaction, however, is likely to be secondary to the ones mediated by the PIP box and the ubiquitin-binding motifs.
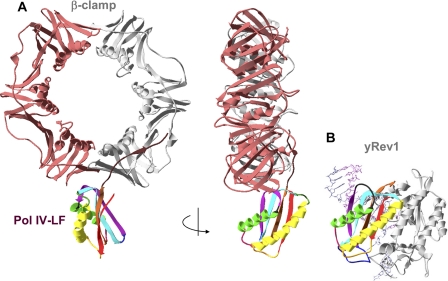
The crystallographic studies of the E. coli Pol IV-LF-β-clamp complex suggested that the interaction of Pol IV-LF at the subunit interface would lock Pol IV in an inactive conformation with the active site away from the primer terminus (32). Comparison of the orientations of the PAD domain in the yeast Rev1-DNA-dCTP ternary complex and the LF domain in the E. coli Pol IV-LF-β-clamp complex (Fig. 5) suggests that the PAD binding at the clamp subunit interface is also likely to keep Rev1 in an inactive conformation. If the mode of interaction of LF/PAD domains and the clamps is conserved between E. coli and eukaryotes, DNA passing through the central hole of the clamp would need to bend almost 180° to enter the active site of Rev1 bound at the subunit interface. It is conceivable that such an inactive conformation could be predominantly adopted by Rev1 that plays an organizing rather than catalytic role in most types of TLS.
FIGURE 5.
Binding of the PAD domain of Rev1 at the clamp subunit interface is likely to keep Rev1 in an inactive conformation. A, orthogonal views of the structure of Pol IV-LF-β-clamp complex (32). B, structure of Rev1-DNA-dCTP ternary complex (50) with the orientation of the PAD domain (rainbow-colored) matching that of the Pol IV-LF in panel A.
Our earlier studies suggested that the PCNA region marked by the pol30-113 mutation also mediates the interaction with Polζ (31). Rev1 and Polζ act in concert during TLS, and the organizing function of Rev1 is required for the vast majority if not all Polζ-dependent processes (3). Thus, Rev1 and Polζ are likely to be present simultaneously at the replication fork. It remains to be determined how the same site at the monomer-monomer interface of PCNA is used for the interaction with both proteins. The presence of three identical monomer-monomer interface sites in the PCNA trimer could potentially allow Rev1 and Polζ to bind simultaneously to different sites. Alternatively, an attractive possibility is that it is Rev1 and not Polζ that predominantly occupies the intermolecular interface site, providing a docking station for the other polymerases. Rev1 then hands over the intermolecular interface site to Polζ as the latter is being recruited to the primer terminus. In this case, Rev1 could remain bound to PCNA through additional interaction sites while Polζ is synthesizing DNA. Consistent with this model, the interaction between Polζ and PCNA has been observed during Polζ-dependent DNA synthesis (14, 31), but has not been reported to occur in the absence of DNA.
Given the plethora of PCNA partners, it is not surprising that the same site on PCNA is used for interaction with more than one protein. This is best illustrated by the wide use of the PIP motif for binding to the interdomain connector loop of PCNA (28). Similarly, the same ∼100 amino acids at the C terminus of Rev1 mediate the interaction with multiple DNA polymerases (3). We have identified the region near the intermolecular interface of PCNA as a site of interaction with both Polζ and the PAD of Rev1. Future studies will help understand the hierarchy and regulation of these interactions. Interestingly, the region of Rev1 containing PAD and the linker connecting PAD to the thumb subdomain was also found to be necessary and sufficient for the interaction with Polη (48) and the Rev7 subunit of Polζ (49). Thus, the PAD may represent another universal module that, in addition to its role in DNA binding, is involved in facilitating DNA polymerase exchange at the primer terminus.
Supplementary Material
Acknowledgments
We thank Peter Burgers for pRS425-GALGST, plasmid, Victoria Liston, Corinn Grabow, and Elizabeth Moore for technical assistance, and Youri Pavlov for helpful discussions.
This work was supported, in whole or in part, by Nebraska DHHS Grant LB506, and National Institutes of Health Grants ES011644 and ES015869 (to P. V. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
D. M. Baitin and P. V. Shcherbakova, unpublished work.
- TLS
- translesion synthesis
- Polζ
- DNA polymerase ζ
- PCNA
- proliferating cell nuclear antigen
- PAD
- polymerase-associated domain
- CC
- catalytic core.
REFERENCES
- 1. Broyde S., Wang L., Rechkoblit O., Geacintov N. E., Patel D. J. (2008) Trends Biochem. Sci. 33, 209–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. McCulloch S. D., Kunkel T. A. (2008) Cell Res. 18, 148–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Waters L. S., Minesinger B. K., Wiltrout M. E., D'Souza S., Woodruff R. V., Walker G. C. (2009) Microbiol. Mol. Biol. Rev. 73, 134–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yang W. (2005) FEBS Lett. 579, 868–872 [DOI] [PubMed] [Google Scholar]
- 5. Nelson J. R., Lawrence C. W., Hinkle D. C. (1996) Nature 382, 729–731 [DOI] [PubMed] [Google Scholar]
- 6. Masuda Y., Takahashi M., Tsunekuni N., Minami T., Sumii M., Miyagawa K., Kamiya K. (2001) J. Biol. Chem. 276, 15051–15058 [DOI] [PubMed] [Google Scholar]
- 7. Masuda Y., Takahashi M., Fukuda S., Sumii M., Kamiya K. (2002) J. Biol. Chem. 277, 3040–3046 [DOI] [PubMed] [Google Scholar]
- 8. Haracska L., Prakash S., Prakash L. (2002) J. Biol. Chem. 277, 15546–15551 [DOI] [PubMed] [Google Scholar]
- 9. Fukuda H., Takamura-Enya T., Masuda Y., Nohmi T., Seki C., Kamiya K., Sugimura T., Masutani C., Hanaoka F., Nakagama H. (2009) J. Biol. Chem. 284, 25585–25592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lin W., Xin H., Zhang Y., Wu X., Yuan F., Wang Z. (1999) Nucleic Acids Res. 27, 4468–4475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang Y., Wu X., Rechkoblit O., Geacintov N. E., Taylor J. S., Wang Z. (2002) Nucleic Acids Res. 30, 1630–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhou Y., Wang J., Zhang Y., Wang Z. (2010) Nucleic Acids Res. 38, 5036–5046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Acharya N., Johnson R. E., Pagès V., Prakash L., Prakash S. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 9631–9636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Garg P., Stith C. M., Majka J., Burgers P. M. (2005) J. Biol. Chem. 280, 23446–23450 [DOI] [PubMed] [Google Scholar]
- 15. Haracska L., Johnson R. E., Unk I., Phillips B., Hurwitz J., Prakash L., Prakash S. (2001) Mol. Cell. Biol. 21, 7199–7206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Haracska L., Johnson R. E., Unk I., Phillips B. B., Hurwitz J., Prakash L., Prakash S. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 14256–14261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Haracska L., Unk I., Johnson R. E., Phillips B. B., Hurwitz J., Prakash L., Prakash S. (2002) Mol. Cell. Biol. 22, 784–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hoege C., Pfander B., Moldovan G. L., Pyrowolakis G., Jentsch S. (2002) Nature 419, 135–141 [DOI] [PubMed] [Google Scholar]
- 19. Kannouche P. L., Wing J., Lehmann A. R. (2004) Mol. Cell 14, 491–500 [DOI] [PubMed] [Google Scholar]
- 20. Stelter P., Ulrich H. D. (2003) Nature 425, 188–191 [DOI] [PubMed] [Google Scholar]
- 21. Haracska L., Torres-Ramos C. A., Johnson R. E., Prakash S., Prakash L. (2004) Mol. Cell. Biol. 24, 4267–4274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bienko M., Green C. M., Crosetto N., Rudolf F., Zapart G., Coull B., Kannouche P., Wider G., Peter M., Lehmann A. R., Hofmann K., Dikic I. (2005) Science 310, 1821–1824 [DOI] [PubMed] [Google Scholar]
- 23. Guo C., Tang T. S., Bienko M., Parker J. L., Bielen A. B., Sonoda E., Takeda S., Ulrich H. D., Dikic I., Friedberg E. C. (2006) Mol. Cell. Biol. 26, 8892–8900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Parker J. L., Bielen A. B., Dikic I., Ulrich H. D. (2007) Nucleic Acids Res. 35, 881–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Plosky B. S., Vidal A. E., Fernández de Henestrosa A. R., McLenigan M. P., McDonald J. P., Mead S., Woodgate R. (2006) EMBO J. 25, 2847–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wood A., Garg P., Burgers P. M. (2007) J. Biol. Chem. 282, 20256–20263 [DOI] [PubMed] [Google Scholar]
- 27. Garg P., Burgers P. M. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 18361–18366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Majka J., Burgers P. M. (2004) Prog. Nucleic Acids Res. Mol. Biol. 78, 227–260 [DOI] [PubMed] [Google Scholar]
- 29. Ross A. L., Simpson L. J., Sale J. E. (2005) Nucleic Acids Res. 33, 1280–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Guo C., Sonoda E., Tang T. S., Parker J. L., Bielen A. B., Takeda S., Ulrich H. D., Friedberg E. C. (2006) Mol. Cell 23, 265–271 [DOI] [PubMed] [Google Scholar]
- 31. Northam M. R., Garg P., Baitin D. M., Burgers P. M., Shcherbakova P. V. (2006) EMBO J. 25, 4316–4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bunting K. A., Roe S. M., Pearl L. H. (2003) EMBO J. 22, 5883–5892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Eissenberg J. C., Ayyagari R., Gomes X. V., Burgers P. M. (1997) Mol. Cell. Biol. 17, 6367–6378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fortune J. M., Stith C. M., Kissling G. E., Burgers P. M., Kunkel T. A. (2006) Nucleic Acids Res. 34, 4335–4341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wach A., Brachat A., Pöhlmann R., Philippsen P. (1994) Yeast 10, 1793–1808 [DOI] [PubMed] [Google Scholar]
- 36. Northam M. R., Robinson H. A., Kochenova O. V., Shcherbakova P. V. (2010) Genetics 184, 27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Johansson E., Garg P., Burgers P. M. (2004) J. Biol. Chem. 279, 1907–1915 [DOI] [PubMed] [Google Scholar]
- 38. Haracska L., Unk I., Prakash L., Prakash S. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 6477–6482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gilljam K. M., Feyzi E., Aas P. A., Sousa M. M., Müller R., Vågbø C. B., Catterall T. C., Liabakk N. B., Slupphaug G., Drabløs F., Krokan H. E., Otterlei M. (2009) J. Cell Biol. 186, 645–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. D'Souza S., Waters L. S., Walker G. C. (2008) DNA Repair 7, 1455–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang H., Gibbs P. E., Lawrence C. W. (2006) Genetics 173, 1983–1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Freudenthal B. D., Gakhar L., Ramaswamy S., Washington M. T. (2009) Acta Crystallogr. D Biol. Crystallogr. 65, 560–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Freudenthal B. D., Ramaswamy S., Hingorani M. M., Washington M. T. (2008) Biochemistry 47, 13354–13361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yang W. (2003) Curr. Opin. Struct. Biol. 13, 23–30 [DOI] [PubMed] [Google Scholar]
- 45. Haracska L., Kondratick C. M., Unk I., Prakash S., Prakash L. (2001) Mol. Cell 8, 407–415 [DOI] [PubMed] [Google Scholar]
- 46. Ogi T., Kannouche P., Lehmann A. R. (2005) J. Cell Sci. 118, 129–136 [DOI] [PubMed] [Google Scholar]
- 47. Vidal A. E., Kannouche P., Podust V. N., Yang W., Lehmann A. R., Woodgate R. (2004) J. Biol. Chem. 279, 48360–48368 [DOI] [PubMed] [Google Scholar]
- 48. Acharya N., Haracska L., Prakash S., Prakash L. (2007) Mol. Cell. Biol. 27, 8401–8408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Acharya N., Haracska L., Johnson R. E., Unk I., Prakash S., Prakash L. (2005) Mol. Cell. Biol. 25, 9734–9740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nair D. T., Johnson R. E., Prakash L., Prakash S., Aggarwal A. K. (2005) Science 309, 2219–2222 [DOI] [PubMed] [Google Scholar]
- 51. Shcherbakova P. V., Kunkel T. A. (1999) Mol. Cell. Biol. 19, 3177–3183 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.