Abstract
Interferon regulatory factor 3 (IRF-3) plays a central role in inducing the expression of cellular antiviral genes, including the interferon-β gene, in response to Pattern Recognition Receptors. IRF-3 is targeted for proteasome-mediated degradation, which modulates the strength and duration of the innate immune responses that depend on it. We have found that caspase-8, which is activated by cytosolic RIG-I-dependent signaling, catalyzes an essential intermediate step in the ubiquitination and proteasome-mediated degradation of IRF-3. Mutation of a consensus cleavage site within IRF-3 generates a form that is not cleaved by caspase-8 and that is protected from ubiquitination and degradation. An in vitro assay confirms the direct action of caspase-8 cleavage on IRF-3. We also show that caspase-8-mediated cleavage of IRF-3 helps to modulate dsRNA-dependent gene induction.
Keywords: Caspase, Double-stranded RNA, Proteasome, Protein Degradation, Ubiquitination, Caspase-8, IRF-3, Proteasomal Degradation, RIG-I
Introduction
A central component of host two-part immune system is the initial rapid innate response that responds to early steps in infection. This response is orchestrated by pattern-recognition receptors (PRRs) that respond to distinct pathogen-associated molecular patterns (PAMPs). Virus-associated PAMPs include their genomic DNAs and RNAs, or double-stranded RNAs (dsRNAs), generated by viral replication. Toll-like receptor-3 (TLR-3),2 an endosomal membrane-bound PRR, recognizes extracellular viral dsRNA to initiate downstream signaling pathways that activate the transcription factors IRF-3, NF-κB, and AP-1. In response to intracellular dsRNA, cytoplasmic helicases (RLHs) that include a caspase-recruiting domain (CARD), such as RIG-I, MDA5, or LGP2, generate similar responses by activating the mitochondrial adaptor protein IPS-1. A crucial aspect of this antiviral system is the synthesis and secretion of the type I interferons, especially IFN-β, to coordinate and initiate adaptive immunity (1, 2). IRF-3, upon activation by the TLR3 or RLH signaling pathways, is phosphorylated, dimerizes, and translocates to the nucleus to transcribe antiviral genes, including the IFN-β gene. The intricacies of IRF-3 activation, modulation, and subsequent degradation are coordinated through myriad post-translational modifications, including phosphorylation, ubiquitination, sumoylation, S-glutathionylation, and ISGylation, which help to shape the strength and duration of its action (3–6). Phosphorylation-dependent ubiquitination terminates IRF-3 function by proteasome-dependent degradation (7, 8). However, the precise events in this pathway are not completely understood, and several cellular proteins have been implicated in the proteasome-mediated degradation of IRF-3 (4, 5, 8–12).
In addition to the induction of antiviral genes, infections by viruses often result in premature cell death by triggering apoptosis in response to activation of caspases, and IRF-3 plays an essential role in mediating virus-induced apoptosis (13, 14). We have discovered a novel apoptotic pathway that is induced by IRF-3-mediated activation of the pro-apoptotic protein BAX (15, 16). Several different caspases are activated during IRF-3-mediated apoptosis. Caspases are also involved in other important aspects of antiviral signaling. FADD, a critical component of interferon response, has been associated with caspase-8 and plays a critical role in antiviral responses and apoptosis. In addition to the production of inflammatory cytokines through IRF-3, the activation of caspases has been implicated in antiviral signaling (9, 17, 18). Caspase-8, which is predominantly involved in triggering cellular apoptosis, also plays additional important roles. Caspase-8 deficiency leads to impaired activation of NF-κB (19). A recent study showed that deficiency of caspase-8 in mouse keratinocytes leads to the hyperactivation of IRF-3 (18). Caspase-8-deficient mice showed up-regulation of pro-inflammatory cytokines. Additional work has revealed that several proteins of antiviral signaling pathways are processed by caspases to attenuate antiviral gene induction. Both of the antiviral adaptor proteins IPS-1 and TRIF are processed in a caspase-dependent manner (20). In addition, the NF-κB activating kinase RIP4 is processed by caspases (21). Various studies have suggested that caspase-mediated processing of antiviral proteins provides potent pro-viral mechanisms for evading the host innate immune responses. Therefore, a critical understanding of the caspase-mediated attenuation of cellular responses is important to develop suitable strategies against viral infections.
In the present investigation, we report that caspase-8 catalyzes cleavage of IRF-3 during dsRNA-dependent signaling. Further, we show that the disruption of caspase-8 activity prevents signal-dependent degradation of IRF-3. Finally, we report that mutation of the caspase cleavage site in IRF-3 inhibits the ubiquitination of IRF-3 and thus disrupts its degradation. Caspase-8-mediated cleavage and degradation of activated IRF-3 is involved in the down-regulation of dsRNA-inducible genes.
EXPERIMENTAL PROCEDURES
Cells and Reagents
HT1080 cells (human fibrosarcoma cells, (22)), HT1080/RIG-Ic (HT1080 cells expressing dominant negative RIG-I mutant, (13)), P2.1 (HT1080 derived chemically mutagenized cells, expressing very low level of endogenous IRF-3, (23)), and ARPE19 (human retinal epithelial cells, (24, 25)) were described previously. P2.1/IRF-3-Flag cells were generated by using lentiviral transduction, as described below. ARPE19/Cas-8 cells were generated by transfecting ARPE19 cells with a wild-type caspase-8 plasmid and selecting stable transfectants with G418. Antibody against human IRF-3 was a gift from Michael David (University of California, San Diego, CA). Antibodies against human P60 and P56 were raised in our laboratory. Antibodies against hemagglutinin (HA), Flag and actin were obtained from Sigma-Aldrich, caspase-8 was from Cell Signaling, cleaved Bid was from Abcam. Poly(I:C) was from GE Healthcare or Sigma-Aldrich. FuGENE 6 and Lipofectamine 2000 were from Roche Diagnostics and Invitrogen, respectively. All caspase inhibitors were obtained from Calbiochem and used according to the manufacturer's protocol. Staurosporine and doxorubicin were from Sigma-Aldrich. Active form of recombinant caspase-8 was obtained from Millipore.
Poly(I:C) Treatment
Stocks were prepared by re-suspending poly(I:C) in PBS and passing it through a 26-gauge needle to shear it. For activation of intracellular receptors, poly(I:C) was transfected into cells by using FuGENE 6. For each milliliter of cell medium, 4 μg of poly(I:C) was combined with 6 μl of FuGENE in 100 μl of OptiMEM (Invitrogen), followed by incubation for 15–30 min before adding the mixture to supernatant media containing serum. For extracellular stimulation of TLR3, 100 μg/ml dsRNA was added directly to cells.
Virus Infections
Sendai virus (SeV) infections were carried out as described before (13). Briefly, cells were washed with serum-free DMEM and infected in DMEM containing 2% serum at MOI:10 for 1 h, with occasional gentle agitation, the virus was removed and cells were washed and placed in complete DMEM for the indicated times. Adenovirus strain Ad5 was a kind gift from Dr. Thomas Shenk (Princeton University). For infection, HT1080 cells were washed twice with virus infection medium (serum-free DMEM) and then placed in a minimal amount of virus infection medium. Adenovirus was used at an MOI of 10. Cells were incubated with virus for 1 h with gentle agitation. The virus was removed, and cells were washed twice with complete medium. The cells were placed in complete medium until they were harvested, as indicated in the figure legends.
Lentiviral Transduction
293T cells were transfected with pWPI-IRF-3-FLAG, pCMV-R8.2 and pVSV-G plasmids using Lipofectamine 2000 reagent (Invitrogen), according to the manufacturer's protocol. Cell supernatant solutions were collected after 36 h, filtered through a 0.45 μm filter, combined with polybrene (4 μg/ml, Sigma-Aldrich), and used to infect P2.1 cells. Serial dilution of supernatant media from virus-infected cells was used, ranging from 1:1 to 1:1000. GFP expression confirmed positive transduction, while Western analysis confirmed the levels of protein expression.
Expression of Caspase-8
ARPE19 cells, naturally deficient in caspase-8, were made to express this enzyme by using the vector pcDNA3-caspase-8 and Lipofectamine 2000. The pcDNA3-caspase-8 plasmid was a kind gift from Dr. Marcus Peter (University of Chicago). After transfection, cells were selected with G418 at 800 μg/ml for 2 weeks. The concentration of G418 was maintained at 400 μg/ml for subsequent cell growth.
Western Analyses and Immunoprecipitations
Western blots were performed as described elsewhere (13). Cells grown in DMEM containing 5% FBS were treated as indicated. The cells were then washed with PBS twice, scraped into PBS and pelleted. The cells were lysed in 50 mm Tris buffer, pH 7.4, containing 150 mm NaCl, 0.1% Triton X-100, 1 mm sodium orthovanadate, 10 mm NaF, 10 mm glycerophosphate, 5 mm sodium pyrophosphate, and protease/phosphatase inhibitors on ice for 20 min. The lysates were clarified by centrifugation at 4 °C, and the supernatant solutions were harvested. Total protein extracts were separated in SDS-polyacrylamide gels (PAGE) followed by transfer and analysis by Western blot.
For IRF-3 immunoprecipitations, cells were lysed as described above. The protein extracts were mixed with Flag-linked beads (Exactacruz, Santa Cruz Biotechnology) for 16 h. The beads were washed in lysis buffer and incubated in SDS protein sample buffer at 100 °C. Beads were pelleted by centrifugation, and the supernatant solution was analyzed by Western blot.
Analysis of Ubiquitination, Degradation, and Cleavage of IRF-3
To analyze the ubiquitination of IRF-3, P2.1 cells expressing IRF-3 were transfected with HA-tagged ubiquitin (HA-Ub) and 48 h later the cells were transfected with poly(I:C) in the presence of a proteasome inhibitor (MG132) for the indicated times; the cell lysates were immunoprecipitated with Flag antibody and the immunoprecipitates were analyzed by Western blot, using anti-HA. The immunoprecipitation was carried out using Exactacruz reagents (Santa Cruz Biotechnology). For analysis of IRF-3 degradation, the proteins in the cell lysates after treatment or transfection of poly(I:C) were resolved by SDS-PAGE and analyzed by Western blot for IRF-3.
The cleavage of IRF-3 was analyzed using the Image J program (26), to quantify the full-length and the cleaved forms of IRF-3. Cleavage was calculated as the amount of cleaved IRF-3 over the total IRF-3 (full-length and cleaved) and presented as % cleavage.
RNA Preparation, Bead Array Gene Expression Analysis and Quantitative Real-time PCR
Total RNA was isolated from P2.1 cells expressing either WT or D121E IRF-3, at 12 and 24 h post-transfection of poly(I:C). RNA extraction was carried out by TRIzol (Invitrogen) according to the manufacturer's instructions. After RNA extraction, samples were treated with recombinant DNase I for 1 h. The enzyme was inactivated, and the RNA was further purified by using the RNAeasy kit (Qiagen). DNase-treated RNA was then analyzed in an Illumina gene array and the data analysis was carried out by using Illumina GenomeStudio V2009.2. Genes shown in Table 1 exhibited at least 1.5-fold induction at 12 h, and trended toward baseline by 20 h in the WT IRF-3-expressing cells.
TABLE 1.
dsRNA-induced genes that show sustained expression in the presence of an IRF-3 mutant protein that is not cleaved by caspase-8
P2.1 cells expressing Wt or D121E IRF-3 were transfected with poly(I:C) for 12 or 20 h. RNA was isolated and analyzed with a microarray. The numbers indicate fold inductions.
| GENES | WT-12h | DE-12h | WT-20h | DE-20h |
|---|---|---|---|---|
| PPP4R4 | 9.6 | 11.3 | 4.0 | 12.3 |
| OTOF | 3.4 | 5.9 | 2.3 | 10.1 |
| VAMP4 | 3.1 | 2.9 | 2.7 | 7.7 |
| KRT34 | 2.3 | 3.0 | 2.0 | 5.1 |
| ATP6V1C1 | 3.0 | 2.1 | 1.6 | 4.0 |
| TRAPPC6B | 8.9 | 5.4 | 4.1 | 6.3 |
| CT45A1 | 1.8 | 2.0 | 1.5 | 3.5 |
| RN7SK | 3.6 | 2.7 | 1.4 | 3.3 |
| CFI | 1.6 | 1.8 | 1.6 | 3.4 |
| PPP3CA | 1.5 | 1.5 | 0.6 | 2.4 |
| CT45A5 | 1.9 | 1.7 | 1.7 | 3.4 |
| GPBP1L1 | 1.7 | 2.1 | 0.9 | 2.6 |
| TROVE2 | 1.6 | 1.7 | 0.8 | 2.4 |
| MGC42630 | 1.6 | 2.2 | 1.1 | 2.6 |
| TESK2 | 2.1 | 2.3 | 1.5 | 2.9 |
| EIF4E3 | 1.7 | 1.7 | 1.1 | 2.5 |
| NNT | 2.2 | 1.6 | 1.2 | 2.7 |
| SRPK2 | 1.6 | 2.0 | 1.1 | 2.5 |
| CD3D | 2.8 | 2.0 | 2.3 | 3.5 |
| KBTBD3 | 2.4 | 1.8 | 1.3 | 2.4 |
| THAP1 | 1.5 | 2.0 | 1.3 | 2.4 |
| GABPB1 | 1.7 | 1.6 | 1.3 | 2.3 |
| SOCS2 | 1.8 | 1.8 | 1.7 | 2.7 |
| GPR56 | 2.1 | 2.0 | 2.0 | 2.9 |
| ZNF77 | 2.3 | 2.2 | 1.7 | 2.6 |
| TRIM9 | 2.0 | 2.3 | 1.5 | 2.4 |
| HSPC159 | 1.8 | 1.8 | 1.4 | 2.3 |
| ETNK1 | 2.1 | 1.7 | 1.4 | 2.3 |
| PMS2CL | 1.5 | 1.8 | 1.1 | 1.8 |
| RPS6KC1 | 1.5 | 1.8 | 1.4 | 2.1 |
| SERPINE1 | 2.2 | 1.6 | 1.8 | 2.6 |
| KIAA0564 | 1.6 | 1.7 | 1.6 | 2.3 |
| GDPD1 | 2.7 | 2.7 | 2.4 | 3.1 |
| ZNF197 | 1.7 | 1.7 | 1.3 | 2.0 |
| IFT52 | 1.9 | 1.6 | 1.6 | 2.3 |
| ETS1 | 1.5 | 1.8 | 1.5 | 2.2 |
| ZNF254 | 1.6 | 1.7 | 1.2 | 1.9 |
| TMEM135 | 2.0 | 1.9 | 1.6 | 2.2 |
| CCNYL1 | 1.8 | 1.9 | 1.5 | 2.1 |
| LOC650898 | 2.1 | 1.6 | 1.8 | 2.4 |
| SNAI2 | 2.2 | 2.0 | 2.0 | 2.5 |
| SLC25A20 | 2.1 | 1.6 | 1.5 | 2.0 |
| SESN3 | 2.0 | 1.5 | 1.9 | 2.2 |
For quantitiative real-time PCR analysis RNA was used from the same samples used for the array analyses. Reverse transcription with random hexamers (ImProm-II, Promega) was performed according to the manufacturer's instructions to produce cDNA. Triplicate cDNA samples of 0.5 ng were used in 384 well-format realtime PCRs in a Roche LightCycler 480 II using Applied Biosystem's SYBR Green PCR core reagents. PCR primers for human ETS1 (AGGAAGAGTGGTGGGTGGT, TGGAGAAGGGAACAAAAGTG) and GPBP1L1 (CCACCGACAGGAAGAGTGAG, AGGGAGGGCAAGACCATTT) and 18 S rRNA (ATTGACGGAAGGGCACCACCAG, CAAATCGCTCCACCAACTAAGAACG) were synthesized by Invitrogen and used according to SYBR Green protocol. The data were analyzed by LightCycler 480 SW 1.5 software and the relative averages of expression levels, normalized to internal 18 S rRNA controls are presented.
In Vitro Cleavage Assay
Full-length IRF-3 was purified by Flag pull-down from P2.1 cells expressing Flag-tagged human IRF-3 and was used for in vitro cleavage assays. A recombinant human active caspase-8 was obtained from Millipore and used for the in vitro cleavage reaction. Increasing amounts of caspase-8 were incubated with full-length IRF-3 in buffer containing 50 mm Hepes, 50 mm NaCl, 0.1% CHAPS, 10 mm EDTA, 5% glycerol, and 10 mm DTT, at 37 °C for the indicated times, and the reactions were terminated by adding SDS-PAGE loading buffer and analyzed by Western blot.
RESULTS
IRF-3 Is Cleaved upon dsRNA Signaling
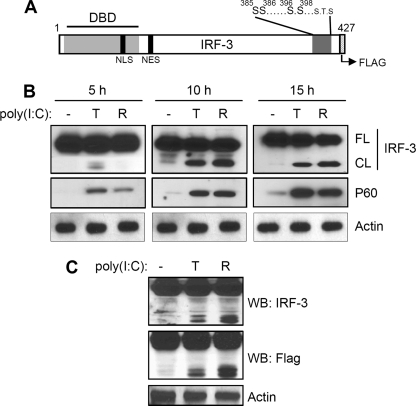
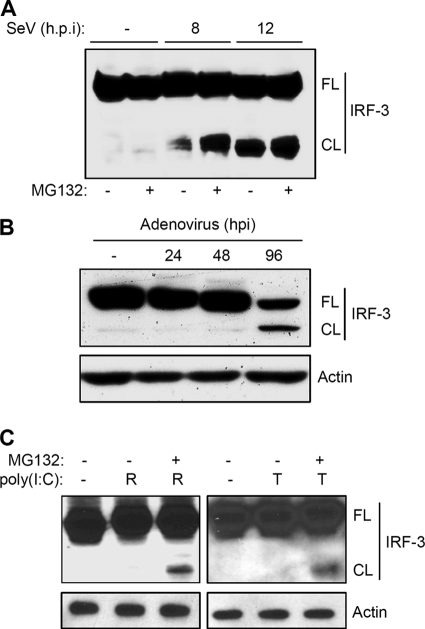
Multiple proteins of host antiviral signaling pathways are proteolytically processed by caspases. This regulatory mechanism attenuates the induction of antiviral genes. To investigate whether such a proteolytic processing mechanism exists for IRF-3, we expressed human IRF-3 (C-terminally Flag-tagged, Fig. 1A) in P2.1 cells which express very low levels of IRF-3. Ectopic expression of IRF-3 in these cells restored the induction of P60, an IRF-3-dependent protein (Fig. 1B). In addition to gene induction, activation of either RLH (R) or TLR3 (T) signaling pathways generated a 40 kDa IRF-3 fragment, which was detected by both IRF-3 and Flag antibodies (Fig. 1, B and C). We therefore concluded that the fragment was generated by the cleavage of the N-terminal DNA binding domain of IRF-3 (Fig. 1C). Both RLH- and TLR3-dependent signaling pathways, activated by dsRNA, triggered the proteolysis that generated the C-terminal fragment of IRF-3; however RLH-dependent signaling induced a somewhat more extensive cleavage, compared with TLR3-dependent signaling (Fig. 1C). To test whether virus infection can similarly induce proteolytic processing of IRF-3, we infected 1080.10 cells (HT1080 cells overexpressing human IRF-3 (27)), with Sendai virus (SeV), an RNA virus that activates the cytosolic RIG-I signaling pathway (13). Infection of SeV, alone or in combination with MG132, an inhibitor of proteasomal degradation, resulted in accumulation of the cleaved fragment of IRF-3 (Fig. 2A). A similar result was obtained when HT1080 cells were infected with adenovirus, a DNA virus that also activates the RIG-I signaling pathway (Fig. 2B, Ref. 14). More importantly, adenovirus-induced cleavage was generated from endogenous IRF-3, supporting the idea that the cleavage of IRF-3 is physiologically relevant. The dsRNA-induced cleaved fragment of endogenous IRF-3 appeared to undergo rapid proteasomal degradation, making it difficult to detect under normal conditions. However, inhibition of proteasomal degradation by the inhibitor MG132 led to the accumulation of the C-terminal cleavage product of IRF-3 (Fig. 2C).
FIGURE 1.
IRF-3 is proteolytically cleaved by dsRNA-dependent signaling. A, schematic representation of human IRF-3, indicating the functional domains. DBD, DNA binding domain; NLS, nuclear localization signal; NES, nuclear export signal. Critical Ser and Thr residues that are phosphorylated in response to virus infection are also shown, as is the C-terminal Flag tag; B, P2.1 cells expressing Flag-tagged IRF-3, were either treated (TLR3 activation, T) or transfected (RLH activation, R) with poly(I:C) for the indicated times; the cell lysates were analyzed by Western blot for IRF-3 (FL, full-length; CL, cleaved) and P60, an IRF-3-dependent gene product; C, P2.1 cells expressing IRF-3 were treated (T) or transfected (R) with poly(I:C), for 15 h, and cell lysates were analyzed by Western blot for Flag and IRF-3.
FIGURE 2.
Virus infection causes cleavage of IRF-3. A, 1080.10 cells (HT1080 cells with overexpression of IRF-3 (27)) were either mock-infected or infected with SeV at MOI of 10 in the absence or the presence of MG132 (10 μm) for the indicated times. The cell lysates were analyzed by Western blot for IRF-3 (FL, full-length; CL, cleaved; h.p.i, hours postinfection); B, HT1080 cells were infected with adenovirus (Ad5) at MOI of 10 for the indicated times, and the cell lysates were analyzed by Western blot for IRF-3 (FL, full-length; CL, cleaved); C, HT1080 cells were either treated (T) or transfected (R) with poly(I:C) in the absence or the presence of MG132 (10 μm) for 8 h, and the cell lysates were analyzed by Western blot for IRF-3.
Caspase-8 Is Involved in IRF-3 Cleavage
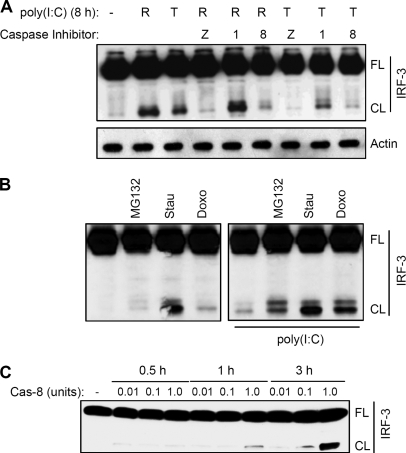
To investigate the biochemical nature of the cleavage, we used a broad spectrum inhibitor of caspases, z-VAD, which inhibited the cleavage of IRF-3, indicating that caspases were involved in the cleavage (Fig. 3A, and supplemental Fig. S1). To identify the specific caspase, we used specific inhibitors. An inhibitor of caspase-8, but not caspase-1, blocked the cleavage of IRF-3, suggesting a role for caspase-8 in the proteolytic processing of IRF-3 (Fig. 3A and supplemental Fig. S1). Caspase-8 is activated by dsRNA-dependent signaling (19); however, it can also be activated by other inducers of apoptosis. Staurosporine and doxorubicin, two strong inducers of caspase-8 activity, also efficiently triggered the cleavage of IRF-3, independently of dsRNA-dependent signaling (Fig. 3B), indicating that the cleavage of IRF-3 can be induced without any prior activation of IRF-3. However, these agents further increased the amount of cleavage product when dsRNA-dependent signaling pathways were activated (Fig. 3B, right panel). In contrast, treatment of cells with MG132 did not induce the cleavage, but led to accumulation of cleaved IRF-3 upon dsRNA signaling (Fig. 3B). To further investigate whether caspase-8 can directly target IRF-3 without any intermediate caspases, we performed in vitro cleavage of IRF-3 by an active form of recombinant caspase-8. The results clearly indicate that caspase-8 alone was able to induce proteolysis of IRF-3, generating the C-terminal fragment of IRF-3 (Fig. 3C).
FIGURE 3.
Caspase-8 activity is essential for the cleavage of IRF-3. A, P2.1 cells expressing IRF-3, were pretreated with the inhibitors of multiple caspases (Z, z-VAD, 10 μm), caspase-1 (z-WEHD, 10 μm), or caspase-8 (z-IETD, 10 μm) for 1 h, when the cells were either treated (T) or transfected (R) with poly(I:C) for 8 h, and cell lysates were analyzed by Western blot for IRF-3; B, P2.1 cells expressing Wt IRF-3 were treated with Staurosporine (Stau, 0.5 μm) or Doxorubicin (Doxo, 1 μm), agents that are known to activate caspase-8, or MG132 (10 μm) in the absence or the presence of transfected poly(I:C), and cell lysates were analyzed for IRF-3 by Western blot; C, IRF-3 was isolated from P2.1 cells expressing Flag-tagged Wt human IRF-3 and subjected to in vitro cleavage assay in the presence of varying amount of active caspase-8 (Cas-8) for the indicated times; the reaction mixtures were analyzed by Western blot for IRF-3, using anti-Flag.
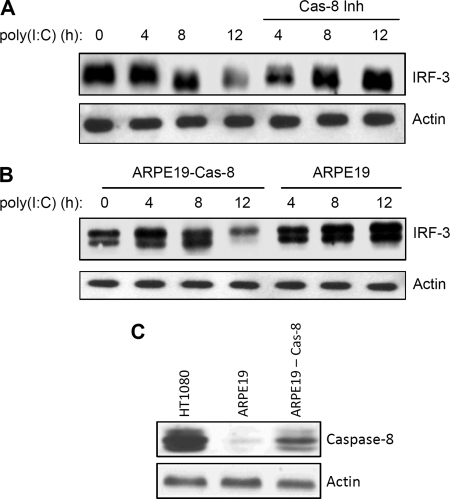
IRF-3, activated by virus infection, undergoes degradation by proteasome-dependent machinery and multiple cellular and viral proteins have been implicated in this process (4, 5, 7, 8, 11, 12, 28, 29). To investigate whether caspase-8-mediated cleavage of IRF-3 contributes to the proteasome-mediated degradation of IRF-3, we monitored its degradation in the presence of a caspase-8 inhibitor. The degradation of IRF-3 in response to RLH-dependent signaling was inhibited by a caspase-8 inhibitor over a period of 12 h after dsRNA stimulation (Fig. 4A). TLR3-dependent signaling, as expected, induced a relatively slower degradation of IRF-3, which was also inhibited by the caspase-8 inhibitor (supplemental Fig. S2). To confirm the specific involvement of caspase-8, we used ARPE19 retinal epithelial cells, which express very low levels of caspase-8 (Fig. 4C). IRF-3 was activated in these cells by RLH-dependent signaling, as indicated by the expression of P60, an IRF-3-dependent gene product (data not shown); however, IRF-3 did not undergo proteasome-mediated degradation in these cells (Fig. 4B). When we expressed caspase-8 in these cells ectopically, IRF-3 degradation was restored upon RLH-dependent signaling (Fig. 4, B and C). Similar results were also obtained for TLR3-dependent signaling, which caused degradation of IRF-3 in ARPE19 cells expressing caspase-8 (supplemental Fig. S3, top panel).
FIGURE 4.
Caspase-8 is required for the proteasome-mediated degradation of IRF-3. A, HT1080 cells were transfected with poly(I:C) in the absence or the presence of an inhibitor of caspase-8 (z-IETD, 10 μm) for the indicated times, and cell lysates were analyzed for IRF-3 by Western blot; B, ARPE19 cells expressing Wt caspase-8 were transfected with poly(I:C) for the indicated times, and cell lysates were analyzed for IRF-3 by Western blot; C, lysates from HT1080, ARPE19, and caspase-8 expressing ARPE19 cells were analyzed for caspase-8 by Western blot.
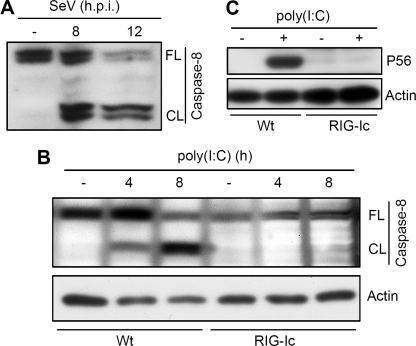
Having established the role of caspase-8 in the proteasome-mediated degradation of IRF-3, we asked how caspase-8 was activated by dsRNA-signaling. Although, extracellular dsRNA can be recognized by endosomal TLR3, multiple receptors have been implicated in cytosolic dsRNA-signaling, including RLR, NLR, and PKR. SeV infection, which is known to trigger RIG-I dependent signaling, caused rapid activation of caspase-8 in HT1080 cells. Caspase activation was analyzed by auto-proteolysis of pro-caspase-8, the precursor form, which generates the active caspase-8 enzyme. As indicated in Fig. 5A, SeV infection resulted in the generation of cleaved caspase-8 fragments, indicating a possible role of RIG-I signaling in the activation of caspase-8. To conclusively determine the requirement of RIG-I in the activation of caspase-8, we tested a human cell line that expresses a dominant negative mutant of RIG-I (RIG-Ic). Cytosolic dsRNA stimulation resulted in the activation of caspase-8 in HT1080 cells (Wt); however, the RIG-Ic expressing cells were incapable of activating caspase-8 (Fig. 5B), suggesting that RIG-I-dependent signaling is essential for cytosolic dsRNA-induced activation of caspase-8. As expected, the RIG-Ic-expressing cells were defective in RIG-I signaling; induction of P56, an IRF-3-dependent gene product, was inhibited in these cells (Fig. 5C).
FIGURE 5.
Caspase-8 is activated by cytosolic RIG-I-dependent signaling. A, 1080.10 cells were infected with SeV at MOI 10 for the indicated times, and the cell lysates were analyzed for the activation of caspase-8 by Western blot (FL, full-length; CL, cleaved; h.p.i., hours postinfection); B, HT1080 (Wt) or HT1080/RIG-Ic cells (expressing a dominant negative mutant of RIG-I) were transfected with poly(I:C) for the indicated time, and cell lysates were analyzed for the activation of caspase-8 by Western blot (FL, full-length; CL, cleaved caspase-8); C, HT1080 and HT1080/RIG-Ic cells were transfected with poly(I:C), and the cell lysates were analyzed for the induction of P56 by Western blot.
Cleavage Deficiency Leads to Impaired Degradation of IRF-3
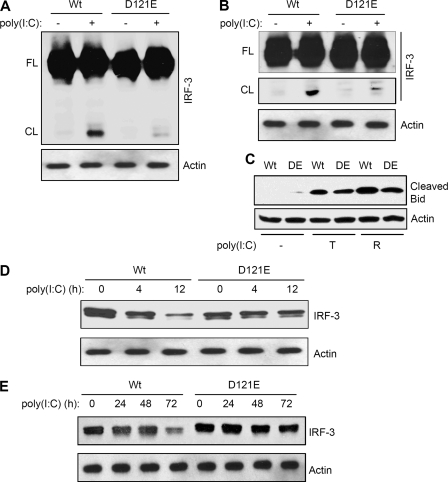
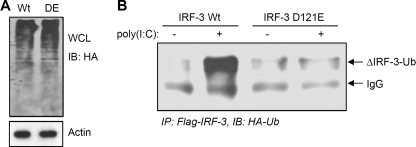
Specific recognition motifs have been identified for the substrates of caspases (30, 31). Several likely caspase recognition sequences were found within IRF-3. Because the caspase-8-mediated cleavage of IRF-3 generated a 40 kDa C-terminal cleaved fragment, we mutated a putative caspase recognition motif that was located close to the end of the N-terminal DNA binding domain; putative cleavage at this site would produce a C-terminal fragment of the observed size. When this sequence was mutated (118SQPD121 to AAAA or D121E), the resultant mutants of IRF-3 were not cleaved by caspase-8 when expressed in P2.1 cells, as indicated by the absence of the cleaved fragment in response RLH- or TLR3-dependent signaling (Fig. 6A and B and supplemental Fig. S4, data not shown). Moreover, the expression of the mutant IRF-3 did not affect the activation of caspase-8; this was tested by the cleavage of Bid, a substrate of caspase-8, in the cells expressing Wt or the mutant forms of IRF-3 (Fig. 6C). To determine whether the cleavage-deficient mutant of IRF-3 (D121E) was protected from proteasome-mediated degradation, we expressed this mutant protein in P2.1 cells and analyzed its degradation in response to dsRNA-dependent signaling. Although, Wt IRF-3 followed degradation by activation of RLH (Fig. 6D and supplemental Fig. S5) or TLR3 (Fig. 6E) pathway, the mutant IRF-3 was not degraded during the course of the experiment. These results clearly indicate that the proteasome-mediated degradation of IRF-3 is at least partly dependent on caspase-8-mediated proteolytic processing. The proteasomal degradation of IRF-3 is known to depend on its poly-ubiquitination, in addition to other necessary modifications (8, 10–12). We tested the relationship between caspase-8-mediated cleavage and the ubiquitination of IRF-3. When Wt or the D121E mutant of IRF-3 was co-expressed with ubiquitin in P2.1 cells (Fig. 7A), the Wt IRF-3 was strongly ubiquitinated in response to dsRNA-dependent signaling but the mutant IRF-3 was not (Fig. 7B). This indicates that the caspase-8-cleaved C-terminal fragment (immunoprecipitated by Flag antibody) of IRF-3 is targeted for polyubiquitination. These results suggest that caspase-8-mediated cleavage of IRF-3 is a prerequisite for its ubiquitination and subsequent proteasome-mediated degradation.
FIGURE 6.
Mutation of a caspase-8 recognition motif leads to impaired cleavage and degradation of IRF-3. P2.1 cells expressing Wt or the D121E (DE) mutant of IRF-3 were used in these experiments (A–C). A, cells were transfected with poly(I:C); cell lysates were analyzed for IRF-3 by Western blot (FL, full-length, CL, cleaved); B, cells were treated with poly(I:C) to activate TLR3-dependent signaling, the full-length (FL) and the cleaved (CL) forms of IRF-3 are shown; C, cell lysates from TLR3 activation (T) or RLH activation (R) reactions were analyzed for cleaved Bid by Western blot; D, IRF-3 degradation was analyzed by Western blot following transfection of poly(I:C) (RLH activation); E, IRF-3 degradation was analyzed by Western blot following treatment with poly(I:C) (TLR3 activation).
FIGURE 7.
IRF-3 ubiquitination is impaired upon mutation of a caspase recognition motif. P2.1 cells expressing Wt or the D121E mutant of IRF-3 were transfected with HA-Ub. A, total cell lysates were analyzed by Western blot for HA-Ub; B, cells were transfected with poly(I:C), and cell lysates were immunoprecipitated with Flag using Exactacruz (Santa Cruz Biotechnology) and analyzed by Western blot for HA-Ub.
Impaired Degradation of IRF-3 Leads to Altered Expression of dsRNA-induced Genes
To investigate whether caspase-8-mediated cleavage and degradation of IRF-3 regulate the expression of the dsRNA-induced genes, we conducted a gene array experiment to compare the expression patterns of these genes in Wt- or D121E IRF-3-expressing cells. We hypothesized that impaired degradation of IRF-3 would lead to the accumulation of active IRF-3 and therefore would contribute to continuous expression of the dsRNA-induced genes. Indeed, the analysis revealed that a subset of dsRNA-induced genes was expressed continuously in the presence of mutant IRF-3 but not Wt IRF-3 (Table 1 and supplemental Fig. S6). The genes that showed accumulated expression, although induced at similar levels by dsRNA initially, were down-regulated when the IRF-3 was degraded (for Wt IRF-3), and their expression was sustained when IRF-3 degradation was impaired (for the D121E mutant). The microarray results were also validated by quantitative realtime PCR for two of the selected genes, which showed sustained expression patterns in the later time point (supplemental Fig. S7). Interestingly, another subset of dsRNA-inducible genes was identified that exhibited similar levels of expression in the presence of both the Wt and the mutant IRF-3 (supplemental Table S1). However, we have not observed the opposite pattern of gene expression under similar analytical conditions. A small group of dsRNA-inducible genes showed sustained expression in Wt IRF-3-expressing cells but their expression was not significantly different from the cells expressing the mutant IRF-3 (data not shown). We therefore conclude that caspase-8-mediated cleavage and degradation of IRF-3 differentially regulates the induction of dsRNA-stimulated genes.
DISCUSSION
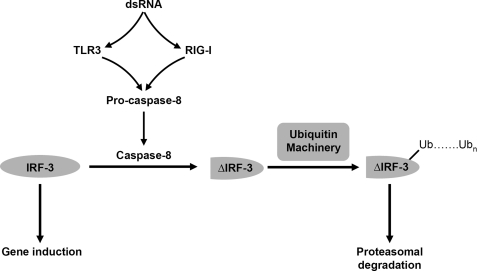
IRF-3 plays a central role in the transcription of cellular antiviral genes, including IFN-β. In uninfected cells, IRF-3 is in the cytosol as inactive monomers. After infection, phosphorylation by the activated kinase TBK1 on C-terminal Ser/Thr residues causes IRF-3 to undergo conformational changes that lead to homodimerization and translocation to the nucleus, where it binds to ISREs (Interferon Stimulated Response Element) in promoters. As a part of the regulation of antiviral signaling, IRF-3 undergoes proteasome-mediated degradation following its ubiquitination. Several cellular ubiquitin ligases are involved in the degradation of IRF-3, including Pin1, RBCK1, Cul-1, and RAUL (5, 8, 11, 32, 33). We show here that caspase-8 catalyzes a specific cleavage of IRF-3, a step that is required for its subsequent proteasome-mediated degradation (Fig. 8). Impaired proteolysis, caused by inhibiting caspase-8 activity or mutating the caspase-recognition motif of IRF-3, leads to the accumulation of full-length IRF-3 and up-regulation of dsRNA-induced gene expression.
FIGURE 8.
Caspase-8-mediated cleavage of IRF-3 is an intermediate step in its proteasome-mediated degradation. IRF-3 undergoes proteasome-mediated degradation in response to dsRNA-dependent signaling. Stimulation of TLR3 or RIG-I signaling by dsRNA activates caspase-8 by proteolysis of inactive procaspase-8. The activated caspase-8 initiates proteolytic processing of IRF-3, generating an intermediate product. The cleaved IRF-3 is then targeted for poly-ubiquitination en route to further degradation.
Termination of signaling is important for maintaining cellular homeostasis. Viruses often activate normal cellular termination mechanisms to evade antiviral response. For example, a protein of bovine viral diarrhea virus triggers the proteasome-mediated degradation of IRF-3 to inhibit interferon production (12). Degradation of IRF-3 is, therefore, an important means of attenuating its transcriptional activity. However, premature degradation of IRF-3 will inhibit the induction of antiviral genes, leading to insufficient host defense against viral infection. The cellular ubiquitination machinery provides a way to dispose the undesired proteins. Ubiquitination is a complex process with diverse implications, leading either to cessation or promotion of signal transduction (34–36). Proteolysis catalyzed by caspases usually inactivates proteins and negatively regulates signaling. Caspase-induced cleavage of TRIF and IPS-1 leads to inhibition of TLR3- and RIG-I-dependent signaling pathways (20). The results presented here demonstrate a connection between the two reactions: a caspase-mediated cleavage is required for the ubiquitination and subsequent proteasome-mediated degradation of IRF-3.
Our results indicate that caspase-8 is activated by RIG-I-dependent signaling. Cells expressing a dominant negative mutant of RIG-I exhibited impaired caspase-8 activation. Caspase-8 is activated by various signaling pathways involving death receptors, TNF, and other apoptotic stimuli. Caspase-8 is activated by auto-proteolysis of procaspase-8 to generate the active enzyme, which triggers the extrinsic apoptotic pathway. Activated caspase-8 also cleaves the pro-apoptotic protein Bid, to mediate crosstalk with the mitochondrial intrinsic apoptotic pathway, resulting in release of cytochrome c into the cytosol and subsequent activation of caspase-9. Our results also indicate that Sendai virus infection induces the activation of caspase-8. Although, caspase-8 was activated by SeV, its role in viral apoptosis is not clear. Induction of viral apoptosis is dependent on caspase-9 activity; however, caspase-8 mediated crosstalk with the mitochondrial pathway may amplify the apoptotic effect. In addition to its primary role in apoptosis, caspase-8 is also associated with dsRNA-dependent signaling. Caspase-8 is recruited to the adaptor proteins IPS-1 and TRIF to promote inflammatory and antiviral responses through the activation of NF-κB and IRFs in response to TLR3- and RIG-I-dependent signaling (37, 38). We have observed the caspase-8-mediated degradation of IRF-3 in both TLR3 and RIG-I signaling pathways, although the degradation of IRF-3 was faster in the case of RIG-I-dependent signaling. Therefore, we hypothesize that procaspase-8, recruited through the RIG-I/IPS-1 complex, is activated by auto-proteolysis to generate active caspase-8. At present, the molecular details of caspase-8 activation by RIG-I signaling remain unclear and additional studies will be necessary to clarify this mechanism.
Our findings help to explain the observation that caspase-8-deficient mice exhibit inflammatory skin disorders due to unregulated activation and accumulation of active IRF-3 (18). IRF-3 activation is kept in check by caspase-8-mediated decay and the absence of caspase-8 leads to aberrant expression of inflammatory genes in the keratinocytes of these mice. The negative regulatory role of caspase-8 in the activation of IRF-3 is also evident from a recent study that demonstrated that RIP1, which is required for RIG-I-mediated activation of IRF-3, is targeted for caspase-8-induced proteolysis (39). IRF-7, a close relative of IRF-3, is also targeted for ubiquitination in virus-infected cells (32), and it remains to be seen whether caspase-mediated regulation is involved in this process. In addition, it is possible that caspases may target IRFs expressed by certain viruses (40), to degrade these proteins as a component of cellular antiviral responses.
Supplementary Material
Acknowledgments
We thank Thomas Shenk, Marcus Peter, and Michael David for important reagents used in this study. We thank the Genomics Core Service at Lerner Research Institute for conducting the microarray.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 AI 073303 (to G. C. S.) and P01 CA 062220 (to G. C. S. and G. R. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7 and Table S1.
- TLR
- Toll-like receptor
- SeV
- Sendai virus.
REFERENCES
- 1. Isaacs A., Lindenmann J. (1957) Proc. R. Soc. Lond. B. Biol. Sci. 147, 258–267 [PubMed] [Google Scholar]
- 2. Génin P., Vaccaro A., Civas A. (2009) Cytokine Growth Factor Rev. 20, 283–295 [DOI] [PubMed] [Google Scholar]
- 3. Lu G., Reinert J. T., Pitha-Rowe I., Okumura A., Kellum M., Knobeloch K. P., Hassel B., Pitha P. M. (2006) Cell. Mol. Biol. 52, 29–41 [PubMed] [Google Scholar]
- 4. Kubota T., Matsuoka M., Chang T. H., Tailor P., Sasaki T., Tashiro M., Kato A., Ozato K. (2008) J. Biol. Chem. 283, 25660–25670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bibeau-Poirier A., Gravel S. P., Clément J. F., Rolland S., Rodier G., Coulombe P., Hiscott J., Grandvaux N., Meloche S., Servant M. J. (2006) J. Immunol. 177, 5059–5067 [DOI] [PubMed] [Google Scholar]
- 6. Prinarakis E., Chantzoura E., Thanos D., Spyrou G. (2008) EMBO J. 27, 865–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lin R., Heylbroeck C., Pitha P. M., Hiscott J. (1998) Mol. Cell. Biol. 18, 2986–2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Saitoh T., Tun-Kyi A., Ryo A., Yamamoto M., Finn G., Fujita T., Akira S., Yamamoto N., Lu K. P., Yamaoka S. (2006) Nat. Immunol. 7, 598–605 [DOI] [PubMed] [Google Scholar]
- 9. Aresté C., Mutocheluh M., Blackbourn D. J. (2009) J. Biol. Chem. 284, 23272–23285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yang K., Shi H. X., Liu X. Y., Shan Y. F., Wei B., Chen S., Wang C. (2009) J. Immunol. 182, 3782–3792 [DOI] [PubMed] [Google Scholar]
- 11. Higgs R., Ní Gabhann J., Ben Larbi N., Breen E. P., Fitzgerald K. A., Jefferies C. A. (2008) J. Immunol. 181, 1780–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen Z., Rijnbrand R., Jangra R. K., Devaraj S. G., Qu L., Ma Y., Lemon S. M., Li K. (2007) Virology 366, 277–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Peters K., Chattopadhyay S., Sen G. C. (2008) J. Virol. 82, 3500–3508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chattopadhyay S., Yamashita M., Zhang Y., Sen G. C. (2011) J. Virol. 85, 3708–3716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chattopadhyay S., Marques J. T., Yamashita M., Peters K. L., Smith K., Desai A., Williams B. R., Sen G. C. (2010) EMBO J. 29, 1762–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. White C. L., Chattopadhyay S., Sen G. C. (2011) J. Virol. 85, 5224–5227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kumar H., Kawai T., Kato H., Sato S., Takahashi K., Coban C., Yamamoto M., Uematsu S., Ishii K. J., Takeuchi O., Akira S. (2006) J. Exp. Med. 203, 1795–1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kovalenko A., Kim J. C., Kang T. B., Rajput A., Bogdanov K., Dittrich-Breiholz O., Kracht M., Brenner O., Wallach D. (2009) J. Exp. Med. 206, 2161–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Takahashi K., Kawai T., Kumar H., Sato S., Yonehara S., Akira S. (2006) J. Immunol. 176, 4520–4524 [DOI] [PubMed] [Google Scholar]
- 20. Rebsamen M., Meylan E., Curran J., Tschopp J. (2008) Cell Death Differ. 15, 1804–1811 [DOI] [PubMed] [Google Scholar]
- 21. Meylan E., Martinon F., Thome M., Gschwendt M., Tschopp J. (2002) EMBO Rep. 3, 1201–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rasheed S., Nelson-Rees W. A., Toth E. M., Arnstein P., Gardner M. B. (1974) Cancer 33, 1027–1033 [DOI] [PubMed] [Google Scholar]
- 23. Leaman D. W., Salvekar A., Patel R., Sen G. C., Stark G. R. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 9442–9447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dunn K. C., Aotaki-Keen A. E., Putkey F. R., Hjelmeland L. M. (1996) Exp. Eye Res. 62, 155–169 [DOI] [PubMed] [Google Scholar]
- 25. Yang P., Peairs J. J., Tano R., Zhang N., Tyrell J., Jaffe G. J. (2007) Invest. Ophthalmol. Vis. Sci. 48, 3341–3349 [DOI] [PubMed] [Google Scholar]
- 26. Collins T. J. (2007) BioTechniques 43, 25–30 [DOI] [PubMed] [Google Scholar]
- 27. Elco C. P., Guenther J. M., Williams B. R., Sen G. C. (2005) J. Virol. 79, 3920–3929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Saira K., Zhou Y., Jones C. (2007) J. Virol. 81, 3077–3086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Barro M., Patton J. T. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 4114–4119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wee L. J., Tan T. W., Ranganathan S. (2007) Bioinformatics 23, 3241–3243 [DOI] [PubMed] [Google Scholar]
- 31. Wee L. J., Tan T. W., Ranganathan S. (2006) BMC Bioinformatics 7, Suppl. 5, S14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yu Y., Hayward G. S. (2010) Immunity 33, 863–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang M., Tian Y., Wang R. P., Gao D., Zhang Y., Diao F. C., Chen D. Y., Zhai Z. H., Shu H. B. (2008) Cell Res. 18, 1096–1104 [DOI] [PubMed] [Google Scholar]
- 34. Kanayama A., Seth R. B., Sun L., Ea C. K., Hong M., Shaito A., Chiu Y. H., Deng L., Chen Z. J. (2004) Mol. Cell 15, 535–548 [DOI] [PubMed] [Google Scholar]
- 35. Zeng W., Xu M., Liu S., Sun L., Chen Z. J. (2009) Mol. Cell 36, 315–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen Z. J., Sun L. J. (2009) Mol. Cell 33, 275–286 [DOI] [PubMed] [Google Scholar]
- 37. Li H. M., Fujikura D., Harada T., Uehara J., Kawai T., Akira S., Reed J. C., Iwai A., Miyazaki T. (2009) Cell Death Differ. 16, 1615–1621 [DOI] [PubMed] [Google Scholar]
- 38. Han K. J., Su X., Xu L. G., Bin L. H., Zhang J., Shu H. B. (2004) J. Biol. Chem. 279, 15652–15661 [DOI] [PubMed] [Google Scholar]
- 39. Rajput A., Kovalenko A., Bogdanov K., Yang S. H., Kang T. B., Kim J. C., Du J., Wallach D. (2011) Immunity 34, 340–351 [DOI] [PubMed] [Google Scholar]
- 40. Lee H. R., Kim M. H., Lee J. S., Liang C., Jung J. U. (2009) J. Interferon Cytokine Res. 29, 621–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.