Background: H. pylori CagA oncoprotein is delivered into gastric epithelial cells, where it interacts with SHP2 oncoprotein and PAR1 kinase.
Results: CagA dimerization stimulates the CagA-SHP2 interaction and thereby causes enhanced SHP2 deregulation.
Conclusion: PAR1-mediated CagA dimerization plays an important role in the oncogenic activity of CagA.
Significance: Inhibition of CagA dimerization has therapeutic value in preventing gastric carcinoma.
Keywords: Bacterial Toxins, Protein Assembly, Protein-Protein Interactions, SH2 Domains, Protein-tyrosine Phosphatase (Tyrosine Phosphatase), CagA, Helicobacter pylori, SHP2, Dimerization, Stomach Cancer
Abstract
Chronic infection with Helicobacter pylori cagA-positive strains is associated with atrophic gastritis, peptic ulceration, and gastric carcinoma. The cagA gene product, CagA, is delivered into gastric epithelial cells via type IV secretion, where it undergoes tyrosine phosphorylation at the EPIYA motifs. Tyrosine-phosphorylated CagA binds and aberrantly activates the oncogenic tyrosine phosphatase SHP2, which mediates induction of elongated cell morphology (hummingbird phenotype) that reflects CagA virulence. CagA also binds and inhibits the polarity-regulating kinase partitioning-defective 1 (PAR1)/microtubule affinity-regulating kinase (MARK) via the CagA multimerization (CM) sequence independently of tyrosine phosphorylation. Because PAR1 exists as a homodimer, two CagA proteins appear to be passively dimerized through complex formation with a PAR1 dimer in cells. Interestingly, a CagA mutant that lacks the CM sequence displays a reduced SHP2 binding activity and exhibits an attenuated ability to induce the hummingbird phenotype, indicating that the CagA-PAR1 interaction also influences the morphological transformation. Here we investigated the role of CagA dimerization in induction of the hummingbird phenotype with the use of a chemical dimerizer, coumermycin. We found that CagA dimerization markedly stabilizes the CagA-SHP2 complex and thereby potentiates SHP2 deregulation, causing an increase in the number of hummingbird cells. Protrusions of hummingbird cells induced by chemical dimerization of CagA are further elongated by simultaneous inhibition of PAR1. This study revealed a role of the CM sequence in amplifying the magnitude of SHP2 deregulation by CagA, which, in conjunction with the CM sequence-mediated inhibition of PAR1, evokes morphological transformation that reflects in vivo CagA virulence.
Introduction
Helicobacter pylori is a Gram-negative bacterium infecting more than half of the global human population. Since its first report in 1984, H. pylori has been shown to cause upper gastrointestinal disorders such as chronic atrophic gastritis and peptic ulcerations. Furthermore, chronic infection with H. pylori strains producing the CagA protein is the highest risk factor for the development of gastric carcinoma (1, 2). CagA is encoded by the cagA gene, which is located in the cag pathogenicity island, a DNA segment that also contains a set of genes encoding components of a bacterial microsyringe termed the type IV secretion system (3).
H. pylori CagA is delivered into gastric epithelial cells via the cag pathogenicity island-encoded type IV secretion system (1). Inside the host cells, CagA underlies tyrosine phosphorylation at the Glu-Pro-Ile-Tyr-Ala (EPIYA) motif, which is present in variable numbers in the C-terminal region, by Src family kinases and/or Abl kinase (4). The C-terminal region of CagA from H. pylori isolated in East Asian countries is composed of EPIYA-A, EPIYA-B, and EPIYA-D segments, each of which contains a single EPIYA motif. Hence, East Asian CagA is structurally defined as ABD-type CagA. On the other hand, the C-terminal region of CagA isolated from the rest of the world (Western CagA) comprises EPIYA-A, EPIYA-B, and a variable number of Western-specific EPIYA-C segments, which also contain a single EPIYA motif. Accordingly, Western CagA is defined structurally as ABCn-type CagA (where n is an arbitrary number) (1, 5).
Tyrosine-phosphorylated CagA acquires the ability to specifically bind to the Src homology-2 (SH2)2-containing protein-tyrosine phosphatase SHP2 (6). SHP2 is expressed in a wide range of cell types, and gain-of-function mutations of SHP2 have been found in a variety of human malignancies, indicating that constitutively activated SHP2 acts as an oncoprotein (7, 8). Physiologically, SHP2 functions as a positive regulator of signals generated by growth factor/cytokine stimuli that promote Erk MAP kinase signaling in both Ras-dependent and Ras-independent manners. More recently, SHP2 was found to activate the nuclear Wnt signal through tyrosine dephosphorylation of parafibromin, a component of the RNA polymerase II-associated factor complex (9). SHP2 possesses two SH2 domains (N-SH2 and C-SH2) at the N-terminal region, a protein-tyrosine phosphatase domain, and a C-terminal tail. The N-SH2 domain interacts with the protein-tyrosine phosphatase domain, which inhibits phosphatase activity. Binding of phosphotyrosyl peptides to the N- and/or C-SH2 domain induces a conformational change in SHP2 that relieves interaction between the protein-tyrosine phosphatase domain and the SH2 domain, resulting in phosphatase activation. The bacterial CagA protein also binds to the SH2 domains of SHP2 and aberrantly activates the phosphatase activity in a manner that is dependent on CagA tyrosine phosphorylation (6). In addition to Erk activation, CagA-deregulated SHP2 dephosphorylates and inactivates focal adhesion kinase (FAK), a tyrosine kinase that controls the turnover of focal adhesion spots (10). As a consequence, CagA induces an elongated cell shape known as the hummingbird phenotype.
In polarized epithelial cells, CagA disrupts the tight junction and causes loss of epithelial cell polarity in a tyrosine phosphorylation-independent manner (11). This CagA activity is mediated by a specific interaction with partitioning-defective 1 (PAR1)/microtubule affinity-regulating kinase (MARK) (12). There are four PAR1 isoforms (PAR1a/MARK3, PAR1b/MARK2, PAR1c/MARK1, and PAR1d/MARK4) in mammalian cells, which redundantly phosphorylate microtubule-associated proteins (MAPs) and thereby regulate stability of microtubules (13). CagA binds to all of the PAR1 family members in a tyrosine phosphorylation-independent manner and inhibits the kinase activity (14). The PAR1-binding region of CagA has been shown to be a C-terminal 16-amino acid sequence, which was originally identified as a CagA multimerization (CM) sequence (12, 15, 16).
The CagA-SHP2 interaction requires both the N-SH2 and the C-SH2 domains of SHP2, whereas CagA proteins possessing a single EPIYA-C or -D segment can form a stable complex with SHP2 (17). Based on this observation, a model in which a CagA dimer simultaneously binds to the two SH2 domains of an SHP2 molecule to make a stable CagA-SHP2 complex was reported (12, 18). Because PAR1b, and probably other PAR1 members as well, is thought to exist as a dimer in cells, two CagA molecules may be passively dimerized through binding with a PAR1 dimer via the CM sequence (12, 19). The model predicts that the CagA CM sequence is essential not only for the CagA-PAR1 interaction but also for stable CagA-SHP2 complex formation.
In this work, we investigated the role of CagA dimerization in the pathophysiological action of CagA using a chemical dimerization technique. We demonstrated that CagA dimerization stabilizes CagA-SHP2 interaction and thereby potentiates deregulation of the SHP2 oncoprotein by CagA.
EXPERIMENTAL PROCEDURES
Expression Vectors
pSP65SRα mammalian expression vectors for hemagglutinin (HA)-tagged wild-type CagA (ABD-type CagA) and the CagA mutant lacking the CM sequence, CagAΔCM, have been described previously (16, 18). A CagA mutant in which the EPIYA motif of the EPIYA-D segment was changed to EPIFA (CagA-ABd) was made by site-directed mutagenesis. The gene encoding CagA-ABd was tagged with the FLAG or HA sequence epitope and was cloned into pSP65SRα. A DNA fragment encoding B-subunit of bacterial DNA gyrase (GyrB)-fused CagAΔCM (GyrB-CagAΔCM) was made from CagAΔCM by introducing a DNA sequence encoding 1–219 amino acids of GyrB into the 5′ end of the cagA gene. The fusion DNA was cloned into pSP65SRα. The DNA fragment encoding FLAG-tagged CagA, FLAG-tagged GyrB-CagAΔCM, or HA-tagged GyrB-CagAΔCM was cloned into pSP65SRα. A pEF mammalian expression vector for T7-tagged human PAR1b/MARK2 has been described previously (12, 14). The cDNA fragment encoding FLAG-tagged human PAR1b/MARK2 was cloned into pSP65SRα.
Antibodies
Anti-HA monoclonal antibody 3F10 (Roche Applied Science) was used as primary antibody for immunoblotting and immunostaining. Anti-FLAG monoclonal antibody M2 (Sigma) was used as primary antibody for immunoprecipitation and immunoblotting. Anti-SHP2 polyclonal antibody C-18 (Santa Cruz Biotechnology), anti-phosphotyrosine monoclonal antibody 4G10 (Millipore), anti-FAK (pY576) phospho-specific polyclonal antibody (BioSource), anti-actin polyclonal antibody C-11 (Santa Cruz Biotechnology), anti-T7 polyclonal antibody (M-21, Santa Cruz Biotechnology), and anti-PAR1b antibody (provided by S. Ohno, Yokohama City University, Yokohama, Japan) were used as primary antibodies for immunoblotting.
Cell Culture and Transfection
AGS human gastric epithelial cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) and at 37 °C in 5% CO2. Cells were transfected with expression vectors or siRNA by using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's protocol.
Immunostaining
Cells were fixed with Mildform 10N (Wako) for 20 min and permeabilized with 0.5% Triton X-100 for 20 min. The fixed cells were then treated with a primary antibody and were visualized with Alexa Fluor 488- or 546-conjugated secondary antibody (Invitrogen). F-actin was stained with Alexa Fluor 546-phalloidin (Invitrogen). The nuclei were stained with 4′,6-diamidino-2-phenylindole dihydrochloride n-hydrate (DAPI). Images were captured by using a confocal microscope system (TCS-SPE, Leica).
Cell Morphological Analysis
The morphology of the AGS cells was observed at 17 h after transfection. AGS cells were transiently transfected with expression vectors. At 8 h after transfection, cells were treated with or without 700 nm coumermycin and were cultured for an additional 9 h. Cell morphology was examined by light microscopy. Cells showing the hummingbird phenotype were counted in 10 different fields in each of three dishes (the area of one field = 0.25 mm2). For detailed cell morphological analysis, the morphology of the AGS cells was captured at 17 h after transfection by using a confocal microscope system (TCS-SPE, Leica). Images of individual cell morphology were analyzed by using the ImageJ software (National Institutes of Health, rsbweb.nih.gov). Hummingbird phenotype was defined as an elongated cell morphology, in which the ratio of the longest protrusion of cell to the shortest cell diameter was more than 2-fold.
Immunoprecipitation and Immunoblotting
AGS cells were harvested at 17 h after transfection and lysed in lysis buffer (50 mm Tris-HCl, pH 7.5, 100 mm NaCl, 5 mm EDTA, 1% Brij-35, 2 mm Na3VO4, 10 mm NaF, 10 mm β-glycerophosphate, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 10 μg/ml trypsin inhibitor, 2 mm PMSF). Total cell lysates and immunoprecipitates were subjected to SDS-polyacrylamide gel electrophoresis (PAGE). Proteins transferred to PVDF membrane filters (Millipore) were incubated in primary antibodies and then visualized by using Western blot chemiluminescence reagent (PerkinElmer Life Sciences). Intensities of chemiluminescence on the immunoblotted filters were quantitated by using a luminescence image analyzer (LAS-4000, Fuji Film).
RNA Interference
PAR1b-specific siRNA-709 has been described previously (14). siRNA was transfected into cells using Lipofectamine 2000 reagent. At 24 h after siRNA treatment, cells were transfected with plasmid vectors. At 17 h after plasmid transfection, cells were subjected to cell morphological analysis.
RESULTS
Construction and Expression of a GyrB-CagA Fusion Protein
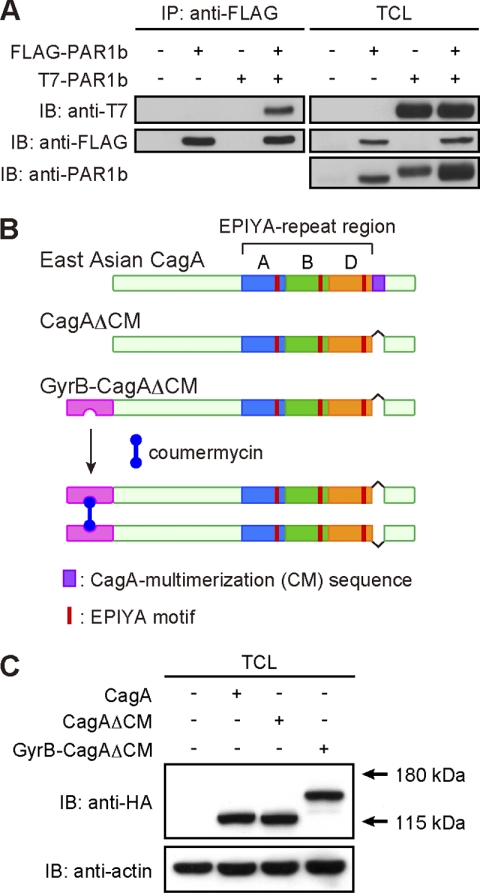
Upon delivery into host gastric epithelial cells, H. pylori CagA is thought to passively homodimerize through its interaction with a PAR1 dimer via the CM sequence (12, 19). To formally proof the presence of a PAR1 dimer in cells, AGS human gastric epithelial cells were transiently transfected with a FLAG-tagged PAR1b vector and/or a T7-tagged PAR1b vector. The cell lysates were immunoprecipitated with an anti-FLAG antibody, and the anti-FLAG immunoprecipitates were subjected to immunoblotting with an anti-T7 antibody. As shown in Fig. 1A, T7-tagged PAR1b was efficiently co-precipitated with FLAG-tagged PAR1b, demonstrating homodimerization of PAR1b in cells. Next, to understand the role of CagA dimerization in its pathophysiological action, we sought to dimerize CagA without the help of PAR1. The PAR1-binding CM sequence is located immediately downstream of the EPIYA-C segment and EPIYA-D segment in the C-terminal region of Western CagA and East Asian CagA, respectively (supplemental Fig. 1) (5, 12, 16). Interestingly, the N-terminal 16-amino acid stretch of the EPIYA-C segment is exactly the same as the CM sequence. Accordingly, Western CagA possesses at least two CM sequences, each of which can independently bind to PAR1 (supplemental Fig. 1) (12). Given this structural difference, we chose East Asian CagA (ABD-type CagA isolated from H. pylori strain F32) containing only a single CM sequence. To generate a CagA mutant that can dimerize in the absence of PAR1, we first deleted the CM sequence from CagA and generated a CagA mutant, CagAΔCM. To conditionally dimerize CagAΔCM, we made use of coumermycin, an antibiotic isolated from Streptomyces rishiriensis, which consists of two identically substituted coumarin rings joined by a methyl pyrrole (20). Coumermycin specifically binds to the N-terminal subdomain (24 kDa) of GyrB with a stoichiometry of 1:2. Because of this unique binding property, the coumermycin-GyrB dimerization strategy has been successfully applied to the artificial homodimerization of proteins of interest (21–25). To chemically dimerize CagA with coumermycin, we fused the GyrB sequence to the N terminus of CagAΔCM and generated GyrB-CagAΔCM (Fig. 1B). The DNA encoding C-terminal HA-tagged CagAΔCM, HA-tagged GyrB-CagAΔCM, or FLAG-tagged GyrB-CagAΔCM was inserted into a pSP65SRα mammalian expression vector.
FIGURE 1.
Generation of conditionally dimerizable CagA. A, homodimerization of PAR1b in cells. Total cell lysates (TCLs) prepared from AGS cells transiently transfected with a FLAG-tagged PAR1b vector and/or a T7-tagged PAR1b vector were immunoprecipitated with an anti-FLAG antibody, and the anti-FLAG immunoprecipitates (IP) were subjected to immunoblotting (IB) with the indicated antibodies. B, strategy for chemical dimerization of CagA using the coumermycin-GyrB system. The 1–219-amino acid stretch of GyrB was fused to the N terminus of CagAΔCM, which lacks the CM sequence and thereby cannot bind to PAR1. Because coumermycin bivalently binds with GyrB, two GyrB-CagAΔCM mutants can be artificially dimerized in the presence of coumermycin. C, AGS cells were transiently transfected with an HA-tagged CagA, CagAΔCM, or GyrB-CagAΔCM expression vector. At 17 h after transfection, cells were harvested and lysed. TCLs were subjected to immunoblotting with the indicated antibodies.
Subcellular Localization and Tyrosine Phosphorylation of CagA Mutants
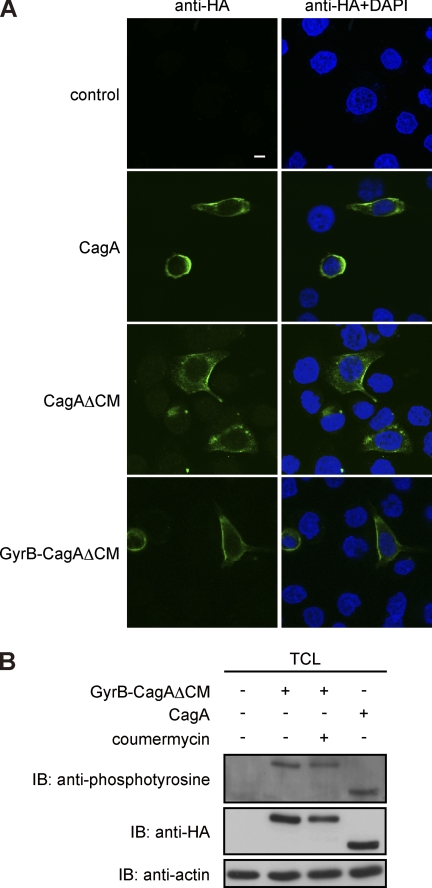
To characterize biological properties of the CagA mutants in cells, AGS cells were transiently transfected with an expression vector for HA-tagged wild-type CagA (ABD-type), HA-tagged CagAΔCM, or HA-tagged GyrB-CagAΔCM. Cell lysates were then prepared and were subjected to immunoblotting analysis using an anti-HA antibody. As expected from the primary sequences, CagAΔCM and GyrB-CagAΔCM were detected as 130-, and 154-kDa proteins, respectively, in AGS cells (Fig. 1C). H. pylori-delivered CagA has been shown to localize to the inner side of the plasma membrane (26, 27). We therefore investigated subcellular distribution of transfected CagA mutants in AGS cells. Immunostaining of cells with an anti-HA antibody showed that both CagAΔCM and GyrB-CagAΔCM were localized to the plasma membrane like wild-type CagA (Fig. 2A). Cell lysates were also prepared from the transfected AGS cells and were subjected to immunoblotting with an anti-phosphotyrosine antibody. The results of the experiment revealed that the GyrB-CagAΔCM mutant underwent tyrosine phosphorylation to levels comparable with that of wild-type CagA in AGS cells (Fig. 2B). The results also showed that coumermycin treatment did not influence the tyrosine phosphorylation level of GyrB-CagAΔCM (Fig. 2B). Hence, both the wild-type CagA and the GyrB-CagAΔCM mutant are localized to the plasma membrane, where they are tyrosine-phosphorylated, in gastric epithelial cells.
FIGURE 2.
Biological properties of conditionally dimerizable CagA in cells. A, subcellular localization of CagA mutants. AGS cells were transfected with an HA-tagged CagA, CagAΔCM, or GyrB-CagAΔCM vector. Cells were fixed at 17 h after transfection and immunostained with an anti-HA antibody (green). Nuclei were stained with DAPI (blue). Confocal x-y images are shown. Scale bar, 10 μm. B, tyrosine phosphorylation of CagA mutants. AGS cells were transiently transfected with an HA-tagged CagA or GyrB-CagAΔCM vector in the presence or absence of 700 nm coumermycin. At 17 h after transfection, cells were harvested and lysed. TCLs were subjected to immunoblotting with the indicated antibodies. IP, immunoprecipitates.
Chemical Dimerization of CagA in Gastric Epithelial Cells
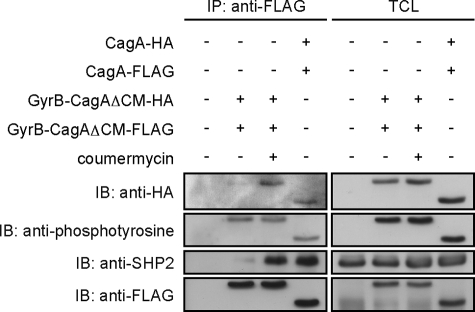
Given the above described observations, we next investigated whether the GyrB-CagAΔCM mutant expressed in AGS cells is capable of specifically dimerizing in a coumermycin-dependent manner. As a positive control experiment, we co-expressed HA-tagged wild-type CagA vector and FLAG-tagged wild-type CagA vector in AGS cells. Total cell lysates prepared from the transfected cells were immunoprecipitated with an anti-FLAG antibody, and the immunoprecipitates were immunoblotted with an anti-HA antibody. As reported previously (16), HA-tagged wild-type CagA was detectable in FLAG-tagged wild-type CagA immunoprecipitates, indicating that CagA is homodimerized in AGS cells (Fig. 3). AGS cells were then co-transfected with an HA-tagged GyrB-CagAΔCM vector and a FLAG-tagged GyrB-CagAΔCM vector and were cultured in the presence or absence of coumermycin at a final concentration of 700 nm, which did not induce any toxicity to AGS cells (data not shown). Total cell lysates prepared were immunoprecipitated with an anti-FLAG antibody, and the immunoprecipitates were immunoblotted with an anti-HA antibody. HA-tagged GyrB-CagAΔCM was also co-immunoprecipitated with FLAG-tagged GyrB-CagAΔCM from lysates prepared from cells treated with coumermycin but not from lysates prepared from cells without coumermycin treatment (Fig. 3). From these observations, we concluded that GyrB-CagAΔCM was conditionally homodimerized by coumermycin in cells.
FIGURE 3.
Chemical dimerization of GyrB-CagAΔCM by coumermycin. AGS cells were transfected with the indicated vectors. At 8 h after transfection, coumermycin was added at a final concentration of 700 nm to the culture, and cells were incubated additional 9 h before harvest. TCLs were prepared and subjected to immunoprecipitation with an anti-FLAG antibody. Immunoprecipitates (IP) and TCLs were subjected to immunoblotting (IB) with the indicated antibodies.
We next investigated whether chemical dimerization of CagA influences the degree of CagA-SHP2 complex formation. Upon coumermycin treatment of AGS cells transfected with the HA-tagged and FLAG-tagged GyrB-CagAΔCM vectors, the amount of SHP2 co-immunoprecipitated with FLAG-tagged GyrB-CagAΔCM was markedly increased (12.8-fold) (Fig. 3, left panel, third row, second and third lanes). Also notably, the level of SHP2 that co-precipitated with chemically dimerized GyrB-CagAΔCM was comparable with that of SHP2 that co-precipitated with wild-type CagA (Fig. 3, left panel, third row, third and fourth lanes). These observations indicated that CagA dimerization plays a critical role in the formation of a stable CagA-SHP2 complex.
Morphogenetic Activity of Monomeric and Dimeric CagA Proteins
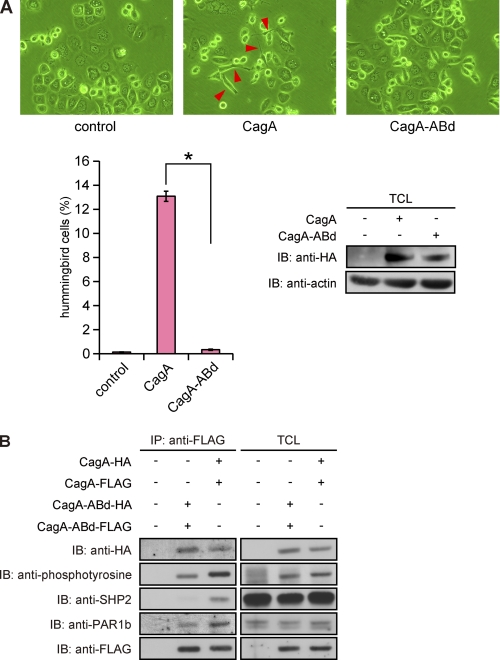
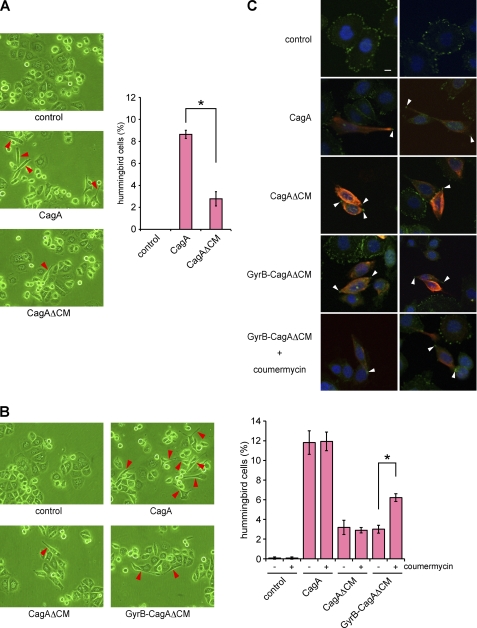
Expression of CagA in AGS cells, either by infection with cagA-positive H. pylori or by transfection of a CagA expression vector, induces an elongated cell morphology termed the hummingbird phenotype. The morphological change, which is characterized by development of one or two long protrusions, requires aberrant activation of SHP2 by CagA that has been tyrosine-phosphorylated at the EPIYA-C or EPIYA-D segment (6, 28). Indeed, a CagA mutant, CagA-ABd, in which the tyrosine residue in the EPIYA-D segment was replaced by non-phosphorylatable phenylalanine, neither bound to SHP2 nor induced the hummingbird phenotype in AGS cells, although it retained the ability to dimerize because of the presence of the PAR1-binding CM sequence (Fig. 4). Notably, the CagA-ABd mutant was tyrosine-phosphorylated in cells because of the presence of the EPIYA-A and -B segments, although the level was less than that of wild-type CagA. We therefore wished to know the contribution of CagA dimerization to induction of the hummingbird phenotype by CagA-activated SHP2. To this end, we examined the hummingbird-inducing potential of a monomeric CagA by transiently transfecting a CagAΔCM vector into AGS cells. In this morphological study, we defined cells showing the hummingbird phenotype (hummingbird cells) as those having cell protrusion, the length of which is more than 2-fold of the length of the shortest diameter of the cell. Although the efficiency was much lower than that induced by wild-type CagA, expression of CagAΔCM was also capable of inducing an elongated cell shape characteristic of the hummingbird phenotype (Fig. 5A). Likewise, expression of GyrB-CagAΔCM in the absence of coumermycin caused the hummingbird phenotype in a very small fraction of transected AGS cells (Fig. 5B). These observations are consistent with the finding that monomeric GyrB-CagAΔCM can still bind to SHP2 in the absence of coumermycin, although the amount of SHP2 bound was much less than that of SHP2 that formed complexes with wild-type CagA (Fig. 3, left panel, third row, compare second and fourth lanes). Because the CagAΔCM mutant has only a single SHP2-binding site (EPIYA-D), the observations also indicate that a monomeric CagA can interact with either the N-SH2 domain or the C-SH2 domain of SHP2 in a tyrosine phosphorylation-dependent manner to form an unstable CagA-SHP2 complex, which could only weakly induce the hummingbird phenotype because of its transient nature.
FIGURE 4.
Failure of hummingbird phenotype induction by CagA lacking a functional SHP2-binding site. A, upper, AGS cells were transfected with an HA-tagged CagA or CagA-ABd vector, and cell morphology was observed by light microscopy at 17 h after transfection. Red arrows indicate hummingbird cells induced by CagA (middle). Lower left, the number of hummingbird cells induced by CagA or CagA-ABd at 17 h after transfection was counted. Error bars, ±S.D. (n = 3), *, p < 0.01, Student's t test. Lower right, at this time point, TCLs were also prepared and were subjected to immunoblotting (IB) with the indicated antibodies. B, AGS cells were co-transfected with CagA-FLAG and CagA-HA vectors or CagA-ABd-FLAG and CagA-ABd-HA vectors. At 17 h after transfection, TCLs were prepared and subjected to immunoprecipitation with an anti-FLAG antibody. The anti-FLAG immunoprecipitates (IP) were separated on SDS-polyacrylamide gel followed by immunoblotting with the indicated antibodies.
FIGURE 5.
Induction of hummingbird phenotype by monomeric and dimeric CagA proteins. A, AGS cells were transfected with a CagA or CagAΔCM vector, and cell morphology was observed by light microscopy at 17 h after transfection. Red arrows indicate hummingbird phenotype induced by CagA (left). The number of hummingbird cells was counted. Error bars, ±S.D. (n = 3), *, p < 0.01, Student's t test (right). B, AGS cells were transfected with a CagA, CagAΔCM, or GyrB-CagAΔCM vector. At 8 h after transfection, coumermycin was added to the culture, and cells were incubated for an additional 9 h before harvest. Cell morphology was analyzed by light microscopy. Red arrows indicate hummingbird cells induced by CagA (left). The number of hummingbird cells was counted. Error bars, ±S.D. (n = 3), *, p < 0.01, Student's t test (right). C, cells were co-stained with anti-active FAK (pY576) (green) and anti-HA (red) antibodies. Nuclei were visualized by DAPI staining (blue). Confocal x-y images are shown. White arrows indicate distal ends of membrane protrusions in cells with the hummingbird phenotype. Scale bar, 10 μm.
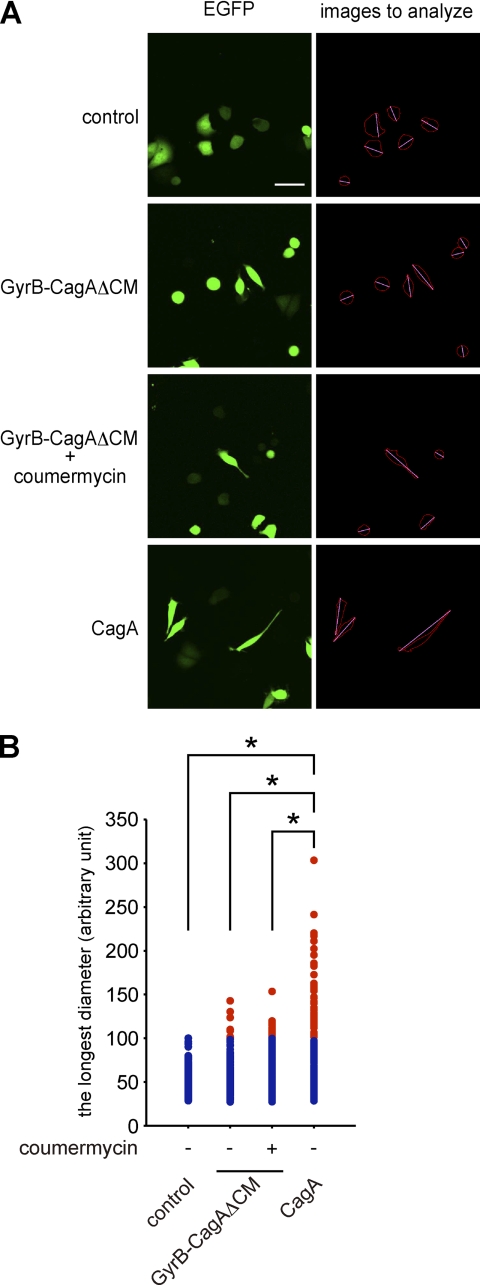
Next, to investigate the effect of CagA dimerization on the morphogenetic activity of CagA, AGS cells were transfected with a GyrB-CagAΔCM vector and were then treated with coumermycin to conditionally dimerize the CagA mutant. The results of cell morphological analysis with a microscope revealed that coumermycin treatment substantially increases the number of cells displaying the hummingbird phenotype (Fig. 5B). In cells showing the hummingbird phenotype, CagA is distributed throughout the cell membrane but is absent in the distal ends of the protrusions. Conversely, active FAK, which has escaped from CagA-deregulated SHP2, is characteristically concentrated at the tips of the cell protrusions in hummingbird cells (10). Accordingly, we stained AGS cells expressing CagAΔCM with an anti-active FAK (anti-FAK pY576) antibody and confirmed specific accumulation of active FAK in the distal ends of cell protrusions. Specific accumulation of active FAK at the tips of the cell protrusions was also observed in hummingbird cells induced by GyrB-CagAΔCM with or without coumermycin (Fig. 5C). Accordingly, CagA dimerization per se enhances the ability of CagA to induce cell elongation, independently of CagA-PAR1 interaction. We then wished to know whether there is any qualitative difference between hummingbird cells induced by wild-type CagA and those induced by artificial dimerization of GyrB-CagAΔCM. To this end, AGS cells were transfected with a CagA or GyrB-CagAΔCM vector together with an EGFP expression vector (at the molar ratio of 10:1). Cells were then cultured in the presence or absence of coumermycin. To qualitatively evaluate the morphology of hummingbird cells, EGFP-positive AGS cells were automatically captured by using ImageJ software (Fig. 6A) (14), and the lengths of cell protrusion were quantitated. Comparison of cell lengths revealed that the cell protrusion induced by chemical dimerization of GyrB-CagAΔCM was significantly shorter than that induced by expression of wild-type CagA (Fig. 6B). These results indicate that CagA dimerization on its own causes an attenuated form of the hummingbird phenotype.
FIGURE 6.
Qualitative analysis of hummingbird cells. A, AGS cells were transfected with an EGFP vector together with a CagA or GyrB-CagAΔCM vector. At 8 h after transfection, coumermycin was added to the culture, and cells were incubated an additional 9 h before analysis. The cell morphological change was observed by a fluorescence microscope (left). The longest diameter was measured by using images analyzed with the ImageJ software (right). Scale bar, 100 μm. B, the longest diameters of cells expressing CagA or GyrB-CagAΔCM, which were calculated from images presented in panel A (right), are shown. Red dots show cells that are longer than the maximum value of control cells. n = 200, *, p < 0.01, Mann-Whitney U test.
Synergism of PAR1 Inhibition in Induction of the Hummingbird Phenotype
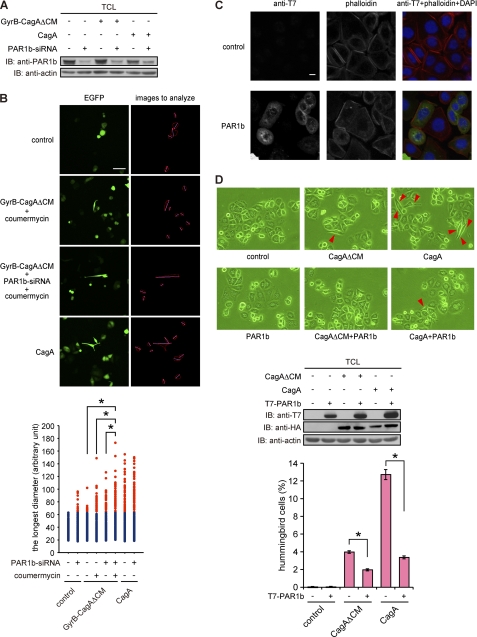
Lastly, we wished to gain insights into the mechanism that directs longer protrusions in hummingbird cells, which were characteristically induced by the expression of wild-type CagA in AGS cells. The CM sequence is not only required for CagA homodimerization but is also critically involved in the inhibition of kinase activity of PAR1 by CagA (12, 16). We therefore assumed that reduced activity of dimerized CagAΔCM in inducing cell elongation is due to failure of the CagA mutant to inhibit PAR1. To address this possibility, we knocked down PAR1b, a major PAR1 isoform expressed in AGS cells, by treating cells with specific siRNA (Fig. 7A). The PAR1b-inhibited AGS cells were then transfected with a wild-type CagA or GyrB-CagAΔCM vector together with EGFP vector, treated with or without coumermycin, and then subjected to morphological analysis using microscopy. The results of the experiment revealed that hummingbird cells induced by chemical dimerization of GyrB-CagAΔCM developed longer cell protrusions when PAR1b expression was simultaneously inhibited (Fig. 7B). In a reciprocal experiment, we ectopically overexpressed PAR1b in AGS cells with or without CagA. In the absence of CagA, AGS cells expressing PAR1b showed a slightly more rounded morphology when compared with the morphology of PAR1b-nonexpressing cells (Fig. 7C). Furthermore, expression of CagA or CagAΔCM together with PAR1b in AGS cells abolished induction of the hummingbird phenotype by CagA (Fig. 7D) (14). Hence, full induction of the hummingbird phenotype by CagA requires two independent functions of the CM sequence: 1) dimerization of CagA that potentiates SHP2 deregulation and 2) inhibition of PAR1 activity.
FIGURE 7.
Effect of PAR1 inhibition on the hummingbird phenotype. A, AGS cells treated with PAR1b-specific siRNA for 24 h were transfected with a CagA or GyrB-CagAΔCM vector. At 17 h after vector transfection, lysates were prepared and were subjected to immunoblotting (IB) with the indicated antibodies. B, AGS cells transfected with PAR1b-specific siRNA for 24 h were co-transfected with CagA or GyrB-CagAΔCM and EGFP vectors. At 8 h after transfection, coumermycin was added to the culture, and cells were incubated for an additional 9 h before analysis. Cell morphology was observed by using a fluorescence microscope (upper). Scale bar, 100 μm. Lower, the longest diameters of cells expressing CagA or GyrB-CagAΔCM, which were calculated from images presented in upper right panels are shown. Red dots show cells that are longer than the maximum value of control cells. n = 200, *, p < 0.01, Mann-Whitney U test (lower). C, AGS cells were transfected with a PAR1b expression vector. At 17 h after transfection, cell morphology was observed by staining cells with an anti-T7 antibody (green), phalloidin (red), or DAPI (blue). Scale bar, 10 μm. D, AGS cells were transfected with a PAR1b expression vector with or without a CagA or CagAΔCM vector. At 17 h after transfection, cell morphology was observed by light microscopy. Red arrows indicate hummingbird phenotype induced by CagA (upper). TCLs were prepared and were subjected to immunoblotting with the indicated antibodies (middle). Lower, the number of hummingbird cells was counted. Error bars, ±S.D. (n = 3), *, p < 0.01, Student's t test.
DISCUSSION
In this work, we generated a CagA mutant that is conditionally homodimerized by a chemical dimerizer in the absence of PAR1 in cells. Upon forced dimerization, the CagA mutant displayed a marked increase in its ability to interact with the SHP2 oncoprotein, which was concomitantly associated with the potentiation of CagA activity to induce an aberrantly elongated cell morphology known as the hummingbird phenotype.
The hummingbird phenotype requires tyrosine phosphorylation of the EPIYA-C or EPIYA-D segment of CagA, to which SHP2 binds (1, 6, 17). Intriguingly, H. pylori strains carrying East Asian CagA (ABD-type CagA) are more closely associated with severe gastric atrophy and gastric carcinoma than are H. pylori strains carrying Western CagA (ABCn-type CagA) (29, 30). Among H. pylori strains carrying Western CagA, those having CagA with a larger number of EPIYA-C segments are more likely to induce intestinal metaplasia and gastric carcinoma than are those having CagA with fewer EPIYA-C segments (31–34). In transgenic mouse studies, East Asian CagA was shown to be more oncogenic than Western CagA (35, 36). In parallel with these observations, East Asian CagA proteins display greater ability to bind SHP2 and exhibit stronger activity to induce the hummingbird phenotype than do Western CagA proteins (17, 37). Likewise, Western CagA proteins with more EPIYA-C segments show greater ability to induce the hummingbird phenotype than do those with fewer EPIYA-C segments (17, 37). Hence, the magnitude of hummingbird induction by individual CagA correlates well with the disease outcome of cagA-positive H. pylori infection, indicating that it reflects the degree of CagA virulence, particularly the oncogenic potential of CagA.
Induction of the hummingbird phenotype by CagA is influenced by the CM sequence, a 16-amino acid stretch originally identified as a sequence that mediates CagA homodimerization (16). Subsequent studies have shown that the CM sequence directly interacts with the catalytic domain of the PAR1 family of polarity-regulating serine/threonine kinases and thereby inhibits their kinase activity (12, 14). Because PAR1 is present as a dimer within the cell (Fig. 1A), two CagA proteins may interact with a PAR1 dimer via the CM sequence and thus be passively homodimerized. The dual functions of the CM sequence in both CagA dimerization and PAR1 inhibition, however, have made it difficult to determine the relative contribution of each of the CM functions to its pathophysiological activity.
To investigate the functional role of CagA dimerization without taking into account the influence of CagA-PAR1 interaction, we used a chemical dimerizer, coumermycin, which enables conditional dimerization of a CagA mutant (CagAΔCM) that lacks the CM sequence and thus cannot bind to PAR1. Coumermycin can specifically dimerize the bacterial GyrB domain, and the coumermycin-GyrB system has been successfully utilized for conditional dimerization of heterologous proteins such as c-Raf-1, ASK1, L-selectin, STAT3, and JAK kinases, demonstrating its versatile application for functional study of protein dimerization (21–25).
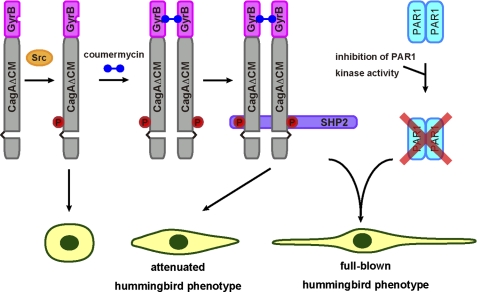
A mutant CagA lacking the CM sequence weakly interacts with SHP2, indicating that a tyrosine-phosphorylated CagA monomer is still capable of binding to one of the two SH2 domains of SHP2 to form an unstable CagA-SHP2 complex. Upon chemical dimerization of CagA, however, the amount of the CagA-SHP2 complex is markedly increased. Hence, simple dimerization of CagA per se appears to be sufficient for stable interaction of CagA with SHP2. This observation supports the previously proposed model that two CagA proteins, each containing a single EPIYA-C or EPIYA-D segment, simultaneously bind to the two SH2 domains of a single SHP2 in an EPIYA phosphorylation-dependent manner to form a stable complex comprising two PAR1 proteins, two CagA proteins, and an SHP2 protein (Fig. 8) (12, 19).
FIGURE 8.
Model for induction of the hummingbird phenotype by chemical dimerization of CagA. As a monomer, GyrB-CagAΔCM undergoes tyrosine phosphorylation (circled P) but barely induces the hummingbird phenotype because of unstable interaction with SHP2. In the presence of coumermycin, dimerized GyrB-CagAΔCM molecules bind to the two SH2 domains of a single SHP2 protein and thereby form a stable CagA-SHP2 complex, which induces an attenuated form of the hummingbird phenotype as a result of SHP2 deregulation. The full-blown hummingbird phenotype by chemically dimerized GyrB-CagAΔCM requires simultaneous inhibition of PAR1 kinase activity. This indicates that the CM sequence has a dual role in mediating both CagA dimerization and PAR1 inhibition in induction of the hummingbird phenotype.
In addition to the enhanced SHP2 interaction, chemical dimerization of CagA substantially increased the number of cells displaying the hummingbird phenotype. This observation is in agreement with the results of previous studies showing that deregulation of SHP2 by CagA is required for induction of the hummingbird phenotype (12, 38). Notably, however, hummingbird cells induced by chemically dimerized CagA were substantially fewer and less elongated than those induced by wild-type CagA. This observation suggests that SHP2 deregulation is not sufficient for induction of the full-blown hummingbird phenotype, which is characterized by the appearance of robustly elongated cells, and suggests a role of CM-mediated PAR1 inhibition in enhancing the hummingbird phenotype. This idea is supported by the observation that hummingbird cells induced by chemical dimerization of CagA, which does not have the CM sequence and thus cannot inhibit PAR1, developed longer protrusions when PAR1 expression was simultaneously inhibited by siRNA. The present study therefore emphasizes the functional integrity of the CagA oncoprotein that enables simultaneous perturbation of two distinct effectors, SHP2 and PAR1, in opposite directions to manifest full virulence potential.
In the present study, CagA was chemically dimerized through the N-terminal region, whereas wild-type CagA was dimerized via the CM sequence located in the C-terminal region. Despite substantial differences in the mode of CagA dimerization, the amount of SHP2 that formed a complex with chemically dimerized CagA was almost comparable with that of SHP2 bound to wild-type CagA (Fig. 3). This observation may be explained by the intrinsically unstructured nature of the C-terminal CagA region that spans the EPIYA segments and the CM sequence (15). A highly dynamic property of the unstructured C-terminal region of CagA may enable efficient interaction with SHP2 once two CagA proteins are juxtaposed through dimerization.
Transgenic mice systemically expressing CagA spontaneously develop gastrointestinal and hematological malignancies (35, 36). In contrast, tumor formation is not observed in transgenic mice expressing phosphorylation-resistant CagA, which cannot bind SHP2 (35). Hence, aberrant activation of SHP2 by CagA plays a pivotal role in the oncogenic transformation of gastric epithelial cells and in the development of gastric adenocarcinoma by cagA-positive H. pylori infection (1, 5, 19, 39). The present study further highlights the importance of CagA dimerization, mediated by a PAR1 dimer, in the deregulation of SHP2 and also the importance of CagA dimerization in the pathogenic action of H. pylori CagA. The results, in turn, indicate a therapeutic application for inhibition of CagA dimerization in preventing H. pylori-associated gastric carcinoma.
Supplementary Material
Acknowledgments
We thank Dr. Roger Perlmutter for providing the GyrB gene and Dr. Shigeo Ohno for providing the PAR1b plasmid and anti-PAR1b antibody. We also thank Ryo Higashi for technical help.
This work was supported by grants-in-aid for scientific research on innovative areas from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- SH2
- Src homology-2
- SHP2
- SH2-containing protein-tyrosine phosphatase
- FAK
- focal adhesion kinase
- PAR1
- partitioning-defective 1
- MARK
- microtubule affinity-regulating kinase
- CM
- CagA multimerization
- GyrB
- B-subunit of bacterial DNA gyrase
- EGFP
- enhanced green fluorescent protein
- TCL
- total cell lysate.
REFERENCES
- 1. Hatakeyama M. (2004) Nat. Rev. Cancer 4, 688–694 [DOI] [PubMed] [Google Scholar]
- 2. Wen S., Moss S. F. (2009) Cancer Lett. 282, 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tegtmeyer N., Wessler S., Backert S. (2011) FEBS J. 278, 1190–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Backert S., Selbach M. (2005) Trends Microbiol. 13, 476–484 [DOI] [PubMed] [Google Scholar]
- 5. Hatakeyama M. (2011) Cancer Sci. 102, 36–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Higashi H., Tsutsumi R., Muto S., Sugiyama T., Azuma T., Asaka M., Hatakeyama M. (2002) Science 295, 683–686 [DOI] [PubMed] [Google Scholar]
- 7. Tartaglia M., Gelb B. D. (2005) Eur. J. Med. Genet 48, 81–96 [DOI] [PubMed] [Google Scholar]
- 8. Mohi M. G., Neel B. G. (2007) Curr. Opin. Genet. Dev. 17, 23–30 [DOI] [PubMed] [Google Scholar]
- 9. Takahashi A., Tsutsumi R., Kikuchi I., Obuse C., Saito Y., Seidi A., Karisch R., Fernandez M., Cho T., Ohnishi N., Rozenblatt-Rosen O., Meyerson M., Neel B. G., Hatakeyama M. (2011) Mol. Cell 43, 45–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tsutsumi R., Takahashi A., Azuma T., Higashi H., Hatakeyama M. (2006) Mol. Cell Biol. 26, 261–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Amieva M. R., Vogelmann R., Covacci A., Tompkins L. S., Nelson W. J., Falkow S. (2003) Science 300, 1430–1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Saadat I., Higashi H., Obuse C., Umeda M., Murata-Kamiya N., Saito Y., Lu H., Ohnishi N., Azuma T., Suzuki A., Ohno S., Hatakeyama M. (2007) Nature 447, 330–333 [DOI] [PubMed] [Google Scholar]
- 13. Ebneth A., Drewes G., Mandelkow E. M., Mandelkow E. (1999) Cell Motil. Cytoskeleton 44, 209–224 [DOI] [PubMed] [Google Scholar]
- 14. Lu H., Murata-Kamiya N., Saito Y., Hatakeyama M. (2009) J. Biol. Chem. 284, 23024–23036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nesić D., Miller M. C., Quinkert Z. T., Stein M., Chait B. T., Stebbins C. E. (2010) Nat. Struct. Mol. Biol. 17, 130–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ren S., Higashi H., Lu H., Azuma T., Hatakeyama M. (2006) J. Biol. Chem. 281, 32344–32352 [DOI] [PubMed] [Google Scholar]
- 17. Higashi H., Tsutsumi R., Fujita A., Yamazaki S., Asaka M., Azuma T., Hatakeyama M. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 14428–14433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lu H. S., Saito Y., Umeda M., Murata-Kamiya N., Zhang H. M., Higashi H., Hatakeyama M. (2008) Cancer Sci. 99, 2004–2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hatakeyama M. (2008) Oncogene 27, 7047–7054 [DOI] [PubMed] [Google Scholar]
- 20. Gilbert E. J., Maxwell A. (1994) Mol. Microbiol. 12, 365–373 [DOI] [PubMed] [Google Scholar]
- 21. Farrar M. A., Alberol-Ila J., Perlmutter R. M. (1996) Nature 383, 178–181 [DOI] [PubMed] [Google Scholar]
- 22. Mizuguchi R., Hatakeyama M. (1998) J. Biol. Chem. 273, 32297–32303 [DOI] [PubMed] [Google Scholar]
- 23. Mohi M. G., Arai K., Watanabe S. (1998) Mol. Biol. Cell 9, 3299–3308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li X., Steeber D. A., Tang M. L., Farrar M. A., Perlmutter R. M., Tedder T. F. (1998) J. Exp. Med. 188, 1385–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Farrar M. A., Tian J., Perlmutter R. M. (2000) J. Biol. Chem. 275, 31318–31324 [DOI] [PubMed] [Google Scholar]
- 26. Higashi H., Yokoyama K., Fujii Y., Ren S., Yuasa H., Saadat I., Murata-Kamiya N., Azuma T., Hatakeyama M. (2005) J. Biol. Chem. 280, 23130–23137 [DOI] [PubMed] [Google Scholar]
- 27. Murata-Kamiya N., Kikuchi K., Hayashi T., Higashi H., Hatakeyama M. (2010) Cell Host Microbe 7, 399–411 [DOI] [PubMed] [Google Scholar]
- 28. Segal E. D., Cha J., Lo J., Falkow S., Tompkins L. S. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 14559–14564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Azuma T., Yamazaki S., Yamakawa A., Ohtani M., Muramatsu A., Suto H., Ito Y., Dojo M., Yamazaki Y., Kuriyama M., Keida Y., Higashi H., Hatakeyama M. (2004) J. Infect. Dis. 189, 820–827 [DOI] [PubMed] [Google Scholar]
- 30. Schmidt H. M., Goh K. L., Fock K. M., Hilmi I., Dhamodaran S., Forman D., Mitchell H. (2009) Helicobacter 14, 256–263 [DOI] [PubMed] [Google Scholar]
- 31. Argent R. H., Kidd M., Owen R. J., Thomas R. J., Limb M. C., Atherton J. C. (2004) Gastroenterology 127, 514–523 [DOI] [PubMed] [Google Scholar]
- 32. Basso D., Zambon C. F., Letley D. P., Stranges A., Marchet A., Rhead J. L., Schiavon S., Guariso G., Ceroti M., Nitti D., Rugge M., Plebani M., Atherton J. C. (2008) Gastroenterology 135, 91–99 [DOI] [PubMed] [Google Scholar]
- 33. Argent R. H., Hale J. L., El-Omar E. M., Atherton J. C. (2008) J. Med. Microbiol. 57, 1062–1067 [DOI] [PubMed] [Google Scholar]
- 34. Batista S. A., Rocha G. A., Rocha A. M., Saraiva I. E., Cabral M. M., Oliveira R. C., Queiroz D. M. (2011) BMC Microbiol. 11, 61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ohnishi N., Yuasa H., Tanaka S., Sawa H., Miura M., Matsui A., Higashi H., Musashi M., Iwabuchi K., Suzuki M., Yamada G., Azuma T., Hatakeyama M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 1003–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Miura M., Ohnishi N., Tanaka S., Yanagiya K., Hatakeyama M. (2009) Int. J. Cancer 125, 2497–2504 [DOI] [PubMed] [Google Scholar]
- 37. Naito M., Yamazaki T., Tsutsumi R., Higashi H., Onoe K., Yamazaki S., Azuma T., Hatakeyama M. (2006) Gastroenterology 130, 1181–1190 [DOI] [PubMed] [Google Scholar]
- 38. Higuchi M., Tsutsumi R., Higashi H., Hatakeyama M. (2004) Cancer Sci. 95, 442–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Saito Y., Murata-Kamiya N., Hirayama T., Ohba Y., Hatakeyama M. (2010) J. Exp. Med. 207, 2157–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.