Abstract
We have recently established the fine structure of the glycan backbone of lipooligosaccharides (LOS-I to LOS-IV) isolated from Mycobacterium marinum, a close relative of Mycobacterium tuberculosis. These studies culminated with the description of an unusual terminal N-acylated monosaccharide that confers important biological functions to LOS-IV, such as macrophage activation, that may be relevant to granuloma formation. It was, however, also suggested that the lipid moiety was required for LOSs to exert their immunomodulatory activity. Herein, using highly purified LOSs from M. marinum, we have determined through a combination of mass spectrometric and NMR techniques, the structure and localization of the fatty acids composing the lipid moiety. The occurrence of two distinct polymethyl-branched fatty acids presenting specific localizations is consistent with the presence of two highly related polyketide synthases (Pks5 and Pks5.1) in M. marinum and presumably involved in the synthesis of these fatty acyl chains. In addition, a bioinformatic search permitted us to identify a set of enzymes potentially involved in the biosynthesis or transfer of these lipids to the LOS trehalose unit. These include MMAR_2343, a member of the Pap (polyketide-associated protein) family, that acylates trehalose-based glycolipids in M. marinum. The participation of MMAR_2343 to LOS assembly was demonstrated using a M. marinum mutant carrying a transposon insertion in the MMAR_2343 gene. Disruption of MMAR_2343 resulted in a severe LOS breakdown, indicating that MMAR_2343, hereafter designated PapA4, fulfills the requirements for LOS acylation and assembly.
Keywords: Cell Wall, Glycolipid Structure, Lipid Synthesis, Mycobacteria, Nuclear Magnetic Resonance, Lipooligosaccharides, Polymethyl-branched Fatty Acid
Introduction
Members of the Mycobacterium genus possess a unique lipid-rich cell wall that confers resistance to many therapeutic agents and contributes to virulence of pathogenic mycobacteria (1, 2). This cell wall consists of numerous ubiquitous or species-specific lipids/glycolipids embedded into a layer of mycolic acids covalently linked to the arabinogalactan-peptidoglycan complex (3, 4). During the infection process, free cell wall lipids/glycolipids are thought to play an immunomodulatory role that leads to physiopathological manifestations and condition the outcome of the infection (5, 6).
Lipooligosaccharides (LOS)3 are among these glycolipids and have been reported to exist in more than 10 mycobacterial species, including the opportunistic pathogens Mycobacterium kansasii, Mycobacterium gastri, and Mycobacterium marinum, as well as the tuberculosis etiological agent Mycobacterium canettii (7–12). LOSs are antigenic compounds containing a α,α′-trehalose unit that is acylated and further glycosylated in a species-specific manner. Although LOSs were discovered 30 years ago, their precise role in virulence and colony morphology remains elusive (13–15). Earlier studies showed that rough variants of M. kansasii, devoid of all LOSs, cause chronic systemic infections in mice, whereas smooth variants bearing LOSs are rapidly cleared from the organs of infected animals (13, 14). It was therefore proposed that LOS might be avirulent factor masking other cell wall-associated lipids/glycolipids that are important virulence determinants. Recent studies suggested that LOSs are also key players contributing to the virulence of M. marinum, a pathogenic mycobacterial species that is closely related to Mycobacterium tuberculosis (16, 17). Initially, four major types of LOSs (LOS-I to LOS-IV) were identified and partially characterized in M. marinum (12). The defect of LOS-IV biosynthesis of a mutant strain was correlated with a lower efficiency to enter and infect macrophages in vitro (12, 18). Furthermore, this result was supported by the finding that a M. marinum isolate (Mma7) unable to produce LOS-IV (19) failed to show signs of infection in adult zebrafish (20).
As a first step to address the role of LOSs in M. marinum virulence, we characterized the glycosidic structures of LOS-I to LOS-IV (19, 21). Structural analyses demonstrated that LOS-I, -II, -III, and -IV share a common glycan core consisting of the acylated trehalose substituted by two β-d-Glcp residues and one 3-O-methylated α-l-Rhap. A residue of α-d-Xylp further substitutes the glycan core in LOS-II, LOS-III, and LOS-IV. In addition, LOS-II contains an α-caryophyllose residue (α-3,6-dideoxy-4-C-(d-altro-1,3,4,5-tetrahydroxyhexyl)-d-xylo-hexopyranose), whereas two α-caryophyllose units are found in LOS-III and LOS-IV (19). In minor species, caryophyllose may be substituted by the related monosaccharide hydroxylated in position C3 (α-6-deoxy-4-C-(d-altro-1,3,4,5-tetrahydroxyhexyl)-d-galactopyranose). Finally, the terminal monosaccharide of LOS-IV substituting the second caryophyllose unit has recently been identified as a unique α-4-amino-4,6-dideoxy-Galp residue N-acylated by a 3-hydroxy-3-methylated-pyrrolidone cycle (21). Importantly, in vitro-based assays using purified LOSs demonstrated their potent anti-inflammatory activity, characterized by the inhibition of TNF-α secretion from LPS-activated macrophages (19). Moreover, only LOS-IV was found to induce secretion of IL-8 as well as cell surface antigen expression such as CD40 and ICAM-1 on macrophages (21). It was therefore proposed that this specific immunostimulatory property was associated to the presence of the unique terminal N-acylated monosaccharide of LOS-IV. However, in vitro assays using deacylated LOSs also indicated that the fatty acyl chains are required for LOSs to exert their immunomodulatory activity (21).
Mycobacterial LOSs possess one to four methyl-branched fatty acids as well as species-specific linear fatty acids. For instance, the major LOS from Mycobacterium smegmatis carries one 2,4-dimethyl-2-eicosenoic acid (C22:1; 2,4-dimethyl), one tetradecanoate/hexadecanoate (C14/C16), and one octanoate (C8), whereas LOSs from M. kansasii are mainly acylated by three 2,4-dimethyltetradecanoic acids (C16:0, 2,4-dimethyl) (9, 22). In M. smegmatis, polymethyl-branched fatty acyl substituents of LOSs are found at single specific positions on the trehalose core (22). In M. canettii, a mixture of LOSs are synthesized, differing by the localization of the polymethyl-branched fatty acyl substituents (23). The biosynthesis and attachment of polymethyl-branched fatty acids of mycobacterial glycolipids require the action of numerous enzymes. First, a fatty acyl-AMP ligase (FadD protein) activates a linear fatty acid to an acyl-adenylate, which is subsequently transferred to a polyketide synthase (Pks) (24). The latter uses methymalonyl-CoA to elongate fatty acids, resulting in the formation of polymethyl-branched fatty acids (25). M. smegmatis Pks5 catalyzes the synthesis of 2,4-dimethyl-2-eicosenoic acid (C22:1; 2,4-dimethyl) of LOS (26). Disruption of the MSMEG_4727 gene encoding Pks5 resulted in the absence of LOS production in M. smegmatis (26). The next biosynthetic step involves an acyltransferase of the Pap (polyketide synthase-associated protein) family, which catalyzes the transfer of the polymethyl-branched fatty acid to a sugar moiety of the glycolipid (27), although such an enzyme has not been described yet for LOSs.
Considering the potential influence of the fatty acyl chains on biosynthesis, antigenicity, and/or biological activity of LOSs, we aimed to precisely define the lipid moiety characterizing each LOS subtype of M. marinum. Herein, we provide evidence that LOSs are bearing three polymethyl-branched fatty acyl chains, one of which is unsaturated. Moreover, we show that these fatty acyl substituents are located on specific positions of the trehalose backbone. Finally, disruption of MMAR_2343/papA4 in M. marinum led to a severe defect in LOS production, thus establishing a key role of PapA4 in LOS acylation.
EXPERIMENTAL PROCEDURES
M. marinum Strains and Growth Culture Conditions
M. marinum strain M, used in this study, was first isolated from a human patient as described previously (16). Bacteria were grown in Sauton broth medium at 30 °C or on plates containing Sauton agar supplemented with 10% oleic acid/albumin/dextrose/catalase enrichment. M. marinum MMAR_2343/papA4::Tn was grown in the presence of 50 μg/ml hygromycin.
Determination of the Transposon Insertion Site
Tn5370 insertion site was determined as described previously (28) with slight modifications. Two rounds of PCR were performed using the Goldstar Mix from Eurogentec on 1 μl of bacterial lysates prepared as followed: 500 μl of culture was centrifuged, and mycobacteria were resuspended in 100 μl of water, incubated 30 min at 80 °C, frozen, and thawed prior to PCR amplification. The first round of PCR was carried out with the transposon-specific primer O84L-R (5′-AACTGGCGCAGTTCCTCTGG-3′) (29) and the arbitrarily degenerated primer RS6–4 (5′-GTAATACGACTCACTATAGGGCNNNNCATG-3′). The second round of PCR was performed with the nested primers SP2 (5′-CTCTTGCTCTTCCGCTTCTTCTCC-3′) and T7 (5′-TAATACGACTCACTATAGGG-3′). Purified PCR fragments were then sequenced using the SP2 primer. Transposon insertion was further confirmed by PCR on genomic DNA using different combinations of primers specific of the transposon, 084L-R and O84R-F (5′-ATACACGCGCACCGGTTCTAGC-3′) (29) and primers specific to MMAR_2343 (5′-AACCTCGAGCTGCGCACCCGCATC-3′ and 5′-ATGTCCATGGCCGCGACCACC-3′). Genomic DNA was prepared as described previously (30).
Polar Lipid Extraction and Analysis
Polar lipids were extracted according to established procedures (12). Polar lipid extracts were examined by two-dimensional TLC on plates of aluminum backed silica Gel 60 (Merck), using chloroform/methanol/water (60:30:6, v/v/v) in the first direction and chloroform/acetic acid/methanol/water (40:25:3:6, v/v/v/v) in the second direction. Glycolipids were visualized by spraying plates with orcinol/sulfuric acid reagent (0.2% (w/v) orcinol in H2SO4/water (1:4, v/v)) followed by charring.
Purification of LOSs
Purification of LOSs was performed as previously described (19). Briefly, extracted polar lipids were dissolved in chloroform/methanol (2:1, v/v) and applied on DEAE cellulose column for purification. LOS-I to -III were eluted by chloroform/methanol (2:1, v/v) from DEAE column. LOS-IV was eluted from DEAE by 40–60 mm of ammonium acetate in chloroform/methanol (2:1, v/v). The presence of LOSs in the eluted fractions was monitored by one-dimensional TLC on glass-backed plates of silica gel 60 (Merck) using chloroform/acetic acid/methanol/water (40:25:3:6, v/v/v/v) as running solvent. LOSs were visualized by spraying the plates with orcinol/sulfuric acid reagent followed by charring. LOS-IV was further purified into individual species by preparative TLC (19, 21).
Gas Chromatography Coupled to Mass Spectrometry
Fatty acids from purified LOSs were released by methanolysis in methanol/HCl 0.5 n at 80 °C for 16 h. After cooling, the reagent was evaporated under a stream of nitrogen, and the sample was dissolved in chloroform prior to GC-MS analysis using a Carlo Erba GC 8000 gas chromatograph equipped with a 25-m × 0.25-mm TR5 low bleed/MS capillary column, 0.25-μm film phase (Thermo Fisher Scientific, Villebon-sur-Yvette, France). Temperature of the Ross injector was 280 °C, and the samples were analyzed using a temperature program starting at 120 °C for 1 min, followed by a temperature ramp of 5 °C min−1 to 280 °C maintained for 20 min. Electron ionization-mass spectra were obtained using a Finnigan Automass II mass spectrometer.
MALDI-MS and -MS/MS
Permethylation of LOSs was performed according to the procedure of Ciucanu and Kerek (31). Briefly, glycolipids were incubated for 2 h in a suspension of 200 mg/ml NaOH in dry Me2SO (300 μl) and iodomethane (200 μl). The methylated LOSs were extracted in chloroform and washed seven times with water. After evaporation of the reagents, the samples were dissolved in methanol prior to mass spectrometry analysis. The molecular masses of permethylated compounds were measured by MALDI-TOF on a Voyager Elite reflectron mass spectrometer (PerSeptive Biosystems, Framingham, MA), equipped with a 337-nm UV laser. Samples were prepared by mixing in a tube 5 μl of diluted permethylated LOS solution in methanol and 5 μl of 2,5-dihydroxybenzoic acid matrix solution (10 mg·ml−1) dissolved in methanol/water (1:1, v/v). The mixtures (2 μl) were then spotted on the target plate. MALDI-MS/MS analyses of native LOSs were performed on a 4800 Proteomics Analyzer (Applied Biosystems, Framingham, MA) mass spectrometer, operated in the reflectron mode. For MS acquisition, 5 μl of diluted samples in CHCl3/CH3OH/H2O (3:1.5:0.25, v/v/v) were mixed with 5 μl of 2,5-dihydroxybenzoic acid matrix solution (10 mg/ml dissolved in CHCl3/CH3OH/H2O (3:1.5:0.25, v/v/v). The mixtures (2 μl) were then spotted on the target plate and air-dried. MS survey data comprise a total of 50 subspectra of 1,500 laser shots. Peaks observed in the MS spectra were selected for further MS/MS. Collision induced dissociation MS/MS data comprises a total of 100 subspectra of 3,000 laser shots. Two or more spectra can be combined after acquisition with mass tolerance set at 0.1 Da to improve the S/N ratio. The potential difference between the source acceleration voltage and the collision cell was set to 1 kV, and argon was used as collision gas.
High Resolution nanoESI-Qh-FT-ICR MS Analysis
LOS-IV was dissolved in methanol/chloroform solution (1:1, v/v) to obtain concentrated 300 pmol/μl stock solution and then diluted in methanol/chloroform solution (2:1, v/v) to obtain the final solution at 500 fmol/μl. To increase the ionization efficiency, lithium chloride was added to the sample (2% LiCl in methanol/chloroform (2:1, v/v)). LOS-IV was analyzed with an Apex Qe 9.4 T Fourier transform ion cyclotron resonance mass spectrometer (Bruker Daltonics, Bremen, Germany). The FT-ICR mass spectrometer was equipped with a nanoelectrospray source. Detection was carried out in positive mode. A potential of 1.6 kV was applied on the needle (PicoTip Emitter; New Objective, Woburn, MA). The detection parameters were as follows: broadband detection, 512 K acquisition size, and start mass at m/z 1000. Ions were accumulated in the storage hexapole during 1 s and in the second hexapole during 0.01 s. The spectra were manually processed using data analysis software (Bruker Daltonics, Bremen, Germany).
NMR Analysis
Solution-state NMR experiments were recorded at 300 K on Bruker Avance 400 and Avance 600 spectrometers equipped with a 5-mm broad band inverse probe and a 5-mm triple resonance cryoprobe, respectively. Prior to NMR spectroscopic analyses, LOSs were repeatedly exchanged in CDCl3/CD3OD (2:1, v/v) (99.97% purity; Eurisotop-Saclay, Saint-Aubin, France) with intermediate drying and then dissolved in CDCl3/CD3OD (2:1, v/v). Chemical shifts are expressed as ppm using the tetramethylsilane signal (0.000 ppm) in CDCl3/CD3OD. All of the experiments were recorded without sample spinning. Pulse programs used in 1H/1H COSY, 1H/1H ROESY, 1H/1H nuclear Overhauser effect spectroscopy, 1H/13C HSQC, and 1H/13C HMBC experiments were extracted from Bruker pulse program library.
RESULTS
Structure of the LOS Fatty Acyl Chains
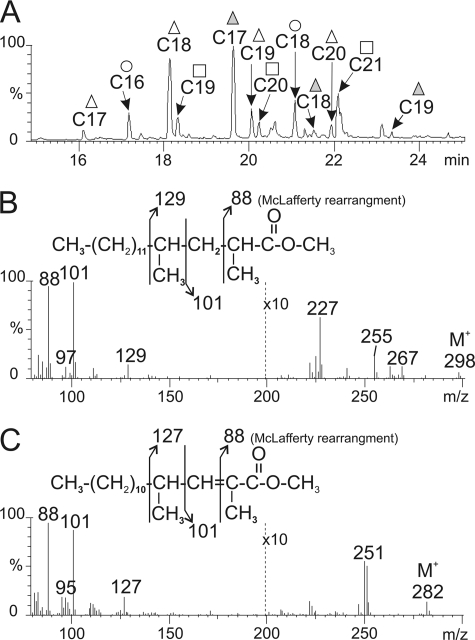
The glycan moiety of the four major LOS subtypes (LOS-I to -IV) of M. marinum has recently been delineated and found to participate in LOS biological functions (19, 21). Additional experiments using deacylated LOSs unraveled a role of the lipid moiety in the LOS biological properties (21), although the fine structure and composition of the acyl chains has not yet been addressed. This prompted us to determine the fatty acid composition and structure of LOS-I to LOS-IV. LOSs were first purified into individual species using a combination of column chromatography and preparative TLC as reported earlier (21). Following purification, fatty acids were released by methanolysis and subsequently analyzed by GC-MS. The chromatogram of fatty acid methyl esters released from LOS-I showed the presence of two prominent peaks with retention times of 18.07 and 19.38 min, respectively (Fig. 1A). Similar chromatograms were obtained for LOS-II, LOS-III and LOS-IV (data not shown), indicating that all LOS subtypes are sharing a similar fatty acid composition. Electron impact EI-MS of the peak at 18.07 min (Fig. 1B) showed a molecular ion (M+) at m/z 298 and (M-31) ion at m/z 267, consistent with a C18-fatty acid methyl ester. The McLafferty rearranged ion at m/z 88 indicated the incorporation of a methyl group into the resulting fragment because of methyl-branched substitution in C2 position of the fatty acid. The additional intense ions at m/z 101, 129, and 97 clearly demonstrated the presence of a second methyl substituent in the C4 position (9, 32, 33). The methyl side chains at C2 and C4 were confirmed by signals at m/z 255 (M-43) and m/z 227 (M-71), resulting from the loss of [CH(CH3)-CH2 + H] and [CH(CH3)-CH-CH(CH3) + H] by the molecular ion (32). The absence of an additional methyl substitution at C6 was reflected by the lack of significant peaks at m/z 171 and 139. Thus, this fatty acid methyl ester was identified as a 2,4-dimethylhexadecanoate (C18:0; 2,4-dimethyl). EI-MS of the peak at 19.38 min (Fig. 1C) showed a molecular ion (M+) at m/z 282 and a (M-31) fragment ion at m/z 251 corresponding to a monounsaturated C17-fatty acid methyl ester. The intense fragment ions at m/z 88, 101, 127, and 95 indicated the presence of two methyl groups in C2 and C4 positions but also the presence of an unsaturation in C2 position. Thus, the fatty acid was characterized as a 2,4-dimethyl-2-pentadecenoate (C17:1; 2,4-dimethyl).
FIGURE 1.
Identification of polymethyl-branched fatty acyl substituents of LOSs by GC-MS. GC-MS analysis of fatty acid methyl esters of LOS-I from M. marinum is shown. A, chromatogram showing presence of 2,4-dimethyl-branched (△), 2,4,6-trimethyl-branched (□), 2-unsaturated 2,4-dimethyl-branched (▴), and linear (○) fatty acids ranging from C16 to C21. B, EI-MS fragmentation of the peak at 18.07 min corresponding to 2,4-dimethylhexadecanoate (C18:0; 2,4-dimethyl). C, EI-MS spectrum of the peak at 19.38 min corresponding to 2,4-dimethyl-2-pentadecenoate (C17:1; 2,4-dimethyl).
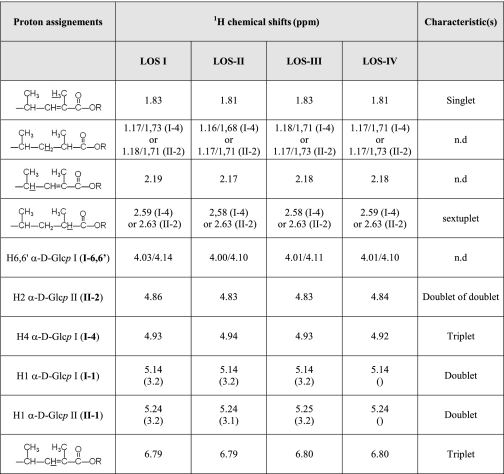
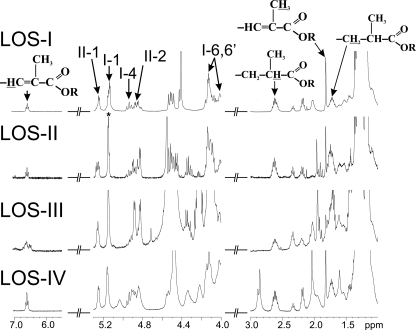
The structures of the two major fatty acids were confirmed by 1H/1H homonuclear NMR analyses of LOS-I to LOS-IV (Table 1). The presence of the saturated 2,4-dimethyl fatty acid was confirmed by the observation of the protons carried by its C2 and C3 at 2.60 and 1.71 ppm, respectively (Fig. 2). Similarly, the presence of 2-unsaturated 2,4-dimethyl fatty acids was confirmed by the identification of the protons associated to its C3 around 6.80 ppm, as well as the protons of the methyl group substituting its C2 at 1.81 ppm (Fig. 2). In addition to these two major fatty acids, minor compounds with sizes ranging from 16 to 21 carbons were also detected on the gas chromatogram (Fig. 1A) and identified by EI-MS as 2,4-dimethyl (C17, C19, and C20), 2,4,6-trimethyl (C19, C20, and C21), 2-unsaturated 2,4-dimethyl (C18 and C19) and linear fatty acids (essentially C16 and C18). Because C16 and C18 linear fatty acids are extremely frequent during lipid analysis, they were considered as ubiquitous contaminants. Relative quantification of the fatty acid methyl esters derived from LOS-I to -IV demonstrated twice as much saturated methyl branch compounds than their 2-unsaturated counterparts (data not shown). Therefore, the major LOS isomer is substituted by one 2-unsaturated 2,4-dimethyl fatty acid and two 2,4-dimethyl and/or 2,3,4-trimethyl fatty acids.
TABLE 1.
Proton chemical shifts (ppm) values of trehalose moiety and fatty acyl substituent from LOS-I to LOS-IV
Measured at 300 K in CDCl3/CD3OD (2:1, v/v) using tetramethylsilane as internal standard. n.d.: not determined.
FIGURE 2.
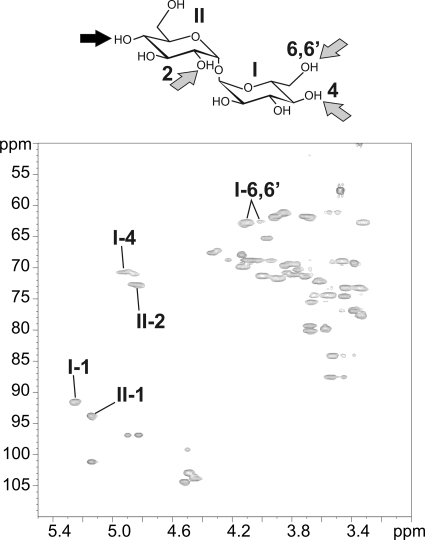
NMR characterization of polymethyl-branched fatty acyl chains of LOSs. Details of the alkenyl, anomer, and methyl/methylene resonances containing regions of the one dimensional 1H NMR spectra of LOS-I to LOS-IV. I-1, I-4, and I-6,6′ indicate H1, H4, and H6,6′ resonances of the first α-Glcp residue of the trehalose moiety of LOSs. II-2 corresponds to the H2 resonance of the second α-Glcp of the trehalose. R stands for carbons on the trehalose moiety. 1H chemical shift values are indicated in Table 1.
Composition of LOS Fatty Acyl Substituents
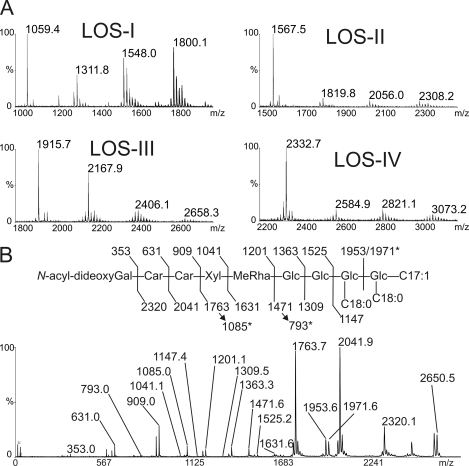
Delineation of the fatty acyl substituents on the trehalose moiety was further confirmed by MALDI-MS analysis of the O-methylated derivatives (22). Under the used condition of per-O-methylation (31), the alkali-labile fatty acyl substituents were incompletely replaced by methyl groups, a fraction of them being retained on the trehalose moiety. Comparison of the resulting molecular ions observed by mass spectrometry thus provides an informative map of the fatty acyl substituents of LOSs (22). MALDI-MS spectrum of each per-O-methylated LOSs demonstrated successive losses of 252, 236, and 252 atomic mass units that correspond to the replacement of two C18 fatty acyl substituents and one C17:1 fatty acyl substituent with O-methyl groups (Fig. 3A). The association of the lipid moiety to the LOSs was confirmed by high resolution mass spectrometry analysis of LOS-IV. Using a nanoESI-Qh-FT-ICR MS, LOS-IV was detected as a doubly charged ion cluster [M+2Li]2+ with a major signal at m/z 1343.26374 consistent with a C128H226O55N2 atomic composition, with a measured error compared with theoretical m/z value below 1 ppm (Δm = −0.7 ppm) (data not shown). Considering that the composition of glycan moiety of predominant LOS-IV isomer was shown to be 4-(2-carbamoyl-3-hydroxy-4-methoxy-1,3-dimethyl-5-oxopyrrolidine-2-carboxylic acid)-4,6-dideoxy-Gal-Car2-Xyl-Me-O-Rha-Glc4 (21), the measured C128H226O55N2 atomic composition of LOS-IV is in agreement with the presence of the three fatty acids identified by GC/MS analysis, i.e. two C18:0 and one C17:1. Altogether, GC and mass spectrometry allowed us to establish that the major form of all LOSs of M. marinum comprises two 2,4-dimethylhexadecanoates (C18:0; 2,4-dimethyl) and one 2,4-dimethyl-2-pentadecenoate (C17:1; 2,4-dimethyl).
FIGURE 3.
Mass spectrometry analysis of LOS. A, MALDI-MS spectra of permethylated derivatives of LOSs showed losses of 236 or 252 atomic mass units corresponding to the replacement of C17:1 or C18 polymethyl-branched fatty acyl chains with O-methyl groups. B, MALDI-MS/MS of the [M+Na]+ molecular ion at m/z 2650 corresponding to the major form of native LOS-IV. The 45-atomic mass unit discrepancy observed between values generated by MALDI and ES-MS originates from the selective decarboxylation of the 2-carbamoyl-3-hydroxy-4-methoxy-1,3-dimethyl-5-oxopyrrolidine-2-carboxylic acid moiety as previously established (19, 21). Glc, glucopyranose; MeRha, 3-O-methyl-rhamnopyranose; Xyl, xylopyranose; Car, caryophyllose; N-acyl-dideoxyGal, 4-(2-carbamoyl-3-hydroxy-4-methoxy-1,3-dimethyl-5-oxopyrrolidine)-4,6-dideoxy-galactopyranose.
Localization of LOS Fatty Acyl Chains
Localization of the various fatty acyl substituents on the LOS oligosaccharide backbone was essentially investigated using a combination of homonuclear 1H/1H COSY, TOCSY, and ROESY and heteronuclear 1H/13C HSQC and HMBC NMR experiments. On the basis of spin systems, chemical shift values, and relevant vicinal coupling constants patterns observed by NMR, we previously characterized all glycosyl residues of de-O-acylated LOS-I to LOS-IV (19, 21). Thus, assignment of NMR signals and identification of 1H/13C parameters of the trehalose moiety in native LOSs were obtained by comparing spectra of native and de-O-acylated LOSs. As shown on the HSQC spectrum of native LOS-II, the deshielded values of H4 and H6,6′ from the terminal α-Glcp (I) of trehalose at 4.92 and 4.10/4.01 compared with 3.64 and 3.85/3.75 ppm for the de-O-acylated residue I (19) typified substitution by acyl groups (Fig. 4 and Table 1). Acylation in position 6 of I was also supported by the H-5 resonance at δ ≥ 4 (data not shown) (23). Similarly, deshielding of H2 from the second α-Glcp unit (II) at 4.84 ppm compared with nonacylated residue II (Δδ +1,15 ppm) established the position of the third acylation (Fig. 4 and Table 1). NMR spectra did not allow the observation of other acylation positions or differences between LOS-I, LOS-II, LOS-III, and LOS-IV (Table 1).
FIGURE 4.
Identification of acylation locations on the trehalose part of LOSs. The 1H-13C HSQC NMR spectrum of LOS-II was taken as an example. Anomeric signal annotated I-1 and II-1 belong to the terminal and second α-Glcp units of the trehalose core, respectively. The deshielding of H4 and H6,6′ of the first α-Glcp residue (I-4 and I-6,6′) and of H-2 of the second α-Glcp residue (II-2) of LOS-II clearly demonstrate substitutions by acyl groups (indicated with gray arrows on the trehalose moiety). The dark arrow indicates the glycan substitution of the trehalose. LOS-I, LOS-III, and LOS-IV were found to be acylated on the same positions than LOS-II as indicated by the proton deshielding of I-4, I-6,6′, and II-2 as observed in Fig. 2. 1H chemical shift values are indicated in Table 1.
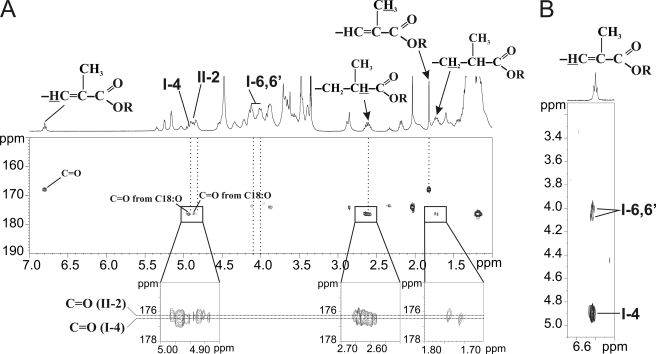
NMR analysis of native LOSs showed that saturated and unsaturated methyl-branched fatty acids occupy specific positions on the trehalose moiety by observing HMBC 3JH,C correlations between fatty acids carbonyl groups and protons of the trehalose substituted positions. Chemical shifts of carbonyl (C=O) groups were first established on HMBC 3JH,C spectrum for 2,4-dimethylhexadecanoate at 176.2 and 176.4 ppm by observing intramolecular 3JH,C/2JH,C correlations between C3/C2 protons and C=O and for 2,4-dimethyl-2-pentadecenoate at 168.1 ppm by observing 3JH,C/2JH,C correlation between CH3 group/CH=C- and C=O (Fig. 5A). Then extraresidue HMBC 3JH,C correlations of C=O from saturated methyl-branched fatty acids at 176.2 and 176.4 ppm with H4 of α-Glcp I and H2 α-Glcp II, respectively, established that these positions were substituted by 2,4-dimethylhexadecanoate (C18:0; 2,4-dimethyl) (Fig. 5A). Because LOSs comprise mainly two 2,4-dimethylhexadecanoate and one 2,4-dimethyl-2-pentadecenoate, we hypothesized that the latter fatty acid substitutes at position 6 of the α-Glcp II of the trehalose. However, we could not observe a clear 3JH,C correlation between carbonyl group of unsaturated methyl-branched fatty acids at 168.1 ppm and H6,6′ of α-Glcp I on HMBC spectra (Fig. 5A). The specific localization of the unsaturated fatty acyl substituent on position 6 of the α-Glcp I residue was supported by the observation of nuclear Overhauser effect correlations on ROESY spectrum between proton of the doubled bound (δ 6.81) of the unsaturated fatty acyl substituent and I-H6,6′ and I-H4 (Fig. 5B). The specific location of fatty acids on the trehalose backbone was confirmed by the fragmentation pattern of LOS-IV by MALDI-MS/MS experiment. As shown on Fig. 3B, fragments ions at m/z 2320 to 1147 originating from the recurrent fragmentation of the oligosaccharide chain supported that the trehalose core was substituted by three fatty acids chains, two C18:0 and one C17:1. Then signals at m/z 1953, 1085 and 793 originating from the primary and secondary cleavages of trehalose glycosidic bond confirmed that the internal glucose residue was substituted by a single C18:0 fatty acid chain, which leaves C18:0 and C17:1 fatty acid chains on the terminal Glc residue, as established by NMR. Altogether, these data established that the trehalose core of the major LOS species is substituted by two 2,4-dimethylhexadecanoate chains in the C4 position of terminal Glc residue and in the C2 position of internal Glc residue and by one 2,4-dimethyl-2-pentadecenoate in the C6 position of the terminal Glc residue.
FIGURE 5.
Specificity of location of families of polymethyl-branched fatty acyl chains. A, 1H-13C HMBC NMR spectrum of LOS-IV showing that position 4 of the first α-Glcp residue (I-4) and position 2 (II-2) of the second Glcp residue are substituted by carbonyl groups (C=O) of the saturated polymethyl-branched fatty acyl chains. B, 1H-1H ROESY NMR spectrum of LOS-IV indicating spatial proximity between H6,6′ of the first α-Glcp residue (I-6,6′) and proton of the doubled bound (δ 6.81) of the unsaturated fatty acyl chain. The spatial proximity strongly suggests the location of unsaturated fatty acyl chain on position C6 of the terminal α-Glcp residue of trehalose.
In Silico Search for Potential Genes Involved in Biosynthesis/Transfer of the Polymethyl-branched Fatty Acids
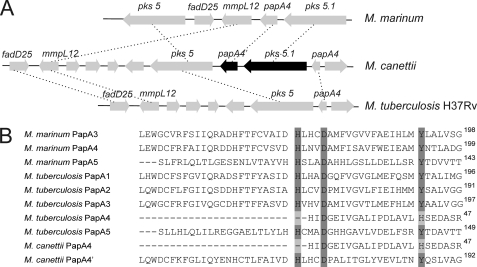
The LOS biosynthetic gene cluster was previously identified in M. marinum encompassing the region spanning from MMAR_2307 to MMAR_2344 (12, 18). Structural characterization of LOSs offered the ability to assign biological functions to several enzymes encoded within this gene cluster and involved in the biosynthesis of the glycan moiety, including glycosyltransferases that transfer caryophyllose(s) on LOS-II to LOS-IV and N-acylated α-4-amino-4,6-dideoxy-Galp in LOS-IV (12, 18, 19, 21, 34). In contrast, restricted information is available regarding the genes involved in the synthesis and/or the transfer of fatty acids on LOSs. A notable exception is MSMEG_4727, recently identified as the Pks5-encoding gene catalyzing the synthesis of 2,4-dimethyl-2-eicosenoic acid (C22:1; 2,4-dimethyl) substituting LOS of M. smegmatis (26). BLAST analysis revealed that the MMAR_2340 and MMAR_2344 translational products in M. marinum share 65 and 63% identity with MSMEG_4727/Pks5 of M. smegmatis, respectively. It is therefore tempting to speculate that both polyketide synthases (annotated MMAR_2340/Pks5 and MMAR_2344/Pks5.1) are involved in the synthesis of polymethyl-branched fatty acids in M. marinum (Table 2 and Fig. 6A). Accordingly, the presence of two distinct Pks5 homologues in M. marinum corroborates with the presence of two types of polymethyl-branched fatty acyl substituents, predominantly represented by the 2,4-dimethylhexadecanoate (C18:0; 2,4-dimethyl) and the 2,4-dimethyl-2-pentadecenoate (C17:1; 2,4-dimethyl). Other genes immediately adjacent to MMAR_2340 and MMAR_2344 are postulated to encode enzymes associated with the Pks in the synthesis and transfer of the methyl-branched fatty acids. These include the fatty acyl-AMP ligase (MMAR_2341/fadD25), the large mycobacterial membrane protein (MMAR_2342/mmpL12), and the polyketide synthase-associated protein (MMAR_2343/papA4) (Fig. 6A). Overall, these data indicate that the LOS biosynthetic cluster (Table 2) strongly resembles that of sulfolipid SL-1 and polyacyltrehalose PAT, which are also trehalose-based glycolipids, specific to pathogenic mycobacteria (35–37).
TABLE 2.
Predictive functions of the genes involved in LOS lipid metabolism
| Open reading frame | Gene name | Number of amino acids in product | Proposed function of product | Sequence similarity or protein family | Identity/similarity (%) |
|---|---|---|---|---|---|
| MMAR_2344 | Pks5.1 | 2084 | Polyketide synthase | MSMEG_4727 (pks5), M. smegmatis | 63/76 |
| Rv1527c (pks5), M. tuberculosis H37Rv | 63/74 | ||||
| contig001063 (139633–145917), M. canettii | 64/74 | ||||
| MMAR_2343 | MMAR_2343 | 478 | Polyketide synthase-associated protein (Pap) | MSMEG_4728 (pap), M. smegmatis | 54/69 |
| Rv1528c (papA4, only 165 amino acids), M. tuberculosis H37Rv | 52/69 | ||||
| contig001063 (146014–147393), M. canettii, (pap) | 48/59 | ||||
| MMAR_2342 | mmpL | 1457 | Conserved transmembrane transport protein, MmpL family | MSMEG_4741 (mmpL), M. smegmatis | 49/69 |
| Rv1522c (mmpL12), M. tuberculosis H37Rv | 55/69 | ||||
| contig001063 (158349–161570), M. canettii | 55/70 | ||||
| MMAR_2341 | fadD25 | 585 | Acyl-CoA synthetase | MSMEG_4731 (fadD), M. smegmatis | 58/72 |
| Rv1521 (fadD), M. tuberculosis H37Rv | 70/80 | ||||
| (163641–161899), M. canettii, contig001063 | 70/80 | ||||
| MMAR_2340 | Pks5 | 2090 | Polyketide synthase | MSMEG_4727 (pks5), M. smegmatis | 65/78 |
| Rv1527c (pks5), M. tuberculosis H37Rv | 69/79 | ||||
| contig001063 (147579–153851), M. canettii | 69/78 |
FIGURE 6.
Genetic organization of the lipid biosynthetic locus of LOSs and sequence comparison of Pap active site. A, part of the LOS biosynthetic locus involved in lipid production of M. marinum and the corresponding regions in M. canettii and M. tuberculosis H37Rv. Block arrows represent open reading frames. Open reading frames connected by dashed lines are conserved genes between the three loci. The open reading frame names are indicated in Table 2. fad, protein belonging to the fatty acyl-AMP ligase family; mmpL, mycobacterial membrane protein large. B, multiple sequence alignments of Pap from M. marinum, M. canettii, and M. tuberculosis H37Rv. Protein names are based on similarity to M. tuberculosis Paps. The HX3DX14Y motif, essential for acyltransferase activity, is conserved in all Paps except in the truncated PapA4 of M. canettii and M. tuberculosis H37Rv.
Inactivation of MMAR_2343 Leads to Decreased LOS Production
The SL-1 locus in M. tuberculosis encodes two acyltransferase genes, papA1 and papA2, which are required for SL-1 biosynthesis (36, 37). The PAT locus contains a gene homologous to both papA1 and papA2, designated papA3, which has been shown to encode an acyltransferase essential for the biosynthesis of PAT (35). Indeed, deletion of papA3 resulted in a total loss of the glycolipid in M. tuberculosis (35). Three homologous PapA-like acyltransferases were found to be encoded in M. marinum: PapA3 (MMAR_2355), PapA4 (MMAR_2343), and PapA5 (MMAR_1768). BLAST analysis revealed that the characteristic acyltransferase motif HX3DX14Y is conserved in the three M. marinum PapA-like acyltransferases, including MMAR_2343 (Fig. 6B). The His and Asp residues within this motif are considered essential for acyltransferase activity (27, 38–40). This suggests that MMAR_2343 is likely to be catalytically active and may fulfill the role of a Pap in the acylation steps during LOS assembly and will be hereafter referred to as PapA4.
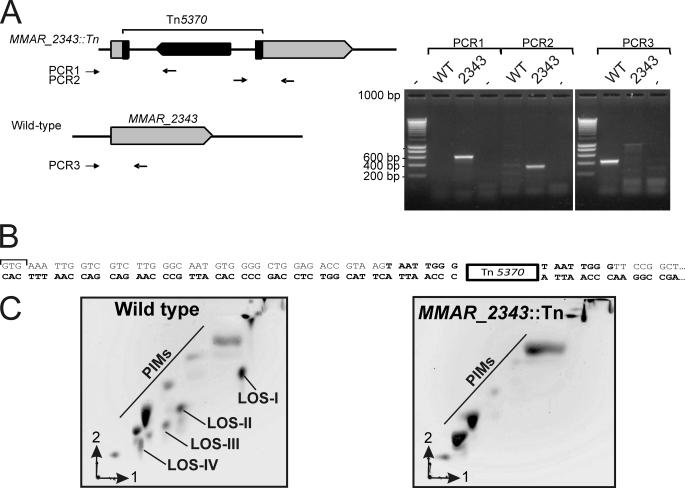
We have previously screened a M. marinum Tn library in Dictyostelium discoideum aiming to identify new cell wall defective mutants (41). During the course of this screening, a M. marinum mutant carrying a Tn insertion in papA4 was identified, designated papA4::Tn. This mutant allowed us to examine the bioinformatic predictions by genetic and biochemical analyses. The Tn insertion was mapped to the 5′ end of the papA4-coding sequence (after the 52nd base with respect to the initiation GTG codon (Fig. 7, A and B). This was further confirmed by PCR and Southern blot analyses (Fig. 7A and data not shown).
FIGURE 7.
LOS content of the MMAR_2343/papA4::Tn mutant. A, schematic representation of the transposon insertion site and of the sets of primers used for PCR analyses. Amplifications were performed on genomic DNA. ∼600- and ∼330-bp fragments were expected for PCR1 and PCR2 amplifications in the papA4::Tn mutant, respectively. A 423-bp fragment (PCR3 amplification) was only expected to occur in the wild-type M. marinum strain. 2343 stands for MMAR_2343::Tn, and − indicates that no template was added. B, localization of the transposon in the MMAR_2343/papA4 gene. The Tn5370 was found to be inserted after the 52nd base relative to the GTG start codon. Duplicated sequences flanking the transposon are shown in bold type. C, two-dimensional TLC patterns of M. marinum polar glycolipids. Following extraction, polar glycolipids were separated on two-dimensional TLC plates using the solvent system chloroform/methanol/water (60:30:6, by v/v/v) in the first direction and chloroform/acetic acid/methanol/water (40:25:3:6, v/v/v/v) in the second direction. Glycolipids were detected with orcinol/sulfuric acid staining. In parallel, TLC plate was stained with iodine vapor, and the glycolipids were individually collected and identified by mass spectrometry.
Polar lipids were then extracted from the parental M. marinum M strain and the papA4::Tn mutant strain. Glycolipid profiles were recorded on two-dimensional TLC and revealed with orcinol/sulfuric acid. The polar glycolipid pattern of the wild-type strain was typically characterized by the presence of PIMs and the different LOS-type species, LOS-I to LOS-IV (Fig. 7C), in agreement with previous studies (12, 19). By comparison, the papA4::Tn mutant displayed a profoundly altered polar lipid pattern, characterized by a dramatic decrease in LOS production. Only traces amounts of LOSs could be observed in the papA4::Tn mutant, thus supporting the view that PapA4 plays a critical role in the acylation step required in assembly of complete LOS. As previously noted for SL-1 and PAT, a defect in the acylation step severely impairs biosynthesis of the corresponding glycolipids. Altogether, these results also indicate that, despite the presence of at least three Pap-like proteins in M. marinum, PapA4 appears specific for LOS synthesis.
DISCUSSION
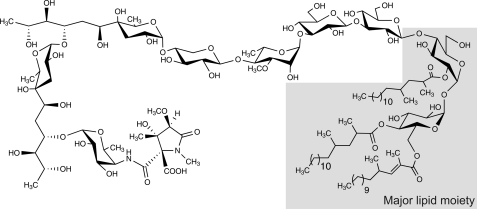
The present data along with our previous results on the glycan moiety allow us to propose a complete and detailed structure of the major LOS-IV of M. marinum (Fig. 8). Here, we showed that LOSs of M. marinum bear two types of fatty acyl chains differing by the presence of unsaturations. To the best of our knowledge, acylation of LOSs by several types of methyl-branched fatty acids were only observed in M. canettii, M. mucogenicum, and M. gordonae (11, 42, 43). As for M. marinum, LOSs of M. mucogenicum are acylated by saturated and unsaturated polymethyl-branched fatty acyl substituents (43), whereas those from M. canettii and M. gordonae are substituted by saturated polymethyl-branched fatty acids and hydroxylated methyl-branched fatty acids (11, 42). Importantly, this study demonstrated that each acylation position of the trehalose moiety was substituted by a specific fatty acid: the 2-unsaturated 2,4-dimethyl-branched fatty acid substitutes at position C6 of the first α-Glcp of the trehalose, whereas the 2,4-dimethyl- or 2,4,6-trimethyl-branched fatty acids acylate positions C4 and C2 of the first and the second α-Glcp units, respectively (Fig. 8). However, one cannot exclude the existence of minor LOSs in which the locations of fatty acids are different from those of major compounds. Indeed, a high degree of heterogeneity exists for other acylated trehalose-based glycolipids such as sulfolipids (44).
FIGURE 8.
Complete structure of M. marinum LOS and localization of the major polymethyl-branched fatty acyl chains. Fine structure of the glycan moiety was previously established by a combination of NMR, mass spectrometry, and gas chromatography (19, 21), whereas composition and localization of the acyl chains on the trehalose core were established in the present study (in gray).
Of all pks genes linked to Pap proteins, pks2 is the only one located next to two pap genes (papA1 and papA2) involved in sulfolipid acylation, whereas papA3 and pks3/4 are involved in the biosynthesis of PAT. PapA5 was reported to participate to PDIM biosynthesis (24, 27, 38) by transferring two mycocerosic acid residues to the hydroxyl groups of a phthiocerol. In contrast, the metabolite synthesized by PapA4 remained unknown. Herein, we provide for the first time evidence that PapA4 is required for the transfer of fatty acids to the trehalose core of LOSs. This conclusion is supported by the presence of the characteristic acyltransferase motif HX3DX14Y found in other Pap enzymes, including the PapA5 prototype (27) and the impairment of LOS biosynthesis in a papA4::Tn mutant (Fig. 7C).
It is noteworthy that this is also the first report of a M. marinum strain profoundly affected in the synthesis of all LOS subspecies, thus providing a very powerful tool for future studies dedicated to address the role of LOS in virulence and in vivo survival of M. marinum in the zebrafish model of infection.
In silico analysis allowed us to identify multiple genes predicted to encode enzymes potentially involved in the synthesis or transfer of the LOS lipid moiety (Table 2). This gene cluster also contains two pks5 genes interspersed by fadD25, mmpL12, and papA4 (Fig. 6A), thus corroborating the presence of polymethyl-branched fatty acids described in the present study. However, it remains to be experimentally established whether Pks5 and/or Pks5.1 are required for LOS biosynthesis.
Interestingly, M. canettii, a LOS-producing species (23), possesses two families of polymethyl-branched fatty acids differing by the presence or absence of an hydroxyl group in the fatty acids. BLAST analysis also revealed the presence of two putative Pks (pks5 and pks5.1) separated by the gene encoding a putative polyketide synthase-associated protein, designated PapA4′ (Fig. 6A and Table 2) (21). Interestingly, both papA4′ and pks5.1 are missing in M. tuberculosis H37Rv, a LOS-deficient strain (Fig. 6A). Moreover, the gene previously assigned as pap (Rv1528c/papA4) is predicted to encode a truncated protein of only 165 amino acids instead of 465 and 480 amino acid residues of the Pap proteins involved in LOS biosynthesis of M. marinum and M. smegmatis, respectively (Table 2). Thus, one can anticipate that the absence of functional PapA4 and Pks5.1 in M. tuberculosis H37Rv is responsible for the lack of LOS production in this species. However, cross-complementation studies are further required to support this hypothesis.
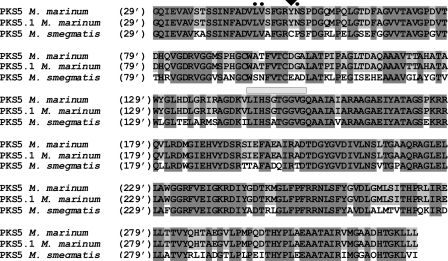
Pks are multimodular multi-enzymatic megasynthases characterized by several functional domains including acyltransferase, ketosynthase, dehydratase, enoylreductase (ER), ketoreductase, and acyl carrier protein. Using methylmalonyl-CoA, all of these enzymatic modules of the Pks are required to elongate linear fatty acids resulting in formation of methyl-branched fatty acids (25). Synthesis of unsaturated methyl-branched fatty acids usually results from either (i) the absence/inactivity of the ER domain in the Pks or (ii) the involvement of a desaturase-type enzyme acting late in the biosynthesis of methyl-branched fatty acids, although such enzymes have not been identified yet in the vicinity of LOS gene cluster. We thus examined and compared the primary sequence of Pks5 and Pks5.1 using the SBSPKS program, and both were found to share acyltransferase, ketosynthase, dehydratase, ketoreductase, acyl carrier protein, and ER domains (data not shown) (45, 46). BLAST analysis reveals that Pks5 and Pks5.1 possess 76% identity, but their two predictive ER domains exhibit 100% identity (Fig. 9). A closer inspection of the ER domains suggested that they were very likely to be catalytically active based on the presence of the invariant LIHSGTGGVG sequence, which corresponds to the conserved NADPH binding motif (47, 48) as well as conserved amino acid residues of active ER domains as described previously (data not shown) (48). Tyr at position 52 of the ER domain of Pks5 and Pks5.1 is predictive for an enoylreduction with (S) configuration (47, 48). From these data, it can then be inferred that all methyl-branched fatty acids in M. marinum LOS are in (S) configuration. However, confirmation of the absolute configuration of C2 and C4 chiral centers can only come from the individual study by NMR or optical rotation values of synthetic standards with defined configurations. Still, the large 1H Δδ values (0.53–0.56 ppm) between HA and HB C3 methylene protons revealed that both CH3 groups in saturated fatty acids presented a syn configuration, meaning that both chiral centers exhibit a similar configuration (S,S or R,R) (49). In contrast to M. marinum, Pks5 of M. smegmatis possesses a Cys residue at position 52, suggesting an (R) configuration for the two methyl branches of 2,4-dimethyl-2-eicosenoic acid (47). Because of the high similarity of M. smegmatis Pks5 with M. marinum MMAR_2340/Pks5, one can speculate that MMAR_2340/Pks5 is responsible for the synthesis of the 2-unsaturated 2,4-dimethyl-branched fatty acids. However, this hypothesis awaits further experimental proof.
FIGURE 9.
Sequence alignments of the ER domain of Pks5 in M. marinum and M. smegmatis. Pks5 from M. smegmatis as well as Pks5 and Pks5.1 form M. marinum comprise a catalytically competent ER domain based on the presence of the conserved NADPH-binding motif (horizontal gray bar) in addition to several residues important for ER activity (dark circles). The Tyr at position 52 (black triangle) present only in the two M. marinum polyketide synthases is predictive of methyl-branched fatty acids with a (S) configuration.
Acknowledgments
We thank S. L. G. Cirillo and J. D. Cirillo for providing the M. marinum Tn library and C. Biot for helpful discussion.
This work was supported by French National Research Agency Grant ANR-05-MIIM-025 (to Y. G. and L. K.), ZebraFlam Grant ANR-10-MIDI 010 (to L. K.), and a grant from the Ministère de l'Enseignement Supérieur (to Y. R.). The 400-MHz facility was funded by the Centre Commun de Mesure RMN (Résonance Magnétique Nucléaire) de l'Université de Lille 1. The 600-MHz facility used in this study was funded by the European Union, Région Nord-Pas de Calais, CNRS, and Institut Pasteur de Lille. The mass spectrometry facilities used for this study are funded by the European community (Fonds Européen de Développement Régional), the Région Nord-Pas de Calais (France), the Infrastructures en Biologie Santé et Agronomie network, the CNRS, the Université Lille 1 Sciences et Technologies, and the TGE FT-ICR (Très Grand Equipement Fourier Transform Ion Cyclotron Resonance).
- LOS
- lipooligosaccharide
- El-MS
- Electronic Impact Mass Spectrometry
- Car
- caryophyllose
- Pks
- polyketide synthase
- HSQC
- heteronuclear single quantum coherence
- ER
- enoylreductase
- ROESY
- rotating frame Overhauser enhancement spectroscopy
- HMBC
- Heteronuclear Multiple Bond Correlation.
REFERENCES
- 1. Jarlier V., Nikaido H. (1990) J. Bacteriol. 172, 1418–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Daffé M., Draper P. (1998) Adv. Microb. Physiol. 39, 131–203 [DOI] [PubMed] [Google Scholar]
- 3. Kremer L., Baulard A. R., Besra G. S. (2000) Molecular Genetics of Mycobacteria (Hatfull G. F., Jacobs W. R. eds) pp. 173–190, ASM Press, Washington, D.C [Google Scholar]
- 4. Brennan P. J., Nikaido H. (1995) Annu. Rev. Biochem. 64, 29–63 [DOI] [PubMed] [Google Scholar]
- 5. Karakousis P. C., Bishai W. R., Dorman S. E. (2004) Cell. Microbiol. 6, 105–116 [DOI] [PubMed] [Google Scholar]
- 6. Neyrolles O., Guilhot C. (2011) Tuberculosis 91, 187–195 [DOI] [PubMed] [Google Scholar]
- 7. McNeil M., Chatterjee D., Hunter S. W., Brennan P. J. (1989) Methods Enzymol. 179, 215–242 [DOI] [PubMed] [Google Scholar]
- 8. Hunter S. W., Fujiwara T., Murphy R. C., Brennan P. J. (1984) J. Biol. Chem. 259, 9729–9734 [PubMed] [Google Scholar]
- 9. Hunter S. W., Murphy R. C., Clay K., Goren M. B., Brennan P. J. (1983) J. Biol. Chem. 258, 10481–10487 [PubMed] [Google Scholar]
- 10. Gilleron M., Vercauteren J., Puzo G. (1993) J. Biol. Chem. 268, 3168–3179 [PubMed] [Google Scholar]
- 11. Daffé M. (1991) Res. Microbiol. 142, 405–410 [DOI] [PubMed] [Google Scholar]
- 12. Burguière A., Hitchen P. G., Dover L. G., Kremer L., Ridell M., Alexander D. C., Liu J., Morris H. R., Minnikin D. E., Dell A., Besra G. S. (2005) J. Biol. Chem. 280, 42124–42133 [DOI] [PubMed] [Google Scholar]
- 13. Collins F. M., Cunningham D. S. (1981) Infect. Immun. 32, 614–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Belisle J. T., Brennan P. J. (1989) J. Bacteriol. 171, 3465–3470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lemassu A., Lévy-Frébault V. V., Lanéelle M. A., Daffé M. (1992) J. Gen. Microbiol. 138, 1535–1541 [DOI] [PubMed] [Google Scholar]
- 16. Stinear T. P., Seemann T., Harrison P. F., Jenkin G. A., Davies J. K., Johnson P. D., Abdellah Z., Arrowsmith C., Chillingworth T., Churcher C., Clarke K., Cronin A., Davis P., Goodhead I., Holroyd N., Jagels K., Lord A., Moule S., Mungall K., Norbertczak H., Quail M. A., Rabbinowitsch E., Walker D., White B., Whitehead S., Small P. L., Brosch R., Ramakrishnan L., Fischbach M. A., Parkhill J., Cole S. T. (2008) Genome Res. 18, 729–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tobin D. M., Ramakrishnan L. (2008) Cell. Microbiol. 10, 1027–1039 [DOI] [PubMed] [Google Scholar]
- 18. Ren H., Dover L. G., Islam S. T., Alexander D. C., Chen J. M., Besra G. S., Liu J. (2007) Mol. Microbiol. 63, 1345–1359 [DOI] [PubMed] [Google Scholar]
- 19. Rombouts Y., Burguière A., Maes E., Coddeville B., Elass E., Guérardel Y., Kremer L. (2009) J. Biol. Chem. 284, 20975–20988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. van der Sar A. M., Abdallah A. M., Sparrius M., Reinders E., Vandenbroucke-Grauls C. M., Bitter W. (2004) Infect. Immun. 72, 6306–6312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rombouts Y., Elass E., Biot C., Maes E., Coddeville B., Burguière A., Tokarski C., Buisine E., Trivelli X., Kremer L., Guérardel Y. (2010) J. Am. Chem. Soc. 132, 16073–16084 [DOI] [PubMed] [Google Scholar]
- 22. Besra G. S., Khoo K. H., Belisle J. T., McNeil M. R., Morris H. R., Dell A., Brennan P. J. (1994) Carbohydr. Res. 251, 99–114 [DOI] [PubMed] [Google Scholar]
- 23. Daffe M., McNeil M., Brennan P. J. (1991) Biochemistry 30, 378–388 [DOI] [PubMed] [Google Scholar]
- 24. Trivedi O. A., Arora P., Sridharan V., Tickoo R., Mohanty D., Gokhale R. S. (2004) Nature 428, 441–445 [DOI] [PubMed] [Google Scholar]
- 25. Chopra T., Gokhale R. S. (2009) Methods Enzymol. 459, 259–294 [DOI] [PubMed] [Google Scholar]
- 26. Etienne G., Malaga W., Laval F., Lemassu A., Guilhot C., Daffé M. (2009) J. Bacteriol. 191, 2613–2621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Onwueme K. C., Ferreras J. A., Buglino J., Lima C. D., Quadri L. E. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 4608–4613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McAdam R. A., Quan S., Smith D. A., Bardarov S., Betts J. C., Cook F. C., Hooker E. U., Lewis A. P., Woollard P., Everett M. J., Lukey P. T., Bancroft G. J., Jacobs W. R., Jr., Duncan K. (2002) Microbiology 148, 2975–2986 [DOI] [PubMed] [Google Scholar]
- 29. Cox J. S., Chen B., McNeil M., Jacobs W. R., Jr. (1999) Nature 402, 79–83 [DOI] [PubMed] [Google Scholar]
- 30. Larsen M. H., Biermann K., Tandberg S., Hsu T., Jacobs W. R., Jr. (2007) Curr. Protoc. Microbiol. Chapter 10, Unit 10A 12 [DOI] [PubMed] [Google Scholar]
- 31. Ciucanu I., Kerek F. (1984) Carbohydr. Res. 131, 209–217 [Google Scholar]
- 32. Ryhage R., Stenhagen E. (1960) J. Lipid Res. 1, 361–390 [PubMed] [Google Scholar]
- 33. Camphausen R. T., McNeil M., Jardine I., Brennan P. J. (1987) J. Bacteriol. 169, 5473–5480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sarkar D., Sidhu M., Singh A., Chen J., Lammas D. A., van der Sar A. M., Besra G. S., Bhatt A. (2011) J. Bacteriol. 193, 2336–2340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hatzios S. K., Schelle M. W., Holsclaw C. M., Behrens C. R., Botyanszki Z., Lin F. L., Carlson B. L., Kumar P., Leary J. A., Bertozzi C. R. (2009) J. Biol. Chem. 284, 12745–12751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bhatt K., Gurcha S. S., Bhatt A., Besra G. S., Jacobs W. R., Jr. (2007) Microbiology. 153, 513–520 [DOI] [PubMed] [Google Scholar]
- 37. Kumar P., Schelle M. W., Jain M., Lin F. L., Petzold C. J., Leavell M. D., Leary J. A., Cox J. S., Bertozzi C. R. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 11221–11226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Buglino J., Onwueme K. C., Ferreras J. A., Quadri L. E., Lima C. D. (2004) J. Biol. Chem. 279, 30634–30642 [DOI] [PubMed] [Google Scholar]
- 39. Bergendahl V., Linne U., Marahiel M. A. (2002) Eur. J. Biochem. 269, 620–629 [DOI] [PubMed] [Google Scholar]
- 40. Lewendon A., Murray I. A., Kleanthous C., Cullis P. M., Shaw W. V. (1988) Biochemistry 27, 7385–7390 [DOI] [PubMed] [Google Scholar]
- 41. Alibaud L., Rombouts Y., Trivelli X., Burguiere A., Cirillo S. L., Cirillo J. D., Dubremetz J. F., Guerardel Y., Lutfalla G., Kremer L. (2011) Mol. Microbiol. 80, 919–934 [DOI] [PubMed] [Google Scholar]
- 42. Besra G. S., McNeil M. R., Khoo K. H., Dell A., Morris H. R., Brennan P. J. (1993) Biochemistry 32, 12705–12714 [DOI] [PubMed] [Google Scholar]
- 43. Muñoz M., Raynaud C., Lanéelle M. A., Julián E., López Marín L. M., Silve G., Ausina V., Daffé M., Luquin M. (1998) Microbiology 144, 137–148 [DOI] [PubMed] [Google Scholar]
- 44. Goren M. B. (1970) Biochim. Biophys. Acta 210, 127–138 [DOI] [PubMed] [Google Scholar]
- 45. Anand S., Prasad M. V., Yadav G., Kumar N., Shehara J., Ansari M. Z., Mohanty D. (2010) Nucleic Acids Res. 38, (suppl.) W487–W496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yadav G., Gokhale R. S., Mohanty D. (2003) J. Mol. Biol. 328, 335–363 [DOI] [PubMed] [Google Scholar]
- 47. Kwan D. H., Sun Y., Schulz F., Hong H., Popovic B., Sim-Stark J. C., Haydock S. F., Leadlay P. F. (2008) Chem. Biol. 15, 1231–1240 [DOI] [PubMed] [Google Scholar]
- 48. Kwan D. H., Leadlay P. F. (2010) ACS Chem. Biol. 5, 829–838 [DOI] [PubMed] [Google Scholar]
- 49. Schmidt Y., Breit B. (2010) Org. Lett. 12, 2218–2221 [DOI] [PubMed] [Google Scholar]