Abstract
Host survival depends on an effective immune system and pathogen survival on the effectiveness of immune evasion mechanisms. Staphylococcus aureus utilizes a number of molecules to modulate host immunity, including the SSL family of which SSL7 binds IgA and inhibits Fcα receptor I (FcαRI)-mediated function. Other Gram-positive bacterial pathogens produce IgA binding proteins, which, similar to SSL7, also bind the Fc at the CH2/CH3 interface (the junction between constant domains 2 and 3 of the heavy chain). The opposing activities of the host FcαRI-IgA receptor ligand pair and the pathogen decoy proteins select for host and pathogen variants, which exert stronger protection or evasion, respectively. Curiously, mouse but not rat IgA contains a putative N-linked glycosylation site in the center of this host receptor and pathogen-binding site. Here, we demonstrate that this site is glycosylated and that the effect of amino acid changes and glycosylation of the CH2/CH3 interface inhibits interaction with the pathogen IgA binding protein SSL7, while maintaining binding of pIgR, essential to the biosynthesis and transport of SIgA.
Keywords: Antibodies, Bacterial Toxins, Evolution, FC Receptors, Staphylococcus aureus, Immunoglobulin A (IgA), Staphylococcal Superantigen-like (SSL) Proteins
Introduction
Many IgA functions are dependent on IgA receptors (1–3). In most mammals but not mice, IgA effector function is dependent on Fcα receptor I (FcαRI2; CD89), the Fc receptor specific for the Fc portion of IgA. Mice lack this receptor gene and a number of other immunoreceptors of the so called leukocyte receptor cluster (4). FcαRI is an activating receptor expressed on a wide variety of innate myeloid immune cells, including neutrophils, monocytes, dendritic cells, eosinophils, and tissue macrophages, including alveolar macrophages and Kuppfer cells (3, 5–9). Thus, through binding IgA, FcαRI couples the IgA immune response with potent cell-based effector systems. Phagocytosis (10), proinflammatory cytokine and chemokine secretion, neutrophil antibody-dependent cellular cytotoxicity (11), and respiratory burst are potently triggered by activation of FcαRI, sometimes more effectively than other Fc receptors (12). Uniquely, neutrophil activation via FcαRI stimulates the production of leukotriene B4, which acts to recruit further neutrophils and so may orchestrate an inflammatory response at the site of failure of the mucosal barrier (13). Furthermore, FcαRI expression on Kuppfer cells has been suggested to integrate with mucosal immunity in providing a second level of IgA mediated protection if the gut mucosal barrier is breached (9). Indeed, FcαRI-dependent IgA-mediated immunity has been shown to be effective in the resistance to a number of pathogens, including Candida albicans (14), Streptococcus pneumonia (15), Porphyromonas gingivalis (16), and Bordetella pertussis (17) (reviewed in Ref. 8). Studies of human FcαRI ex vivo and in transgenic mice demonstrated the importance of this effector arm in resistance to mucosal infection by tuberculosis (18).
Despite the importance of IgA and FcαRI in immunity in such model systems, IgA deficiency is the most common immunodeficiency with a relatively mild phenotype, which includes susceptibility to allergy and recurrent enteric and respiratory infections (19). It may be that the supersufficiency of the immune system, by having multiple effective protective mechanisms, has allowed marked co-evolution of and even loss of the FcαRI effector arm. Notably, mice, rabbits, and dogs lack a functional IgA/FcαRI effector arm by loss of the receptor genes or its presence as pseudogenes (20). A detailed phylogenetic analysis found the contact residues of the FcαRI ectodomain 1, and the IgA-Fc have been subject to strong positive selection. This selection is postulated to arise from Gram-positive streptococci and Staphylococcus aureus, important human pathogens that produce proteins that bind the IgA-Fc to evade IgA-mediated immunity (20).
We have focused on the staphylococcal superantigen-like (SSL) protein SSL7, which binds IgA, blocking the interaction with FcαRI (21). Previously, we determined the structure of the complex of SSL7 and human IgA1-Fc by x-ray crystallography (22). SSL7 binding to IgA was centered on the CH2/CH3 domain interface in the Fc (21, 23), which is predominantly the same site recognized by the granulocyte IgA receptor, FcαRI (i.e. CD89) (24). Consequently, SSL7 directly blocks IgA-dependent triggering of FcαRI and its activation of neutrophil anti-microbial activity.
The SSL7 protein and other pathogen IgA-binding proteins have exerted selective pressure on this important FcαRI-IgA interaction. In this study, we have examined critical differences in human, rat, and mouse IgA and its human and rat receptor FcαRI. These differences, including glycosylation of the mouse IgA-Fc alter binding interactions and are consistent with selection by pathogen IgA-binding decoy proteins.
MATERIALS AND METHODS
DNA Constructs
Restriction enzymes and DNA-modifying enzymes were all from New England Biolabs except for PCR applications, which used Pfx (Invitrogen) with standard mutagenesis methods employed as described previously (25).
IgA-Fc Constructs
The expression of the N terminus and transmembrane region of the human transferrin receptor (TfR) fused to the human IgA-Fc has been described previously (23). Fusion proteins of TfR and rat and mouse IgA-Fc, which lacked the tail piece region and had C-terminal hexahistidine tags, were produced similarly using TfR and IgA-Fc DNAs amplified from Rattus norvegicus IgA heavy chain cDNA (clone IMAGE 7375116) or mus musculus IgA heavy chain cDNA (clone IMAGE 6477061) templates subcloned into a derivative of pENTR1A (Invitrogen) encoding a C-terminal hexahistidine tag.
Rat FcαRI-IgG2b-Fc
A DNA encoding the normal ectodomains of rat FcαRI was amplified from cloned cDNA (gift from Dr Masanori Kasahara (4)) and ligated with a mouse IgG2b-Fc region DNA from pBAR225 (23) and subcloned in pENTR1A. Expression constructs were produced using the LR clonase reaction to transfer the DNAs to gateway reading frame A cassette adapted pCR3 (Invitrogen).
Intact Mouse IgA
The mouse HyHEL-10 IgA WT heavy and light chain expression constructs in pcDNA3 and the N442S mutant heavy chain construct were used in transient co-transfection of Chinese hamster ovary cells expressing polyoma large T antigen (CHOP) to produce WT and N442S mutant IgA with anti-hen egg lysozyme specificity. Construct sequences were confirmed using BigDye3.1 (Applied Biosystems, Inc.).
Proteins
Biotinylated recombinant SSL7 and human FcαRI ectodomains fused to the Fc region of mouse IgG2b (hu-FcαRI-Fcγ2b) were prepared, and transient transfection of CHOP cells with FcαRI-Fcγ2b expression constructs were performed as described previously (21).
Expression and Binding Analysis of TfR-IgA-Fc Fusion Proteins
Transient expression in CHOP cells using Lipofectamine 2000 reagent (Invitrogen) was largely as described previously (8) except here, 0.04 μg of pEGFP-N1 (Clontech) was mixed with the expression plasmid DNA (0.8 μg) in each transfection. After 48 or 72 h, the expression of TfR-rodent IgA-Fc was measured by incubating cells (50 μl, 105cells) 1 h on ice with 1/400 anti-polyhistidine mAb, (clone HIS-1; Sigma-Aldrich) After incubation, the cells were resuspended in PBS containing 0.1% BSA, centrifuged (1000 rpm for 5 min), and the cells were incubated in 1:400 dilution of Alexa Fluor® 633 goat anti-mouse IgG (heavy and light chains) (Invitrogen) for 1 h on ice. Cells were resuspended in PBS containing 0.1% BSA, centrifuged (1000 rpm for 5 min), and the collected cells were analyzed. Human FcαRI-Ig and rat FcαRI-Ig binding activities of the various of IgA-Fc were measured by incubating cells (105) 1 h on ice with 50 μl of transfectant cell supernatant and subsequently with 1:400 dilution of Alexa Fluor® 633 goat anti-mouse IgG as above. Staining for the SSL7 binding activities of the various IgA-Fc proteins was as described previously (8). Analysis by flow cytometry used a FACSCantoII (Becton Dickinson, Melbourne, Australia) with post acquisition gating on EGFP+ cells to enrich for transfected cells. Binding data were analyzed by one-way analysis of variance with Newman-Keuls post test (GraphPad Prism version 5, GraphPad Software, Inc.).
Immunoprecipitation and Digestion with N-Glycanase
The TfR-mouse IgA-Fc and the TfR-mouse N442S IgA-Fc fusion proteins were transiently expressed in CHOP cells (3 × 10 cm2 wells), and at 48 h, cells were lysed with 1 ml of lysis buffer (10 mm Tris, pH 7.4, 150 mm NaCl, 0.5% Brij-96 and one mini-complete protease tablet without EDTA (Roche Applied Science) per 10 ml). The lysate was clarified by centrifugation at 10,000 rpm for 10 min at 4 °C, and the supernatant was bound to Talon NTA-cobalt-agarose (Clontech) and incubated at 4 °C for 30 min. Beads were washed with lysis buffer (five washes), with PBS, pH 8.5, containing 5 mm imidazole and eluted with PBS, pH 8.5, containing 500 mm imidazole, and the eluate was dialyzed against 100 mm phosphate, pH 7.5. Samples were denatured and digested with N-glycanase (Genzyme, Cambridge, MA) and analyzed by SDS-PAGE and semidry transfer to polyvinylidene difluoride membrane. Immunodetection of the hexahistidine tagged TfR-IgA-Fc proteins used 1/3000 anti-polyhistidine mAb, (Sigma-Aldrich) followed by HRP-conjugated anti-mouse Ig (Amersham Biosciences), diluted 1/10,000, and lastly, ECL reagent (PerkinElmer Life Sciences).
ELISA
ELISA plates (Maxisorb F96 Nunc) were coated with hen egg lysozyme (50 μg/ml), blocked with PBS containing 1% BSA (Sigma), and incubated with serial dilutions of the recombinant IgA transfection supernatants. Bound IgA was detected by sequential incubations with biotin-labeled anti-mouse IgA at 1/3000 dilution (Pharmingen), 1/6000 dilution of HRP-conjugated streptavidin (Amersham Biosciences), and 3,3′,5,5′-tetramethylbenzidine substrate (Zymed Laboratories Inc., San Francisco, CA). Similarly, to determine galectin 3 binding activity, biotin-conjugated galectin 3 was used at 1/150 dilution. Galectin 3 was purified from mouse macrophage J774 lysates by lactose affinity chromatography as described in Ref. 26 and biotinylated according to (21). Similarly, rat FcαRI binding activity was measured by ELISA using incubation in a mixture of rat FcαRI-IgG2b supernatant with 1/1600 HRP-conjugated streptavidin and 1/13000 biotin-conjugated anti-IgG2b (Pharmingen).
Modeling
Homology models of rat and mouse IgA-Fc (CH2-CH3 domains lacking the tailpiece peptide) were generated using crystal structures as templates: PDB code 2QEJ to assess SSL7 interactions and PDB code 1OW0 for FcαRI interactions as well as for modeling of the rat FcαRI ectodomains. The Modeler (27) algorithm was used to generate optimized homology models in Discovery Studio (version 1.6; Accelrys, San Diego, CA) as described previously (28). Using the coordinates of the IgA-Fc proteins, N-glycans were added by the GlyProt server using SWEET II (29) to model and attach core branched oligosaccharides to accessible asparagine residues within Nx(T/S) motifs.
RESULTS
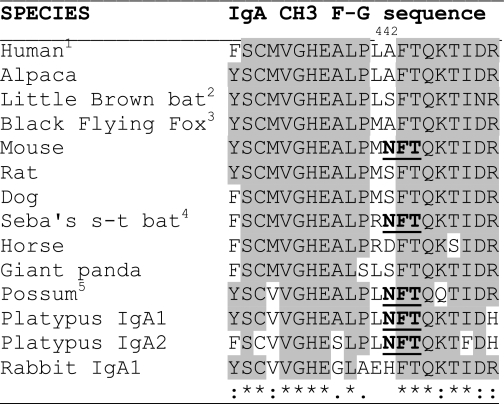
The last loop (connecting the F and G β-strands) of the CH3 domain of the IgA-Fc plays a key role in the binding of both the granulocyte receptor FcαRI and also bacterial pathogen IgA binding proteins. This region in monotreme, marsupial, and all strains of mouse IgA contains a putative N-linked glycosylation site (NFT, 442–444) that is not present in primate, cow, sheep, pig, dog, or rat IgA (Table 1). Expression of a WT mouse IgA-Fc and a N442S mutant Fc was used to determine whether the sequence NFT (residues 442–444) formed a functional glycosylation site. These Fc fusion proteins lack the tailpiece and rodent IgA-Fc lacks the equivalent of Asn-263 of human IgA so that Asn-442 forms the only possible N-linked glycosylation site in this recombinant mouse IgA-Fc (Fig. 1A). These IgA-Fc fragments were expressed in CHOP cells as hexahistidine-tagged TfR fusion proteins, purified by binding to NTA-cobalt agarose, and digested with endoglycosidase F. Western blot analysis detected a single band for the endoglycosidase F-treated WT mouse IgA-Fc and both the treated and untreated mutant N442S Fc, which corresponded with an expected molecular mass for the polypeptide of 34.8 kDa. The untreated WT Fc consisted of the ∼35 kDa band and an additional ∼37 kDa band, indicating NFT (442–444) is a functional glycosylation site. The presence of both the 35 and 37 kDa bands suggests partial or heterogeneous glycosylation of Asn-442 (Fig. 1B).
TABLE 1.
A sequence alignment of the F-G strands from the CH3 domains of mammalian IgA heavy chains
The putative Asn-442-linked glycosylation site is shown in boldface.
1 The sequences of human, chimpanzee, crab-eating macaque, Rhesus monkey, Northern white-cheeked gibbon, white-tufted ear marmoset, bovine, sheep, and Pig IgA are identical in this region.
2 Little brown bat and big brown bat.
3 Black Flying fox and Indian short-nosed fruit bat.
4 Seba's short-tailed bat.
5 Brush-tailed possum and Grey opossum. Fc residues are numbered according to their equivalent residue in human IgA Fc.
FIGURE 1.
Glycosylation of mouse IgA-Fc. A, schematic diagram of transferrin receptor fusion proteins with mouse and rat IgA-Fc. B, mouse IgA-Fc is N-glycosylated on residue Asn-442. Transfectant cell lysates containing mouse TfR-IgA-Fc WT and N442S mutant fusion proteins were reacted with NTA-agarose. Purifed proteins were treated or untreated with N-glycanase and IgA polypeptide revealed by Western blot.
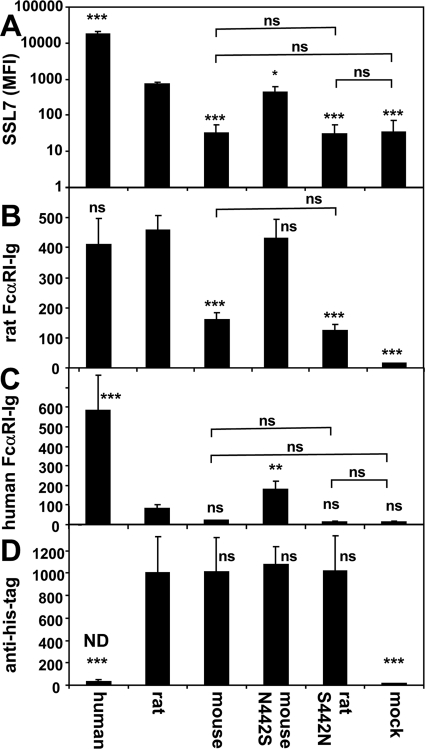
The human, rat, and mouse WT IgA-Fc proteins were expressed on CHOP cells, and the interactions with the staphylococcal IgA binding decoy protein SSL7 were analyzed by flow cytometry. These Fc fragments are highly homologous, but differ in residues defined in our previous work as human IgA contacts for FcαRI and SSL7 (22, 23). The rat IgA-Fc (PMSFT, 440–444) was ∼25-fold less active in binding SSL7 (MFI 730 c.f. 18,000) than the human IgA-Fc (PLAFT, 440–444), indicating that the amino acid differences in this key binding region affect SSL7 binding affinity (Fig. 2A). However, the mouse IgA-Fc showed no detectable binding of SSL7 above background staining (MFI 32 ± 21 c.f. mock 34 ± 36). Because the mouse IgA-Fc may be heterogeneously glycosylated (Fig. 1B), the amino acid differences as well as N-glycosylation of these residues (MN, 441–442), could abolish SSL7 binding. The pattern of poor SSL7 binding to the rat and mouse IgA underscores that S. aureus is well adapted to its human host, and this particular interaction itself shows strong species selectivity. Importantly, the N442S mutation of the mouse IgA-Fc restored SSL7 binding to almost that of the rat IgA-Fc (Fig. 2A).
FIGURE 2.
SSL7 and FcαRI binding activities of human, mouse, and rat IgA. Transfected cells expressing human, mouse, and rat IgA-Fc on the cell surface were analyzed by flow cytometry for co-expressed EGFP and for binding of SSL7 (A), rat FcαRI-Ig (B), human FcαRI-Ig (C), and anti-poly-histidine antibody (D). Note that the human IgA-Fc fusion protein was not hexahistidine-tagged and hence not detectable (ND). Newman keuls post-comparison tests to rat IgA-Fc binding data (log10 for SSL7 binding) are shown for each column compared with binding to rat IgA-Fc. Other post-comparison test pairs are shown by linking bars. ***, p < 0.001; **, p < 0.01; *, p < 0.05; ns, not significant. Data are from three independent experiments comprising four replicates.
The analysis of rat FcαRI-Ig binding to the IgA-Fc proteins indicated equivalent activities for the human (MFI 410 ± 90) and rat (MFI 460 ± 50) WT IgA-Fc proteins (Fig. 2B). The activity of the mouse IgA-Fc (MFI 160 ± 25) was 3-fold less than that of the rat IgA-Fc for binding rat FcαRI. Importantly, the N442S mutant mouse Fc had binding activity for rat FcαRI equivalent to the rat Fc. In accordance with Asn-442 being primarily responsible for reduced binding of mouse IgA-Fc, the reciprocal mutation S442N in the rat Fc reduced rat FcαRI binding activity to the level of the WT mouse Fc.
Human FcαRI-Ig bound well to human IgA-Fc (MFI 580 ± 180) but relatively poorly to rat WT IgA-Fc (MFI 81 ± 19), indicating the sequence differences at this key site (PMSF c.f. PLAF) affect binding of human FcαRI (Fig. 2C) but not the binding of rat FcαRI (Fig. 2B). The mouse IgA-Fc did not show significant binding to human FcαRI. Notably, the N442S mutated mouse IgA-Fc showed a gain (MFI 179 ± 40) of human FcαRI binding activity, still 3-fold less than the activity of the human IgA-Fc. The negative affect of Asn-442 and/or the N-glycan on FcαRI binding to IgA-Fc was further confirmed by the S442N mutation of rat IgA-Fc, which reduced its binding activity to that of background. Staining the transfected cells with anti-polyhistidine mAb showed equivalent expression of the WT and mutant rodent IgA-Fc fragments (Fig. 2D).
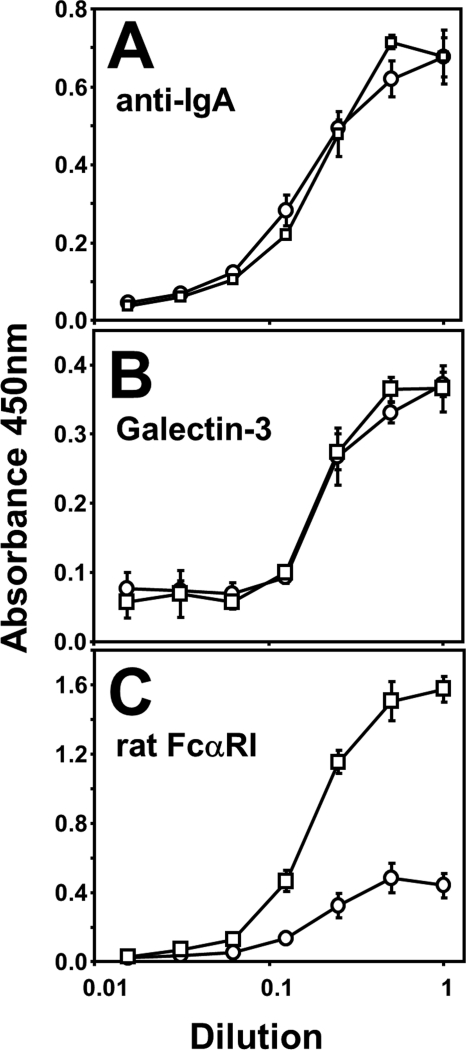
The previous experiments using recombinant Fc fusion proteins demonstrated a role for mouse IgA-Fc Asn-442, and probably the attached N-glycan, in blocking SSL7 and FcαRI binding. Recombinant mouse anti-hen egg lysozyme (HEL) IgA and its N442S mutant were used to confirm these findings in intact IgA antibody. Both the normal IgA and the mutant IgA bound antigen, HEL, with equivalent activities (Fig. 3A), and these HEL bound IgAs also had equivalent binding activities for galectin 3 (Fig. 3B), an IgA binding protein in the mouse (26). In contrast, the N442S IgA had enhanced binding activity for rat FcαRI-Ig with ∼3.5-fold greater saturable binding in the ELISA than normal mouse IgA (Fig. 3C) in agreement with the enhanced activity of the N442S mutant Fc (Fig. 3B). Hence, in both recombinant Fc and whole IgA, Asn-442 or its N-linked glycan inhibits binding by FcαRI.
FIGURE 3.
N442S mutant mouse IgA has enhanced FcαRI binding activity. Recombinant anti-HEL mouse IgA, WT (○), and N442S mutant (□) were analyzed by ELISA for binding to HEL antigen (A), murine galectin-3 (B), and rat FcαRI-Ig (C). Data represent the average ± S.D. (n = 6 replicates) representative of three experiments.
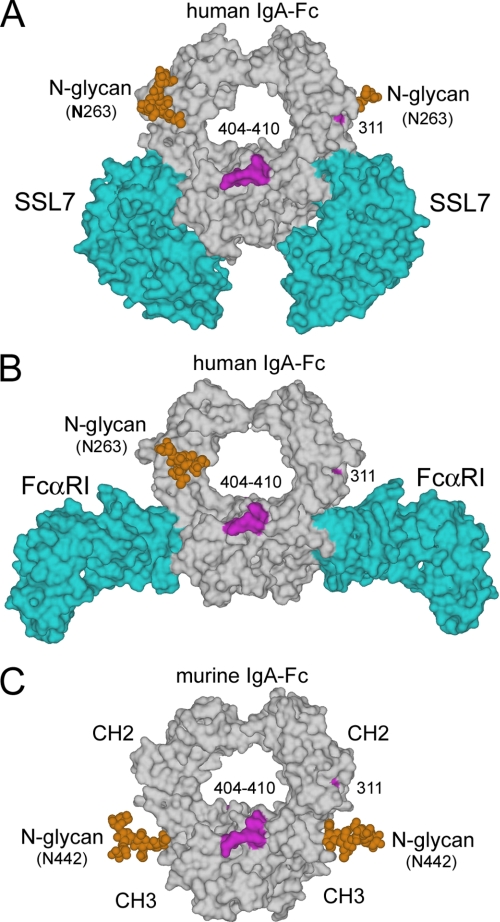
We generated homology models of the rat FcαRI ectodomains and the rodent IgA-Fc fragments from the crystallographic structures of the human proteins (PDB codes 1OW0 and 2QEJ) and analyzed their various complexes between IgA-Fc, FcαRI, and SSL7 (Fig. 4). The different binding activities of the human IgA-Fc and the rat IgA-Fc result from two amino acid residue differences (441 and 442) in the binding site for the receptor and three differences (317, 441, and 442) in the binding site for SSL7. Residues 441 and 442 are key as they occupy the center of the FcαRI and SSL7 binding sites. In human IgA, Leu-441 protrudes prominently from the FG loop contributing strongly to interactions of the Fc complexes with receptor and SSL7 (Fig. 4, A and B). Moreover, the rat IgA-Fc (Met-441–Ser-442) or the mouse N442S IgA-Fc mutant (Met-441-N442S) compared with the human IgA-Fc (Leu-441–Ala-442) both have ≥25-fold reduced binding activity toward SSL7, whereas binding to the human FcαRI is reduced 7- or 3-fold, respectively (Fig. 2B). The greater effect of these amino acid differences on the SSL7 binding rather than the human FcαRI binding may result from the SSL7 binding with a higher affinity and shape complementarity to the core of the CH2/CH3 interface, an interaction that is therefore less accommodating of changes in the central contact residues. Further diversity in the capacity to tolerate amino acid differences in the CH2/CH3 interface is seen with the binding of FcαRI. Although the human FcαRI binds weakly (∼8-fold less) to the rat IgA-Fc (Fig. 2C), the rat FcαRI binds well to both the rat and human IgA-Fc (Fig. 2B). In FcαRI, seven of ten residues contacting IgA differ between the human and rat receptors reflecting the extensive co-adaption of receptor and IgA under evolutionary selection (20). This divergence of the human and rat receptors explains their differing tolerance of amino acid differences in the CH2/CH3 interface.
FIGURE 4.
Structural comparisons of key host-pathogen and N-glycan sites on IgA-Fc. A, crystal structure of the human IgA1-Fc in complex with S. aureus protein SSL7 (PDB code 2QEJ) (22). B, crystal structure of human IgA1-Fc in complex with FcαRI (PDB code 1OW0) (36). C, homology model of murine IgA-Fc based on the IgA1-Fc·SSL7 complex. The IgA-Fc N-glycans (orange CPK space-filling spheres) for all structures were modeled as the core branched oligosaccharide (see “Materials and Methods”). Sites for interacting with the pIgR, as identified previously by mutagenesis, residues 404–410 (39) and Cys-311 (40, 46) are highlighted (magenta). Solvent accessible surfaces were generated using DS Visualizer (version 2.0 (Accelrys)). Glycosylation of asparagine 442 in the mouse IgA-Fc effectively blocks access to this key site for the interaction of host and pathogen IgA binding proteins. Mouse IgA-Fc lacks the equivalent Asn-263 glycan in the CH2 of human IgA (Fig. 4B). The C-terminal tail piece, which is N-glycosylated in both mouse and human IgA, is not represented in this model of the IgA-Fc (CH2 and CH3 domains).
DISCUSSION
Amino acid differences in the CH2/CH3 domain interface of the IgA-Fc occur between human, rat, and mouse IgA in the FG loop of CH3. This region is critical in binding the host receptor, FcαRI, and pathogen IgA binding proteins, such as SSL7. Divergent CH3 FG loops, PLAFT in humans, PMSFT in the rat, and PMNFT in the mouse, account for the species differences in IgA binding to SSL7 and FcαRI. S. aureus is a highly adapted pathogen to its human host and reflective of this SSL7 binding is reduced to IgA-Fc that differs from the human sequence. The binding activity of rat IgA is ∼25-fold less than that of human IgA and mouse IgA is unable to bind SSL7. Also human FcαRI has ∼8-fold lower binding activity on the rat IgA and negligible binding to mouse IgA. Mice lack FcαRI as the gene for FcαRI, and many related immunoreceptors, are absent (4). Asn-442 in the mouse IgA-Fc is notable as the only residue different to that of the rat IgA-Fc and being a bulkier sidechain when compared with serine in the rat Fc, it may protrude into the CH2/CH3 interface reducing binding (3-fold) to rat FcαRI and abrogating binding to the human receptor.
Furthermore, N-glycanase treatment and mutagenesis analysis demonstrated N-glycosylation of the PMNFT site of mouse IgA-Fc. Glycan analysis by mass spectroscopy has previously identified N-linked glycosylation of mouse IgA at two sites, one in the CH1 domain and one in the tail piece (30). This is to our knowledge the first report of glycosylation at Asn-442 in the FG loop of CH3. The glycosylation of this site may provide a general block to pathogen IgA binding proteins that interact primarily with the CH2/CH3 interface. The CH2/CH3 interfaces in the Fc of antibodies are promiscuous binding interfaces (31), and the glycosylation of this site in mouse IgA is an effective block to undesirable pathogen interactions. In mouse IgA, this acquisition is perhaps a reversion to a more ancient solution to blocking pathogen IgA-binding proteins as the monotremes and marsupials lack FcαRI and their IgAs are also potentially glycosylated at this site (Table 1).
Streptococcal IgA binding proteins (32–34) and SSL7 of S. aureus have multiple binding activities, so they may provide additional benefit to pathogen fitness beyond blocking FcαRI-mediated immunity. For example, SSL7 also binds and inhibits the function of complement C5 (21). The simultaneous binding of IgA has a role in this complement inhibitory activity (35). In fact, antibodies generally are targeted at this interface in the Fc by pathogen binding proteins with binding specificities for other host immune molecules and may form large molecular assemblies (31). Because the host receptor FcαRI is not present in the mouse genome, the selection of IgA variants in the CH2/CH3 interface that inhibit binding of pathogen IgA binding proteins is unconstrained by positive selection maintaining the interaction with FcαRI (20). Indeed, an N-linked glycan at Asn-442 of mouse IgA-Fc fully impedes access to the CH2/CH3 interface abrogating its potential for protein-protein interactions (Fig. 4). FcαRI, binds edge on to the Fc at the CH2/CH3 interface (36) with a 2:1 stoichiometry (28, 37). The S. aureus protein SSL7 forms a complex with IgA-Fc of flat planar topology centered on the CH2/CH3 interface (22). Thus, Asn-442 glycosylation is incompatible with binding either the receptor FcαRI or other host or pathogen proteins that bind centrally to this interface.
Transcytosis and formation of SIgA is dependent on the interaction of dimeric IgA and pIgA with the pIgR (38). The critical function of SIgA in mucosal immunity will exert strong selection of IgA-Fc variants. Mutagenesis studies of dimeric IgA demonstrate that pIgR interacts mostly with a ridge on the face of the IgA-Fc, residues Pro-404 to Thr-410 and the adjacent residues 411–414 (39, 40). The CH3 FG loop residues Pro-440 to Phe-443 form the periphery of the pIgR interactive site with mutations L441M/A442N and A442R of human dimeric-IgA modestly reducing pIgR binding ∼2-fold (40). These residues are, however, central to the interaction of SSL7 and FcαRI, which interact edge on with the CH2/CH3 (22, 36). A recent solution structure of human SIgA proposes the secretory component is located along an edge of the dimeric IgA in a near to planar topology (41), but the SC domains appear displaced sufficiently from the central CH2/CH3 interface to accommodate an N-glycan at residue 442. Thus, glycosylation of Asn-442 in mouse IgA would appear an elegant adaption that blocks IgA binding pathogen proteins, which react with the central CH2/CH3 interface, but still permits interaction of pIgR to preserve SIgA mediated mucosal immunity.
Glycosylation is often used by pathogens to mask epitopes to prevent their recognition by the immune system. Neutralizing antibodies to HIV gp120 (42), influenza HA (43), and hepatitis C E2 glycoprotein (44) drive the selection of acquisition of glycosylation variants by which carbohydrate shields important antigenic features. The mucin domain of Ebola virus glycoprotein forms a glycan umbrella that generally masks the infected cell surface (45). In this study, the glycosylation of CH2/CH3 interface of murine IgA is an example of the adaption of the mammalian immune system whereby acquisition of a glycan shields a key host protein from pathogen interactions.
Acknowledgment
We greatly appreciate the gift of rat FcαRI cloned cDNA from Dr Masanori Kasahara.
This work was supported by a National Health and Medical Research Council project grant.
- FcαRI
- Fcα receptor I
- CH
- heavy chain constant domain
- PDB
- Protein Data Bank
- TfR
- transferrin receptor
- pIgR
- polymeric Ig receptor
- MFI
- mean fluorescent intensity
- SC
- secretory component
- SIgA
- secretory IgA
- SSL
- staphylococcal superantigen-like
- NTA
- nitrilotriacetic acid.
REFERENCES
- 1. Monteiro R. C., Van De Winkel J. G. (2003) Annu. Rev. Immunol. 21, 177–204 [DOI] [PubMed] [Google Scholar]
- 2. Wines B. D., Hogarth P. M. (2006) Tissue Antigens 68, 103–114 [DOI] [PubMed] [Google Scholar]
- 3. Woof J. M., Kerr M. A. (2006) J. Pathol. 208, 270–282 [DOI] [PubMed] [Google Scholar]
- 4. Maruoka T., Nagata T., Kasahara M. (2004) Immunogenetics 55, 712–716 [DOI] [PubMed] [Google Scholar]
- 5. Maliszewski C. R., March C. J., Schoenborn M. A., Gimpel S., Shen L. (1990) J. Exp. Med. 172, 1665–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Otten M. A., van Egmond M. (2004) Immunol. Lett. 92, 23–31 [DOI] [PubMed] [Google Scholar]
- 7. Pleass R. J., Dunlop J. I., Woof J. M. (1997) Biochem. Soc. Trans. 25, 327S [DOI] [PubMed] [Google Scholar]
- 8. van Egmond M., Damen C. A., van Spriel A. B., Vidarsson G., van Garderen E., van de Winkel J. G. (2001) Trends Immunol. 22, 205–211 [DOI] [PubMed] [Google Scholar]
- 9. van Egmond M., van Garderen E., van Spriel A. B., Damen C. A., van Amersfoort E. S., van Zandbergen G., van Hattum J., Kuiper J., van de Winkel J. G. (2000) Nat. Med. 6, 680–685 [DOI] [PubMed] [Google Scholar]
- 10. Shen L. (1992) Immunol. Res. 11, 273–282 [DOI] [PubMed] [Google Scholar]
- 11. Stockmeyer B., Dechant M., van Egmond M., Tutt A. L., Sundarapandiyan K., Graziano R. F., Repp R., Kalden J. R., Gramatzki M., Glennie M. J., van de Winkel J. G., Valerius T. (2000) J. Immunol. 165, 5954–5961 [DOI] [PubMed] [Google Scholar]
- 12. Otten M. A., Rudolph E., Dechant M., Tuk C. W., Reijmers R. M., Beelen R. H., van de Winkel J. G., van Egmond M. (2005) J. Immunol. 174, 5472–5480 [DOI] [PubMed] [Google Scholar]
- 13. van der Steen L., Tuk C. W., Bakema J. E., Kooij G., Reijerkerk A., Vidarsson G., Bouma G., Kraal G., de Vries H. E., Beelen R. H., van Egmond M. (2009) Gastroenterology 137, 2018–2029 [DOI] [PubMed] [Google Scholar]
- 14. van Spriel A. B., van den Herik-Oudijk I. E., van Sorge N. M., Vilé H. A., van Strijp J. A., van de Winkel J. G. (1999) J. Infect. Dis. 179, 661–669 [DOI] [PubMed] [Google Scholar]
- 15. van der Pol W., Vidarsson G., Vilé H. A., van de Winkel J. G., Rodriguez M. E. (2000) J. Infect. Dis. 182, 1139–1145 [DOI] [PubMed] [Google Scholar]
- 16. Kobayashi T., Takauchi A., van Spriel A. B., Vilé H. A., Hayakawa M., Shibata Y., Abiko Y., van de Winkel J. G., Yoshie H. (2004) Vaccine 23, 585–594 [DOI] [PubMed] [Google Scholar]
- 17. Hellwig S. M., van Spriel A. B., Schellekens J. F., Mooi F. R., van de Winkel J. G. (2001) Infect Immun. 69, 4846–4850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Balu S., Reljic R., Lewis M. J., Pleass R. J., McIntosh R., van Kooten C., van Egmond M., Challacombe S., Woof J. M., Ivanyi J. (2011) J. Immunol. 186, 3113–3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yel L. (2010) J. Clin. Immunol. 30, 10–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Abi-Rached L., Dorighi K., Norman P. J., Yawata M., Parham P. (2007) J. Immunol. 178, 7943–7954 [DOI] [PubMed] [Google Scholar]
- 21. Langley R., Wines B., Willoughby N., Basu I., Proft T., Fraser J. D. (2005) J. Immunol. 174, 2926–2933 [DOI] [PubMed] [Google Scholar]
- 22. Ramsland P. A., Willoughby N., Trist H. M., Farrugia W., Hogarth P. M., Fraser J. D., Wines B. D. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 15051–15056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wines B. D., Willoughby N., Fraser J. D., Hogarth P. M. (2006) J. Biol. Chem. 281, 1389–1393 [DOI] [PubMed] [Google Scholar]
- 24. Pleass R. J., Dunlop J. I., Anderson C. M., Woof J. M. (1999) J. Biol. Chem. 274, 23508–23514 [DOI] [PubMed] [Google Scholar]
- 25. Wines B. D., Hulett M. D., Jamieson G. P., Trist H. M., Spratt J. M., Hogarth P. M. (1999) J. Immunol. 162, 2146–2153 [PubMed] [Google Scholar]
- 26. Reljic R., Crawford C., Challacombe S., Ivanyi J. (2004) Immunol. Lett. 93, 51–56 [DOI] [PubMed] [Google Scholar]
- 27. Sali A., Potterton L., Yuan F., van Vlijmen H., Karplus M. (1995) Proteins 23, 318–326 [DOI] [PubMed] [Google Scholar]
- 28. Wines B. D., Sardjono C. T., Trist H. H., Lay C. S., Hogarth P. M. (2001) J. Immunol. 166, 1781–1789 [DOI] [PubMed] [Google Scholar]
- 29. Bohne A., Lang E., von der Lieth C. W. (1999) Bioinformatics 15, 767–768 [DOI] [PubMed] [Google Scholar]
- 30. Kragten E. A., Bergwerff A. A., van Oostrum J., Müller D. R., Richter W. J. (1995) J. Mass Spectrometry 30, 1679–1686 [Google Scholar]
- 31. Wines B. D., Trist H. M., Farrugia W., Ngo C., Trowsdale J., Areschoug T., Lindahl G., Fraser J. D., Ramsland P. A. (2011) Adv. Exp. Med. Biol., in press [DOI] [PubMed] [Google Scholar]
- 32. Areschoug T., Carlsson F., Stålhammar-Carlemalm M., Lindahl G. (2004) Vaccine 22, S9–S14 [DOI] [PubMed] [Google Scholar]
- 33. Areschoug T., Stålhammar-Carlemalm M., Karlsson I., Lindahl G. (2002) J. Biol. Chem. 277, 12642–12648 [DOI] [PubMed] [Google Scholar]
- 34. Pleass R. J., Areschoug T., Lindahl G., Woof J. M. (2001) J. Biol. Chem. 276, 8197–8204 [DOI] [PubMed] [Google Scholar]
- 35. Laursen N. S., Gordon N., Hermans S., Lorenz N., Jackson N., Wines B., Spillner E., Christensen J. B., Jensen M., Fredslund F., Bjerre M., Sottrup-Jensen L., Fraser J. D., Andersen G. R. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 3681–3686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Herr A. B., Ballister E. R., Bjorkman P. J. (2003) Nature 423, 614–620 [DOI] [PubMed] [Google Scholar]
- 37. Herr A. B., White C. L., Milburn C., Wu C., Bjorkman P. J. (2003) J. Mol. Biol. 327, 645–657 [DOI] [PubMed] [Google Scholar]
- 38. Mostov K. E., Blobel G. (1982) J. Biol. Chem. 257, 11816–11821 [PubMed] [Google Scholar]
- 39. Hexham J. M., White K. D., Carayannopoulos L. N., Mandecki W., Brisette R., Yang Y. S., Capra J. D. (1999) J. Exp. Med. 189, 747–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lewis M. J., Pleass R. J., Batten M. R., Atkin J. D., Woof J. M. (2005) J. Immunol. 175, 6694–6701 [DOI] [PubMed] [Google Scholar]
- 41. Bonner A., Almogren A., Furtado P. B., Kerr M. A., Perkins S. J. (2009) Mucosal Immunol. 2, 74–84 [DOI] [PubMed] [Google Scholar]
- 42. Wei X., Decker J. M., Wang S., Hui H., Kappes J. C., Wu X., Salazar-Gonzalez J. F., Salazar M. G., Kilby J. M., Saag M. S., Komarova N. L., Nowak M. A., Hahn B. H., Kwong P. D., Shaw G. M. (2003) Nature 422, 307–312 [DOI] [PubMed] [Google Scholar]
- 43. Wei C. J., Boyington J. C., Dai K., Houser K. V., Pearce M. B., Kong W. P., Yang Z. Y., Tumpey T. M., Nabel G. J. (2010) Sci. Transl. Med. 2, 24ra21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Falkowska E., Kajumo F., Garcia E., Reinus J., Dragic T. (2007) J. Virol. 81, 8072–8079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Francica J. R., Varela-Rohena A., Medvec A., Plesa G., Riley J. L., Bates P. (2010) PLoS Pathog. 6, e1001098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chintalacharuvu K. R., Tavill A. S., Louis L. N., Vaerman J. P., Lamm M. E., Kaetzel C. S. (1994) Hepatology 19, 162–173 [PubMed] [Google Scholar]