Abstract
Allergic asthma is a chronic airway inflammatory disease in which exposure to allergens causes intermittent attacks of breathlessness, airway hyper-reactivity, wheezing, and coughing. Allergic asthma has been called a “syndrome” resulting from a complex interplay between genetic and environmental factors. Worldwide, >300 million individuals are affected by this disease, and in the United States alone, it is estimated that >35 million people, mostly children, suffer from asthma. Although animal models, linkage analyses, and genome-wide association studies have identified numerous candidate genes, a solid definition of allergic asthma has not yet emerged; however, such studies have contributed to our understanding of the multiple pathways to this syndrome. In contrast with animal models, in which T-helper 2 (TH2) cell response is the dominant feature, in human asthma, an initial exposure to allergen results in TH2 cell-dependent stimulation of the immune response that mediates the production of IgE and cytokines. Re-exposure to allergen then activates mast cells, which release mediators such as histamines and leukotrienes that recruit other cells, including TH2 cells, which mediate the inflammatory response in the lungs. In this minireview, we discuss the current understanding of how associated genetic and environmental factors increase the complexity of allergic asthma and the challenges allergic asthma poses for the development of novel approaches to effective treatment and prevention.
Keywords: Gene Expression, Immunology, Inflammation, Innate Immunity, Interleukin, Allergic Asthma
Allergic Asthma: A Complex Syndrome
Asthma is a highly prevalent (330 million people affected worldwide) chronic inflammatory disease of the conducting airways of the respiratory system (American Academy of Allergy, Asthma and Immunology and reviewed in Ref. 2). It is a complex syndrome in which allergen exposure often induces intermittent attacks of breathlessness, airway hyper-reactivity, wheezing, and coughing (3). During the past 6 decades, the worldwide incidence and severity of asthma have steadily increased. Allergic asthma is one aspect of atopic disease, which is also increasing. This disease has become an expanding burden on public health services in both industrialized and developing countries (1–6). It is estimated that >35 million people in the United States alone develop asthma during their lifetime, and more than three-fourths of these individuals suffer from allergies (1, 5). During the past 3 decades, much has been learned about the pathogenesis of allergen-induced airway inflammation, which is recognized as one of the predominant underlying causes of allergic asthma. Currently, it is recognized that allergic asthma is a complex disease that results from interactions between multiple genetic and environmental factors (7). Moreover, the advances in molecular genetics and the use of animal models have advanced our understanding of the pathogenic mechanism(s) of various aspects of this complex disease, and it is expected that further advance in this area will facilitate the development of novel and more effective therapeutic approaches. It should be noted, however, that the mouse models of allergic asthma do not exactly replicate the human disease. In contrast to the mouse models, in which T-helper 2 (TH2)2 cell response is the dominant feature, the pathogenesis of human allergic asthma involves an initial exposure to an allergen that results in TH2 cell-dependent stimulation of the immune response that mediates the production of IgE and cytokines. Repeated exposure to allergen then activates mast cells, which release mediators that facilitate recruitment of other cell types, including TH2 cells, which mediate the inflammatory response in the airways (reviewed in Ref. 8). Thus, although the TH2 axis plays a critical role in initiating the IgE response and airway inflammation, the presence of allergen-specific IgE does not necessarily culminate in asthma. In this minireview, we discuss what makes allergic asthma a complex disease in which genetic and environmental factors merge to cause pathogenesis.
Current Understanding of Allergic Asthma Pathogenesis
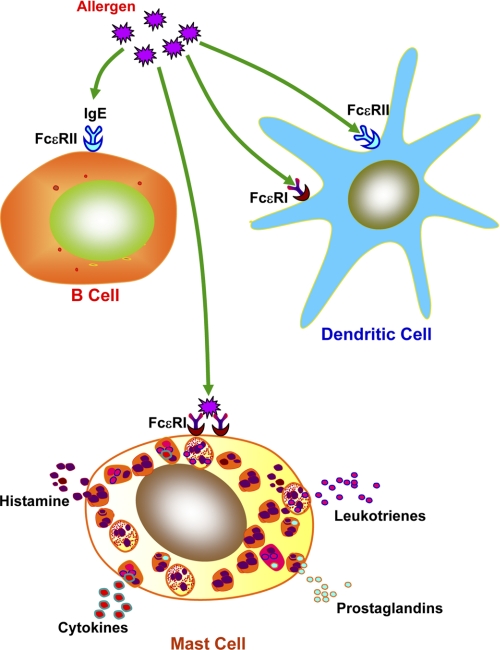
Although several types of asthma have been recognized clinically, allergic asthma is the most common form of the disease (reviewed in Ref. 9). In susceptible individuals, the initiation of allergen sensitivity occurs at the mucosal surfaces, where environmental allergens meet the mucosal epithelia. The interaction of inhaled allergen(s) with sensitized immune cells in the airway results in allergic asthma. Allergic rhinitis, atopic dermatitis, and asthma, which constitute atopic conditions, occur in individuals with markedly increased levels of IgE antibodies (10). The expression of high-affinity (FcϵRI) and low-affinity (FcϵRII; CD23) IgE receptors occurs in a wide variety of cell types, including dendritic cells (DCs) and B cells (Fig. 1). IgE bound to these receptors on B cells and DCs facilitates the uptake of allergen by these cells, promoting allergen presentation to T cells, which mediate secondary immune responses. The majority of IgE is bound by FcϵRI on mast cells as well as basophils, and IgE-bound FcϵRI cross-linking by a specific antigen mediates the release of inflammatory mediators (e.g. histamine and leukotrienes) by mast cells (Fig. 1), leading to the inflammatory response. The regulation of IgE production and its relationship to the development of TH2 cells that drive IgE responses have been reviewed (8). An enhanced tendency toward airway hyper-reactivity (AHR) culminating in bronchial smooth muscle contraction, characteristically found in patients with asthma, is often linked to high IgE levels (11). Moreover, it has been reported that, in cohorts of children with asthma and physiologic evidence of AHR, high serum levels of IgE are detectable (12). Although, in both animal models and humans, some component of asthmatic pathophysiology, especially acute reactions to allergen, may be IgE-mediated, the other features of this disease may arise independently of IgE. Thus, in atopic families, inhalation of allergens and subsequent production of IgE are associated with predisposition to allergic asthma. Furthermore, it is likely that IgE participates in triggering mast cell-mediated acute- and late-phase airflow obstruction (10). Allergen exposure also triggers activation of bone marrow-derived and non-bone marrow-derived cells of the innate immune system, which eventually leads to the secretion of various cytokines (3). The recruitment of antigen-processing cells, most likely monocyte-derived DCs, initiates the pathway to inflammation. Recently, it was reported that basophils may also be involved in certain situations. Moreover, it has been reported that, in the airways of patients with asthma and especially in those patients suffering from allergic asthma, allergen-specific TH2 cells are readily detectable (13). Recently, several excellent reviews were published on TH2 cell differentiation (14–17), which may be referred to for detail knowledge on this subject. The TH2 cells secrete cytokines, which promote the synthesis of allergen-specific IgE. These cytokines also promote presentation of antigens (allergens) to CD4+ T cells, which facilitate both DCs and T cells in eliciting TH2 cell responses (18, 19). In addition, activated TH2 cells also secrete the cytokines IL-5, IL-9 and IL-13, which facilitate the recruitment of eosinophils and promote the growth of mast cells, respectively, ultimately stimulating AHR, characteristically found in asthma (9, 20). However, recent experiments indicate that TH2 cells fail to produce IL-4, IL-5, or IL-13 without CD11c+ DCs (9, 21). Interestingly, adoptively transferred, bone marrow-derived, antigen-loaded DCs or DCs in lungs can induce the TH2 cell response (22), suggesting that lung DCs are the major antigen-presenting cells (APCs) and are essential for TH2 cell response during allergen-mediated airway inflammation.
FIGURE 1.
IgE-mediated allergic response. A wide variety of immune cells such as DCs and B cells express the low-affinity IgE receptor, FcϵRII (CD23). Uptake of allergen is mediated via IgE-bound FcϵRI and FcϵRII on APCs, augmenting secondary immune responses. The mast cells and basophils express the high-affinity IgE receptor, FcϵRI, which binds IgE, and the cross-linking of IgE-bound FcϵRI on these cells mediates release of proinflammatory mediators such as histamine, prostaglandins, leukotrienes, cytokines, and enzymes that lead to biological manifestation of allergy (8).
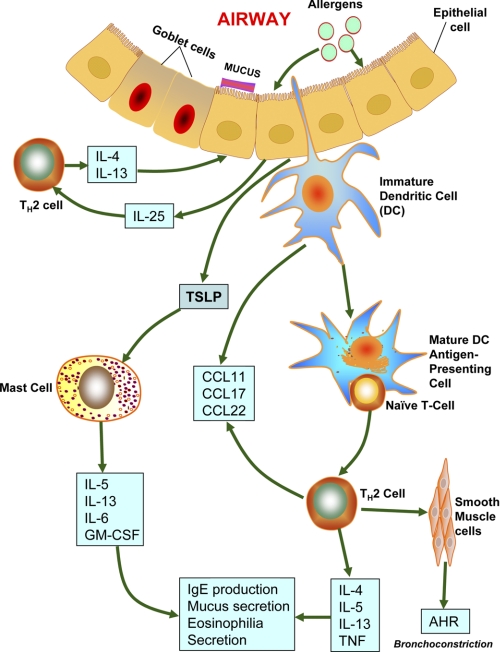
In addition to the recruitment of TH2 cells, allergen challenge also facilitates the recruitment of other inflammatory cells such as mast cells, basophils, and eosinophils. However, a recent report indicates that, in addition to being inflammatory, these cells also participate as APCs and initiate or enhance TH2 cell responses (9). Among the three cell types, mast cells can initiate immediate hypersensitivity responses by releasing histamines in response to both IgE-mediated adaptive and innate immune responses. Moreover, mast cells can be activated via cross-linking of allergen-specific IgE (23) or via Toll-like receptor ligands or cytokines such as IL-33 (24). Furthermore, in addition to releasing histamines and cysteinyl leukotrienes, mast cells secrete various cytokines such as IL-1, IL-3, IL-4, IL-5, IL-6, IL-8, IL-10, IL-13, IL-16, TGF-β, and TNF-α and chemokines such as TCA-3, RANTES, MCP-1, and MIP-1α (reviewed in Ref. 25). In Fig. 2, the pathways that lead to allergic asthma are summarized.
FIGURE 2.
Possible pathways to allergic asthma. Allergens reaching the airways via inhaled air are taken up and processed by DCs that are primed by thymic stromal lymphopoietin (TSLP) secreted by airway epithelial cells. These allergens also cause the mast cells to release CCL17 and CCL22. CCL17 and CCL22 act on CCR4 (CC chemokine receptor 4), which mediates chemotactic migration of TH2 cells. TH2 cells play critical roles in orchestrating the allergen-induced inflammatory response by releasing IL-4 and IL-13. These interleukins also stimulate IgE production by B cells. These activated B cells also produce IL-5 (required for eosinophilic inflammation) and IL-9 (stimulator of mast cell proliferation). Airway epithelial cells release CCL11, stimulating recruitment of eosinophils via CCR3. Individuals suffering from allergic asthma may have defective Treg cells, which favor further TH2 cell proliferation and differentiation. Allergens also stimulate activation of sensitized mast cells by cross-linking surface-bound IgE molecules. In turn, activated mast cells secrete mediators of bronchoconstriction such as histamines, prostaglandin D2, and cysteinyl leukotrienes (2).
Genetic Susceptibility to Allergy and Asthma
As stated above, asthma and asthma-related syndromes are complex diseases in which an interplay of strong genetic and environmental components leads to pathogenesis (reviewed in Refs. 7 and 26–32). It is well known that many individuals are predisposed to developing allergic reactions to substances that do not generally elicit immune response. These atopic individuals are thought to be genetically predisposed to develop hypersensitivity to substances such as pollens, antibiotics, and perfumes. Numerous studies have shown that atopy is familial in nature. Elevated levels of IgE have been detected in patients with allergic diseases (7), and IgE production is tightly controlled. Recently, it was reported that NF-IL3, a transcriptional regulator, may control IgE production (33). Early studies reported the prevalence of allergic disease in first degree relatives of affected individuals (34, 35). Subsequently, studies in monozygotic and dizygotic twins have shown a positive correlation with regard to allergic disease traits such as total serum IgE levels, methacholine sensitivity in the lungs, and skin test results being 2-fold higher among monozygotic twins than dizygotic twins. Moreover, children of asthmatic parents are more likely to develop asthma than those of parents without any history of atopy. Studies on atopic disease in human and animal populations have facilitated the identification of susceptibility genes encoding class II MHC, FcϵRIβ, RANTES, IL-4 receptor α, β-adrenergic receptor, T cell receptor α, and mast cell chymase. Furthermore, evidence suggests that genetic loci on human chromosomes 5, 6, and 11 are likely to harbor atopy genes. A thorough review of the molecular genetics of allergic diseases provides insight into the complexity of the atopic disease and why the identification of specific atopy genes remains challenging (reviewed in Ref. 36).
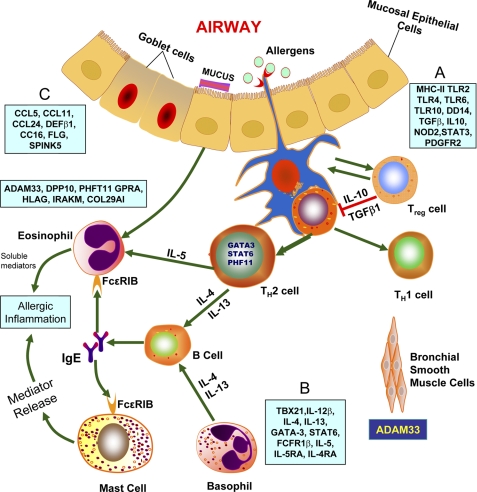
During the past decade, knowledge on asthma genetics has progressed significantly, and several genes or gene loci associated with asthma- and/or atopy-related syndromes have been identified. However, most of these genes have only modest effects, and the majority of these genes have not been systematically tested to determine whether the results are replicable in different populations. Several excellent reviews on the genetics of allergy and asthma have been published (2, 7, 26–32). In this minireview, we discuss only those genes that have shown functional and immunological links for susceptibility to asthma and allergy. In general, asthma susceptibility genes are classified into four main groups (reviewed in Refs. 7, 26, and 32): (i) innate immunity and immunoregulatory genes; (ii) genes associated with TH2 cell differentiation and effector functions; (iii) genes associated with mucosal immunity and epithelial biology; and (iv) genes linked to lung function, airway remodeling, and severity of the disease (7, 26, 32). The interactions among the genes in the first three groups and those identified by positional cloning have been reviewed in detail (32) and are summarized in Fig. 3. The genes included in the first group have been discovered through association studies and are thought to be involved in triggering the immune response and stimulating differentiation of CD4+ T helper cells. These genes encode pattern recognition receptors CD14 (monocyte differentiation antigen 14), NOD1 (nucleotide-binding, oligomerization domain-containing 1), NOD2, TLR2 (Toll-like receptor 2), TLR4, TLR6, and TLR10; cytokines regulating immune responses such as TGF-β1 and IL-10; the transcription factor STAT3 (signal transducer and activator of transcription 3); molecules that facilitate antigen presentation such as the HLA-DR, HLA-DP, and HLA-DQ alleles; and the prostaglandin receptor. Asthma susceptibility genes that belong to the second group regulate TH2 cell differentiation and TH2 cell effector functions (26): FCER1B, GATA3 (GATA-binding protein 3), IL4, IL4RA (interleukin-4 receptor α-chain), IL5, IL5RA, IL12B, IL13, STAT6, and TBX21 (T-box 21; also known as T-bet). Several genes are expressed in epithelial cells and are included in the third group. These genes encode chemokines CCL5 (CC chemokine ligand 5), CCL11, CCL24, and CCL26; factors involved in maintaining the integrity of the epithelial cell barrier (FLG (filaggrin) and SPINK5 (serine protease inhibitor Kazal-type 5)); antimicrobial peptide DEFB1 (defensin β1); and anti-inflammatory protein CC10/CC16 (Clara cell-specific 16 kDa protein; also known as SCGB1A1 and uteroglobin). The fourth group of asthma susceptibility genes has been identified by positional cloning. It includes ADAM33 (a disintegrin and metalloproteinase domain 33), COL29A1 (collagen type XXIX α-1; also called COL6A5 (collagen type VI α-5)), DPP10 (dipeptidyl peptidase 10), GPRA (G-protein-coupled receptor for asthma susceptibility; also known as NPSR1 and GPRA154), HLA-G, IRAKM (interleukin-1 receptor-associated kinase M), and PHF11 (plant homeodomain finger protein 11), which are expressed in the epithelium and/or smooth muscle cells.
FIGURE 3.
Genes identified by association studies or positional cloning. The genes that are associated with asthma/atopy are divided into four groups (26). A, the first group of genes is associated with triggering the allergic response via differentiation of CD4+ T helper cells. This group includes genes encoding CD14, TLR2, TLR4, TLR6, TLR10, NOD1, and NOD2, which are known as pattern recognition receptors. This group also includes genes that encode immunoregulatory cytokines such as IL-10 and TGF-β1; the transcription factor STAT3; antigen-presenting facilitator genes such as the HLA-DR, HLA-DQ, and HLA-DP alleles; and prostaglandin E receptor 2. B, the second group of genes includes GATA3, TBX21, IL4, IL13, IL4RA, FCER1B, IL5, IL5RA, STAT6, and IL12B, which regulate TH2 cell differentiation and effector functions. C, the third group includes genes encoding chemokines CCL5, CCL11, CCL24, and CCL26; antimicrobial peptide DEFB1; anti-inflammatory protein CC16 (also called UG); and factors responsible for maintaining the epithelial cell barrier such as SPINK5 and FLG. The positional cloning method has been used to identify the following genes expressed in the epithelia and smooth muscles: ADAM33, COL29A1, DPP10, GPRA, HLA-G, IRAKM, and PHF11 (26).
Several genes (including ADAM33, DPP10, PHF11, TIM-1, GPRA, OPN3, ORMDL3, and PDE4D) that are associated with atopy and asthma have been identified via association studies and positional cloning (37). The genes encoding the T cell immunoglobulin domain, the mucin-like domain (TIM) family (38, 39), and ADAM33 on human chromosome 20p13 are associated with asthma and AHR (40). In addition to association studies and positional cloning, genome-wide association studies have also been carried out to identify asthma susceptibility genes (27–32). This method utilizes SNPs for screening across the whole genome to identify novel disease susceptibility genes without the bias of prior knowledge. However, because of the lack of biological correlates, in some instances, the genes identified by this method have raised questions about the true relevance of these genes to the disease (32). Three independent reports were published recently (41–43) in which asthma susceptibility genes were identified utilizing knowledge from asthma genetics as well as asthma biology. As stated above, most of the identified genes have not been rigorously tested as to whether the results are replicable in different populations.
Gene Expression in Allergic Airway Inflammation and Allergic Asthma
Our knowledge of the allergen-mediated inflammatory response in airways has been greatly advanced by the availability of mouse models of allergic asthma. The hallmark of allergic asthma immunopathology is the infiltration of the airways by eosinophils and TH2 cells (44). Recruitment of TH2 cells to the lungs mediates the development of eosinophilic inflammation and AHR (9). Mice can be sensitized to foreign antigens (allergens), including ovalbumin (OVA), by introducing this antigen combined with an adjuvant such as alum (45). Immunization with OVA in alum causes IgE and IgG1 production. When subjected to repeated exposure to the allergen, OVA-sensitized mice develop airway eosinophilia, hyperplasia of mucus-secreting goblet cells, and AHR, which are some of the characteristic features of allergic asthma. When these changes occur chronically, they lead to airway remodeling (46), which is also characteristically found in allergic asthma. It should be noted that, although allergen-specific TH2 cells can be induced in mice (26) and in non-human primates (47), there are substantial differences between human and animal models of allergic airway disease (reviewed in Ref. 48).
To characterize gene expression in allergic airway inflammation, gene microarrays have been used (49, 50). The profiling of gene expression by microarray has been used to characterize asthma-related genes in both humans and mice (see Table 1 in Ref. 51). The Gene Expression Omnibus (GEO) Database (www.ncbi.nlm.nih.gov/geo/) contains data sets on gene expression in allergic asthma models, intratracheal treatment with IL-13 (called allergen-induced goblet cells), T cells from atopic and asthma patients, allergen provocation in IL-13 knock-out mice, asthma exacerbation factors, and other studies related to allergy.
The allergen-induced responses in allergic asthma are driven predominantly by effector T cells, mostly by TH2 cells, which are regulated by regulatory T (Treg) cells, and thus, both TH2 and Treg cells have been studied by gene expression profiling. Microarray analyses of TH1 and TH2 cells differentiated from cord blood T lymphocytes have enabled Rogge et al. (52) to demonstrate that 215 of 6000 genes studied were differentially expressed among the two T cell subsets. Studies with CD4+ T cells from patients with atopy or allergic asthma activated in vitro have shown differential gene expression in the two types of allergic disorders (53). Similar studies have facilitated discovery of a family of genes called ephrins, which are associated with allergic asthma pathology (54). Gene expression profiling using Treg cells facilitated the discovery that IL-10 and ICOS (inducible costimulator) promote regulation of effector cells (55). Other investigations related to allergic inflammation have provided gene expression profiles using eosinophils (56) and basophils (57). Similar studies were conducted on a transcriptome level to determine the differentiation of DCs by contact allergens that can also trigger airway inflammation and AHR (58). It should be noted, however, that, although a vast amount of data from investigations on gene expression profiles have been amassed, only a small portion of this information has been used to advance our knowledge on the molecular mechanism(s) of allergic asthma. This may be due to the fact that the roles of numerous transcription factors, signaling molecules, and enzymes in allergic asthma have not yet been fully elucidated. Thus, many recent studies have focused on gene expression that could be associated with specific functions such as chemotaxis, inflammation, and cytokine regulation.
One of the puzzling phenomena in allergic diseases, especially allergic asthma, is that, although our airways are exposed to myriads of allergens on a daily basis, only a small percentage of the world's population suffers from allergic asthma. Thus, although the incidence of allergic asthma continues to rise, the majority of the world's population manages to avert allergic inflammatory diseases, including allergic asthma. Therefore, it is likely that some genes constitutively expressed in the airways maintain homeostasis in the mammalian respiratory system, which suppresses the allergen-mediated inflammatory response. An example of such a gene is uteroglobin (UG) (59), also known as CC10 (60). UG is a secretory protein that manifests immunomodulatory, anti-chemotactic, and anti-inflammatory properties (reviewed in Ref. 61). This protein is produced and secreted by the mucosal epithelia of all organs that communicate with the external environment. Moreover, it is constitutively expressed at a high level in the airway epithelial cells, and UG knock-out mice (62) develop increased susceptibility to allergen-induced airway inflammation. Recently, it was reported that UG suppresses the allergen (OVA)-induced airway inflammatory response by blocking prostaglandin D2 receptor-mediated function (63). This protein has also been reported to inhibit TH2 cell differentiation by acting on DCs (64, 65). Interestingly, human orthologs of murine scca2/serpinb3a (squamous cell carcinoma antigen 2), a serine protease inhibitor associated with allergic inflammation, are overexpressed in the airways of asthma patients (reviewed in Ref. 66). Most interestingly, scca2 is expressed at high levels in the airways of UG knock-out mice and is further augmented by allergen challenge; treatment of these mice with recombinant UG abrogates the allergen-induced elevation of scca gene expression (67). Furthermore, UG also inhibits allergen-induced TH2 cell differentiation by down-regulating the expression of serum amyloid A and the SOCS-3 (suppressor of cytokine signaling 3) gene (68), which play critical regulatory roles in the initiation and maintenance of the TH2 cell-mediated allergic airway inflammatory response (69).
Studies in humans and mouse models of allergic asthma have shown that the TIM family of genes is associated with allergen-mediated diseases (38). It has also been reported that monoclonal antibodies directed against mouse TIM-1 suppress allergen-mediated inflammation in various organs, including the lungs (70–73). More recently, using a humanized mouse model generated by injecting peripheral blood monocytes from asthma patients into immunocompromised severe combined immunodeficient mice, Sonar et al. (74) reported that monoclonal antibody against TIM-1 suppressed the production of proinflammatory TH2 cytokines and prevented AHR. These results clearly demonstrated that the TIM-1 pathway plays a critical role in allergic asthma, and more importantly, antagonizing TIM-1 may provide therapeutic benefit to patients with allergic asthma and other immune-mediated inflammatory disorders.
Most intriguingly, the discovery of a mutant mouse strain called “flaky tail” in the Jackson Laboratory in 1958 and the strain reported in 1972 (75) may provide new insight into the pathogenesis of atopic dermatitis (eczema) and allergic asthma. Recently, Fallon et al. (76) reported that flaky tail mice carry a homozygous frameshift mutation in the gene encoding filaggrin, a large structural protein that facilitates terminal differentiation of the epidermis and forms the skin barrier. Interestingly, this mutation closely resembles human FLG variants that cause ichthyosis vulgaris (77), a skin disorder that is a genetic risk for atopic dermatitis and associated allergic asthma (78, 79). Although the association between FLG mutation and atopic dermatitis is strong, it has been reported that nearly 40% of individuals carrying a null mutation of this gene do not develop the skin disease (80). This suggests that both genetic and environmental modifiers may be involved in the pathogenesis of atopic dermatitis and associated allergic asthma (81).
Environmental Factors Interacting with Genetic Factors in Allergic Asthma
As discussed above, although allergic diseases such as allergic asthma have predisposing genetic factors, the interaction of environmental factors in the pathogenesis of allergic diseases is compelling. Environmental factors (both ingested and inhaled) have been suggested to contribute to allergic asthma pathogenesis (reviewed in Ref. 27). Examples of the environmental factors include air pollutants, respiratory viruses, tobacco smoke, endotoxin, allergens in the air, and diet. Occupational environment can also trigger asthma in genetically susceptible individuals (82). Thus, studies on gene-environment interactions may advance our understanding of the complex mechanism(s) of allergic asthma (83–86). Genes encoding pattern recognition receptors such as TLR4 and CD14 are reported to recognize and clear bacterial endotoxin and LPS, and SNPs in these genes may initiate asthma pathology in early development (27). SNPs in CD14, TLR4, and other TLR genes have been found to have a modest risk of asthma susceptibility in atopic individuals (87). Similarly, SNPs in TGFB1 (88), IL10 (89), DCNP1 (dendritic cell-associated nuclear protein 1) (90), and inflammatory cytokines and enzymes (91) have been reported to alter gene-environment interaction in allergic asthma. Polymorphisms in the TNF-α gene (92) and the risk of childhood asthma in relation to environmental tobacco smoke and SNPs in the q21 region of human chromosome 17 (93) also point to gene-environment interaction. The subject of gene-environment interactions in the pathogenesis of respiratory diseases has been recently reviewed (1, 7, 94).
Concluding Remarks and Perspectives
During the past decade, major advances have been made in our understanding of the immune mechanisms of allergic diseases, including allergic asthma. However, these advances have not yet resulted in the development of effective new therapeutic approaches. It is possible that our knowledge of interacting genetic and environmental factors affecting the pathogenesis of this disease remains incomplete. Thus, further advance in the genetics of allergic asthma and the interaction of candidate asthma genes with environmental factors may facilitate the development of novel therapies. One of the puzzling aspects of allergic asthma pathogenesis is the fact that, despite our exposure to myriads of antigens (allergens), only a relatively small portion of the total human population suffers from allergic diseases. Recent efforts to treat allergic diseases have been focused on the application of biological agents such as monoclonal antibodies to target specific cytokines and cell surface proteins associated with the allergic inflammatory response. For example, it has been demonstrated that blocking TIM-1 using specific monoclonal antibodies may prevent allergic inflammatory responses. These are welcome developments, although we should be cautious that these therapeutic approaches do not compromise the many facets of immune responses (some of which are poorly understood) that are essential for maintaining homeostasis in the human body.
Acknowledgments
We thank I. Owens, J. Y. Chou, and S. W. Levin for critical review of the manuscript and helpful suggestions.
This work was supported by the National Institutes of Health Intramural Research Program of Eunice Kennedy Shriver NICHD. This is the second article in the Thematic Minireview Series on Molecular Bases of Disease: Asthma. This minireview will be reprinted in the 2011 Minireview Compendium, which will be available in January, 2012.
- TH2
- T-helper 2
- FcϵR
- Fcϵ receptor
- DC
- dendritic cell
- AHR
- airway hyper-reactivity
- APC
- antigen-presenting cell
- OVA
- ovalbumin
- Treg
- regulatory T
- UG
- uteroglobin.
REFERENCES
- 1. Garantziotis S., Schwartz D. A. (2010) Annu. Rev. Public Health 31, 37–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barnes P. J. (2008) Nat. Rev. Immunol. 8, 183–192 [DOI] [PubMed] [Google Scholar]
- 3. Locksley R. M. (2010) Cell 140, 777–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mannino D. M., Buist A. S. (2007) Lancet 370, 765–773 [DOI] [PubMed] [Google Scholar]
- 5. Pearce N., Aït-Khaled N., Beasley R., Mallol J., Keil U., Mitchell E., Robertson C., and the ISAAC Phase Three Study Group (2007) Thorax 62, 758–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Akinbami L. J. (2006) Advance Data from Vital and Health Statistics, pp. 1–24, National Center for Health Statistics, Hyattsville, MD [Google Scholar]
- 7. Holloway J. W., Yang I. A., Holgate S. T. (2010) J. Allergy Clin. Immunol. 125, S81–S94 [DOI] [PubMed] [Google Scholar]
- 8. Geha R. S., Jabara H. H., Brodeur S. R. (2003) Nat. Rev. Immunol. 3, 721–732 [DOI] [PubMed] [Google Scholar]
- 9. Kim H. Y., DeKruyff R. H., Umetsu D. T. (2010) Nat. Immunol. 11, 577–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Oettgen H. C., Geha R. S. (2001) J. Allergy Clin. Immunol. 107, 429–440 [DOI] [PubMed] [Google Scholar]
- 11. Postma D. S., Bleecker E. R., Amelung P. J., Holroyd K. J., Xu J., Panhuysen C. I., Meyers D. A., Levitt R. C. (1995) N. Engl. J. Med. 333, 894–900 [DOI] [PubMed] [Google Scholar]
- 12. Sears M.R., Burrows B., Flannery E. M., Herbison G. P., Hewitt C. J., Holdaway and M. D. (1991) N. Engl. J. Med. 325, 1067–1071 [DOI] [PubMed] [Google Scholar]
- 13. Robinson D. S., Hamid Q., Ying S., Tsicopoulos A., Barkans J., Bentley A. M., Corrigan C., Durham S. R., Kay A. B. (1992) N. Engl. J. Med. 326, 298–304 [DOI] [PubMed] [Google Scholar]
- 14. Zhu J., Yamane H., Paul W. E. (2010) Annu. Rev. Immunol. 28, 445–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Paul W. E., Zhu J. (2010) Nat. Rev. Immunol. 10, 225–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Amsen D., Spilianakis C. G., Flavell R. A. (2009) Curr. Opin. Immunol. 21, 153–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sokol C. L., Medzhitov R. (2010) Curr. Opin. Immunol. 22, 73–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Saenz S. A., Taylor B. C., Artis D. (2008) Immunol. Rev. 226, 172–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Finkelman F. (2001) in Samter's Immunologic Diseases (Austen K. F., Frank M. M., Atkinson J. P., Cantor H. eds) 6th Ed., pp. 111–126, Lippincott Williams & Wilkins, Philadelphia [Google Scholar]
- 20. Holgate S. T., Polosa R. (2008) Nat. Rev. Immunol. 8, 218–230 [DOI] [PubMed] [Google Scholar]
- 21. van Rijt L. S., Jung S., Kleinjan A., Vos N., Willart M., Duez C., Hoogsteden H. C., Lambrecht B. N. (2005) J. Exp. Med. 201, 981–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lambrecht B. N., De Veerman M., Coyle A. J., Gutierrez-Ramos J. C., Thielemans K., Pauwels R. A. (2000) J. Clin. Invest. 106, 551–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Prussin C., Metcalfe D. D. (2006) J. Allergy Clin. Immunol. 117, S450–S456 [DOI] [PubMed] [Google Scholar]
- 24. Silver M. R., Margulis A., Wood N., Goldman S. J., Kasaian M., Chaudhary D. (2010) Inflamm. Res. 59, 207–218 [DOI] [PubMed] [Google Scholar]
- 25. Barrett N. A., Austen K. F. (2009) Immunity 31, 425–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vercelli D. (2008) Nat. Rev. Immunol. 8, 169–182 [DOI] [PubMed] [Google Scholar]
- 27. Ober C., Hoffjan S. (2006) Genes Immun. 7, 95–100 [DOI] [PubMed] [Google Scholar]
- 28. Wills-Karp M., Ewart S. L. (2004) Nat. Rev. Genet. 5, 376–387 [DOI] [PubMed] [Google Scholar]
- 29. Cookson W. (2004) Nat. Rev. Immunol. 4, 978–988 [DOI] [PubMed] [Google Scholar]
- 30. Guerra S., Martinez F. D. (2008) Annu. Rev. Med. 59, 327–341 [DOI] [PubMed] [Google Scholar]
- 31. Bossé Y., Hudson T. J. (2007) Annu. Rev. Med. 58, 171–184 [DOI] [PubMed] [Google Scholar]
- 32. Vercelli D. (2010) J. Allergy Clin. Immunol. 125, 347–348 [DOI] [PubMed] [Google Scholar]
- 33. Rothman P. B. (2010) Trans. Am. Clin. Climatol. Assoc. 121, 156–171 [PMC free article] [PubMed] [Google Scholar]
- 34. Sibbald B., Horn M. E., Brain E. A., Gregg I. (1980) Thorax 35, 671–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Taylor B, Broom B. C. (1981) Ann. Allergy 47, 197–199 [PubMed] [Google Scholar]
- 36. Ono S. J. (2000) Annu. Rev. Immunol. 18, 347–366 [DOI] [PubMed] [Google Scholar]
- 37. Agrawal D. K., Shao Z. (2010) Curr. Allergy Asthma Rep. 10, 39–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. McIntire J. J., Umetsu S. E., Akbari O., Potter M., Kuchroo V. K., Barsh G. S., Freeman G. J., Umetsu D. T., DeKruyff R. H. (2001) Nat. Immunol. 2, 1109–1116 [DOI] [PubMed] [Google Scholar]
- 39. McIntire J. J., Umetsu D. T., DeKruyff R. H. (2004) Springer Semin. Immunopathol. 25, 335–348 [DOI] [PubMed] [Google Scholar]
- 40. Van Eerdewegh P., Little R. D., Dupuis J., Del Mastro R. G., Falls K., Simon J., Torrey D., Pandit S., McKenny J., Braunschweiger K., Walsh A., Liu Z., Hayward B., Folz C., Manning S. P., Bawa A., Saracino L., Thackston M., Benchekroun Y., Capparell N., Wang M., Adair R., Feng Y., Dubois J., FitzGerald M. G., Huang H., Gibson R., Allen K. M., Pedan A., Danzig M. R., Umland S. P., Egan R. W., Cuss F. M., Rorke S., Clough J. B., Holloway J. W., Holgate S. T., Keith T. P. (2002) Nature 418, 426–430 [DOI] [PubMed] [Google Scholar]
- 41. Wu H., Romieu I., Shi M., Hancock D. B., Li H., Sienra-Monge J. J., Chiu G. Y., Xu H., del Rio-Navarro B. E., London S. J. (2010) J. Allergy Clin. Immunol. 125, 321–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mathias R. A., Grant A. V., Rafaels N., Hand T., Gao L., Vergara C., Tsai Y. J., Yang M., Campbell M., Foster C., Gao P., Togias A., Hansel N. N., Diette G., Adkinson N. F., Liu M. C., Faruque M., Dunston G. M., Watson H. R., Bracken M. B., Hoh J., Maul P., Maul T., Jedlicka A. E., Murray T., Hetmanski J. B., Ashworth R., Ongaco C. M., Hetrick K. N., Doheny K. F., Pugh E. W., Rotimi C. N., Ford J., Eng C., Burchard E. G., Sleiman P. M., Hakonarson H., Forno E., Raby B. A., Weiss S. T., Scott A. F., Kabesch M., Liang L., Abecasis G., Moffatt M. F., Cookson W. O., Ruczinski I., Beaty T. H., Barnes K. C. (2010) J. Allergy Clin. Immunol. 125, 336–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li X., Howard T. D., Zheng S. L., Haselkorn T., Peters S. P., Meyers D. A., Bleecker E. R. (2010) J. Allergy Clin. Immunol. 125, 328–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kay A. B. (2001) N. Engl. J. Med. 344, 30–37 [DOI] [PubMed] [Google Scholar]
- 45. Finkelman F. D., Wills-Karp M. (2008) J. Allergy Clin. Immunol. 121, 603–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Holgate S. T., Roberts G., Arshad H. S., Howarth P. H., Davies D. E. (2009) Proc. Am. Thorac. Soc. 6, 655–659 [DOI] [PubMed] [Google Scholar]
- 47. Zou J., Young S., Zhu F., Gheyas F., Skeans S., Wan Y., Wang L., Ding W., Billah M., McClanahan T., Coffman R. L., Egan R., Umland S. (2002) Genome Biol. 2002, 3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Finkelman F. D., Boyce J. A., Vercelli D., Rothenberg M. E. (2010) J. Allergy Clin. Immunol. 125, 312–318 [DOI] [PubMed] [Google Scholar]
- 49. Sandler N. G., Mentink-Kane M. M., Cheever A. W., Wynn T. A. (2003) J. Immunol. 171, 3655–3667 [DOI] [PubMed] [Google Scholar]
- 50. Chtanova T., Kemp R. A., Sutherland A. P., Ronchese F., Mackay C. R. (2001) J. Immunol. 167, 3057–3063 [DOI] [PubMed] [Google Scholar]
- 51. Schmidt-Weber C. B. (2006) Chem. Immunol. Allergy 91, 188–194 [DOI] [PubMed] [Google Scholar]
- 52. Rogge L., Bianchi E., Biffi M., Bono E., Chang S. Y., Alexander H., Santini C., Ferrari G., Sinigaglia L., Seiler M., Neeb M., Mous J., Sinigaglia F., Certa U. (2000) Nat. Genet. 25, 96–101 [DOI] [PubMed] [Google Scholar]
- 53. Wohlfahrt J. G., Kunzmann S., Menz G., Kneist W., Akdis C. A., Blaser K., Schmidt-Weber C. B. (2003) Int. Arch. Allergy Immunol. 131, 272–282 [DOI] [PubMed] [Google Scholar]
- 54. Wohlfahrt J. G., Karagiannidis C., Kunzmann S., Epstein M. M., Kempf W., Blaser K., Schmidt-Weber C. B. (2004) J. Immunol. 172, 843–850 [DOI] [PubMed] [Google Scholar]
- 55. Herman A. E., Freeman G. J., Mathis D., Benoist C. (2004) J. Exp. Med. 199, 1479–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Brenner V., Lindauer K., Parkar A., Fordham J., Hayes I., Stow M., Gama R., Pollock K., Jupp R. (2001) J. Immunol. Methods 250, 15–28 [DOI] [PubMed] [Google Scholar]
- 57. Saito H., Nakajima T., Matsumoto K. (2001) Int. Arch. Allergy Immunol. 125, 1–8 [DOI] [PubMed] [Google Scholar]
- 58. Ryan C. A., Gildea L. A., Hulette B. C., Dearman R. J., Kimber I., Gerberick G. F. (2004) Toxicol. Lett. 150, 301–316 [DOI] [PubMed] [Google Scholar]
- 59. Beier H. M. (1968) Biochim. Biophys. Acta 160, 289–291 [DOI] [PubMed] [Google Scholar]
- 60. Singh G., Katyal S. L. (2000) Ann. N.Y. Acad. Sci. 923, 43–58 [DOI] [PubMed] [Google Scholar]
- 61. Mukherjee A. B., Zhang Z., Chilton B. S. (2007) Endocr. Rev. 28, 707–725 [DOI] [PubMed] [Google Scholar]
- 62. Zhang Z., Kundu G. C., Yuan C. J., Ward J. M., Lee E. J., DeMayo F., Westphal H., Mukherjee A. B. (1997) Science 276, 1408–1412 [DOI] [PubMed] [Google Scholar]
- 63. Mandal A. K., Zhang Z., Ray R., Choi M. S., Chowdhury B., Pattabiraman N., Mukherjee A. B. (2004) J. Exp. Med. 199, 1317–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hung C. H., Chen L. C., Zhang Z., Chowdhury B., Lee W. L., Plunkett B., Chen C. H., Myers A. C., Huang S. K. (2004) J. Allergy Clin. Immunol. 114, 664–670 [DOI] [PubMed] [Google Scholar]
- 65. Johansson S., Wennergren G., Aberg N., Rudin A. (2007) (2007) J. Allergy Clin. Immunol. 120, 308–314 [DOI] [PubMed] [Google Scholar]
- 66. Silverman G. A., Bird P. I., Carrell R. W., Church F. C., Coughlin P. B., Gettins P. G., Irving J. A., Lomas D. A., Luke C. J., Moyer R. W., Pemberton P. A., Remold-O'Donnell E., Salvesen G. S., Travis J., Whisstock J. C. (2001) J. Biol. Chem. 276, 33293–33296 [DOI] [PubMed] [Google Scholar]
- 67. Ray R., Choi M., Zhang Z., Silverman G. A., Askew D., Mukherjee A. B. (2005) J. Biol. Chem. 280, 9761–9764 [DOI] [PubMed] [Google Scholar]
- 68. Ray R., Zhang Z., Lee Y. C., Gao J. L., Mukherjee A. B. (2006) FEBS Lett. 580, 6022–6026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Seki Y., Inoue H., Nagata N., Hayashi K., Fukuyama S., Matsumoto K., Komine O., Hamano S., Himeno K., Inagaki-Ohara K., Cacalano N., O'Garra A., Oshida T., Saito H., Johnston J. A., Yoshimura A., Kubo M. (2003) Nat. Med. 9, 1047–1054 [DOI] [PubMed] [Google Scholar]
- 70. Encinas J. A., Janssen E. M., Weiner D. B., Calarota S. A., Nieto D., Moll T., Carlo D. J., Moss R. B. (2005) J. Allergy Clin. Immunol. 116, 1343–1349 [DOI] [PubMed] [Google Scholar]
- 71. Sizing I. D., Bailly V., McCoon P., Chang W., Rao S., Pablo L., Rennard R., Walsh M., Li Z., Zafari M., Dobles M., Tarilonte L., Miklasz S., Majeau G., Godbout K., Scott M. L., Rennert P. D. (2007) J. Immunol. 178, 2249–2261 [DOI] [PubMed] [Google Scholar]
- 72. Fukushima A., Sumi T., Fukuda K., Kumagai N., Nishida T., Akiba H., Okumura K., Yagita H., Ueno H. (2007) Biochem. Biophys. Res. Commun. 353, 211–216 [DOI] [PubMed] [Google Scholar]
- 73. Feng B. S., Chen X., He S. H., Zheng P. Y., Foster J., Xing Z., Bienenstock J., Yang P. C. (2008) J. Allergy Clin. Immunol. 122, 55–61 [DOI] [PubMed] [Google Scholar]
- 74. Sonar S. S., Hsu Y. M., Conrad M. L., Majeau G. R., Kilic A., Garber E., Gao Y., Nwankwo C., Willer G., Dudda J. C., Kim H., Bailly V., Pagenstecher A., Rennert P. D., Renz H. (2010) J. Clin. Invest. 120, 2767–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lane P. W. (1972) J. Hered. 63, 135–140 [DOI] [PubMed] [Google Scholar]
- 76. Fallon P. G., Sasaki T., Sandilands A., Campbell L. E., Saunders S. P., Mangan N. E., Callanan J. J., Kawasaki H., Shiohama A., Kubo A., Sundberg J. P., Presland R. B., Fleckman P., Shimizu N., Kudoh J., Irvine A. D., Amagai M., McLean W. H. (2009) Nat. Genet. 41, 602–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Smith F. J., Irvine A. D., Terron-Kwiatkowski A., Sandilands A., Campbell L. E., Zhao Y., Liao H., Evans A. T., Goudie D. R., Lewis-Jones S., Arseculeratne G., Munro C. S., Sergeant A., O'Regan G., Bale S. J., Compton J. G., DiGiovanna J. J., Presland R. B., Fleckman P., McLean W. H. (2006) Nat. Genet. 38, 337–342 [DOI] [PubMed] [Google Scholar]
- 78. Palmer C. N., Irvine A. D., Terron-Kwiatkowski A., Zhao Y., Liao H., Lee S. P., Goudie D. R., Sandilands A., Campbell L. E., Smith F. J., O'Regan G. M., Watson R. M., Cecil J. E., Bale S. J., Compton J. G., DiGiovanna J. J., Fleckman P., Lewis-Jones S., Arseculeratne G., Sergeant A., Munro C. S., El Houate B., McElreavey K., Halkjaer L. B., Bisgaard H., Mukhopadhyay S., McLean W. H. (2006) Nat. Genet. 38, 441–446 [DOI] [PubMed] [Google Scholar]
- 79. Baurecht H., Irvine A. D., Novak N., Illig T., Bühler B., Ring J., Wagenpfeil S., Weidinger S. (2007) J. Allergy Clin. Immunol. 120, 1406–1412 [DOI] [PubMed] [Google Scholar]
- 80. O'Regan G. M., Sandilands A., McLean W. H., Irvine A. D. (2008) J. Allergy Clin. Immunol. 122, 689–693 [DOI] [PubMed] [Google Scholar]
- 81. Vercelli D. (2009) Nat. Genet. 41, 512–513 [DOI] [PubMed] [Google Scholar]
- 82. Mapp C. E. (2005) Curr. Opin. Allergy Clin. Immunol. 5, 113–118 [DOI] [PubMed] [Google Scholar]
- 83. Cookson W. (1999) Nature 402, B5–B11 [DOI] [PubMed] [Google Scholar]
- 84. Marks G. B. (2006) Clin. Exp. Pharmacol. Physiol. 33, 285–289 [DOI] [PubMed] [Google Scholar]
- 85. Yang I. A., Holloway J. W. (2007) Clin. Exp. Allergy 37, 1264–1266 [DOI] [PubMed] [Google Scholar]
- 86. Miller R. L., Ho S. M. (2008) Am. J. Respir. Crit. Care Med. 177, 567–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Smit L. A., Siroux V., Bouzigon E., Oryszczyn M. P., Lathrop M., Demenais F., Kauffmann F. (2009) Am. J. Respir. Crit. Care Med. 179, 363–368 [DOI] [PubMed] [Google Scholar]
- 88. Sharma S., Raby B. A., Hunninghake G. M., Soto-Quirós M., Avila L., Murphy A. J., Lasky-Su J., Klanderman B. J., Sylvia J. S., Weiss S. T., Celedón J. C. (2009) Am. J. Respir. Crit. Care Med. 179, 356–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Hunninghake G. M., Soto-Quirós M. E., Lasky-Su J., Avila L., Ly N. P., Liang C., Klanderman B. J., Raby B. A., Gold D. R., Weiss S. T., Celedón J. C. (2008) J. Allergy Clin. Immunol. 122, 93–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Kim Y., Park C. S., Shin H. D., Choi J. W., Cheong H. S., Park B. L., Choi Y. H., Jang A. S., Park S. W., Lee Y. M., Lee E. J., Park S. G., Lee J. Y., Lee J. K., Han B. G., Oh B., Kimm K. (2007) Genes Immun. 8, 369–378 [DOI] [PubMed] [Google Scholar]
- 91. Yang I. A., Fong K. M., Zimmerman P. V., Holgate S. T., Holloway J. W. (2008) Thorax 63, 555–563 [DOI] [PubMed] [Google Scholar]
- 92. Wu H., Romieu I., Sienra-Monge J. J., del Rio-Navarro B. E., Anderson D. M., Dunn E. W., Steiner L. L., Lara-Sanchez Idel C., London S. J. (2007) Environ. Health Perspect. 115, 616–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Bouzigon E., Corda E., Aschard H., Dizier M. H., Boland A., Bousquet J., Chateigner N., Gormand F., Just J., Le Moual N., Scheinmann P., Siroux V., Vervloet D., Zelenika D., Pin I., Kauffmann F., Lathrop M., Demenais F. (2008) N. Engl. J. Med. 359, 1985–1994 [DOI] [PubMed] [Google Scholar]
- 94. Schwartz D. A. (2010) Proc. Am. Thorac. Soc. 7, 123–125 [DOI] [PubMed] [Google Scholar]