Abstract
Recently, we identified extracellular ubiquitin as an endogenous CXC chemokine receptor (CXCR) 4 agonist. However, the receptor selectivity and molecular basis of the CXCR4 agonist activity of ubiquitin are unknown, and functional consequences of CXCR4 activation with ubiquitin are poorly defined. Here, we provide evidence that ubiquitin and the cognate CXCR4 ligand stromal cell-derived factor (SDF)-1α do not share CXCR7 as a receptor. We further demonstrate that ubiquitin does not utilize the typical two-site binding mechanism of chemokine-receptor interactions, in which the receptor N terminus is important for ligand binding. CXCR4 activation with ubiquitin and SDF-1α lead to similar Gαi-responses and to a comparable magnitude of phosphorylation of ERK-1/2, p90 ribosomal S6 kinase-l and Akt, although phosphorylations occur more transiently after activation with ubiquitin. Despite the similarity of signal transduction events after activation of CXCR4 with both ligands, ubiquitin possesses weaker chemotactic activity than SDF-lα in cell migration assays and does not interfere with productive entry of HIV-1 into P4.R5 multinuclear activation of galactosidase indicator cells. Unlike SDF-1α, ubiquitin lacks interactions with an N-terminal CXCR4 peptide in NMR spectroscopy experiments. Binding and signaling studies in the presence of antibodies against the N terminus and extracellular loops 2/3 of CXCR4 confirm that the ubiquitin CXCR4 interaction is independent of the N-terminal receptor domain, whereas blockade of extracellular loops 2/3 prevents receptor binding and activation. Our findings define ubiquitin as a CXCR4 agonist, which does not interfere with productive cellular entry of HIV-1, and provide new mechanistic insights into interactions between CXCR4 and its natural ligands.
Keywords: Chemokines, G Protein-coupled Receptors (GPCR), HIV, Inflammation, Ubiquitin
Introduction
Ubiquitin is a post-translational protein modifier in all eukaryotic cells (1). Besides important intracellular roles, ubiquitin functions as an endogenous immune modulator with anti-inflammatory properties when it is released into the extracellular space (2, 3). In addition, clinical association studies suggest that systemic ubiquitin release after injury is protective and correlates inversely with the degree of organ dysfunction (4). As administration of exogenous ubiquitin has been shown to reduce inflammation and tissue injury in various disease models (5–11), identification of the mechanism of action of extracellular ubiquitin provides the opportunity to develop novel therapeutic strategies.
Previously, we demonstrated that extracellular ubiquitin can be taken up into monocytes and quantification of its uptake kinetics suggested a receptor-mediated process (12). Subsequently, we identified a novel and unforeseen biological function of ubiquitin and showed that it is a natural agonist of the G protein coupled receptor (GPCR)2 CXC chemokine receptor (CXCR) 4 (fusin, CD184) (13, 14).
CXCR4 and its cognate ligand stromal cell-derived factor-1α (SDF-1α) (chemokine (C-X-C motif) ligand 12) are known to be important for normal development and hematopoiesis, and play pleiotropic roles in the immune system and during tissue repair processes (15–20). Furthermore, the SDF-1α/CXCR4 axis gained particular attention as a drug target through its involvement in cancer metastases and HIV infection (21–23). However, the molecular mechanisms underlying CXCR4 agonist activity of ubiquitin are unknown, it remains to be determined whether SDF-1α and ubiquitin show functional selectivity on CXCR4 and whether both ligands also share CXCR7 as a common receptor (24).
Here, we provide evidence that ubiquitin is a CXCR4, but not a CXCR7 ligand. Comparison of CXCR4-mediated chemotaxis and intracellular signaling events after stimulation with ubiquitin and SDF-1α and infection studies with X4 tropic HIV-1, which utilize CXCR4 as a co-receptor, demonstrate that ubiquitin functions as a CXCR4 agonist that does not interfere with HIV-1 infectivity. NMR spectroscopy with the extracellular N-terminal domain of CXCR4, receptor binding, and signaling studies in the presence of antibodies directed against the N terminus and extracellular loops (ECL) 2/3 of CXCR4 demonstrate distinct and ligand-specific CXCR4 interactions of ubiquitin and SDF-1α. These studies suggest that the ubiquitin interaction with CXCR4 is independent of the N-terminal domain of CXCR4, thus suggesting a novel binding mechanism of a natural chemokine receptor ligand.
EXPERIMENTAL PROCEDURES
Proteins, Peptides, and Reagents
Ubiquitin, BSA, zidovudine, AMD3100, and forskolin were obtained from Sigma. N-terminal fluorescein-labeled ubiquitin (FITC-ubiquitin) was purchased from Boston Biochem. Recombinant human SDF-1α was obtained from Peprotech. For NMR spectroscopy studies SDF-1α and the [U-15N]CXCR4-(1–38) peptide were expressed in Escherichia coli and purified as previously described (25).
HA-tagged CXCR4 and CXCR7 Transfections
DNA encoding HA-tagged CXCR4 was as previously described (26). DNA encoding CXCR7 (GenBankTM accession number NP_064707) was purchased from Open Biosystems. CXCR7 was amplified by PCR using the following primers containing XhoI and XbaI restriction enzyme sites, respectively, forward, 5′-ATATCTCGAGTGGATCTGCATCTCTTCGACTAC; reverse, 5′-ATATTCTAGATCATTTGGTGCTCTGCTCCAAG. The PCR product was digested with XhoI and XbaI and ligated into HA-pcDNA3 cassette vector in which the CXCR4 fragment was removed and replaced with CXCR7. The sequence of the clone was verified by dideoxysequencing. DNA encoding each tagged G protein-coupled receptor and empty vector (pcDNA3) was transiently transfected into HEK 293 cells grown on 10-cm tissue culture dishes using TransIT-LT1 transfection reagent (Mirus Bio), according to the manufacturer's recommendation. Forty-eight hours later, cells were harvested and used for Western blotting, flow cytometry, and ubiquitin binding assays.
HIV-1 Production and Infection Assays
Laboratory adapted viruses (R9 (X4); R9BaL (R5)) and pseudotyped virions (HXB2 (X4), JRFL (R5)) were as described (27, 28). Viral stocks were generated from HEK293T cells via polyethylenimine (molecular weight 25,000; Polysciences) using a polyethylenimine:DNA ratio of 2:1, as described (28). Pseudotyped virions were generated by cotransfecting 12 μg of R7Δenv GFP with 8 μg of HXB2 or JRFL envelope expression plasmid. 48 h after transfection, virus was harvested and passed through a 0.45-μm filter. Virus infectivity was assessed as described (29). P4.R5 multinuclear activation of galactosidase indicator (MAGI) cells were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, National Institutes of Health, from Dr. Nathaniel Landau. One hour before infection, P4.R5 MAGI cells were treated with the indicated concentration of AMD3100, ubiquitin, or SDF-1α. Cells were then infected with serial dilutions of viral supernatant, beginning at a multiplicity of infection of ∼0.5. Eighteen hours post-infection, medium was replaced with medium containing 200 μm zidovudine. Thirty-six hours post-infection cells were assayed for β-galactosidase expression by monitoring the cleavage of the colorimetric β-gal substrate o-nitrophenyl β-d-galactopyranoside at 405 nm.
Ubiquitin Binding Assays
Ubiquitin binding assays were performed with HEK293, P4.R5 MAGI, and THP-1 cells, as described (13). In brief, cells were washed with ice-cold PBS and 105 cells were suspended in 100 μl of cold (4 °C) PBS, 1% BSA, 0.01% sodium azide. FITC-ubiquitin was added and incubated for 1 min at 4 °C. Cells were washed twice and the fluorescence intensities were measured (λexcitation/emission: 485/528 nm). Nonspecific binding was assessed as binding of FITC-ubiquitin in the presence of 300 μm native ubiquitin. Where applicable, cells were preincubated for 60 min at 37 °C with 0.2 μg/ml of anti-CXCR4 N-terminal amino acids 1–14 (anti-CXCR4-(1–14), Abcam), anti-CXCR4 amino acids 176–293 (anti-CXCR4-(176–293), Santa Cruz), or anti-rabbit IgG (Sigma).
Calcium Assay
Intracellular calcium was measured using the Fluo-4 NW Calcium Assay Kit (Molecular Probes), as described (13). Where applicable, AMD3100 (10 μm) was added to the cells 5 min before stimulation with ubiquitin or SDF-1α.
cAMP Assay
Quantitative determination of cAMP levels was performed in forskolin-treated cells (5 μm, 10 min, 37 °C) using the cAMP complete enzyme immunoassay kit (Assay Designs), acetylated format, as described (13). Where applicable, AMD3100 (10 μm) was added to the cell cultures 5 min before stimulation with ubiquitin or SDF-1α, and anti-CXCR4-(1–14), anti-CXCR4-(176–293), or anti-rabbit IgG (0.2 μg/ml) were added 60 min before cells were stimulated with ubiquitin or SDF-1α.
Protein Kinase Phosphorylation Array
Screening of the phosphorylation status of various protein kinases after ubiquitin and SDF-1α stimulation was performed in THP-1 cells using a human phospho MAPK antibody array (R&D Systems). Cells were stimulated with 1 μm ubiquitin or SDF-1α for 10 min at 37 °C. Unstimulated cells served as controls in parallel experiments. The array was performed according to the manufacturer's instructions. In brief, 107 THP-1 cells were lysed in 1 ml of lysis buffer (R&D Systems) on ice for 30 min, centrifuged (14,000 × g, 5 min), and the supernatant (=lysate) was collected. 250 μl of the cell lysate was incubated with a pre-wet array membrane for 15 h at 4 °C. After three washing steps at room temperature, array membranes were incubated with the diluted antibody mixture (2 h at room temperature), washed, and incubated with strepavidin-HRP solution for 30 min at room temperature. After washing the array membranes, chemiluminescence substrate (SuperSignal West Dura, Thermo Scientific) was added and the chemiluminescence signal was detected (Chemi Doc XRS with Quantity One version 4.5.2 software (Bio-Rad Laboratories)). Chemiluminescence signals were analyzed with the ImageQuant TL software (Amersham Biosciences).
Western Blots
Western blotting was performed as described (13, 30). Mouse monoclonal anti-HA (Covance) in combination with anti-mouse IgG HRP-linked whole antibody (Amersham Biosciences) were used for detection of HA-tagged CXCR4 and CXCR7 in THP-1 lysates after transfection. Anti-phospho p90 ribosomal S6 kinase (RSK1) (Ser-380) rabbit IgG, anti-phospho-ERK1 (Thr-202/Tyr-204)/ERK2 (Thr-185/Tyr-187) rabbit-IgG, anti-phospho-Akt pan (Ser-437) rabbit IgG (all from R&D Systems) and mouse anti-GAPDH (Applied Biosystems) were used in combination with anti-rabbit or mouse IgG HRP-linked whole antibody (Amersham Biosciences).
Chemotaxis Assays
Cell migration was assessed using the ChemoTx 96-well cell migration system (30-μl well plate, 8 μm filter pore size; Neuroprobe). The bottom wells were filled with 30 μl of test solutions (10 μm AMD3100, ubiquitin, and SDF-1α in PBS). The microplate was then covered with the ChemoTX filter, 25 μl of cell suspension containing 5 × 104 THP-1 cells in PBS were pipetted onto the filter over each well and incubated for 2 h at 37 °C, 5% CO2. After incubation, cells that transmigrated through the filter were stained on the lower filter surface with Accustain Wright-Giemsa stain (Sigma). The average number of cells on the lower filter surface was determined by counting the number of cells in three random non-overlapping high power (×400) fields by light microscopy. The average cell count on the lower filter surface in the presence of PBS in the lower compartment (=control) was 3.2 ± 2.5 per high power field. The chemotactic index was calculated as the ratio of cells that transmigrated through the filter in the presence versus absence (=PBS/control) of the test solutions.
FACS Analyses
FACS was used to analyze cell surface expression of HA-tagged CXCR4 and CXCR7, to quantify CXCR4 cell surface expression, assess FITC-ubiquitin binding, and assess the interactions between ubiquitin, SDF-1α, anti-CXCR4-(1–14), and anti-CXCR4-(176–293). For the analyses of HA-tagged CXCR4/7 expression, cells were labeled with monoclonal mouse anti-HA in combination with anti-mouse Alexa Fluor 488 goat IgG (Invitrogen). Rabbit IgG (R&D Systems) in combination with FITC-conjugated anti-rabbit goat IgG (Abcam) was used as a negative control. For the quantification of CXCR4 expression, cells were labeled with anti-human CXCR4 FITC-conjugated IgG (R&D Systems). FITC-conjugated IgG2A (R&D Systems) was used as a negative control under identical conditions. Binding of anti-CXCR4-(1–14) and anti-CXCR4-(176–293) was detected with FITC-conjugated anti-rabbit (Abcam). Cells were analyzed with a FACSAria flow cytometer (BD Biosciences). The fluorescence intensities of at least 105 cells were recorded and analyzed using the FloJo software (Tree Star).
NMR Spectroscopy
NMR experiments were performed on a Bruker DRX 600 equipped with a 1H/15N/13C Cryoprobe. NMR samples contained: 250 μm [U-15N]CXCR4-(1–38), 90% (v/v) H2O, 10% (v/v) D2O, 0.02% NaN3, and 25 mm deuterated MES buffer (pH 6.8). 1H and 15N chemical shift assignments for CXCR4-(1–38) were obtained from previously published spectra (25). CXCR4-(1–38) was titrated with incremental additions of SDF-1α or ubiquitin and monitored by two-dimensional 1H-15N heteronuclear single quantum coherence. Combined 1H/15N chemical shift perturbations were calculated as [(5ΔδNH)2 + (ΔδN)2]0.5, where ΔδNH and ΔδN are the changes in backbone amide 1H and 15N chemical shifts in ppm, respectively. CXCR4-(1–38) 1H-15N heteronuclear nuclear Overhauser effect values were determined in the absence and presence of 325 μm SDF-1α and ubiquitin.
Statistics
Data are expressed as mean ± S.E. of n independent experiments that were performed on different days. Differences between the various cell culture conditions were compared using Student's t test or analyses of variance with Tukey post test to control for multiple testing. A two-tailed p < 0.05 was considered significant. Data were analyzed using GraphPad Prism 5 software.
RESULTS
Receptor Selectivity of Ubiquitin
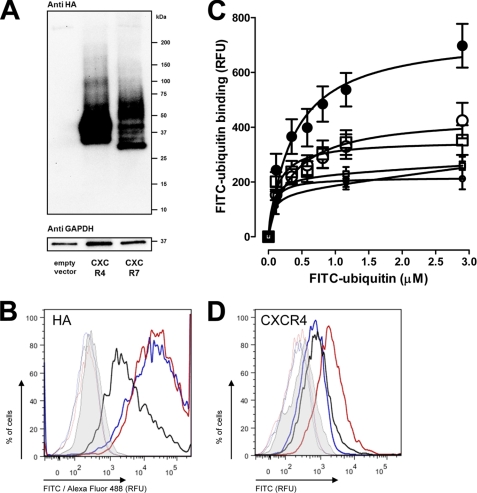
To delineate whether ubiquitin and SDF-1α also share CXCR7 as a common receptor, we transfected human HEK293 cells with HA-tagged CXCR4 and CXCR7 expression vectors. To confirm expression of the chemokine receptors after transfection, whole cell extracts were analyzed by Western blotting with anti-HA. We detected the expected pattern of bands after transfection of the cells with the receptor cDNA clones, as compared with cells that were transfected with the empty vector (Fig. 1A). The multiple bands that were detectable can be likely attributed to the heterogeneity of receptor species that are generated by differential glycosylation in the Golgi apparatus (31). The band at ∼37 kDa in cells transfected with CXCR7 may represent unprocessed receptor or a proteolytically processed fragment not observed with CXCR4. Nevertheless, the most abundant CXCR7 species, which likely represents a fully processed receptor, runs as a broad band at ∼50 kDa. This is slightly larger than the most abundant CXCR4 species, which runs as a broad band at ∼48 kDa. This difference is consistent with their predicted and modestly different molecular masses. Flow cytometry analyses then documented that HA-tagged CXCR4 and CXCR7 were expressed on the cell surface (Fig. 1B). FITC-ubiquitin binding studies showed increased specific FITC-ubiquitin binding after transfection with CXCR4 (Fig. 1C). FITC-ubiquitin binding to cells transfected with CXCR7 was indistinguishable from binding to cells transfected with the empty vector. Quantification of the specific FITC-ubiquitin binding signal showed a 3.3-fold increase in cells transfected with CXCR4 (Bmax, 578 ± 63 RFU), as compared with cells transfected with CXCR7 or empty vector (Bmax, 176 ± 18 RFU). Quantification of CXCR4 cell surface expression by flow cytometry with an anti-CXCR4 antibody (Fig. 1D) confirmed that the degree of CXCR4 overexpression is reflected by a comparable increase of the FITC-ubiquitin binding signal (mean RFU; cells transfected with CXCR4, 1747 ± 176; cells transfected with CXCR7, empty vector or not transfected, 600 ± 100; cells stained with control antibody, 200 ± 3). This demonstrates that ubiquitin binding correlates to the number of receptors expressed on the cell surface and suggests that ubiquitin does not bind to CXCR7.
FIGURE 1.
CXCR4, but not CXCR7 overexpression increases ubiquitin receptor binding. A, HA-tagged open reading frame cDNA clones of CXCR4 and CXCR7 were transfected into HEK293 cells followed by immunoblotting of whole cell lysates with anti-HA and anti-GAPDH. B, quantification of HA expression by flow cytometry after transfection as in A. Thick lines, cells labeled with mouse anti-HA/anti-mouse Alexa Fluor 488 goat IgG. Thin lines, control; cells labeled with rabbit IgG/anti-rabbit FITC goat IgG. Gray, unstained cells. Red, cells transfected with HA-tagged CXCR4. Blue, cells transfected with HA-tagged CXCR7. Black, cells transfected with empty plasmid. C, FITC-ubiquitin binding (1 min, 4 °C) after transfection as in A. ●, CXCR4. □, CXCR7. ○, empty vector. ●, nonspecific binding (NSB)-CXCR4. □, NSB-CXCR7. ●, NSB-empty vector; n = 4. D, quantification of CXCR4 expression by flow cytometry after transfection as in A. Thick lines, cells labeled with anti-human CXCR4 FITC-conjugated IgG. Thin lines, control, cells labeled with FITC-conjugated IgG2A. Gray, unstained cells. Red, cells transfected with HA-tagged CXCR4. Blue, cells transfected with HA-tagged CXCR7. Black, cells transfected with empty plasmid.
CXCR4-induced Protein Kinase Phosphorylation
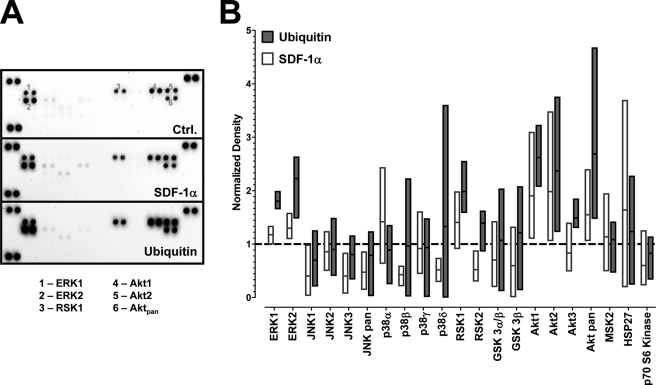
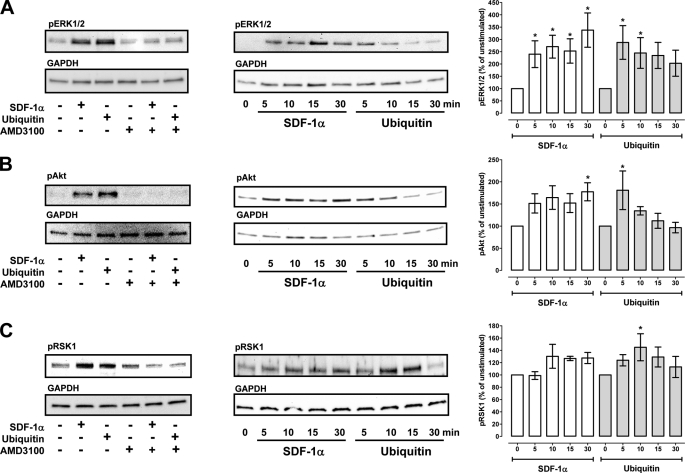
We have shown previously that CXCR4 activation with both agonists results in comparable effects on intracellular Ca2+ flux and cAMP levels in THP-1 cells (13). To further compare subsequent signal transduction events after ubiquitin and SDF-1α activation of CXCR4, we performed a screening of the phosphorylation status of numerous protein kinases and functionally related molecules after CXCR4 stimulation of THP-1 cells utilizing a membrane-based proteome array. A typical image of the array membranes and densitometric quantification of the spot densities from four independent experiments are shown in Fig. 2. Although there was considerable variation among the individual experiments for some of the protein kinases, ubiquitin and/or SDF-1α stimulation (1 μm, 10 min) consistently increased the density of the spots corresponding to phosphorylated ERK1/2, RSK1, and Akt, as compared with untreated cells. Therefore, we selected these protein kinases to determine the effects of both CXCR4 agonists on their phosphorylation status by Western blotting (Fig. 3). Pretreatment of the cells with the selective CXCR4 antagonist AMD3100 prevented the increase in phosphorylation of ERK1/2, Akt, and RSK1 after ubiquitin and SDF-1α stimulation and confirmed that these effects are mediated through CXCR4 (Fig. 3, left panels). Measurements of the time progression of the phosphorylation status of these protein kinases then showed that CXCR4 activation with both ligands results in a comparable increase in phosphorylation. With ubiquitin activation, increased phosphorylation occurred transiently and declined within 30 min. In contrast, increased phosphorylation was sustained for 30 min with SDF-1α activation (Fig. 3, center and right panels), suggesting differential signaling properties of both ligands.
FIGURE 2.
Phosphorylation of mitogen-activated protein kinases after stimulation with SDF-1α and ubiquitin. THP-1 cells were stimulated with 0 or 1 μm ubiquitin or SDF-1α for 10 min at 37 °C. Whole cell lysates were probed for protein kinase phosphorylations utilizing a proteome array. A, proteome array membranes showing the spot densities in untreated (ctrl.), ubiquitin-treated and SDF-1α-treated cells. The numbers on the array membrane correspond to the spot positions for phosphorylated ERK1 (1), ERK2 (2), RSK1 (3), Akt1 (4), Akt2 (5), and Akt pan (6). B, densitometric quantification of the spot densities after treatment as in A, n = 4. Spot densities are given as normalized pixel densities (1 = unstimulated cells, dashed line). The bars (white, SDF-1α treatment; gray, ubiquitin treatment) extend from the minimum to the maximum, the horizontal line shows the mean.
FIGURE 3.
CXCR4-induced protein kinase phosphorylation. Western blot analyses of MAPK phosphorylation after stimulation of THP-1 cells with ubiquitin and SDF-1α. A, phospho-ERK1/2. B, phospho-Akt. C, phospho-RSK1. Left panels, cells were pretreated with or without AMD3100 and stimulated with 1 μm ubiquitin or SDF-1α for 10 min at 37 °C. Center panels, time course of MAPK phosphorylations after stimulation of cells with 1 μm ubiquitin or SDF-1α. Right panels, quantification of the chemiluminescence signals after cell stimulation as in B. White bars, SDF-1α stimulation. Gray bars, ubiquitin stimulation, n = 5–10. *, p < 0.05 versus unstimulated cells.
CXCR4-induced Chemotaxis
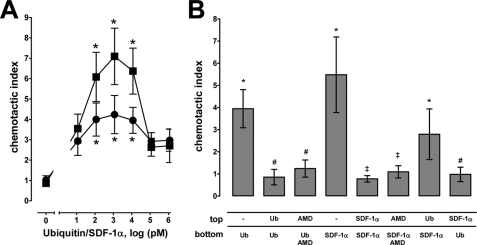
The regulation of cell trafficking is considered as a key function of the SDF-1α/CXCR4 axis. Therefore, we used the chemotactic response of THP-1 cells as a functionally relevant read-out for CXCR4 agonist activity of ubiquitin. As shown in Fig. 4A, THP-1 cells migrated dose dependently toward ubiquitin in filter migration assays. When compared with SDF-1α, cell migration toward ubiquitin was detectable at similar concentrations. However, the chemotactic index at concentrations that induced maximal cell migration was lower with ubiquitin (chemotactic index, 4.2 ± 0.9 with ubiquitin versus 7.1 ± 1.4 with SDF-1α; p < 0.05). Induction of cell migration by ubiquitin and SDF-1α required a concentration gradient and AMD3100 prevented cell migration (Fig. 4B). This suggests that both molecules possess chemotactic activity, which is mediated through CXCR4. The finding that a SDF-1α concentration gradient induced a chemotactic response in the presence of ubiquitin, whereas a ubiquitin concentration gradient did not produce chemotactic movements in the presence of SDF-1α (Fig. 4B), is consistent with the weaker chemotactic activity of ubiquitin that we determined in the dose-response experiments.
FIGURE 4.
CXCR4-mediated chemotaxis. A, dose-dependent migration of THP-1 cells toward a ubiquitin (●) and SDF-1α (■) gradient; n = 7. *, p < 0.05 versus cells in the presence of PBS in the lower compartment. B, migration of THP-1 cells in the presence or absence of ubiquitin, SDF-1α, or AMD3100 (AMD, 10 μm) in the upper (top) and lower (bottom) compartment, as indicated in the graph (n = 4). Ubiquitin and SDF-1α were used at concentrations (two experiments with 1 nm, two experiments with 10 nm) that showed maximal chemotactic activity, as determined in A. *, p < 0.05 versus cells in the presence of PBS in the upper and lower compartment. #, p < 0.05 versus cells in the presence of PBS in the upper compartment and ubiquitin in the lower compartment. ‡, p < 0.05 versus cells in the presence of PBS in the upper compartment and SDF-1α in the lower compartment.
Ubiquitin Is a CXCR4 Agonist That Does Not Affect HIV-1 Infection
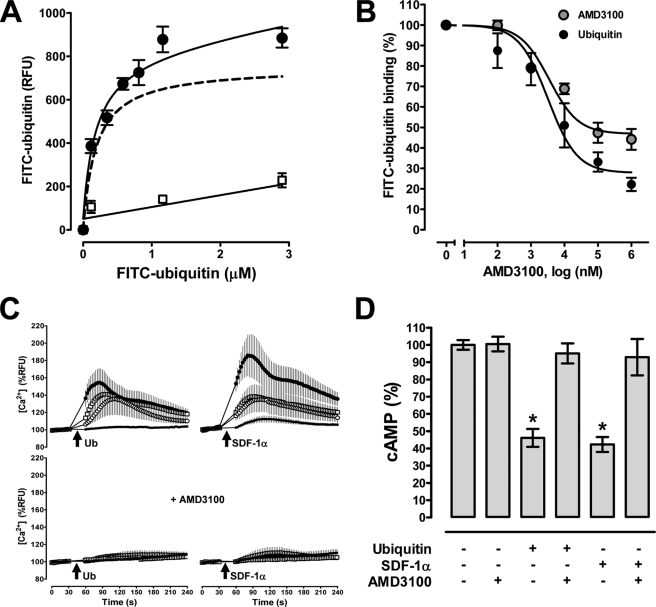
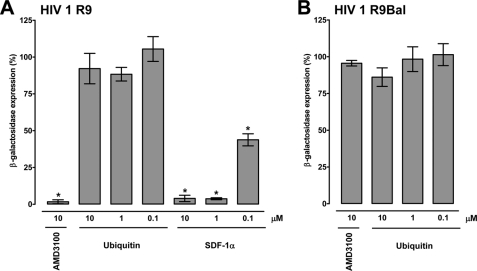
SDF-1α and AMD3100 have been shown to reduce X4 tropic HIV-1 entry into the cell (32–35). Therefore, we tested whether ubiquitin also reduces infectivity of X4 tropic HIV-1 in vitro utilizing the MAGI (29). In this assay, P4.R5 MAGI cells (HeLa CD4-LTR/β-gal indicator cells), which express CXCR4, C-C chemokine receptor type 5, and CD4 on the cell surface and are stably transformed with β-galctosidase under the control of the HIV-1 long terminal repeat promoter, are employed to assess viral infectivity. To confirm that ubiquitin functions as a CXCR4 agonist in P4.R5 MAGI cells, we first determined the CXCR4 binding properties of ubiquitin and confirmed its biological activity (Fig. 5, A–D). Saturation binding experiments with FITC-ubiquitin at 4 °C documented typical receptor binding characteristics (Fig. 5A). The determined Kd from saturation binding experiments was 156 ± 27 nm. Consistent with ubiquitin binding to CXCR4, we detected that native ubiquitin and the CXCR4 antagonist AMD3100 competed with FITC-ubiquitin for receptor binding (Fig. 5B). Next, we compared CXCR4 agonist activity of SDF-1α and ubiquitin using intracellular Ca2+ flux and cellular cAMP levels as read-outs for typical Gαi responses. As shown in Fig. 5C, stimulation of P4.R5 MAGI cells with ubiquitin and SDF-1α dose-dependently promoted intracellular Ca2+ fluxes. Although SDF-1α induced a stronger Ca2+ response than ubiquitin at a supraphysiological concentration (1.16 μm), the abilities of ubiquitin and SDF-1α to promote Ca2+ fluxes were comparable at lower concentrations. Pretreatment of P4.R5 cells with AMD3100 inhibited ubiquitin and SDF-1α promoted Ca2+ fluxes (Fig. 5C, bottom panels). Similarly, ubiquitin and SDF-1α stimulation reduced cellular cAMP levels in forskolin-treated P4.R5 MAGI cells and this effect could also be inhibited with AMD3100 (Fig. 5D). However, when P4.R5 MAGI cells were infected with X4 tropic HIV-1 R9, AMD3100 and SDF-1α reduced HIV-1 infectivity, whereas ubiquitin did not (Fig. 6A). AMD3100 and ubiquitin did not affect infectivity of R5 tropic (C-C chemokine receptor type 5) using HIV-1 R9Bal (control, Fig. 6B). Infectivity assays with pseudotyped X4 (HXB2) and R5 (JRFL) tropic virions showed identical results (not shown). These findings demonstrate that in contrast to SDF-1α, ubiquitin is a CXCR4 agonist that fails to block X4 tropic HIV-1 entry into the cell.
FIGURE 5.
Ubiquitin functions as a CXCR4 agonist in P4.R5 MAGI cells. A, FITC-ubiquitin binding (1 min, 4 °C). ●, FITC-ubiquitin; □, nonspecific binding. Dashed line, specific binding curve (=total FITC-ubiquitin binding − nonspecific binding); n = 6. B, competition binding (1 min, 4 °C) curve for unlabeled ubiquitin (n = 6, ●) and AMD3100 (n = 5, ●) with 1.16 μm FITC-ubiquitin. FITC-ubiquitin binding is expressed as % of the fluorescence signal measured in the absence of unlabeled ubiquitin (=100%). C, top, ubiquitin (left panels) and SDF-1α (right panels) induced Ca2+ flux. Bottom, cells were pretreated with AMD3100 (10 μm); n = 3. Arrows indicate the time point when ubiquitin or SDF-1α was added (●, 1.16 μm; □, 116 nm; ○, 16 nm; ■, 1.6 nm). D, AMD3100 (10 μm) abolishes ubiquitin and SDF-1α (116 nm) induced reduction of cAMP levels in forskolin-stimulated cells; n = 4. Data are expressed as % of untreated cells (=100%). *, p < 0.05 versus untreated cells.
FIGURE 6.
Ubiquitin does not affect HIV-1 infection. Effects of CXCR4 ligands on X4 tropic HIV-1 R9 (A) and R5 tropic HIV-1 R9BaL (B) infection in P4.R5 MAGI cells (n = 3). One h before infection with 0.5 multiplicity of infection of virus, P4.R5 MAGI cells were treated with the indicated concentrations of CXCR4 ligands. Thirty-six hours postinfection cells were assayed for β-galactosidase expression. Experiments with lower multiplicity of infection (0.25 and 0.125) and X4 and R5 tropic pseudotyped virions showed identical results (not shown). *, p < 0.05 versus cells cultured in the absence of CXCR4 ligands.
Ubiquitin-CXCR4 Interaction
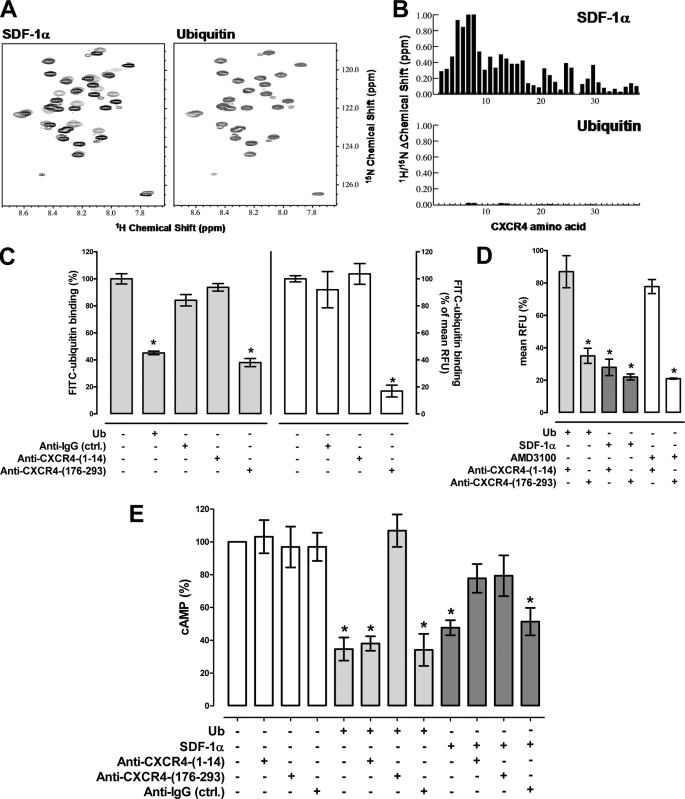
The differential effects of ubiquitin and SDF-1α on protein kinase phosphorylation and HIV-1 infectivity suggested that both ligands may function through distinct interactions on CXCR4. As NMR studies provided direct evidence for the interaction of SDF-1α with the N-terminal domain of CXCR4 (25), we assessed whether ubiquitin also interacts with the N-terminal CXCR4-(1–38) peptide using NMR spectroscopy (Fig. 7, A and B). Two-dimensional 1H-15N heteronuclear single quantum coherence NMR experiments provide atom-specific information and are sensitive to micromolar or weaker interactions. In contrast to SDF-1α, ubiquitin did not induce chemical shift changes of CXCR4-(1–38). 15N-1H heteronuclear nuclear Overhauser effect values further indicated that CXCR4-(1–38) is disordered and does not adopt a stable secondary or tertiary structure in the presence of ubiquitin (data not shown), suggesting that ubiquitin lacks a physicochemical interaction with the N-terminal peptide of CXCR4.
FIGURE 7.
The ubiquitin CXCR4 interaction is independent of the N-terminal receptor domain. A, ubiquitin does not bind CXCR4-(1–38). 15N-1H heteronuclear single quantum coherence of 250 μm [U-15N]CXCR4-(1–38) in the absence (black) and presence (gray) of 325 μm SDF-1α (left) or ubiquitin (right); the chemical shift of CXCR4-(1–38) residues change in the presence of SDF-1α, whereas they are unperturbed by ubiquitin. B, combined 1H/15N shift perturbations of SDF1α (top) and ubiquitin (bottom) plotted as a function of the CXCR4-(1–38) residue. Tyr-7 and Thr-8 were not present at the end of titration with SDF-1α due to line broadening. Shift changes for Pro-27 were not measured because it does not contain an amide proton. C, FITC-ubiquitin binding (1.16 μm) to THP-1 cells after labeling of cells with anti-CXCR4-(1–14), anti-CXCR4-(176–293), or anti-IgG. Gray bars (left y axis), RFU from ubiquitin binding assays. Open bars (right y axis), mean RFU from FACS analyses. Ub, ubiquitin, 30 μm. Data are expressed as % of the RFU after incubation with FITC-ubiquitin alone (=100%); n = 3. *, p < 0.05 versus cells incubated with FITC-ubiquitin alone. D, THP-1 cells were coincubated with each of the CXCR4 ligands (116 nm for ubiquitin (light gray bars) and SDF-1α (dark gray bars), 10 μm for AMD3100 (open bars)) and anti-CXCR4-(1–14) or anti-CXCR4-(176–293) at 4 °C. Antibody binding was detected by FACS and mean RFU (% of max) were quantified; n = 3. Data are expressed as % of the RFU after incubation with antibody alone. *, p < 0.05 versus cells after incubation with antibody alone. E, cAMP levels in forskolin (5 μm)-treated THP-1 cells 15 min after ubiquitin or SDF-1α (116 nm) stimulation in the presence or absence of anti-CXCR4-(1–14), anti-CXCR4-(176–293), or anti-IgG, n = 3. Data are expressed as % of untreated cells (=100%). White bars, cells were incubated with antibodies alone. Light gray bars, coincubations with antibodies and ubiquitin. Dark gray bars, coincubations with antibodies and SDF-1α. *, p < 0.05 versus untreated cells.
To confirm this observation, we then evaluated whether antibodies directed against the N-terminal domain (CXCR4-(1–14)) or ECL2/3 (CXCR4-(176–293)) of CXCR4 interfere with FITC-ubiquitin binding to THP-1 cells. Anti-CXCR4-(176–293) was used because ECL2/3 of CXCR4 are known to be important for binding of SDF-1α and AMD3100 (36, 37). FACS analyses documented that both antibodies bind to the cell surface of THP-1 cells (not shown). When cells were labeled with anti-CXCR4-(1–14), subsequent FITC-ubiquitin binding was comparable with unlabeled cells (Fig. 7C). In contrast, in cells labeled with anti-CXCR4-(176–293), the binding of FITC-ubiquitin was reduced by more than 60% when assessed in FITC-ubiquitin binding assays (Fig. 7C, left panel) and by more than 80% when assessed by FACS analyses (Fig. 7C, right panel). These data confirmed our observations from NMR spectroscopy experiments and further suggested that blocking ECL2/3 of CXCR4 with anti-CXCR4-(176–293) prevents FITC-ubiquitin binding. Therefore, we next performed competition binding experiments to assess whether both antibodies may also interfere with the binding of SDF-1α and AMD3100 to CXCR4. In these experiments cells were coincubated with the antibodies and ligands for 15 min at 4 °C to prevent receptor internalization during coincubation with SDF-1α and ubiquitin. Antibody binding to the cell surface was then quantified by FACS analyses using FITC-labeled secondary antibodies (Fig. 7D). Binding of anti-CXCR4-(1–14) to the cell surface of THP-1 cells was not affected by ubiquitin and reduced by SDF-1α. Binding of anti-CXCR4-(176–293) was reduced when coincubated with either ubiquitin or SDF-1α. Similar to ubiquitin, the CXCR4 antagonist AMD3100 did not interfere with the binding of anti-CXCR4-(1–14) to THP-1 cells and reduced the binding of anti-CXCR4-(176–293).
To determine whether both antibodies also interfere with CXCR4-mediated signaling upon stimulation with ubiquitin and SDF-1α, we tested whether labeling of THP-1 cells with either antibody influences effects of ubiquitin and SDF-1α on cAMP levels in forskolin-stimulated THP-1 cells (Fig. 7E). Anti-CXCR4-(1–14) did not affect reduction of intracellular cAMP levels by ubiquitin, whereas anti-CXCR4-(176–293) was able to prevent this effect. Both antibodies partially attenuated SDF-1α-induced reduction of cAMP levels in parallel experiments.
DISCUSSION
In the present study, we demonstrate that extracellular ubiquitin binds to and signals via CXCR4 through a unique binding mechanism, independent of the N-terminal domain of CXCR4. Our findings define ubiquitin as a CXCR4 agonist, which does not interfere with productive cellular entry of HIV-1. Furthermore, we provide evidence that ubiquitin and SDF-1α display distinct receptor selectivity.
Chemokine receptors belong to the G protein-coupled receptor superfamily, promote Ca2+ flux, reduce cAMP levels, and share >20% sequence identity (38). In general, chemokine receptors and their ligands are highly promiscuous, being able to bind multiple receptors/ligands (15, 38). The currently known endogenous CXCR4 ligands are SDF-1α, macrophage migration inhibitory factor, and ubiquitin (13, 15, 24, 38, 39). Although SDF-1α also binds to CXCR7 (24), macrophage migration inhibitory factor has also been reported as a ligand of CD74/invariant chain and CXCR2 (39, 40). As ubiquitin receptor binding could not be increased when CXCR7 was overexpressed on the cell surface of HEK293 cells, these findings suggest that ubiquitin and SDF-1α do not share CXCR7 as another common receptor. This implies that the natural CXCR4 ligands fulfill, in part, specific biological functions through their actions on distinct cell surface receptors.
The affinity of ubiquitin for CXCR4 that we determined from saturation binding experiments in P4.R5 cells in the present study is consistent with its affinity that we reported previously utilizing THP-1 cells and primary human monocytes (13, 14). Comparison of cellular Gαi responses in P4.R5 MAGI cells and chemotactic movements of THP-1 cells upon exposure to ubiquitin and SDF-1α, and inhibition of these effects by AMD3100, further confirmed CXCR4 agonist activity of ubiquitin (13–15).
Our finding that SDF-1α and ubiquitin displayed chemotactic activity at concentrations between 0.1 and 10 nm is in agreement with the wide range of SDF-1α concentrations that have been reported to induce chemotactic movements previously (41, 42). This range of concentrations corresponds to the affinity of SDF-1α for CXCR4 (Kd: 1.5–24 nm), which has been reported in studies with human peripheral blood monocytes, T-cells, and T-cell lines (39, 43–46). However, this range of concentrations is 10–1000-fold below the affinity of ubiquitin for CXCR4, which we determined previously in THP-1 cells (13). This suggests that occupancy of only a small fraction of receptors by ubiquitin evokes a cellular response. Such a dose-response relationship has been described for other G protein-coupled receptors, which showed half-maximal and maximal responses at receptor occupancies of 0.13 and 0.8%, respectively (47, 48). In addition, our finding corresponds to the previous observation that intramuscular injection of ubiquitin led to the accumulation of large numbers of lymphocytes without inducing cytotoxic effects in the C2C12 myoblast cell line (49), which resembles the effects of SDF-1α after subcutaneous injection (50).
Although CXCR4-mediated downstream signaling events upon SDF-1α stimulation have been previously studied in various cell types, ubiquitin-induced cell signaling events are largely unknown. Side-by-side comparison of MAPK phosphorylations after ubiquitin and SDF-1α stimulation of THP-1 cells showed that both ligands produced similar patterns in a membrane array and confirmed phosphorylation of ERK-1/2, RSK-1, and Akt in response to CXCR4 activation (51–54). The similarity of CXCR4-mediated signaling events upon activation with both ligands argues against biased agonism directed signaling, also referred to as functional selectivity, of CXCR4 (55, 56). However, given that MAPK phosphorylations occurred more transiently after activation of CXCR4 with ubiquitin and that ubiquitin induced weaker chemotactic responses than SDF-1α, SDF-1α appears to be a more efficacious CXCR4 agonist than ubiquitin.
In contrast to the subtle differences in strength of signal of CXCR4-mediated signaling events upon stimulation of cells with ubiquitin and SDF-1α and the CXCR4-mediated chemotactic activity of both ligands, HIV-1 infectivity studies demonstrated the inability of ubiquitin to interfere with the productive cellular entry of X4 tropic virus. This finding provided initial evidence that ubiquitin does not resemble all SDF-1α effects on CXCR4 and suggested that the mechanism of ubiquitin binding to CXCR4 is distinct from SDF-1α.
The interaction between chemokines and their receptors, including CXCR4 and SDF-1α, is thought to follow a two-site binding model (25, 36, 57–61). In this model, the N terminus of the chemokine receptor is important for initial binding of the ligand. Interactions of the ligand with ECL2/3 and transmembrane regions of the receptor are then required to elicit the activation signal. Although complete details of the SDF-1α:CXCR4 interface remain to be determined, the NMR structure of a soluble complex between dimeric SDF-1α and a CXCR4 fragment defined specific binding determinants in the receptor N terminus (42). In the present study, NMR spectroscopy experiments with an N-terminal CXCR4-(1–38) peptide, receptor binding, and signaling studies in the presence of antibodies against the CXCR4 N terminus and ECL2/3 provided multiple layers of evidence that the interaction between ubiquitin and CXCR4 is independent of the N terminus of the receptor.
It has been reported that AMD3100 binds to only three acidic anchor-point residues located in transmembrane regions IV, VI, and VII of CXCR4, which form the main ligand binding pocket (37). Furthermore, AMD3100 was able to displace the N terminus of SDF-1α from the receptor without displacing the SDF-1α core domain from the CXCR4 N terminus (60). Our finding that AMD3100 displaced ubiquitin from CXCR4 in P4.R5 cells is consistent with previous observations that SDF-1α and AMD3100 also displaced ubiquitin from CXCR4 in THP-1 cells (13). Furthermore, anti-ECL2/3 inhibited ubiquitin binding to CXCR4, interfered with ubiquitin and SDF-1α induced CXCR4 signaling, and anti-CXCR4 ECL2/3 binding to the cell surface was reduced in the presence of ubiquitin, SDF-1α, and AMD3100. This suggests that ECL2/3 and adjacent transmembrane domains of CXCR4 contain important binding sites that are required for its interaction with all ligands. This further implies that the CXCR4 binding site for ubiquitin is probably in close proximity to the binding sites of SDF-1α and AMD3100.
Previous studies suggested that cellular entry of X4 tropic HIV-1 and the interaction between CXCR4 and HIV-1 glycoprotein gp120 depend on the CXCR4 N terminus and ECL2/3 (36, 58, 62). Interestingly, studies with SDF-1α analogues showed that the ability of SDF-1α to block HIV-1 entry does not depend on its signaling properties (58), which are due to interactions with ECL2/3. This indicates that the N-terminal receptor domain is of importance for the anti-HIV-1 effect of SDF-1α and may explain the inability of ubiquitin to affect HIV-1 entry. On the other hand, AMD3100 does not bind to the N terminus of the receptor but displays anti-HIV-1 activity. Thus, the ubiquitin binding site in the ECL2/3 domain of CXCR4 is probably not identical with the binding sites for AMD3100, and unlikely to interfere with the HIV-1 contact sites in the ECL2/3 region. SDF-1α binding to CXCR4 is followed by a rapid agonist induced internalization of the receptor-ligand complex, which does not occur after AMD3100 binding to CXCR4 (63). Reduction of the number of available CXCR4 cell surface receptors by SDF-1α has been proposed to be another component of its anti-HIV-1 activity (64). Thus, the lack of anti-HIV-1 activity of ubiquitin could also correspond to reduced receptor internalization or enhanced recycling of CXCR4 to the cell surface after ubiquitin binding. Further studies are required to address these hypotheses.
Conclusively, our data suggest a novel binding mechanism of a natural ligand of CXCR4. Despite distinct receptor binding mechanisms of SDF-1α and ubiquitin, our findings imply that stimulation of CXCR4 with ubiquitin and SDF-1α is coupled to the same intracellular signaling pathways and results in comparable effects on cell function. Although the findings of the present study exclude ubiquitin as an anti-HIV-1 agent, they support the notion that the anti-inflammatory and organ protective effects of SDF-1α, the SDF-1α peptide analog CTCE-0214, and ubiquitin, which have been observed after administration in various models of infectious and noninfectious inflammation (5–11, 65–68), have a common molecular basis. These data further support the concept that endogenous extracellular ubiquitin may function to limit exuberant inflammation induced by damage-associated molecular pattern molecules (3, 69, 70). Besides providing new mechanistic insights into the biology of CXCR4-mediated cellular events, our findings have implications for the development of novel anti-inflammatory compounds, which may either target the signaling pathways that are activated after CXCR4 stimulation or agonist-specific CXCR4 binding sites.
Acknowledgments
We thank Debby Wyatt and Jacqueline Romero for technical assistance and Ravi Shankar for help with the chemotaxis experiments.
This work was supported in part by a grant that was awarded and administered by the U.S. Army Medical Research and Materiel Command (USAMRMC) and the Telemedicine and Advanced Technology Research Center (TATRC), at Fort Detrick, MD, under Contract Number W81XWH1020122 (to M. M.). This work was also supported in part by National Institutes of Health Grant GM075159 (to A. M.). The therapeutic use of ubiquitin has been patented (US patent #7,262,162) and M. M. is an inventor. M. M. has not received any income related to the patent.
- GPCR
- G-protein coupled receptor
- CXCR
- CXC chemokine receptor
- ECL
- extracellular loops
- RSK
- p90 ribosomal S6 kinase
- SDF
- stromal cell-derived factor
- RFU
- relative fluorescence units
- MAGI
- multinuclear activation of galactosidase indicator.
REFERENCES
- 1. Hershko A., Ciechanover A. (1998) Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 2. Majetschak M., Krehmeier U., Bardenheuer M., Denz C., Quintel M., Voggenreiter G., Obertacke U. (2003) Blood 101, 1882–1890 [DOI] [PubMed] [Google Scholar]
- 3. Majetschak M. (2011) J. Leukocyte Biol. 89, 205–219 [DOI] [PubMed] [Google Scholar]
- 4. Majetschak M., Zedler S., Hostmann A., Sorell L. T., Patel M. B., Novar L. T., Kraft R., Habib F., de Moya M. A., Ertel W., Faist E., Schade U. (2008) J. Trauma 64, 586–596; discussion 596–598 [DOI] [PubMed] [Google Scholar]
- 5. Majetschak M., Cohn S. M., Nelson J. A., Burton E. H., Obertacke U., Proctor K. G. (2004) Surgery 135, 536–543 [DOI] [PubMed] [Google Scholar]
- 6. Majetschak M., Cohn S. M., Obertacke U., Proctor K. G. (2004) J. Trauma 56, 991–999; discussion 999–1000 [DOI] [PubMed] [Google Scholar]
- 7. Earle S. A., El-Haddad A., Patel M. B., Ruiz P., Pham S. M., Majetschak M. (2006) Transplantation 82, 1544–1546 [DOI] [PubMed] [Google Scholar]
- 8. Earle S. A., Proctor K. G., Patel M. B., Majetschak M. (2005) Surgery 138, 431–438 [DOI] [PubMed] [Google Scholar]
- 9. Garcia-Covarrubias L., Manning E. W., 3rd, Sorell L. T., Pham S. M., Majetschak M. (2008) Crit. Care Med. 36, 979–982 [DOI] [PubMed] [Google Scholar]
- 10. Griebenow M., Casalis P., Woiciechowsky C., Majetschak M., Thomale U. W. (2007) J. Neurotrauma 24, 1529–1535 [DOI] [PubMed] [Google Scholar]
- 11. Ahn H. C., Yoo K. Y., Hwang I. K., Cho J. H., Lee C. H., Choi J. H., Li H., Cho B. R., Kim Y. M., Won M. H. (2009) Exp. Neurol. 220, 120–132 [DOI] [PubMed] [Google Scholar]
- 12. Majetschak M., Ponelies N., Hirsch T. (2006) Immunol. Cell Biol. 84, 59–65 [DOI] [PubMed] [Google Scholar]
- 13. Saini V., Marchese A., Majetschak M. (2010) J. Biol. Chem. 285, 15566–15576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Saini V., Romero J., Marchese A., Majetschak M. (2010) Commun. Integr. Biol. 3, 608–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Busillo J. M., Benovic J. L. (2007) Biochim. Biophys. Acta 1768, 952–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nagasawa T., Hirota S., Tachibana K., Takakura N., Nishikawa S., Kitamura Y., Yoshida N., Kikutani H., Kishimoto T. (1996) Nature 382, 635–638 [DOI] [PubMed] [Google Scholar]
- 17. Tachibana K., Hirota S., Iizasa H., Yoshida H., Kawabata K., Kataoka Y., Kitamura Y., Matsushima K., Yoshida N., Nishikawa S., Kishimoto T., Nagasawa T. (1998) Nature 393, 591–594 [DOI] [PubMed] [Google Scholar]
- 18. Karin N. (2010) J. Leukocyte Biol. 88, 463–473 [DOI] [PubMed] [Google Scholar]
- 19. Zaruba M. M., Franz W. M. (2010) Expert Opin. Biol. Ther. 10, 321–335 [DOI] [PubMed] [Google Scholar]
- 20. Li M., Yu J., Li Y., Li D., Yan D., Ruan Q. (2010) Cell Reprogram. 12, 405–415 [DOI] [PubMed] [Google Scholar]
- 21. Nagasawa T., Tachibana K., Kishimoto T. (1998) Semin. Immunol. 10, 179–185 [DOI] [PubMed] [Google Scholar]
- 22. Veldkamp C. T., Ziarek J. J., Peterson F. C., Chen Y., Volkman B. F. (2010) J. Am. Chem. Soc. 132, 7242–7243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Teicher B. A., Fricker S. P. (2010) Clin. Cancer Res. 16, 2927–2931 [DOI] [PubMed] [Google Scholar]
- 24. Balabanian K., Lagane B., Infantino S., Chow K. Y., Harriague J., Moepps B., Arenzana-Seisdedos F., Thelen M., Bachelerie F. (2005) J. Biol. Chem. 280, 35760–35766 [DOI] [PubMed] [Google Scholar]
- 25. Veldkamp C. T., Seibert C., Peterson F. C., Sakmar T. P., Volkman B. F. (2006) J. Mol. Biol. 359, 1400–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Malik R., Marchese A. (2010) Mol. Biol. Cell 21, 2529–2541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bobardt M. D., Saphire A. C., Hung H. C., Yu X., Van der Schueren B., Zhang Z., David G., Gallay P. A. (2003) Immunity 18, 27–39 [DOI] [PubMed] [Google Scholar]
- 28. Campbell E. M., Nunez R., Hope T. J. (2004) J. Virol. 78, 5745–5755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kimpton J., Emerman M. (1992) J. Virol. 66, 2232–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bhandari D., Trejo J., Benovic J. L., Marchese A. (2007) J. Biol. Chem. 282, 36971–36979 [DOI] [PubMed] [Google Scholar]
- 31. Soto A. G., Trejo J. (2010) J. Biol. Chem. 285, 18781–18793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bleul C. C., Farzan M., Choe H., Parolin C., Clark-Lewis I., Sodroski J., Springer T. A. (1996) Nature 382, 829–833 [DOI] [PubMed] [Google Scholar]
- 33. Oberlin E., Amara A., Bachelerie F., Bessia C., Virelizier J. L., Arenzana-Seisdedos F., Schwartz O., Heard J. M., Clark-Lewis I., Legler D. F., Loetscher M., Baggiolini M., Moser B. (1996) Nature 382, 833–835 [DOI] [PubMed] [Google Scholar]
- 34. Steen A., Schwartz T. W., Rosenkilde M. M. (2009) Mini Rev. Med. Chem. 9, 1605–1621 [DOI] [PubMed] [Google Scholar]
- 35. Kuritzkes D. R. (2009) Curr. Opin. HIV AIDS 4, 82–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Doranz B. J., Orsini M. J., Turner J. D., Hoffman T. L., Berson J. F., Hoxie J. A., Peiper S. C., Brass L. F., Doms R. W. (1999) J. Virol. 73, 2752–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rosenkilde M. M., Gerlach L. O., Jakobsen J. S., Skerlj R. T., Bridger G. J., Schwartz T. W. (2004) J. Biol. Chem. 279, 3033–3041 [DOI] [PubMed] [Google Scholar]
- 38. Murphy P. M., Baggiolini M., Charo I. F., Hébert C. A., Horuk R., Matsushima K., Miller L. H., Oppenheim J. J., Power C. A. (2000) Pharmacol. Rev. 52, 145–176 [PubMed] [Google Scholar]
- 39. Bernhagen J., Krohn R., Lue H., Gregory J. L., Zernecke A., Koenen R. R., Dewor M., Georgiev I., Schober A., Leng L., Kooistra T., Fingerle-Rowson G., Ghezzi P., Kleemann R., McColl S. R., Bucala R., Hickey M. J., Weber C. (2007) Nat. Med. 13, 587–596 [DOI] [PubMed] [Google Scholar]
- 40. Leng L., Metz C. N., Fang Y., Xu J., Donnelly S., Baugh J., Delohery T., Chen Y., Mitchell R. A., Bucala R. (2003) J. Exp. Med. 197, 1467–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Struyf S., Noppen S., Loos T., Mortier A., Gouwy M., Verbeke H., Huskens D., Luangsay S., Parmentier M., Geboes K., Schols D., Van Damme J., Proost P. (2009) J. Immunol. 182, 666–674 [DOI] [PubMed] [Google Scholar]
- 42. Veldkamp C. T., Seibert C., Peterson F. C., De la Cruz N. B., Haugner J. C., 3rd, Basnet H., Sakmar T. P., Volkman B. F. (2008) Sci. Signal. 1, ra4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fricker S. P., Anastassov V., Cox J., Darkes M. C., Grujic O., Idzan S. R., Labrecque J., Lau G., Mosi R. M., Nelson K. L., Qin L., Santucci Z., Wong R. S. (2006) Biochem. Pharmacol. 72, 588–596 [DOI] [PubMed] [Google Scholar]
- 44. Hesselgesser J., Liang M., Hoxie J., Greenberg M., Brass L. F., Orsini M. J., Taub D., Horuk R. (1998) J. Immunol. 160, 877–883 [PubMed] [Google Scholar]
- 45. Loetscher P., Gong J. H., Dewald B., Baggiolini M., Clark-Lewis I. (1998) J. Biol. Chem. 273, 22279–22283 [DOI] [PubMed] [Google Scholar]
- 46. Di Salvo J., Koch G. E., Johnson K. E., Blake A. D., Daugherty B. L., DeMartino J. A., Sirotina-Meisher A., Liu Y., Springer M. S., Cascieri M. A., Sullivan K. A. (2000) Eur. J. Pharmacol. 409, 143–154 [DOI] [PubMed] [Google Scholar]
- 47. Gifford A. N., Bruneus M., Gatley S. J., Lan R., Makriyannis A., Volkow N. D. (1999) J. Pharmacol. Exp. Ther. 288, 478–483 [PubMed] [Google Scholar]
- 48. Ethier M. F., Schaefer O. P., Samant N., Yamaguchi H., Madison J. M. (1996) Am. J. Physiol. Lung Cell. Mol. Physiol. 270, L199–L207 [DOI] [PubMed] [Google Scholar]
- 49. Cai D., Lee K. K., Li M., Tang M. K., Chan K. M. (2004) Arch. Biochem. Biophys. 425, 42–50 [DOI] [PubMed] [Google Scholar]
- 50. Bleul C. C., Fuhlbrigge R. C., Casasnovas J. M., Aiuti A., Springer T. A. (1996) J. Exp. Med. 184, 1101–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nishio M., Endo T., Tsukada N., Ohata J., Kitada S., Reed J. C., Zvaifler N. J., Kipps T. J. (2005) Blood 106, 1012–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. O'Hayre M., Salanga C. L., Kipps T. J., Messmer D., Dorrestein P. C., Handel T. M. (2010) PLoS One 5, e11716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lee Y., Gotoh A., Kwon H. J., You M., Kohli L., Mantel C., Cooper S., Hangoc G., Miyazawa K., Ohyashiki K., Broxmeyer H. E. (2002) Blood 99, 4307–4317 [DOI] [PubMed] [Google Scholar]
- 54. Ganju R. K., Brubaker S. A., Meyer J., Dutt P., Yang Y., Qin S., Newman W., Groopman J. E. (1998) J. Biol. Chem. 273, 23169–23175 [DOI] [PubMed] [Google Scholar]
- 55. Hudson B. D., Hébert T. E., Kelly M. E. (2010) Mol. Pharmacol. 77, 1–9 [DOI] [PubMed] [Google Scholar]
- 56. Bosier B., Muccioli G. G., Hermans E., Lambert D. M. (2010) Biochem. Pharmacol. 80, 1–12 [DOI] [PubMed] [Google Scholar]
- 57. Gupta S. K., Pillarisetti K., Thomas R. A., Aiyar N. (2001) Immunol. Lett 78, 29–34 [DOI] [PubMed] [Google Scholar]
- 58. Crump M. P., Gong J. H., Loetscher P., Rajarathnam K., Amara A., Arenzana-Seisdedos F., Virelizier J. L., Baggiolini M., Sykes B. D., Clark-Lewis I. (1997) EMBO J. 16, 6996–7007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wu B., Chien E. Y., Mol C. D., Fenalti G., Liu W., Katritch V., Abagyan R., Brooun A., Wells P., Bi F. C., Hamel D. J., Kuhn P., Handel T. M., Cherezov V., Stevens R. C. (2010) Science 330, 1066–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kofuku Y., Yoshiura C., Ueda T., Terasawa H., Hirai T., Tominaga S., Hirose M., Maeda Y., Takahashi H., Terashima Y., Matsushima K., Shimada I. (2009) J. Biol. Chem. 284, 35240–35250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gozansky E. K., Louis J. M., Caffrey M., Clore G. M. (2005) J. Mol. Biol. 345, 651–658 [DOI] [PubMed] [Google Scholar]
- 62. Brelot A., Heveker N., Montes M., Alizon M. (2000) J. Biol. Chem. 275, 23736–23744 [DOI] [PubMed] [Google Scholar]
- 63. Hatse S., Princen K., Bridger G., De Clercq E., Schols D. (2002) FEBS Lett. 527, 255–262 [DOI] [PubMed] [Google Scholar]
- 64. Amara A., Gall S. L., Schwartz O., Salamero J., Montes M., Loetscher P., Baggiolini M., Virelizier J. L., Arenzana-Seisdedos F. (1997) J. Exp. Med. 186, 139–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Meiron M., Zohar Y., Anunu R., Wildbaum G., Karin N. (2008) J. Exp. Med. 205, 2643–2655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Shyu W. C., Lin S. Z., Yen P. S., Su C. Y., Chen D. C., Wang H. J., Li H. (2008) J. Pharmacol. Exp. Ther. 324, 834–849 [DOI] [PubMed] [Google Scholar]
- 67. Hu X., Dai S., Wu W. J., Tan W., Zhu X., Mu J., Guo Y., Bolli R., Rokosh G. (2007) Circulation 116, 654–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Fan H., Wong D., Ashton S. H., Borg K. T., Halushka P. V., Cook J. A. (2011) Inflammation [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lotze M. T., Zeh H. J., Rubartelli A., Sparvero L. J., Amoscato A. A., Washburn N. R., Devera M. E., Liang X., Tör M., Billiar T. (2007) Immunol. Rev. 220, 60–81 [DOI] [PubMed] [Google Scholar]
- 70. Oppenheim J. J., Yang D. (2005) Curr. Opin Immunol. 17, 359–365 [DOI] [PubMed] [Google Scholar]