Abstract
The classical cannabinoid agonist HU210, a structural analog of (−)-Δ9-tetrahydrocannabinol, binds to brain cannabinoid (CB1) receptors and activates signal transduction pathways. To date, an exact molecular description of the CB1 receptor is not yet available. Utilizing the minor binding pocket of the CB1 receptor as the primary ligand interaction site, we explored HU210 binding using lipid bilayer molecular dynamics (MD) simulations. Among the potential ligand contact residues, we identified residues Phe-1742.61, Phe-1772.64, Leu-1933.29, and Met-3636.55 as being critical for HU210 binding by mutational analysis. Using these residues to guide the simulations, we determined essential cannabinoid-binding domains in the CB1 receptor, including the highly sought after hydrophobic pocket important for the binding of the C3 alkyl chain of classical and nonclassical cannabinoids. Analyzing the simulations of the HU210-CB1 receptor complex, the CP55940-CB1 receptor complex, and the (−)-Δ9-tetrahydrocannabinol-CB1 receptor complex, we found that the positioning of the C3 alkyl chain and the aromatic stacking between Trp-3566.48 and Trp-2795.43 is crucial for the Trp-3566.48 rotamer change toward receptor activation through the rigid-body movement of H6. The functional data for the mutant receptors demonstrated reductions in potency for G protein activation similar to the reductions seen in ligand binding affinity for HU210.
Keywords: G Protein-coupled Receptors (GPCR), Membrane Proteins, Molecular Dynamics, Mutagenesis Mechanisms, Protein Drug Interactions, Receptor Structure-Function
Introduction
Brain cannabinoid (CB1)2 receptors (Fig. 1A) (1) are rhodopsin class G protein-coupled receptors (GPCRs) (2). Multiple chemical classes, including classical and nonclassical cannabinoids (e.g. (−)-11-hydroxydimethylheptyl-Δ8-tetrahydrocannabinol (HU210), (−)-Δ9-tetrahydrocannabinol (Δ9-THC), and (−)-3-[2-hydroxyl-4-(1,1-dimethylheptyl)phenyl]-4-[3-hydroxylpropyl]cyclohexan-1-ol (CP55940)) (Fig. 1B), aminoalkylindoles (e.g. (R)-(+)-[2,3-dihydro-5-methyl-3-[(4-morpholinyl)methyl]pyrrolo[1,2,3-de]-1,4-benzoxazin-6-yl](1-naphthalenyl)methanone (WIN55212-2)), and endocannabinoids (e.g. N-arachidonoylethanolamine and 2-arachidonoylglycerol), bind to the CB1 receptor and activate signal transduction pathways (3) in an agonist-specific manner (4). According to the two-state model of GPCR activation (5), these agonists stabilize the receptor in its active state, whereas an inverse agonist N-(piperidin-1-yl)-5-(4-chlorophenyl)-1-(2,4-dichlorophenyl)-4-methyl-1H-pyrazole-3-carboxamide(SR141716A) (6, 7) stabilizes the receptor in its inactive state.
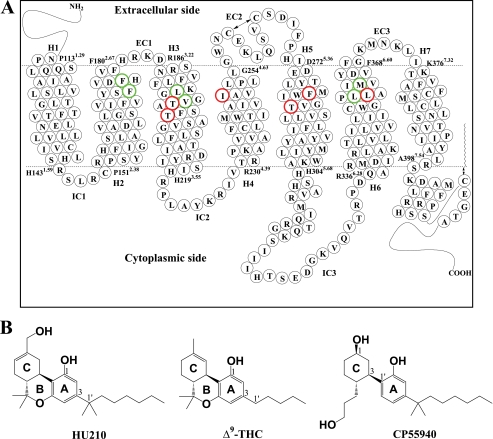
FIGURE 1.
CB1 receptor. A, human CB1 receptor in two-dimensional representation. Seven TM helices (H1 through H7), three extracellular loops (EC1, EC2, and EC3), and three intracellular loops (IC1, IC2, and IC3) are presented. Putative TM helical boundaries (H1, Pro-1.29–His-1431.59; H2, Pro-1512.38–Phe-1802.67; H3, Arg-1863.22–His-2193.55; H4, Arg-2304.39–Gly-2544.63; H5, Asp-2725.36–His-3045.68; H6, Arg-3366.28–Phe-3686.60; and H7, Lys-3767.32–Ala-3987.54) (49) are represented by dotted lines. The EC2 intra-loop disulfide linkage between Cys-257EC2 and Cys-264EC2 and the palmitoylation of the C-terminal Cys-415 are represented by a two-sided arrow. Amino acid residues examined in the present mutation studies are depicted in boldface type; residues critical for classical and nonclassical cannabinoid ligand binding (Phe-1742.61, Phe-1772.64, Leu-1933.29, Val-1963.32, Leu-3596.51, and Met-3636.55) are in green and residues noncritical for classical and nonclassical cannabinoid ligand binding (Thr-1973.33, Thr-2013.37, Ile-2474.56, Phe-2785.42, Thr-2835.47, and Leu-3606.52) are in red. The details of the binding pocket are represented in Fig. 5. B, molecular structures of the classical (ABC-tricyclic) cannabinoids HU210 (left) and Δ9-THC (middle), and the nonclassical (AC-bicyclic) cannabinoid CP55940 (right).
The CB1 receptor is a valuable therapeutic target for a number of disorders, including the treatment of anorexia in patients who suffer from AIDS wasting syndrome, reducing nausea and vomiting associated with chemotherapy treatment (8), reducing spasticity in multiple sclerosis patients (9), the treatment of neurodegenerative disease (10), and relief of neuropathic pain in multiple sclerosis (11). The accumulated mutational and structure-activity relationship data for the CB1 receptor enable us to better understand the receptor-ligand interactions, as an exact molecular description of the CB1 receptor is not yet available.
It has been proposed that there exists a hydrophobic binding pocket that interacts with the C3 alkyl chain of classical and nonclassical cannabinoids (12–16), a key pharmacophoric element for the CB1 receptor (17, 18). In this regard, the identification of Cys-3556.47, located next to Trp-3566.48 of the highly conserved CWXP motif, as a binding contact for the C3 alkyl chain of a nonclassical cannabinoid (19) is highly informative.3,4 The finding that Cys-2856.47 of the β2 adrenergic receptor (β2AR) became accessible to a thiol-reactive reagent only when the receptor was activated (20) suggests the corresponding residue of the CB1 receptor, Cys-3556.47, unavailable for ligand binding in the inactive state, becomes available in the active state as the Cys-3556.47 moves into the binding core as a result of an anticlockwise rigid-body rotation of the transmembrane (TM) helix 6 (H6) (21). Thus, it appears that the hydrophobic pocket that interacts with the C3 alkyl chain of cannabinoids forms dynamically as the receptor shifts its equilibrium toward the active state. No residues, other than Cys-3556.47, of the CB1 receptor have been reported to exhibit contact to the C3 alkyl chain of classical or nonclassical cannabinoids.
The recently determined x-ray structures of GPCRs both in the inactive state (22–25) and in the active state (26–30) together with the biophysical data (21, 31–36) have provided insights into understanding ligand-receptor interactions. The x-ray structures of GPCRs with diffusible ligands have shown that the location of ligand binding is slightly more extracellular than that of the covalently bound retinal in rhodopsin and that the binding pocket is open or partially open, possibly for easy access of the ligand to the binding cavity (23). In GPCRs, the region formed by the extracellular ends of H2/H3/H7 has been proposed to contain a cleft that forms the minor binding pocket (37). In the x-ray structure of the A2A receptor (PDB code 3EML) (25), the major binding pocket was occupied by an antagonist, whereas the minor binding pocket was occupied by a series of coordinated water molecules. Participating in H-bonding with these is His-2787.43, which is equivalent to Lys-2967.43 in rhodopsin which forms a covalent bond to the cis-retinal ligand. Interestingly, Ser-3837.39 and Cys-3867.42, in close proximity to the 7.43 position, in the CB1 receptor have been reported as residues important for ligand binding (38, 39). Combining these results with the results of other mutation studies, including Asp-184EC1 (40), Phe-1893.25 (40), and Lys-1923.28 (41, 42), the minor binding pocket appears important for the binding of classical and nonclassical cannabinoids. This is supported by the notion that the H2/H3/H6/H7 region of the CB1 receptor is important for classical and nonclassical cannabinoid interactions (39, 43).
The starting point of this study was our observation in the ligand-unbound CB1 receptor model (44) that a group of water molecules were networked through H-bonding to Ser-1732.60 in the minor binding pocket and extended to Tyr-2755.39 in the major binding pocket. We utilized the minor binding pocket of the CB1 receptor as the primary ligand interaction site to explore HU210 binding within the TM binding pore using lipid bilayer molecular dynamics (MD) simulations. We identified residues Phe-1742.61, Phe-1772.64, Leu-1933.29, and Met-3636.55 as being critical for HU210 by mutational analysis and determined essential cannabinoid-binding domains by the MD simulations. Functional data for the mutant receptors demonstrated a diminished potency for G protein activation suggesting that the hydrophobic binding pocket is crucial for binding classical and nonclassical cannabinoids and for G protein coupling.
EXPERIMENTAL PROCEDURES
Generation of CB1 Receptor Mutants
Site-directed mutagenesis (QuikChange; Stratagene, La Jolla, CA) was performed using the human CB1 cDNA cloned into pcDNA3.1. The presence of the mutations was confirmed by DNA sequencing.
CB1 Expression and Membrane Preparation
HEK293T cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 3.5 mg/ml glucose at 37 °C in 5% CO2. To transiently transfect human embryonic kidney (HEK293T) cells, cells were seeded at ∼1.0 × 106 cells/100-mm dish 1 day prior to transfection via calcium phosphate precipitation (45). Approximately 24 h after transfection, the cells were harvested and washed twice with phosphate-buffered saline (PBS) and membranes prepared as described previously (46). Cells were resuspended in PBS containing a protease inhibitor mixture (4-(2-aminoethyl)benzenesulfonylfluoride hydrochloride, pepstatin A, E-64, bestatin, leupeptin, and aprotinin) (Sigma) and then lysed by nitrogen cavitation at 750 p.s.i. for 5 min using a Parr cell disruption bomb. Cell debris and nuclei were pelleted at 4 °C at 500 × g for 10 min. The resulting supernatant was spun at 100,000 × g for 45 min at 4 °C. The membrane-containing pellet was then resuspended via homogenization in Tris/Mg2+/EDTA (TME) buffer (25 mm Tris-HCl, 5 mm MgCl2, and 1 mm EDTA, pH 7.4) containing 7% sucrose (w/v). Protein concentration was determined by the Bradford assay (47), and the membrane preparation was stored at −70 °C.
Ligand Binding
Ligand binding assays were performed as described previously (46) with minor modifications. For saturation binding assays, ∼4–10 μg of membrane was incubated with nine concentrations of [3H]SR141716A (43 Ci/mmol; PerkinElmer Life Sciences) between 0.24 and 20 nm for 60 min at 30 °C in TME containing 0.1% fatty acid-free BSA (w/v) in a final volume of 200 μl. For competition binding assays, ∼4–10 μg of membrane was incubated with 2 or 4 nm [3H]SR141716A and typically nine concentrations of displacing ligand between 10 pm and 31.6 μm for 60 min at 30 °C in TME containing 0.1% fatty acid-free BSA (w/v) in a final volume of 200 μl. The addition of 250 μl of chilled TME + 5% BSA was used to terminate the reaction before separating bound from unbound ligand by filtering with a 24-manifold Brandel cell harvester (Brandel, Gaithersburg, MD). Nonspecific binding was determined with 1 μm of unlabeled ligand. Filters were washed four times with cold TME buffer. Scintillation counting was then used to determine the bound radioactivity. Assays were performed at least three times in duplicate.
GTPγS Binding Assay
GTPγS binding assays were performed by incubating ∼10 μg of membrane in GTPγS binding buffer (50 mm Tris-HCl, pH 7.4, 3 mm MgCl2, 0.2 mm EGTA, and 100 mm NaCl) with at least nine concentrations of unlabeled HU210 (ranging from 10 pm to 10 μm), 10 μm GDP, 0.1% fatty acid-free BSA (w/v), and 0.1 nm [35S]GTPγS (1,250 Ci/mmol; PerkinElmer Life Sciences) in a final volume of 200 μl for 60 min at 30 °C. Nonspecific binding was determined with 10 μm of unlabeled GTPγS (Sigma). The reaction was terminated by separating bound from unbound [35S]GTPγS by filtering with a 24-manifold Brandel cell harvester (Brandel, Gaithersburg, MD). Filters were washed four times with cold TME buffer. The bound radioactivity was then determined by scintillation counting. Assays were performed two times in duplicate.
Data Analysis
The data are presented as the mean of two or three independent experiments performed in duplicate with the 95% confidence intervals given in parentheses. For ligand binding assays, IC50 values were calculated by nonlinear regression (fit to a one-site competition model) using GraphPad Prism (GraphPad Software Inc., San Diego). The Cheng-Prusoff equation (48) was used to calculate Ki values using the Kd values for the tracer obtained from saturation binding analyses. For GTPγS binding assays, the EC50 values were determined using a sigmoidal dose-response relationship. Analysis of variance followed by Dunn's post hoc test was used to compare the wild-type Ki and EC50 values to those of the mutant receptors. p values of < 0.05 were considered statistically significant.
Simulation in a 1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) Bilayer
Simulations were performed, as described in our previous studies (49), by the NAMD simulation package (version 2.6b2 for Linux-Power-MPI) (50, 51), using CHARMM22 force field parameters with the φ/ψ angle cross-term map correction for the protein (52, 53) and the TIP3 water model (54, 55), and CHARMM27 force field parameters for the lipids (56). The topology definitions and the parameters for the palmitoylated Cys residue, including the parameters around the bond connecting Cys-415 of the CB1 receptor and the carbonyl carbon of the palmitoyl moiety, as used in the literature (57), were found in the NAMD Parameter Topology Repository site. The temperature was maintained at 310 K through the use of Langevin dynamics (59) with a damping coefficient of 1/ps. The pressure was maintained at 1 atm by using the Nosé-Hoover method (60) with the modifications as described in the NAMD user's guide. The van der Waals interactions were switched at 10 Å and zero smoothly at 12 Å. Electrostatic interactions were treated using the Particle Mesh Ewald method (62). A pair list for calculating the van der Waals and electrostatic interactions was set to 13.5 Å and updated every 10 steps. A multiple time-stepping integration scheme, the impulse-based Verlet-I reversible reference system propagation algorithm method (63), was used to efficiently compute full electrostatics. The time step size for integration of each step of the simulation was 1 fs.
HU210 Docking
Initially homology-built using the inactive state crystal structure of β2AR (PDB code 2RH1) (23) as the template (49), our ligand-unbound CB1 receptor model was recently refined by determining the second extracellular loop (EC2) structure (44). In preparation of the receptor for HU210 docking, using the snapshot of the refined CB1 receptor model with the EC2 intra-loop disulfide linkage at 53 ns of the simulation, in which the system appeared converged before the rotamer change (see below), the system size of the ligand-unbound CB1 receptor model was significantly increased. The dimensions were set to 115 × 150 × 220 Å3 to reduce the artifacts related to the finite size of the membrane systems, and the resulting system was simulated at 310 K for 10 ns in the constant pressure (NPT) ensemble. After 2,500 steps of minimization, the receptor was extracted and used to explore HU210 docking modes by employing the genetic algorithm-based flexible docking program GOLD (64, 65). For the ligand, an initial structure of HU210 was built by the Build Fragment tool in Discovery Studio (Accelrys, San Diego). The lowest energy conformation of HU210 obtained by the Generate Conformations tool, using the BEST conformation algorithm as implemented in Discovery Studio, was used for docking. All the receptor residues within 20 Å of Phe-3797.35, whose aromatic ring was positioned toward the TM core, were defined as the binding site. This region adequately covered the putative ligand binding region, including the innermost binding core region near Trp-3566.48. For the GOLD docking experiment, we used the pre-defined default GOLD generic algorithm settings. GOLDScore was used for evaluating HU210 docking modes. The GOLD cavity detection algorithm was used to locate the ligand within the TM binding core of the protein. No ligand bumping to any part of the protein was allowed. We performed the docking experiment to retain 50 docking poses from 10,000 docking runs and repeated the same docking experiment several times to obtain convergent binding poses. Among a diverse set of HU210 docking poses, we selected high score docking poses such that the ABC-ring moiety was placed in the minor binding pocket, and the hydrophobic C3 side chain moiety was located in the major binding pocket. Possible docking poses were further screened using H-bonding of the ABC-ring to polar residues; a few distinct poses were identified that satisfied all criteria, and these were placed into the hydrated lipid bilayer. Any water molecule within 1.0 Å of the newly introduced ligand was removed.
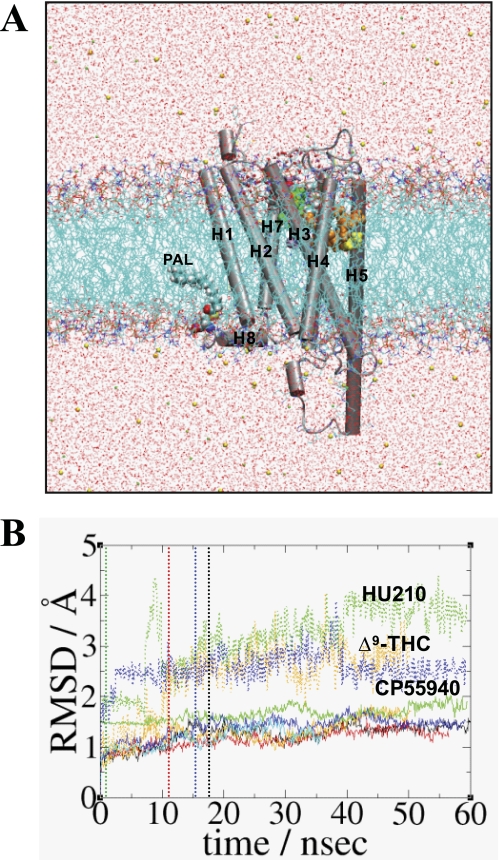
The HU210-bound CB1 receptor embedded in a hydrated POPC bilayer, resulted in a total of ∼380,000 atoms (Fig. 2A). To relax the protein in the presence of the tightly fitted ligand in the cavity, the system was simulated at 310 K for a few ns while constraining a few H-bonds between the ligand and the protein, using adaptive biasing force (66) as implemented in NAMD (67). Then the whole system was simulated at 310 K for 60 ns in the NPT ensemble without constraint. We defined this simulation system HU210-CB1y1 (i.e. HU210-bound to the CB1 receptor and yes to the Trp-3566.48 rotamer χ1 angle changed from g+ to trans (see below)). Repeating similar procedures, we obtained the following: HU210-CB1y2 with a different ligand binding mode; CP55940-CB1y or Δ9-THC-CB1n by replacing HU210 in HU210-CB1y2 after a few nanoseconds of the simulation by CP55940 or Δ9-THC; HU210-CB1y3 by replacing CP55940 in CP55940-CB1y after a few nanoseconds of the simulation by HU210; and finally CB1n by removing HU210 in HU210-CB1y1 at 10 ns of the simulation. The structures taken every 100 ps of these simulations were used for the analysis. The validity of the modeled receptor structures was examined by WHAT IF (68, 69). All the simulations performed are summarized in Table 2.
FIGURE 2.
Simulation systems of the CB1 receptor in the present study. A, typical simulation system consisting of the CB1 receptor, ∼440 POPC molecules, ∼107,800 water molecules, ∼140 sodium ions, and ∼160 chloride ions. Palmitoyl moiety that is covalently bonded to Cys-415 is represented by a space-filling model. Lipids and water molecules are represented as lines and ions are represented as balls. Lipid hydrogen atoms are omitted for clarity. The system at 50 ns of HU210-CB1y3 is shown. Color coding is as follows: carbon, cyan; oxygen, red; nitrogen, blue; phosphorus, orange; sodium ion, green; and chloride ion, yellow. Hydrogen atoms are omitted for clarity. B, r.m.s.d. values (RMSD) of the proteins in HU210-CB1y1 (in black), HU210-CB1y2 (in red) HU210-CB1y3 (in green), CP55940-CB1y (in blue), Δ9-THC-CB1n (in orange), and CB1n (in cyan), calculated by root mean square fitting to the initial coordinates with respect to the backbone heavy atoms of the TM helical residues of the CB1 receptor, are represented by continuous lines, whereas the r.m.s.d. values of the ligands in HU210-CB1y3 (in green), CP55940-CB1y (in blue), and Δ9-THC-CB1n (in orange), calculated with respect to the initial coordinates after fitting the proteins based upon the backbone atoms of TMHs, are represented by dotted lines. The time when the rotamer change occurred is represented by a dotted line: for HU210-CB1y1, in black; for HU210-CB1y2, in red; for HU210-CB1y3, in green; and for CP55940-CB1y, in blue.
TABLE 2.
MD simulations performed in the present study
| Systema | Binding mode in agreement with mutational datab | Rotamer changec | Receptor activationd | MD simulation |
|
|---|---|---|---|---|---|
| Duration | Time for rotamer changee | ||||
| ns | |||||
| HU210-CB1y1 | No | Yes | No | 60 | ∼17 |
| HU210-CB1y2 | Yes | Yes | No | 56 | ∼11 |
| HU210-CB1y3 | Yes | Yes | Yes | 60 | Immediatef |
| CP55940-CB1y | Yes | Yes | Yes | 59 | ∼15 |
| Δ9-THC-CB1n | Yes | No | No | 50 | |
| CB1n | No | No | 36 | ||
a All simulations were performed in a fully hydrated lipid-bilayer using POPC (see “Experimental Procedures”).
b Judged by the interaction between the C3 alkyl chain and the ligand-contacting residues, Leu-1933.29, Val-1963.32, Leu-3596.51, and Met-3636.55.
c Judged by the Trp-3566.48 χ1 angle: if trans (i.e. +120° < χ1 < +240°) yes; otherwise, no.
d Judged by the orientation of the Trp-3566.48 side chain indole ring: if the indole ring faces toward H5 (91), yes; otherwise, no.
e The time taken for the rotamer change as judged by the Trp-3566.48 χ1 angle.
f In this case, the initial structure included the Trp-3566.48 rotamer change (see text).
CHARMM Parameterization
To describe HU210, CP55940, and Δ9-THC in the MD simulations using the CHARMM force field, we determined the missing parameters. To determine the atomic charges of each ligand atom, charges from electrostatic potentials using a grid-based method (70) at the ab initio RHF/6–31G* level were computed using the Gaussian program (71). The charge from electrostatic potentials using a grid-based method at every atom was averaged over five representative conformations from a short (500 ps) MD simulation in a water box (40 × 40 × 40 Å3). To minimize any inconsistencies with the existing CHARMM parameters, most of the missing parameters were borrowed, if possible, from the parameter values of chemically relevant structures. If necessary, employing ab initio RHF/6–31G* and MP2/6–31G* level calculations (71), we adjusted the values of bonds, angles, and torsion angles preferentially over adjustments to the values of force constants. We checked the validity of the newly determined CHARMM parameters for describing HU210, CP55940, and Δ9-THC by comparing molecular geometries from the MD simulations and key torsional energy barriers by CHARMM with those obtained by ab initio RHF/6–31G* and MP2/6–31G* level calculations (supplemental Table 1 and supplemental Fig. 1).
Estimation of Nonbonding Interaction Energy Values
The nonbonding interaction energy is defined as the summation of electrostatic energy and van der Waals energy. The nonbonding interaction energy values between any two residues of the protein and between a residue of the protein and the ligand were estimated using the NAMD energy plugin in VMD (72). A smooth switching function was activated at the distance of 10 Å to truncate the nonbonding interaction energies smoothly at the cutoff distance of 12 Å. Favorable interaction energies have negative signs, and unfavorable interaction energies have positive signs.
RESULTS
Potential Contact Residues for the Binding of Cannabinoids to the CB1 Receptors
Residues Phe-1742.61, Phe-1772.64, Leu-1933.29, and Met-3636.55 of the CB1 receptor were identified as being critical for the binding of the classical cannabinoid agonist HU210. These residues were individually substituted with alanine, and the mutant receptors were evaluated for ligand binding with cannabinoids. Radiolabeled SR141716A was used as the tracer in all experiments due the reduction in affinity observed for the other ligands tested. [3H]SR141716A saturation binding assays suggested all the mutant receptors were expressed at levels similar to the wild-type receptor (WT Bmax = 5,200 fmol/mg). Inhibition constants were determined for HU210 binding to all cannabinoid receptor mutants. The mutant receptors, F1742.61A, F1772.64A, L1933.29A, and M3636.55A, all exhibited diminished HU210 binding relative to the wild-type receptor (Table 1). The results indicate the greatest decrease in HU210 binding affinity for this series was observed for the F1772.64A mutant receptor that bound HU210 with an affinity ∼700-fold lower than the wild-type receptor (WT HU210 Ki = 0.23 nm). In comparison, the F1742.61A, and L1933.29A mutant receptors had somewhat less substantial losses in HU210 binding affinity (with 146- and 105-fold losses in affinity, respectively). HU210 binding was least sensitive to the M3636.55A mutation with a 15-fold loss in binding affinity relative to the wild-type receptor.
TABLE 1.
Binding and G protein activation by ligands to CB1 receptors with substitutions in potential contact residues
| Receptor | HU210 |
CP55940, Kia | Δ9-THC, Kia | |
|---|---|---|---|---|
| Kia | GTPγS EC50b | |||
| nm | nm | nm | nm | |
| WT | 0.23(0.16–0.33) | 0.66(0.39–1.10) | 13.5(7.71–23.6) | 24.6(17.1–35.5) |
| F1742.61A | 33.6*(6.19–182) | 20.5*(11.8–35.4) | 294*(199–434) | 59%* displacement at 3.16 μm |
| F1772.64A | 160*(102–252) | 116*(36.3–370) | 5564*(3241–9553) | 34%* displacement at 3.16 μm |
| L1933.29A | 24.2*(13.2–44.1) | 10.6*(4.08- 27.7) | 1049*(623–1765) | 258*(152–440) |
| M3636.55A | 3.37*(1.87–6.07) | 4.23*(2.69–6.66) | 52.2*(35.1–77.8) | 16.0(10.8–23.7) |
a Data are the means of three independent experiments performed in duplicate, and the 95% confidence intervals are given in parentheses. Ki values were determined using [3H]SR141716A.
b Data are the means of two independent experiments performed in duplicate, and the 95% confidence intervals are given in parentheses.
* indicates statistically significant differences from wild-type (p < 0.05) using analysis of variance followed by Dunn's post hoc test.
To assess the effect of the mutations on receptor-mediated G protein activation, HU210-induced GTPγS binding was examined. The data (Table 1) demonstrate that the F174A2.61, F177A2.64, L193A3.29, and M363A6.55 mutant receptors exhibited the same rank order loss in HU210 potency for G protein activation, relative to the wild-type receptor (WT HU210 EC50 = 0.66 nm), as was observed for HU210 binding affinity (Table 1). The comparable Ki and EC50 values for HU210 ligand binding and G protein activation suggest a similar relative activity of HU210 for these receptors. Moreover, the ability of these mutant receptors to exhibit similar Emax values to the wild-type receptor (wild-type Emax = 29.8 fmol/mg) (data not shown) suggest these mutations do not affect the global conformation of the receptors.
To gain insight of the possible role of Phe-1742.61, Phe-1772.64, Leu-1933.29, and Met-3636.55 in incorporating the B-ring of HU210 at the CB1 receptor-binding site, the B-ring lacking CP55940 binding affinity was determined for the mutated cannabinoid receptors. The fold loss in binding affinity for CP55940 was comparable with that of HU210 for the F1772.64A and L1933.29A mutant CB1 receptors relative to the wild-type receptor. F1742.61A and M3636.55A receptors exhibited a smaller reduction in binding affinity relative to the wild-type receptor for CP55940 than for HU210 (F1742.61A exhibited a 146- and 22-fold loss in binding affinity and the M3636.55A receptor exhibited a 15- and 4-fold loss in binding affinity, for HU210 and CP55940, respectively).
Ki values for Δ9-THC were not obtained for the F1742.61A and F1772.64A mutant receptors because just 59 and 34% of the tracer was displaced by 3.16 μm Δ9-THC, respectively. These results suggest there is a marked loss in Δ9-THC binding (substantially greater than 100-fold relative to the wild-type receptor) as was observed for HU210 binding affinity by these receptors. In contrast, the near wild-type binding affinity of Δ9-THC by the M3636.55A receptor suggests this residue may be more involved in enabling the binding of agonists with alkyl tails containing a dimethyl group such as HU210 and CP55940. Likewise, L1933.29A bound Δ9-THC with a smaller decrease in binding affinity than it bound HU210 and CP55940 (10-, 105-, and 78-fold losses, respectively).
MD Simulations of the CB1 Receptor in the Presence of HU210, CP55940, and Δ9-THC
We performed several MD simulations, including HU210-CB1y1, HU210-CB1y2, HU210-CB1y3, CP55940-CB1y, Δ9-THC-CB1n, and CB1n (Table 2). As shown in Fig. 2B, all the receptor systems, showing some fluctuations, appeared to be converged with the r.m.s.d. values <2 Å with respect to the Cα atoms of the receptor TM helical bundle, indicating the CB1 receptor models were stable. Despite high r.m.s.d. values (> 2 Å) for the ligands, which suggested some noticeable changes in position within the binding site, all of the tested ligands appeared converged at the end of the simulations (Fig. 2B). This indicates the ligands were stable and maintained their binding to the receptor. High fluctuations shown in the ligand r.m.s.d. values were due to the fluctuation of the ligand C3 alkyl chain.
To determine the binding mode of HU210, we explored many possible modes, including one with the ABC-ring hydroxyl groups buried inside the hydrophobic core region (e.g. HU210-CB1y1) and another with the ABC-ring hydroxyl groups positioned toward the hydrophilic extracellular surface (e.g. HU210-CB1y2 and HU210-CB1y3) (Fig. 3A). In HU210-CB1y1, the binding contacts of HU210 were in poor agreement with the mutational data (Table 1). A potential alternative binding mode, in which the C3 alkyl chain of HU210 was inserted into a hydrophobic crevice formed by H5/H6, was identified during an early stage of the simulation that appeared in agreement with the present mutational data, but it was dropped due to the poor stability of the ligand in maintaining its binding to this region of receptor. In HU210-CB1y3, two noticeable changes in the ligand r.m.s.d. values were caused by the adjustment of the ABC-ring within the minor binding pocket as follows: its orientation from parallel to perpendicular to the membrane surface at 10 ns of the simulation, and its position from the middle of the core to toward H2 at 40 ns of the simulation. These results suggested that the MD simulation approach was suitable to sample the conformational space within the ligand-binding site. This permitted adjustment of the thermal equilibrium of the whole system, as small local energy barriers were overcome. During this process, the binding modes of structurally different ligands could be uniquely optimized, as shown in Fig. 3B of the ligand binding modes in HU210-CB1y3, Δ9-THC-CB1n, and CP55940-CB1y.
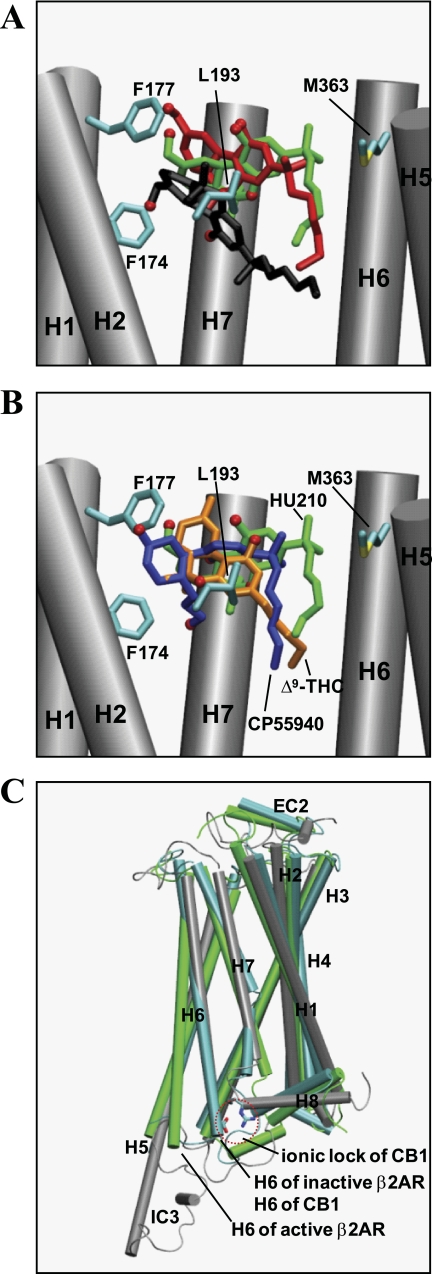
FIGURE 3.
Superposition of the ligands and the receptors. A, superposition of different binding modes of HU210 in HU210-CB1y1 (in black), HU210-CB1y2 (in red), and HU210-CB1y3 (in green) at the end of the simulations, with respect to the backbone atoms of TMHs. Phe-1742.61, Phe-1772.64, Leu-1933.29, and Met-3636.55 are represented by stick (in atom type: carbon, cyan; and sulfur, yellow). Only the side chains without hydrogen atoms of these residues are represented for clarity. H3 and H4 are omitted for clarity. B, superposition of HU210 in HU210-CB1y3 (in green), Δ9-THC, in Δ9-THC-CB1n (in orange), and CP55940 in CP55940-CB1y (in blue), with respect to the backbone atoms of TMHs. Phe-1742.61, Phe-1772.64, Leu-1933.29, and Met-3636.55 are represented by stick (in atom type: carbon, cyan; and sulfur, yellow). Only the side chains without hydrogen atoms of these residues are represented for clarity. H3 and H4 are omitted for clarity. C, superposition of HU210-CB1y3 (in gray) on the x-ray structures of the inactive form (PDB code 2RH1) (23) (in cyan) and the active form (PDB code 3P0G) (28) (in green) of β2AR, with respect to the backbone atoms of TMHs except H5 and H6. The ionic lock Arg-2143.50/Asp-3386.30 of the CB1 receptor is also presented by using only the side chains without hydrogen atoms within a red dotted circle.
Both in HU210-CB1y2 and in CP55940-CB1y, the ligand binding contacts were good, but only CP55940-CB1y proceeded toward receptor activation, as judged by the distance between the ligand and Cys-3556.47 (see below) (19). In Δ9-THC-CB1n, the ligand binding contacts were good, but the rotamer change was not observed. In CB1n, the rotamer change also was not observed. In all receptor systems analyzed, the salt bridge Arg-2143.50/Asp-3386.30, retaining the receptor in the inactive state (73), was maintained (Fig. 3C).
HU210-CB1y2 Showed the Rotameric Change but Did Not Proceed toward Receptor Activation
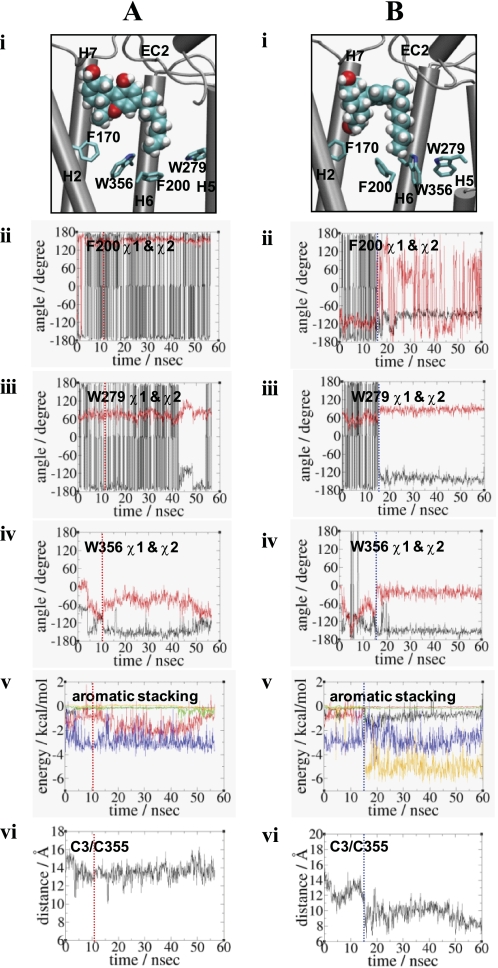
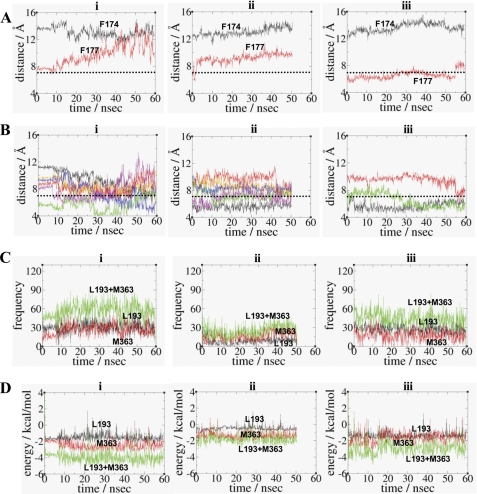
Similar to Trp-2866.48/Phe-2906.52 of β2AR (74), the Trp-3566.48/Phe-2003.36 pair of the CB1 receptor has been proposed as a rotamer switch for receptor activation (75). By definition, the rotameric angles are divided into three angle categories as follows: g−, 0° to 120°; trans, 120° to 270° and g+, 240° to 360° (74). Thus, the Phe-2003.36 χ1 trans/Trp-3566.48 χ1 g+ conformation in the inactive receptor changes to the Phe-2003.36 χ1 g+/Trp-3566.48 χ1 trans conformation in the active receptor. In support, the F2003.36A mutation of the CB1 receptor resulted in higher constitutive activity compared with the wild-type receptor (76). In HU210-CB1y2, the χ1 angle of Trp-3566.48 changed from g+ to trans at 10.7 ns of the simulation and maintained the trans conformation throughout the duration of the simulation (Fig. 4A). In HU210-CB1y2, the χ1 angle of Trp-3566.48 changed from g+ to trans at 10.7 ns of the simulation and was maintained during the rest of the simulation (Fig. 4A). Interestingly, both Phe-2003.36 and Trp-2795.43 almost maintained the χ1 angle of trans throughout the duration of the simulation. Our interaction energy analysis revealed that Trp-3566.48 interacted little with Trp-2795.43 although it interacted closely with Phe-2003.36 (Fig. 4A). Overall, HU210-CB1y2, where Trp-3566.48 was isolated from Trp-2795.43 and at the same time locked by Phe-2003.36, failed to move toward the active state despite the Trp-3566.48 rotamer change. In HU210-CB1y2, we observed no decrease in the distance between the ligand's C3 alkyl terminal carbon and Cys-3556.47, as Trp-3566.48 projected away from H5, unfavorable for initiating the rigid body motion of H6 (Fig. 4A, panel vi, and supplemental Fig. 2A, panel ii). Our cross-correlation analysis for HU210-CB1y2 showed that coupling between H5 and H6 was impaired (supplemental Fig. 2A, panel iii).
FIGURE 4.
Comparison of the Trp-3566.48 rotamer changes in HU210-CB1y2 (A) and in CP55940-CB1y (B) at the end of the simulations. Panel i, binding of the ligand (in space filling) with respect to the arrangement of Phe-1702.57, Phe-2003.36, Trp-2795.43, and Trp-3566.48 (in stick) is shown. Only the side chains without hydrogen atoms of residues are shown. H3 and H4 are omitted for a clearer view. Color coding: carbon, cyan; oxygen, red; and hydrogen, white. Panels ii–iv, rotameric angles, the χ1 angles (in black) and the χ2 angles (in red), of Phe-2003.36, Trp-2795.43, and Trp-3566.48 during the simulations. Panel v, estimated nonbonding interaction energy values between two aromatic residues, including Phe-1702.57/Phe-2003.36 (in black), Phe-1702.57/Trp-3566.48 (in red), Phe-2003.36/Trp-2795.43 (in green), Phe-2003.36/Trp-3566.48 (in blue), and Trp-2795.43/Trp-3566.48 (in orange). Only the side chains of residues were considered in calculating the nonbonding energy values. For HU210-CB1y2, the energy values (in kcal/mol), with the standard deviation in parentheses, averaged over the last 20.0 ns of the simulation are as follows: Phe-1702.57/Phe-2003.36, −0.13(0.02); Phe-1702.57/Trp-3566.48, −1.47(0.71); Phe-2003.36/Trp-2795.43, −0.33(0.24); Phe-2003.36/Trp-3566.48, −3.03(0.55); and Trp-2795.43/Trp-3566.48, −0.08(0.12). For CP55940-CB1y, the energy values (in kcal/mol), with the standard deviation in parentheses, averaged over the last 20.0 ns of the simulation are as follows: Phe-1702.57/Phe-2003.36, −0.68(0.32); Phe-1702.57/Trp-3566.48, −0.07(0.02); Phe-2003.36/Trp-2795.43, −0.17(0.04); Phe-2003.36/Trp-3566.48, −2.79(0.69); and Trp-2795.43/Trp-3566.48, −5.06(0.69). Panel vi, distance between the end carbon atom of the C3 alkyl chain of the ligand and the side chain sulfur atom of Cys-3556.47. In panels ii–vi, the time when the rotamer change occurred is represented by a dotted line: for HU210-CB1y2, in red and for CP55940-CB1y, in blue.
CP55940-CB1y Showed the Rotameric Change and Proceeded toward Receptor Activation
In CP55940-CB1y, the χ1 angles changed from trans to g+ for both Phe-2003.36 and Trp-2795.43 in 15 ns of the simulation, by which time the Trp-3566.48 rotamer change was completely established (Fig. 4B, panel iv). As a result, Trp-3566.48 faced toward H5 and Trp-2795.43 toward the TM core region and together these exhibited aromatic stacking. At the same time, Phe-2003.36 maintained aromatic stacking with Trp-3566.48 and also formed new aromatic stacking with Phe-1702.57 (Fig. 4B, panel v). Thus, our interaction energy analysis revealed both Phe-1702.57/Phe-2003.36 and Trp-2795.43/Trp-3566.48 aromatic stacking interactions, in addition to Phe-2003.36/Trp-3566.48, contributed to the Trp-3566.48 rotameric change. In CP55940-CB1y, we observed decreases in the distance between the ligand's C3 alkyl terminal carbon and Cys-3556.47, as Trp-3566.48 became favorably positioned toward H5 for initiating the rigid-body motion of H6 (Fig. 4B, panel vi, and supplemental Fig. 2B, panel ii). Our cross-correlation analysis for CP55940-CB1y showed that coupling between H5 and H6 was strong (supplemental Fig. 2B, panel iii).
HU210 Binding Mode to the CB1 Receptor
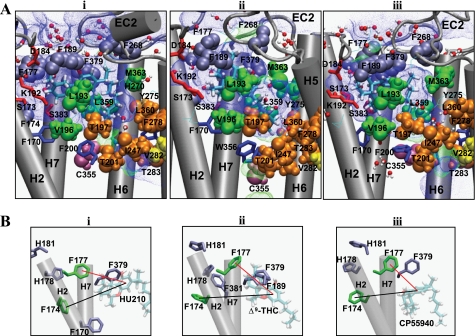
Realizing that it was necessary to obtain a HU210-CB1 system that shows the Trp-3566.48 rotameric change and proceeds toward receptor activation, as seen in CP55940-CB1y, we unsuccessfully tried many other HU210-CB1 systems, including HU210-CB1y2. Instead, we used CP55940-CB1y at the early stage of the simulation and obtained HU210-CB1y3. In this system, with the binding mode in agreement with the mutational data, HU210 binding and the Trp-3566.48 rotameric change were fully established by the simulation. The HU210 binding mode in HU210-CB1y3 (Fig. 5A, panel i) is described below.
FIGURE 5.
CB1 receptor interactions with HU210, Δ9-THC, and CP55940. A, binding modes of HU210 in HU210-CB1y3 (panel i), Δ9-THC in Δ9-THC-CB1n (panel ii), and CP55940 in CP55940-CB1y (panel iii) at the end of the simulations. The ligand is represented by stick (in atom type). Polar/charged residues Ser-1732.60, Asp-184EC1, and Lys-1923.28, and Ser-3837.39 that interact with the ligand's ring moiety are represented by stick (in red). Aromatic residues Phe-1772.64, Phe-1893.25, Phe-268EC2, and Phe-3797.45 that interact with the ligand's ring moiety are represented by space-filling (in ice-blue). Hydrophobic binding pocket residues Leu-1933.29, Val-1963.33, Leu-3596.51, and Met-3636.55 that interact with the ligand's C3 alkyl chain are represented by space-filling (in green). Residues Thr-1973.33, Thr-2013.37, Ile-2474.56, Phe-2785.42, Thr-2835.47, and Leu-3606.52 that exhibited no ligand contacts are represented by space-filling (in orange). Cys-3556.47 known to interact with the C3 alkyl chain of the cannabinoids (19) is represented by space-filling (in purple), whereas Val-2825.46 thought to interact little with the C3 alkyl chain of the cannabinoids (81) is represented by space-filling (in yellow). Any additional residues within 4 Å of the ligand are also presented by line (in atom type). For the amino acids, only the side chains of the amino acids are represented. H3 and H4 are omitted for a better view. The volume in the space-filling representation is scaled down to 70%. The solvent-accessible pore (in blue dots for low radius surface and in green dots for mid radius surface) was created by using HOLE (58) at the ligand binding core region at the end of the MD simulations. B, aromatic stacking of Phe-1742.61 and Phe-1772.64 with neighboring aromatic residues (in purple) and with the A-ring of the ligands in HU210-CB1y3 (panel i), Δ9-THC-CB1n (panel ii), and CP55940-CB1y (panel iii). For clarity, only H2 and H7 of the receptor are represented.
Minor Binding Pocket Region Was Important for the ABC-ring of HU210
(i) Covered by EC2, the A-ring phenolic hydroxyl oxygen atom of HU210 formed an H-bond directly to the backbone amide oxygen atom of Ile-267EC2, indicating that the A-ring hydroxyl is quite close to the extracellular membrane surface, in agreement with the observation for CP55940 in a POPC bilayer by solid-state NMR (77). (ii) The A-ring was in close proximity to a group of aromatic residues, including Phe-1893.25, Phe-268EC2, and Phe-3797.35, suggesting that aromatic stacking is crucial for HU210 binding. (iii) The C-ring hydroxyl oxygen atom formed a tight H-bond to one of the side chain oxygen atoms of Asp-184EC1 to which Ser-1732.60 formed water-mediated H-bonds. Considering little alteration in HU210 binding by the S1732.60A mutation (39), the role of Ser-1732.60 appears to stabilize the receptor. Overall, the ABC-ring of HU210 tightly binds to the polar/charged/aromatic residues in the minor binding pocket, as shown in the present simulations.
Phe-1742.61 and Phe-1772.64 Stabilize the Minor Binding Pocket through Aromatic Stacking
Significant decreases in ligand binding by the F174A2.61 and F177A2.64 mutations were observed for all the tested ligands (Table 1), suggesting important roles for Phe-1742.61 and Phe-1772.64 in classical and nonclassical cannabinoid binding. Detailed distance analyses showed that both Phe-1742.61 and Phe-1772.64 were not close enough for aromatic stacking with any of the tested nonclassical and classical cannabinoid ligands (Fig. 6A). Phe-1742.61 and Phe-1772.64, instead, were heavily involved in aromatic stacking with the neighboring aromatic residues (Fig. 5B and Fig. 6B). Thus, it appears that both Phe-1742.61 and Phe-1772.64 are indirectly involved in ligand binding by stabilizing the minor binding pocket through aromatic stacking. In support, our simulations of the F1772.64A and F1742.61A mutant receptors show a noticeable change (∼2 Å) in the aromatic ring centroid distance between the A-ring of HU210 and Phe-1742.61 by the F1772.64A mutation or between the A-ring of HU210 and Phe-1772.64 by the Phe-1742.61A mutation (supplemental Fig. 3B, panel i).
FIGURE 6.
Interactions of Phe-1742.61, Phe-1772.64, Leu-1933.29, and Met-3636.55 in the binding pocket. A, aromatic ring centroid distance between the A-ring of the ligand and Phe-1742.61 (in black) or Phe-1772.64 (in red). B, aromatic ring centroid distances between Phe-1742.61 or Phe-1772.64 and the neighboring aromatic residues in HU210-CB1y3 (panel i), Δ9-THC-CB1n (panel ii), and CP55940-CB1y (panel iii). Color coding in panel i: Phe-1742.61/Phe-1702.57, black; Phe-1742.61/Phe-1772.64, red; Phe-1742.61/His-1782.65, green; Phe-1772.64/His-1782.65, blue; Phe-1772.64/His-1812.68, orange; and Phe-1772.64/Phe-3797.35, magenta. Color coding in panel ii: Phe-1742.61/His-1782.65, black; Phe-1742.61/Phe-3817.37, red; Phe-1772.64/His-1782.65, green; Phe-1772.64/His-1812.68, blue; Phe-1772.64/Phe-1893.25, orange; and Phe-1772.64/Phe-3797.35, magenta. Color coding in panel iii: Phe-1742.61/His-1782.65, black; Phe-1772.64/His-1812.68, red; and Phe-1772.64/Phe-3797.35, green. A and B, the upper limit of the distance for aromatic stacking (7.0 Å) (61) is represented by the dotted line. C, contact numbers of the C3 alkyl chain of HU210 (panel i), Δ9-THC (panel ii), or CP55940 (panel iii) with Leu-1933.29 (in black), Met-3636.55 (in red), and Leu-1933.29/Met-3636.55 (in green). Only the side chains of residues were considered in calculating the contact numbers. A criterion of 4.0 Å was used between nonbonded atoms The contact numbers, with the standard deviation in parentheses, averaged over the last 20.0 ns of the simulation are as follows: 31(10), 26(9), and 57(14) for HU210; 10(4), 21(7), and 31(9) for Δ9-THC; and 26(8), 17(8), and 43(12) for CP55940. D, estimated nonbonding interaction energy values of the C3 alkyl chain of HU210 (panel i), Δ9-THC (panel ii), or CP55940 (panel iii) with Leu-1933.29 (in black), Met-3636.55 (in red), and Leu-1933.29/Met-3636.55 (in green). Only the side chains of residues were considered in calculating the nonbonding energy values. The energy values (in kcal/mol), with the standard deviation in parentheses, averaged over the last 20.0 ns of the simulation are as follows: −1.61(0.40), −2.46(0.58), and −4.07(0.73) for HU210; −0.52(0.30), −1.32(0.46), and −1.84(0.53) for Δ9-THC; and −1.28(0.46), −1.54(0.55), and −2.82(0.72) for CP55940.
Leu-1933.29 and Met-3636.55 Form the Hydrophobic Pocket Important for the C3 Alkyl Chain of HU210
The greatest number of C3 alkyl chain contacts with Leu-1933.29 and Met-3636.55, for the classical and nonclassical cannabinoid ligands tested, is HU210 > CP55940 > Δ9-THC (Fig. 6C). The order of favorable interactions between the C3 alkyl chain of the ligand and Leu-1933.29 and Met-3636.55 are as follows: HU210 (−4.07 kcal/mol) > CP55940 (−2.82 kcal/mol) > Δ9-THC (−1.84 kcal/mol) (Fig. 6D). These data correlated well with the observed order of binding potency of these ligands (Table 1), and these results suggest that Leu-1933.29 and Met-3636.55 are important for classical and nonclassical cannabinoid binding, particularly for the C3 alkyl chain of the ligand. As shown in Fig. 5A, the ligand binding modes deduced here reveal a rather flexible hydrophobic binding pocket; the key ligand contact residues, Leu-1933.29 and Met-3636.55, form the wall of the pocket on each side and the proposed toggle switch Phe-2003.36/Trp-3566.48 (75) on the floor of the pocket.
The dimethyl group on the C1′ atom of the C3 alkyl chain was covered in part by the EC2 C-terminal residues known to be important for HU210 binding (78). The C3 alkyl chain of HU210 interacted closely with Leu-1933.29 and Met-3636.55, which is in excellent agreement with the present mutational results. Met-3636.55 appeared to stabilize Tyr-2755.39 at the bottom of the hydrophobic binding pocket and His-270EC2 at the side of the binding pocket. The interactions between sulfur-containing residues (Met and Cys) and aromatic residues are well documented (79). Both residues His-270EC2 and Tyr-2755.39 have been reported to be important for cannabinoid binding (78, 80).
Our mutational analysis indicated that Thr-1973.33, Thr-2013.37, Ile-2474.56, Phe-2785.42, Thr-2835.47, and Leu-3606.52 were not HU210 contacts (data not shown). Similarly, a mutational study by Song et al. (81) indicated that Val-2825.46 was not an HU210 contact. In agreement, these residues are positioned away from the ligand in the present HU210 binding mode (Fig. 5A).
DISCUSSION
Differential Influence of the F1742.61A, F1772.64A, L1933.29A, and M3636.55A Mutations on the Binding Affinity of HU210, Δ9-THC, and CP55940
According to our mutational data, both the F1742.61A and F1772.64A mutations were more detrimental to classical cannabinoid binding than to nonclassical cannabinoid binding (Table 1). As shown in Fig. 6B, our aromatic-aromatic distance analysis indicated that Phe-1742.61 and Phe-1772.64 in HU210-CB1y3 and in Δ9-THC-CB1n were highly involved in aromatic stacking with the neighboring aromatic residues compared with the same residues in CP55940-CB1y, potentially contributing to the stabilization of the ligand binding pocket geometry. In agreement, it was shown in the simulations of the F1742.61A and F1772.64A mutant receptors that H2 moved toward the central core, pushing the bound ABC-ring moiety of the ligand completely out of the minor binding pocket and displacing the C3 alkyl chain in an unfavorable position within the hydrophobic pocket (supplemental Fig. 3).
It was also observed in our mutational analysis that the F1772.64A mutation was more detrimental than the F1742.61A mutation for cannabinoid binding. A possible explanation is that Phe-1772.64, positioned toward the central TM core, would have a larger role than Phe-1742.61, oriented away from the TM core, in stabilizing the minor binding pocket. In support, the backbone r.m.s.d. of the F1772.64A mutant receptor (2.17 Å) was slightly higher than the backbone r.m.s.d. of the F1742.61A mutant receptor (2.01 Å), relative to the wild-type receptor (supplemental Fig. 3A, panels ii and iii). Moreover, it was noted that the F1772.64A mutation precluded aromatic stacking between Phe-1742.61 and Phe-1702.57 (supplemental Fig. 3B, panel iii).
Our mutational data that ligand binding was less sensitive to the M3636.55A mutation than to the L1933.29A mutation suggest Leu-1933.29 may be more important than Met-3636.55 in the receptor-ligand interaction. According to the present ligand binding modes (Fig. 5A), Leu-1933.29, located in the central binding region and connecting the hydrophobic pocket with the minor binding pocket, interacts not only with the C3 alkyl chain but also with the ABC- or AC-ring moiety, whereas Met-3636.55, located in the marginal binding region, interacts only with the C3 alkyl chain. Thus, it is expected that the L1933.29A mutation is more detrimental than the M3636.55A mutation to ligand binding.
Our ligand contact and nonbonding interaction energy analyses show that HU210 has a stronger interaction with Met-3636.55 than CP55940, whereas both ligands have similar interactions with Leu-1933.29 (Fig. 6, C and D), which is in agreement with our mutational data that the M3636.55A receptor exhibited a smaller reduction in binding affinity relative to the wild-type receptor for CP55940 than for HU210 (the M3636.55A receptor exhibited a 15- and 4-fold loss in binding affinity, for HU210 and CP55940, respectively).
Our mutational data that the near wild-type binding affinity by the M3636.55A receptor and a smaller decrease in binding affinity by the L1933.29A receptor when these mutants bound Δ9-THC than when they bound HU210 and CP55940 (Table 1) suggest these residues may be more involved in enabling the binding of agonists with the C3 alkyl chain containing a dimethyl group. According to the present binding modes (Fig. 5A), Δ9-THC showed the lowest number of contact points with Leu-1933.29 and Met-3636.55 and the highest nonbonding interaction energy values with these residues.
Overall, the influence on ligand binding by the F1742.61A and F1772.64A mutations appears larger than the L1933.29A and M3636.55A mutations, mainly because of the involvement of the former mutations in the alteration of the binding pocket geometry, as opposed to the involvement of the latter mutated residues in direct ligand contact only.
Identification of the Hydrophobic Pocket Crucial for the C3 Alkyl Chain of Classical and Nonclassical Cannabinoids
We identified the long sought after hydrophobic binding pocket of the CB1 receptor important for the C3 alkyl chain of classical and nonclassical cannabinoid ligands. According to the present binding mode of HU210, the C3 alkyl chain interacted closely with the hydrophobic binding pocket, in which Leu-1933.29 and Met-3636.55 formed the wall of the pocket on each side, and the Trp-3566.48 rotamer switch of the highly conserved CWXP motif was at the bottom of the binding pocket (82). Combining the reported cannabinoid contact residues (39–41, 83) with the presently identified contact residues, an emerging picture of the binding pocket for HU210 is that it is in an L-shape as follows: horizontally, the minor binding pocket region formed by H2/H3/H7 interacts with the ABC-ring; and vertically, the inner hydrophobic core region formed by H3/H5/H6 interacts with the C3 alkyl chain (Fig. 5A).
Prior to this study, Kapur et al. (39) reported the binding mode of HU210. The model of Kapur et al. (39) is a static model obtained by simple energy minimization in an implicit solvation model, whereas the present model is a dynamic model obtained by time-evolved simulation in a fully hydrated lipid bilayer, an environment mimicking the physiological condition. Our binding mode of HU210 is similar to the model of Kapur et al. (39) with respect to the location of the ABC-ring in the region formed by H2/H3/H7 but different in terms of the orientation of the ABC-ring hydroxyl groups. In our model, these hydroxyl groups are close to the extracellular surface (Fig. 5), whereas in the model of Kapur et al. (39), they appear buried inside the hydrophobic core region, which might cause unfavorable binding interactions. In addition, the C3 alkyl chain in our model is positioned in the region formed by H3/H5/H6, whereas it appears positioned near H7 in the model of Kapur et al. (39). In HU210-CB1y1, similar to the model of Kapur et al. (39) in terms of the orientation of the ABC-ring hydroxyl groups, the C3 alkyl chain was outside the hydrophobic pocket (Fig. 3A), thereby precluding receptor activation. A significant finding of the present model is that the bound ligand, with the positioning of the C3 alkyl chain in agreement with the present mutation studies, was able to induce the Trp-3566.48 rotamer change during the simulation.
Why Does HU210 Have Higher Affinity than Δ9-THC?
As shown in Fig. 5, the binding modes of HU210, Δ9-THC, and CP55940 are very similar with respect to the orientations of the bond connecting the A-ring to the C-ring and the bond connecting the A-ring and the C3 alkyl chain, which are parallel to the membrane surface, in agreement with the observation for CP55940 in a POPC bilayer by solid-state NMR (77). However, the positioning of the C3 alkyl chain of the cannabinoids for the interaction with the hydrophobic binding pocket appears to be important for ligand affinity. As shown in Fig. 5A, the C3 alkyl chains of HU210 and CP55940 located between Leu-1933.29 and Met-3636.55 interacted closely with these residues. In contrast, the C3 alkyl chain of Δ9-THC located below Leu-1933.29 and Met-3636.55 interacted poorly with these residues, possibly because of the absence of the dimethyl group on the C1′ position of the C3 alkyl chain. As a result, Δ9-THC was not able to maintain a binding mode similar to HU210 in the hydrophobic binding pocket. This explains, in part, why Δ9-THC exhibits weak binding to the receptor compared with CP55940 or HU210 (Table 1).
Why Does HU210 Bind with Higher Affinity than CP55940?
Similar to the results reported by other laboratories (41, 84), we observed that HU210 was ∼60-fold more potent than CP55940 in receptor binding (Table 1). Assuming that the C3 alkyl chains of both HU210 and CP55940 have similar binding interactions with the receptor, it is likely that the high binding affinity of HU210 is attributed to its ABC-ring that fits better than the AC-ring of CP55940 in satisfying the size requirement for the maximum hydrophobic interactions (85). Here, we discuss why HU210 binds better than CP55940. (i) The rigid nature of the fused ABC-ring. HU210 will dock preferentially over CP55940 upon initial contact with the receptor, for the rigid ABC-ring, which removes the rotational freedom by fusing the C-ring to the A-ring, is entropically less penalized than the flexible AC-ring of CP55940. (ii) For the position of the A-ring hydroxyl, it has been estimated that the cost of burying a hydroxyl group of Tyr is high (∼4.5 kcal/mol) in a lipophilic environment like inside the lipid bilayer (86). Because of the perpendicular orientation of the A-ring of HU210, its hydroxyl group lies in the binding pore more extracellularly than the same group in CP55940, parallel to the membrane surface (Fig. 3B). Thus, the A-ring hydroxyl group of HU210, located in a less hydrophobic environment, is less penalized than the same group of CP55940, located in a more hydrophobic environment, upon ligand binding. This might explain why an HU210 analog without the A-ring hydroxyl retained ligand binding similar to HU210, whereas a CP55940 analog without the A-ring hydroxyl had significantly reduced ligand binding (87). (iii) For the aromatic stacking, the A-ring of HU210, perpendicular to the membrane surface, will more closely interact with the aromatic residues in the minor binding pocket near the extracellular surface than the A-ring of CP55940, parallel to the membrane surface. In agreement, the interaction energy values between the ligand and aromatic residues Phe-1893.25 and Phe-3797.35 forming aromatic stacking interactions with the ligand's A-ring at the end of the simulation showed that aromatic stacking in HU210 was a few kcal/mol more favorable than in CP55940 (supplemental Fig. 4).
Distance between the C3 Alkyl Chain and Cys-3556.47 Is an Indicator of Receptor Activation
We report in this study the Trp-3566.48 toggle switch followed by the distance between CP55940 and Cys-3556.47 (Fig. 4B, panel vi and supplemental Fig. 2B, panel ii) as supporting indications of receptor activation. We successfully observed a decrease in the distance between the end of the C3 alkyl chain and Cys-3556.47, known as a contact residue of CP55940 (19), in the simulation of HU210-CB1y3 or CP55940-CB1y, suggesting that the observed decrease in this distance was due to a small anti-clockwise rigid-body rotation or inward movement, as observed in the x-ray structure of β2AR in the active state (28), of H6 that caused Cys-3556.47 to move toward the ligand in the TM core. Interestingly, this distance dropped significantly at the beginning of the simulation as shown in Fig. 4B, panel vi, but changed little throughout the later stages of the MD simulation. This suggests other toggle switches, such as a series of micro-switches around the highly conserved NPXXY motif, which eventually lead to the cleavage of the ionic lock (88, 89), would be required for any further decrease in this distance.
Role of the Ligand in CB1 Receptor Activation
The agonist bound to the receptor appears to play a crucial role in initiating receptor conformational changes toward receptor activation. We found that the positioning of the C3 alkyl chain, as in HU210-CB1y3 or CP55940-CB1y, as opposed to that in HU210-CB1y1 or Δ9-THC-CB1n, was crucial for the rotamer change toward receptor activation (Fig. 4B). Previously, we observed the rotamer change in the CB1 receptor model in the absence of ligand (44). However, its rotamer change, as seen in HU210-CB1y1 or HU210-CB1y2, failed to proceed toward receptor activation (Fig. 4A). Similarly, no rotamer change was observed by up to >50 ns of the simulation of CB1n. Thus, it appears that the interaction between the C3 alkyl chain of a classical or nonclassical cannabinoid and the hydrophobic pocket is important for the rotamer change. Bound optimally in the hydrophobic binding pocket between Leu-1933.29 and Met-3636.55, the C3 alkyl chain of HU210 or CP55940 appears to be more effective than that of Δ9-THC in activating the Trp-3566.48 toggle switch. It is likely that the fluctuation of the C3 alkyl chain of a cannabinoid triggers the rearrangement of the aromatic residues around the Phe-2003.36/Trp-3566.48 toggle switch (90). Our functional data of the mutant receptors, showing the impairment of G protein activation, suggest that the hydrophobic binding pocket is crucial for the potency of HU210.
Identification of Trp-2795.43 as Crucial for the Rotamer Change in the CB1 Receptor
Holst et al. (91) reported a putative active conformation of the rotamer switch in the ghrelin receptor with the indole ring of Trp-6.48 toward H5 and proposed that Phe-5.47 serves as another rotameric switch for receptor activation by the formation of aromatic stacking with Trp-6.48. In support, Phe-5.47 mutant receptors impaired both constitutive and the agonist-induced receptor signaling without altering agonist affinity (91). The CB1 receptor lacks the corresponding aromatic residues at the 5.47 position but has Trp-2795.43 one helical turn down from the 5.47 position. In this study, it was shown that a critical event was centered on the movement of Trp-2795.43 that changed its χ1 angle from trans to g+, resulting in Trp-3566.48 being rearranged and favorably positioned toward H5 for the formation of aromatic stacking with Trp-2795.43 (Fig. 3B). The stabilization by Trp-2795.43/Trp-3566.48 in CP55940-CB1y was apparent. It was also shown in CP55940-CB1y that Phe-2003.36/Trp-3566.48 aromatic stacking was not abrogated but rather rearranged for new aromatic stacking between Trp-3566.48 and Trp-2795.43. It appears that Trp-2795.43 plays a crucial role in the Trp-3566.48 rotamer change toward the direction necessary for receptor activation through the rigid-body motion of H6. As evidence, the W2795.43A mutation of the CB1 receptor showed a significant decrease in potency (75), despite different effects on ligand binding (83).
Supplementary Material
Acknowledgments
J.-Y. S. thanks Drs. L. Pedersen and L. Perera for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants 5K01 DA020663 (to J.-Y. S.) and DA020763 (to D. A. K.). This work was also supported in part by the National Science Foundation through TeraGrid resources provided by TACC and Indiana University.

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures,” “Results,” Figs. 1–5, Table 1, and additional references.
The Ballesteros-Weinstein numbering is used (in superscript) to indicate relative position of amino acid residues within the TM helical bundle; for loop residues, only the loop positions are indicated.
Because the CB1 mutational data cross-species (e.g. human and mouse), the numbering scheme for mutated residues is translated into the number that it would be in the human.
- CB1 receptor
- brain cannabinoid receptor
- GPCR
- G protein-coupled receptor
- Δ9-THC
- (−)-Δ9-tetrahydrocannabinol
- TM
- transmembrane
- H
- TM helix
- EC
- extracellular loop
- β2AR
- β2 adrenergic receptor
- MD
- molecular dynamics
- TME
- Tris/Mg2+/EDTA
- POPC
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- r.m.s.d.
- root-mean-square deviation
- GTPγS
- guanosine 5′-3-O-(thio)triphosphate
- PDB
- Protein Data Bank.
REFERENCES
- 1. Devane W. A., Breuer A., Sheskin T., Järbe T. U., Eisen M. S., Mechoulam R. (1992) J. Med. Chem. 35, 2065–2069 [DOI] [PubMed] [Google Scholar]
- 2. Gether U. (2000) Endocr. Rev. 21, 90–113 [DOI] [PubMed] [Google Scholar]
- 3. Howlett A. C., Blume L. C., Dalton G. D. (2010) Curr. Med. Chem. 17, 1382–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Glass M., Northup J. K. (1999) Mol. Pharmacol. 56, 1362–1369 [DOI] [PubMed] [Google Scholar]
- 5. Samama P., Cotecchia S., Costa T., Lefkowitz R. J. (1993) J. Biol. Chem. 268, 4625–4636 [PubMed] [Google Scholar]
- 6. Bouaboula M., Perrachon S., Milligan L., Canat X., Rinaldi-Carmona M., Portier M., Barth F., Calandra B., Pecceu F., Lupker J., Maffrand J. P., Le Fur G., Casellas P. (1997) J. Biol. Chem. 272, 22330–22339 [DOI] [PubMed] [Google Scholar]
- 7. Meschler J. P., Kraichely D. M., Wilken G. H., Howlett A. C. (2000) Biochem. Pharmacol. 60, 1315–1323 [DOI] [PubMed] [Google Scholar]
- 8. Walsh D., Nelson K. A., Mahmoud F. A. (2003) Support Care Cancer 11, 137–143 [DOI] [PubMed] [Google Scholar]
- 9. Zajicek J., Fox P., Sanders H., Wright D., Vickery J., Nunn A., Thompson A. (2003) Lancet 362, 1517–1526 [DOI] [PubMed] [Google Scholar]
- 10. Scotter E. L., Abood M. E., Glass M. (2010) Br. J. Pharmacol. 160, 480–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rahn E. J., Hohmann A. G. (2009) Neurotherapeutics 6, 713–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Adams R., Harfenist M., Lowe S. (1949) J. Am. Chem. Soc. 71, 1624–1628 [Google Scholar]
- 13. Loev B., Bender P. E., Dowalo F., Macko E., Fowler P. J. (1973) J. Med. Chem. 16, 1200–1206 [DOI] [PubMed] [Google Scholar]
- 14. Howlett A. C., Johnson M. R., Melvin L. S., Milne G. M. (1988) Mol. Pharmacol. 33, 297–302 [PubMed] [Google Scholar]
- 15. Busch-Petersen J., Hill W. A., Fan P., Khanolkar A., Xie X. Q., Tius M. A., Makriyannis A. (1996) J. Med. Chem. 39, 3790–3796 [DOI] [PubMed] [Google Scholar]
- 16. Martin B. R., Jefferson R., Winckler R., Wiley J. L., Huffman J. W., Crocker P. J., Saha B., Razdan R. K. (1999) J. Pharmacol. Exp. Ther. 290, 1065–1079 [PubMed] [Google Scholar]
- 17. Rapaka R. S., Makriyannis A. A. (1987) NIDA Res. Monogr., Vol. 79, National Institute on Drug Abuse, Rockville, MD [Google Scholar]
- 18. Melvin L. S., Milne G. M., Johnson M. R., Subramaniam B., Wilken G. H., Howlett A. C. (1993) Mol. Pharmacol. 44, 1008–1015 [PubMed] [Google Scholar]
- 19. Picone R. P., Khanolkar A. D., Xu W., Ayotte L. A., Thakur G. A., Hurst D. P., Abood M. E., Reggio P. H., Fournier D. J., Makriyannis A. (2005) Mol. Pharmacol. 68, 1623–1635 [DOI] [PubMed] [Google Scholar]
- 20. Javitch J. A., Fu D., Liapakis G., Chen J. (1997) J. Biol. Chem. 272, 18546–18549 [DOI] [PubMed] [Google Scholar]
- 21. Farrens D. L., Altenbach C., Yang K., Hubbell W. L., Khorana H. G. (1996) Science 274, 768–770 [DOI] [PubMed] [Google Scholar]
- 22. Okada T., Sugihara M., Bondar A. N., Elstner M., Entel P., Buss V. (2004) J. Mol. Biol. 342, 571–583 [DOI] [PubMed] [Google Scholar]
- 23. Cherezov V., Rosenbaum D. M., Hanson M. A., Rasmussen S. G., Thian F. S., Kobilka T. S., Choi H. J., Kuhn P., Weis W. I., Kobilka B. K., Stevens R. C. (2007) Science 318, 1258–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Warne T., Serrano-Vega M. J., Baker J. G., Moukhametzianov R., Edwards P. C., Henderson R., Leslie A. G., Tate C. G., Schertler G. F. (2008) Nature 454, 486–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jaakola V. P., Griffith M. T., Hanson M. A., Cherezov V., Chien E. Y., Lane J. R., Ijzerman A. P., Stevens R. C. (2008) Science 322, 1211–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Park J. H., Scheerer P., Hofmann K. P., Choe H. W., Ernst O. P. (2008) Nature 454, 183–187 [DOI] [PubMed] [Google Scholar]
- 27. Scheerer P., Park J. H., Hildebrand P. W., Kim Y. J., Krauss N., Choe H. W., Hofmann K. P., Ernst O. P. (2008) Nature 455, 497–502 [DOI] [PubMed] [Google Scholar]
- 28. Rasmussen S. G., Choi H. J., Fung J. J., Pardon E., Casarosa P., Chae P. S., Devree B. T., Rosenbaum D. M., Thian F. S., Kobilka T. S., Schnapp A., Konetzki I., Sunahara R. K., Gellman S. H., Pautsch A., Steyaert J., Weis W. I., Kobilka B. K. (2011) Nature 469, 175–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Choe H. W., Kim Y. J., Park J. H., Morizumi T., Pai E. F., Krauss N., Hofmann K. P., Scheerer P., Ernst O. P. (2011) Nature 471, 651–655 [DOI] [PubMed] [Google Scholar]
- 30. Standfuss J., Edwards P. C., D'Antona A., Fransen M., Xie G., Oprian D. D., Schertler G. F. (2011) Nature 471, 656–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cai K., Itoh Y., Khorana H. G. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 4877–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ghanouni P., Steenhuis J. J., Farrens D. L., Kobilka B. K. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 5997–6002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Klein-Seetharaman J., Yanamala N. V., Javeed F., Reeves P. J., Getmanova E. V., Loewen M. C., Schwalbe H., Khorana H. G. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 3409–3413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yao X., Parnot C., Deupi X., Ratnala V. R., Swaminath G., Farrens D., Kobilka B. (2006) Nat. Chem. Biol. 2, 417–422 [DOI] [PubMed] [Google Scholar]
- 35. Altenbach C., Kusnetzow A. K., Ernst O. P., Hofmann K. P., Hubbell W. L. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 7439–7444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hornak V., Ahuja S., Eilers M., Goncalves J. A., Sheves M., Reeves P. J., Smith S. O. (2010) J. Mol. Biol. 396, 510–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rosenkilde M. M., Benned-Jensen T., Frimurer T. M., Schwartz T. W. (2010) Trends Pharmacol. Sci. 31, 567–574 [DOI] [PubMed] [Google Scholar]
- 38. Fay J. F., Dunham T. D., Farrens D. L. (2005) Biochemistry 44, 8757–8769 [DOI] [PubMed] [Google Scholar]
- 39. Kapur A., Hurst D. P., Fleischer D., Whitnell R., Thakur G. A., Makriyannis A., Reggio P. H., Abood M. E. (2007) Mol. Pharmacol. 71, 1512–1524 [DOI] [PubMed] [Google Scholar]
- 40. Murphy J. W., Kendall D. A. (2003) Biochem. Pharmacol. 65, 1623–1631 [DOI] [PubMed] [Google Scholar]
- 41. Song Z. H., Bonner T. I. (1996) Mol. Pharmacol. 49, 891–896 [PubMed] [Google Scholar]
- 42. Chin C. N., Lucas-Lenard J., Abadji V., Kendall D. A. (1998) J. Neurochem. 70, 366–373 [DOI] [PubMed] [Google Scholar]
- 43. Shim J. Y. (2010) Curr. Top. Med. Chem. 10, 779–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shim J. Y., Rudd J., Ding T. T. (2011) Proteins 79, 581–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen C., Okayama H. (1987) Mol. Cell. Biol. 7, 2745–2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Abadji V., Lucas-Lenard J. M., Chin C., Kendall D. A. (1999) J. Neurochem. 72, 2032–2038 [DOI] [PubMed] [Google Scholar]
- 47. Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 48. Cheng Y., Prusoff W. H. (1973) Biochem. Pharmacol. 22, 3099–3108 [DOI] [PubMed] [Google Scholar]
- 49. Shim J. Y. (2009) Biophys. J. 96, 3251–3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stone J. E., Phillips J. C., Freddolino P. L., Hardy D. J., Trabuco L. G., Schulten K. (2007) J. Comput. Chem. 28, 2618–2640 [DOI] [PubMed] [Google Scholar]
- 51. Phillips J. C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R. D., Kalé L., Schulten K. (2005) J. Comput. Chem. 26, 1781–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chen J., Im W., Brooks C. L., 3rd (2006) J. Am. Chem. Soc. 128, 3728–3736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Buck M., Bouguet-Bonnet S., Pastor R. W., MacKerell A. D., Jr. (2006) Biophys. J. 90, L36–L38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Brooks B. R., Bruccoleri R. E., Olafson B. D., States D. J., Swaminathan S., Karplus M. (1983) J. Comp. Chem. 4, 187–217 [Google Scholar]
- 55. MacKerell A. D., Jr., Bashford D., Bellott M., Dunbrack R. L., Jr., Evanseck J., Field M. J., Fischer S., Gao J., Guo H., Ha S., Joseph D., Kuchnir L., Kuczera K., Lau F. T. K., Mattos C., Michnick S., Ngo T., Nguyen D. T., Prodhom B., Reiher I. W., Roux B., Schlenkrich M., Smith J., Stote R., Straub J., Watanabe M., Wiorkiewicz-Kuczera J., Yin D., Karplus M. (1998) J. Phys. Chem. B 102, 3586–3616 [DOI] [PubMed] [Google Scholar]
- 56. Feller S., MacKerell A. D., Jr. (2000) J. Phys. Chem. B 104, 7510–7515 [Google Scholar]
- 57. Saam J., Tajkhorshid E., Hayashi S., Schulten K. (2002) Biophys. J. 83, 3097–3112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Smart O. S., Goodfellow J. M., Wallace B. A. (1993) Biophys. J. 65, 2455–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Feller S. E., Zhang Y., Pastor R. W., Brooks B. R. (1995) J. Chem. Phys. 103, 4613–4621 [Google Scholar]
- 60. Hoover W. G. (1985) Phys. Rev. A 31, 1695–1697 [DOI] [PubMed] [Google Scholar]
- 61. Burley S. K., Petsko G. A. (1985) Science 229, 23–28 [DOI] [PubMed] [Google Scholar]
- 62. Essmann U., Perera L., Berkowitz M. L., Darden T., Lee H., Pedersen L. G. (1995) J. Chem. Phys. 103, 8577–8593 [Google Scholar]
- 63. Tuckerman M., Berne B. J., Martyna G. (1992) J. Chem. Phys. 97, 1990–2001 [Google Scholar]
- 64. Jones G., Willett P., Glen R. C. (1995) J. Mol. Biol. 245, 43–53 [DOI] [PubMed] [Google Scholar]
- 65. Verdonk M. L., Cole J. C., Hartshorn M. J., Murray C. W., Taylor R. D. (2003) Proteins 52, 609–623 [DOI] [PubMed] [Google Scholar]
- 66. Darve E., Pohorille A. (2001) J. Chem. Phys. 115, 9169–9183 [Google Scholar]
- 67. Hénin J., Chipot C. (2004) J. Chem. Phys. 121, 2904–2914 [DOI] [PubMed] [Google Scholar]
- 68. Vriend G. (1990) J. Mol. Graph. 8, 29, 52–56 [DOI] [PubMed] [Google Scholar]
- 69. Hooft R. W., Vriend G., Sander C., Abola E. E. (1996) Nature 381, 272 [DOI] [PubMed] [Google Scholar]
- 70. Breneman C. M., Wiberg K. B. (1990) J. Comp. Chem. 11, 361–373 [Google Scholar]
- 71. Frisch M. J., Trucks G. W., Schlegel H. B., Scuseria G. E., Robb M. A., Cheeseman J. R., Scalmani G., Barone V., Mennucci B., Petersson G. A., Nakatsuji H., Caricato M., Li X., Hratchian H. P., Izmaylov A. F., Bloino J., Zheng G., Sonnenberg J. L., Hada M., Ehara M., Toyota K., Fukuda R., Hasegawa J., Ishida M., Nakajima T., Honda Y., Kitao O., Nakai H., Vreven T., Montgomery J. A., Jr., Peralta J. E., Ogliaro F., Bearpark M., Heyd J. J., Brothers E., Kudin K. N., Staroverov V. N., Kobayashi R., Normand J., Raghavachari K., Rendell A., Burant J. C., Iyengar S. S., Tomasi J., Cossi M., Rega N., Millam J. M., Klene M., Knox J. E., Cross J. B., Bakken V., Adamo C., Jaramillo J., Gomperts R., Stratmann R. E., Yazyev O., Austin A. J., Cammi R., Pomelli C., Ochterski J. W., Martin R. L., Morokuma K., Zakrzewski V. G., Voth G. A., Salvador P., Dannenberg J. J., Dapprich S., Daniels A. D., Farkas O., Foresman J. B., Ortiz J. V., Cioslowski J., Fox D. J. (2009) Gaussian 09, Revision A. 02, Gaussian, Inc., Wallingford, CT [Google Scholar]
- 72. Humphrey W., Dalke A., Schulten K. (1996) J. Mol. Graphics 14, 33–38 [DOI] [PubMed] [Google Scholar]
- 73. Ballesteros J. A., Jensen A. D., Liapakis G., Rasmussen S. G., Shi L., Gether U., Javitch J. A. (2001) J. Biol. Chem. 276, 29171–29177 [DOI] [PubMed] [Google Scholar]
- 74. Shi L., Liapakis G., Xu R., Guarnieri F., Ballesteros J. A., Javitch J. A. (2002) J. Biol. Chem. 277, 40989–40996 [DOI] [PubMed] [Google Scholar]
- 75. McAllister S. D., Hurst D. P., Barnett-Norris J., Lynch D., Reggio P. H., Abood M. E. (2004) J. Biol. Chem. 279, 48024–48037 [DOI] [PubMed] [Google Scholar]
- 76. Shen C. P., Xiao J. C., Armstrong H., Hagmann W., Fong T. M. (2006) Eur. J. Pharmacol. 531, 41–46 [DOI] [PubMed] [Google Scholar]
- 77. Kimura T., Cheng K., Rice K. C., Gawrisch K. (2009) Biophys. J. 96, 4916–4924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ahn K. H., Bertalovitz A. C., Mierke D. F., Kendall D. A. (2009) Mol. Pharmacol. 76, 833–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Meyer E. A., Castellano R. K., Diederich F. (2003) Angew. Chem. Int. Ed. Engl. 42, 1210–1250 [DOI] [PubMed] [Google Scholar]
- 80. McAllister S. D., Tao Q., Barnett-Norris J., Buehner K., Hurst D. P., Guarnieri F., Reggio P. H., Nowell Harmon K. W., Cabral G. A., Abood M. E. (2002) Biochem. Pharmacol. 63, 2121–2136 [DOI] [PubMed] [Google Scholar]
- 81. Song Z. H., Slowey C. A., Hurst D. P., Reggio P. H. (1999) Mol. Pharmacol. 56, 834–840 [PubMed] [Google Scholar]
- 82. Hanson M. A., Stevens R. C. (2009) Structure 17, 8–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. McAllister S. D., Rizvi G., Anavi-Goffer S., Hurst D. P., Barnett-Norris J., Lynch D. L., Reggio P. H., Abood M. E. (2003) J. Med. Chem. 46, 5139–5152 [DOI] [PubMed] [Google Scholar]
- 84. Ashton J. C., Wright J. L., McPartland J. M., Tyndall J. D. (2008) Curr. Med. Chem. 15, 1428–1443 [DOI] [PubMed] [Google Scholar]
- 85. Kuntz I. D., Chen K., Sharp K. A., Kollman P. A. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 9997–10002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. MacCallum J. L., Bennett W. F., Tieleman D. P. (2008) Biophys J. 94, 3393–3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Huffman J. W., Yu S., Showalter V., Abood M. E., Wiley J. L., Compton D. R., Martin B. R., Bramblett R. D., Reggio P. H. (1996) J. Med. Chem. 39, 3875–3877 [DOI] [PubMed] [Google Scholar]
- 88. Nygaard R., Frimurer T. M., Holst B., Rosenkilde M. M., Schwartz T. W. (2009) Trends Pharmacol. Sci. 30, 249–259 [DOI] [PubMed] [Google Scholar]
- 89. Ahuja S., Smith S. O. (2009) Trends Pharmacol. Sci. 30, 494–502 [DOI] [PubMed] [Google Scholar]
- 90. Shim J. Y., Howlett A. C. (2004) J. Chem. Inf. Comput. Sci. 44, 1466–1476 [DOI] [PubMed] [Google Scholar]
- 91. Holst B., Nygaard R., Valentin-Hansen L., Bach A., Engelstoft M. S., Petersen P. S., Frimurer T. M., Schwartz T. W. (2010) J. Biol. Chem. 285, 3973–3985 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.