Abstract
Maraviroc is a nonpeptidic small molecule human immunodeficiency virus type 1 (HIV-1) entry inhibitor that has just entered the therapeutic arsenal for the treatment of patients. We recently demonstrated that maraviroc binding to the HIV-1 coreceptor, CC chemokine receptor 5 (CCR5), prevents it from binding the chemokine CCL3 and the viral envelope glycoprotein gp120 by an allosteric mechanism. However, incomplete knowledge of ligand-binding sites and the lack of CCR5 crystal structures have hampered an in-depth molecular understanding of how the inhibitor works. Here, we addressed these issues by combining site-directed mutagenesis (SDM) with homology modeling and docking. Six crystal structures of G-protein-coupled receptors were compared for their suitability for CCR5 modeling. All CCR5 models had equally good geometry, but that built from the recently reported dimeric structure of the other HIV-1 coreceptor CXCR4 bound to the peptide CVX15 (Protein Data Bank code 3OE0) best agreed with the SDM data and discriminated CCR5 from non-CCR5 binders in a virtual screening approach. SDM and automated docking predicted that maraviroc inserts deeply in CCR5 transmembrane cavity where it can occupy three different binding sites, whereas CCL3 and gp120 lie on distinct yet overlapped regions of the CCR5 extracellular loop 2. Data suggesting that the transmembrane cavity remains accessible for maraviroc in CCL3-bound and gp120-bound CCR5 help explain our previous observation that the inhibitor enhances dissociation of preformed ligand-CCR5 complexes. Finally, we identified residues in the predicted CCR5 dimer interface that are mandatory for gp120 binding, suggesting that receptor dimerization might represent a target for new CCR5 entry inhibitors.
Keywords: Allosteric Regulation, Docking, G-protein-coupled Receptors (GPCR), HIV-1, Molecular Modeling, Mutagenesis
Introduction
CC chemokine receptor 5 (CCR5) is a G-protein-coupled, high affinity receptor for the chemokines macrophage inflammatory protein 1α (MIP1α)/CCL3, MIP1β/CCL4, regulated upon activation, normal T-cell expressed and secreted/CCL5, and monocyte chemoattractant protein-2 (MCP2)/CCL8 (1) and serves as a CD4 coreceptor for R5-tropic human immunodeficiency virus type 1 (HIV-1) entry into activated memory CD4+ T-lymphocytes and monocyte-derived macrophages (2). The importance of CCR5 in HIV-1 transmission and propagation is indicated by the observation that naturally occurring polymorphisms of the CCR5 gene confer resistance to infection (for review see Ref. 3). This has stimulated the development of CCR5 ligands to prevent HIV infection, including chemically modified chemokines (4, 5), monoclonal antibodies (6, 7), and nonpeptidic low molecular weight compounds (8), among which maraviroc (MVC)3 (from Pfizer) (Fig. 1) has just been approved for the treatment of patients infected with only R5 viruses (9, 10).
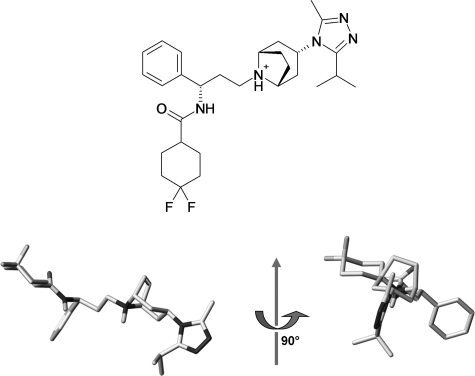
FIGURE 1.
Chemical structure of maraviroc (top) and presumed active three-dimensional structure of maraviroc (bottom).
Experimental evidence suggests that the latter class of molecules prevents chemokines and the surface subunit of the HIV-1 envelope glycoprotein (gp120) from binding to and signaling through CCR5 and inhibits viral entry by means of an allosteric mechanism. First of all, an orthosteric mode of action is not supported by the fact that small molecule CCR5 inhibitors and gp120 or chemokines have large size differences as well as by data showing that interaction of these large size proteins with CCR5 is multivalent (11–13). In further support of the notion that these entry inhibitors are CCR5 allosteric modulators are data showing that these molecules can differentially affect the binding of different chemokines or antibodies to CCR5 (8, 14) and alter different functions of the receptor to different extents (14, 15). As a paradigmatic example of this property, the entry inhibitor aplaviroc permits CCL5 binding to CCR5 and some CCL5-mediated signaling but fully prevents calcium mobilization in response to the chemokine (14, 15). Recently, we have reported that the small molecule HIV-1 entry inhibitors TAK779 and MVC accelerate dissociation of CCL3 and gp120 from CCR5, thus implying that TAK779 and MVC could bind to CCL3- or gp120-occupied CCR5 thereby suggesting that these molecules and the chemokine or the viral glycoprotein have different binding sites in CCR5 (16). Interestingly, although MVC has a lower capacity to dissociate gp120 compared with TAK779, we found that it is 100-fold more potent at inhibiting infection, suggesting that MVC can alter distinct steps of CCR5 usage during viral entry. Unfortunately, the lack of CCR5 crystal structure made it difficult to provide further explanations on how MVC works.
Site-directed mutagenesis (SDM) and molecular modeling agreed on the view that small molecule CCR5 inhibitors bind to a hydrophobic cavity located within the transmembrane domain of CCR5 (17–20), thus presumably far from the proposed binding sites for chemokines and gp120 at the receptor surface. In two recent works, binding of MVC to CCR5 mutants was evaluated by using displacement experiments of [125I]CCL5 binding to receptors (21) and cell-cell fusion assays (22). In both cases, data were interpreted in the light of a rhodopsin-based homology model of CCR5.
In this study, different crystal structures of G-protein-coupled receptors were evaluated for their suitability for CCR5 modeling. Seven CCR5 models were built and compared for their ability to agree with the effects of 48 mutations in the CCR5 transmembrane and extracellular regions on binding of CCL3, gp120, and MVC as well as to discriminate between CCR5 and non-CCR5 ligands in a virtual screening approach. In this regard, we found that the CCR5 model based on the recently published (23) crystal structure of the other HIV-1 coreceptor CXCR4 bound to the peptide CVX15 (PDB code 3OE0) is the most accurate model. Docking MVC into this model and SDM data provided with new insights into the mechanisms whereby MVC allosterically regulates chemokine and gp120 binding to CCR5. Results also point to a role for CCR5 dimerization in interaction of gp120 with the coreceptor, suggesting that this process might represent a therapeutic target for inhibition of HIV infection.
EXPERIMENTAL PROCEDURES
Site-directed Mutagenesis and Transfections
The human CCR5 coding sequence cloned in the HIV-1-based lentiviral pTRIP vector described previously (24) was amplified by PCR using forward (5′-CACCCGTACGATGGATTATCAAGTGTCAAGTCCAATCTATGAC-3′) and reverse (5′-GGCGCGCTCACAAGCCCACAGATATTTCCTGCTCC-3′) primers with the expanded high fidelity PCR system (Roche Applied Science). The PCR products were then cloned directionally into the pcDNA3.1D/V5-His-TOPO plasmid (Invitrogen) according to the manufacturer's instructions. Point mutations were introduced into the CCR5 sequence using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). All mutations were confirmed by DNA sequence analysis (Genome Express). HEK 293T cells, which were cultured as described previously (16), were transiently transfected with the receptor-encoding plasmids using the calcium phosphate-DNA coprecipitation method of transfection. Forty eight hours after transfection, cell-surface expression levels of receptors were measured by flow cytometry analysis using the phycoerythrin-conjugated anti-CCR5 mAbs 2D7 and CTC5 (BD Biosciences). Analysis was performed on a FACSCalibur flow cytometer (BD Biosciences).
Radioligand Binding Assays
Protocols for membrane preparations from the transfected HEK 293T cells, displacement experiments of 0.1 nm [125I]CCL3 (PerkinElmer Life Sciences) binding to wild-type and mutant CCR5-expressing membranes by MVC, and measurement of specific binding of 10 nm [35S]gp120 from the R5-tropic HIV-1 primary strain Bx08 in the presence of 30 nm recombinant soluble human CD4 (Protein Sciences Corp., Meriden, CT) were described previously (16). Saturation binding experiments of [3H]MVC (specific activity 16 Ci/mmol, kindly provided by Pfizer, Sandwich, UK) were performed in 96-well basic flash plates (PerkinElmer Life Sciences). Membrane aliquots containing 2.5 μg of proteins and concentrations of [3H]MVC ranging from 0.5 up to 32 nm were incubated for 1 h at room temperature in a 0.2-ml final volume of assay buffer (50 mm HEPES, pH 7.4, 1 mm CaCl2, 5 mm MgCl2, 0.5% BSA). Nonspecific binding was measured in the presence of 2 μm of unlabeled MVC (obtained from the AIDS Research and Reference Reagent Program, National Institutes of Health). Incubations were then stopped by centrifugation (800 × g for 10 min at 4 °C) and removal of supernatants. Plates were counted in a Wallac 1450 MicroBeta TriLux® (PerkinElmer Life Sciences). Data were fitted to a one-site binding model using the Prism software (GraphPad Software Inc., San Diego).
Modeling the CCR5 Three-dimensional Structure
The seven transmembrane domain (7TM) of CCR5 was modeled using the GPCRmod modeling package (25). A separate model was generated for each of the following templates: inactivate bovine rhodopsin (PDB code 1U19), active bovine opsin (PDB code 3DQB), human adrenergic receptor α2 (PDB code 2RH1), and human adenosine receptor (PDB code 3EML). Each transmembrane helix (TM) was considered individually in the sequence alignment. Alignments were based on the amino acids motifs characteristic of the GPCR class A (or rhodopsin-like class), to which the target and the template both belong. The four three-dimensional models were built using the three-dimensional template backbone and libraries of side chain rotamers and then were subjected to energy minimization using AMBER8 to remove steric clashes.
A fifth three-dimensional model of the 7TM was built by combining ab initio and homology models. The TM2 and TM3 structures were built as ideal α-helices, which were linked by the four residues of the first extracellular loop (ECL). The TM1 and TM4–7 structures are threading-generated helices based on bovine rhodopsin crystal structure (PDB code 1F88). The inter-helix interface of the 7TM was optimized by molecular dynamics simulations (26). The model was refined to accommodate five known modulators (45) and, in a subsequent stage, to improve the aliphatic and aromatic packing of residues and to establish hydrogen bonds (H-bonds) between H-bond donor atoms and H-bond acceptor atoms in apolar environments. Practically, the structure was edited using Sybyl-X (Tripos, Inc., St. Louis, MO), and the TM5 was rotated 10° clockwise along its helical axis (observer at the top of the TM N terminus) and the rotameric state chosen in the Lovell library. The 177TCS179 peptide of the second extracellular loop (ECL2) was built ab initio, with χ1(Cys-178) set to −168, χ1(Cys-101) set to 62.1, and the torsion angle of the disulfide bridge (Cβ-Sγ-Sγ-Cβ) set to 58.5. The three-dimensional model was last subjected to sequential energy minimizations with constraints fixing intra-helical H-bonds.
Finally, two models of the full CCR5 were built by homology to the CXCR4 receptor in complex with the small molecule IT1t (PDB code 3ODU) and with the cyclic peptide CVX15 (PDB code 3OE0). Sequence alignment and homology modeling were performed using MOE 2009.10 (Chemical Computing Group Inc., Montreal, Canada). The three template sequences (PDB code 3ODU, chains A and B, and code 3OE0, chain A) were edited to remove the lysozyme insert and then aligned to the target sequence (MOE default settings). A constraint was placed on both CCR5 Trp-190 and CXCR4 Trp-195 to avoid the introduction of a gap in the N-terminal region of TM5. The coordinates of CCR5 were modeled using MOE defaults settings, except for the AMBER99 force field and R-field implicit solvation model. The two models built from 3OE0 and 3ODU templates consist of a single chain and two chains, respectively. They were subjected to sequential energy minimization in vacuum using AMBER8 with the parm98 parameter set. Four cycles of 2500 steps of steepest descent followed by 2500 steps of conjugated gradient were applied (root mean square gradient of the potential energy in the 0.05 kcal/mol/Å range). The α carbons were constrained using a harmonic constant of 50, 25, and 10 kcal/mol/Å in the first, second, and third cycles, respectively. All sequence alignments are given in the supplemental material (see supplemental Fig. S1).
Docking
The three-dimensional structure of MVC was generated using Corina version 3.1 (Molecular Network GmbH, Erlangen, Germany) from a two-dimensional sketch drawn using Marvinsketch (MarvinBeans version 5.3.01, ChemAxon, Budapest, Hungary). A positive charge was assigned to the ternary amine group. Only the presumed active conformer was considered (i.e. R-exo chair, Fig. 1) (27) The three-dimensional structure was stored in MOL2 file format.
Docking was carried out using five different programs as follows: GOLD (28); SURFLEX (29); PLANTS (30); FRED (OpenEye Scientific Software); and FLEXX (31). For GOLD docking, we used the 4.2.1 version of the program with the automatic settings of the genetic algorithm. The binding site consists of all residues less than 15 Å from the 7TM cavity center (defined as the mass center of Glu-283, Thr-105, and Tyr-108 residues). We submitted 10 independent jobs for each ligand-protein complex, and we allowed the early termination of the algorithm when the top three poses were within 1.5 Å r.m.s.d. Poses were scored with the GOLD fitness score (GOLDScore). For SURFLEX docking, we used the 2.4.2 version of the program with the automatic settings of the docking algorithm. The protomol was generated based on the automatically detected cavity (proto_thres set to 0.5 and proto_bloat set to 1). We scored and output the 10 final poses (ndock_final set to 10, div_rms set to 0.1). For PLANTS docking, we used the 1.1 version of the program with the automatic settings of the ACO optimizer. The binding site consists of all residues closer than 15 Å from the 7TM cavity center (as defined for GOLD docking). The final 10 poses were scored with the Chemplp function. For FRED docking, we used the 2.2.5 version of the program with the default settings (exhaustive search using Chemgauss3, rigid body optimization of poses using Chemgauss3, and best pose selected using consensus PLP-Chemgauss3-OEChemscore scoring). The binding site was defined with a box enclosing all residues closer than 15 Å from the TM cavity center (as defined for GOLD docking). A library of 500 MVC conformers was created using Omega 2 with the default settings (32). For FLEXX docking, we used the 3.1.4 version of the program with the default settings as defined in LeadIt version 1.1 (Triangle matching for base placement). The binding site (as defined for GOLD docking) was tagged in the input MOL2 file of the receptor.
Virtual Screening
We designed four compound libraries to evaluate the CCR5 three-dimensional models as follows. 1) The MVC-like library is made of nine molecules in the MVC chemical series. All molecules are high affinity CCR5 binders, with the functional effect at nanomolar concentrations. The direct edition of the MVC three-dimensional structure followed by energy minimized using Sybyl-X (Tripos, Inc., St. Louis, MO) yielded the 14 entries of the library, including R-exo boat and R-exo chair diastereoisomers. 2) The DIVERSE-active library is made of 17 high affinity CCR5 binders, whose scaffolds are chemically diverse and differ from maraviroc structure. The three-dimensional structure of the library entries was obtained as described above for maraviroc. 3) The FOCUS-inactive library is made of 87 compounds that do not bind CCR5 at 100 μm concentration in vitro, although they meet some of the structural or pharmacophoric requirements for CCR5 binding (e.g. all compounds possess a positively charged nitrogen atom). The three-dimensional structure generation using Corina version 3.1 yielded 178 entries, including all possible enantiomers and diastereoisomers. 4) The ChemoR-decoys library is made of 117 known binders of five members of the chemokine receptor family. The three-dimensional structure generation using Corina version 3.1 yielded 117 entries. The composition of the four libraries is given in supplemental Fig. S2.
The virtual screening of libraries was performed by SURFLEX docking into the CCR5 7TM cavity. The docking settings were identical to that described above for MVC docking, except that only the best scored pose was output for each ligand (ndock_final set to 1). Docking results were processed to identify inter-molecular interactions. The in-house program IFP (33) detected whether an ionic bond was predicted between a positively charged group of the docked ligand and the Glu-283 carboxylate group of the CCR5 receptor (the maximal distance between two charges was set to 5.5 Å). It also evaluated the number of π-π interactions between aromatic rings in the docked ligand and the side chain of residues Tyr-37, Trp-86, Tyr-108, Phe-109, Phe-112, Trp-248, and Tyr-251 in the CCR5 receptor (the maximal distance between two aromatic rings was set to 4 Å, with no requirements for the angle between the ring planes). A Pipeline Pilot protocol (version 4.1, SciTegic 2009) analyzed for each entry the SURFLEX scores (pKi, crash, and polar) and identified interactions in the docked complex to produce hit lists.
RESULTS
Site-directed Mutagenesis of CCR5
We performed single-site mutagenesis of residues within the extracellular loops of CCR5 as well as its transmembrane domain surrounding the proposed hydrophobic binding pocket for small molecule CCR5 inhibitors (Fig. 2), which corresponds to the binding site for retinal in rhodopsin (18, 21). Results are reported in Table 1 and Fig. 4.
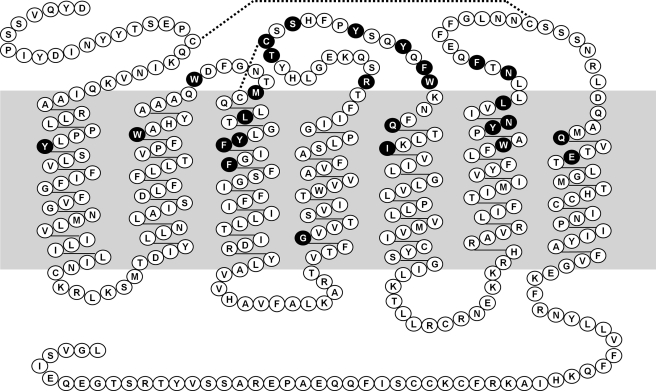
FIGURE 2.
Snake plot of human CCR5 sequence. Residues tested in this study are highlighted with a black background.
TABLE 1.
Site-directed mutagenesis data
The figures for binding and displacement experiments are written in boldface type if significant changes were observed upon mutation of the receptor.
| CCR5 mutantsa | 2D7 geometric MFI (% WT-CCR5) | CTC5 geometric MFI (% WT-CCR5) | [125I]CCL3 binding (% WT -CCR5) | [35S]gp120 binding (% WT-CCR5) | MVC |
||
|---|---|---|---|---|---|---|---|
| [3H]MVC binding KDb | Displacement of [125I]CCL3 IC50b | ||||||
| TM1 | Y37A | 115 ± 13 | 106 ± 6.9 | 14 ± 1.1 (13)c | 41 ± 7.5 (37)c | 0.80 | 2.2 ± 1.3 |
| TM1 | Y37F | 86 ± 28 | 103 ± 11 | 78 ± 10 (88) | 53 ± 1.5 (59) | 1.10 | 0.9 ± 0.2 |
| TM2 | W86A | 117 ± 0.4 | 112 ± 1.6 | 40 ± 2.1 (35) | <10 | 42.9 ± 13 | |
| ECL1 | W94A | 140 ± 15 | 121 ± 7.3 | <10 | <10 | 0.79 ± 0.3 | |
| ECL1 | W94M | 103 ± 7.5 | 108 ± 3.4 | <10 | <10 | 1.18 ± 0.1 | |
| ECL1 | M100A | 114 ± 14 | 122 ± 17 | 95 ± 1.0 (82) | 127 ± 21 (107) | 1.0 ± 0.1 | |
| ECL1 | M100T | 79 ± 9.3 | 86 ± 10 | 42 ± 4.1 (52) | 53 ± 1.5 (65) | 0.63 | 1.3 ± 0.3 |
| TM3 | L104A | 131 ± 18 | 113 ± 1.7 | 81 ± 6.3 (67) | 127 ± 19 (97) | 1.07 | 0.9 ± 0.2 |
| TM3 | L104F | 128 ± 17 | 107 ± 8.2 | 43 ± 3.8 (37) | 53 ± 21 (46) | 2.05 | 1.2 ± 0.2 |
| TM3 | Y108A | 65 ± 21 | 92 ± 8.0 | 44 ± 13 (49) | <10 | 75.5 ± 6.4 | |
| TM3 | F109A | 91 ± 7.3 | 82 ± 6.8 | 159 ± 1.0 (186) | 23 ± 11 (27) | 1.43 | 1.4 ± 0.2 |
| TM3 | F109H | 106 ± 6.1 | 105 ± 18 | 179 ± 23 (174) | 24 ± 0.7 (23) | 1.5 ± 0.1 | |
| TM3 | F112A | 21 ± 1.5 | 28 ± 2.3 | 23 ± 7.6 (101) | 31 ± 7.6 (121) | 0.4 ± 0.2 | |
| TM3 | F112Y | 125 ± 25 | 112 ± 31 | 104 ± 19 (89) | 169 ± 3.6 (115) | 1.1 ± 0.3 | |
| TM4 | G145A | 137 ± 2.7 | 130 ± 6.1 | 103 ± 11 (78) | 150 ± 17 (113) | 0.60 | 1.3 ± 0.3 |
| ECL2 | R168A | 77 ± 8.7 | 67 ± 11 | 47 ± 8.5 (64) | 45 ± 11 (72) | 1.3 ± 0.5 | |
| ECL2 | T177A | 92 ± 2.2 | 118 ± 4.3 | 13 ± 4.4 (13) | 8.1 ± 6.1 (7.6) | 1.70 | 1.4 ± 0.5 |
| ECL2 | T177Q | 82 ± 13 | 98 ± 9.2 | 13 ± 1.6 (15) | 18 ± 6.7 (22) | 0.79 | 1.3 ± 0.2 |
| ECL2 | C178A | 0 ± 0 | 74 ± 37 | <10 | <10 | ||
| ECL2 | S180A | 124 ± 1.1 | 120 ± 13 | 85 ± 14 (69) | 139 ± 19 (115) | 1.10 | 0.9 ± 0.0 |
| ECL2 | S180P | 85 ± 1.7 | 94 ± 2.0 | 58 ± 8.9 (65) | <10 | 0.56 | 0.9 ± 0.1 |
| ECL2-TM5 | Y184A | 160 ± 37 | 134 ± 23 | 33 ± 7.7 (23) | 133 ± 27 (122) | 2.1 ± 0.7 | |
| ECL2-TM5 | Y187A | 88 ± 10 | 112 ± 8.5 | <10 | 144 ± 8.0 (161) | 1.19 ± 0.5 | |
| ECL2-TM5 | F189A | 57 ± 3.9 | 73 ± 4.4 | 71 ± 2.6 (110) | <10 | 1.5 ± 0.0 | |
| ECL2-TM5 | W190A | 22 ± 13 | 84 ± 46 | 16 ± 0.4 (19) | <10 | 1.6 ± 0.8 | |
| ECL2-TM5 | Q194A | 105 ± 4.5 | 110 ± 2.0 | 84 ± 5.8 (78) | 162 ± 40 (150) | 0.43 | 0.6 ± 0.2 |
| ECL2-TM5 | Q194H | 104 ± 1.4 | 113 ± 4.7 | 85 ± 6.4 (78) | 184 ± 40 (169) | 2.40 | 5.6 ± 0.9 |
| TM5 | I198A | 124 ± 14 | 118 ± 12 | 130 ± 4.7 (109) | 49 ± 0.8 (41) | 17.40 | 131 ± 29 |
| TM6 | W248A | 3.6 ± 2.3 | 14 ± 0.4 | <10 | <10 | ||
| TM6 | Y251A | 10 ± 2.4 | 31 ± 0.1 | 13 ± 4.8 (64) | <10 | 7.7 ± 2.3 | |
| TM6 | Y251F | 68 ± 0.8 | 80 ± 3.6 | 88 ± 7.8 (120) | 38 ± 9.3 (52) | 3.3 ± 1.2 | |
| TM6 | Y251I | 4.8 ± 2.1 | 20 ± 1.8 | 14 ± 2.2 (119) | <10 | 19.9 ± 2.1 | |
| TM6 | N252A | 101 ± 2.5 | 86 ± 3.9 | 46 ± 5.2 (49) | 92 ± 29 (98) | 0.80 | 1.1 ± 0.4 |
| TM6 | N252I | 141 ± 14 | 137 ± 8.4 | 16 ± 0.8 (12) | 147 ± 6.2 (106) | 0.44 | 1.2 ± 0.3 |
| TM6 | L255A | 39 ± 2.5 | 41 ± 0.5 | 49 ± 0.1 (123) | 30 ± 2.5 (76) | 2.3 | 0.9 ± 0.5 |
| TM6 | N258A | 91 ± 5.5 | 86 ± 8.6 | 68 ± 4.5 (77) | 123 ± 5.9 (140) | 0.48 | 0.4 ± 0.3 |
| TM6-ECL3 | N258M | 85 ± 6.0 | 87 ± 0.3 | 71 ± 0.4 (82) | 85 ± 3.3 (98) | 1.27 | 0.7 ± 0.2 |
| TM6-ECL3 | N258Q | 138 ± 2.9 | 141 ± 7.2 | 81 ± 2.4 (58) | 149 ± 28 (107) | 1.10 | 1.2 ± 0.2 |
| TM6-ECL3 | F260A | 40 ± 5.1 | 60 ± 15 | 46 ± 3.8 (97) | 13 ± 13 (32) | 0.8 ± 0.0 | |
| TM7 | Q280A | 106 ± 6.2 | 110 ± 2.0 | 84 ± 6.6 (78) | 128 ± 21 (119) | 0.74 | 1.1 ± 0.1 |
| TM7 | E283A | 70 ± 4.3 | 75 ± 8.1 | 21 ± 3.7 (30) | <10 | undd | >10,000 |
| TM7 | E283Q | 92 ± 3.2 | 99 ± 6.6 | 33 ± 11 (35) | 26 ± 12 (28) | undd | >10,000 |
a The seven transmembrane helices of CCR5 are defined as follows: TM1 from Ile-28 to Asn-57; TM2 from Met-64 to Gln-93; TM3 from Asn-98 to Val-130; TM4 from Thr-143 to Ile-165; TM5 from Lys-191 to Gly-216; TM6 from Arg-230 to Leu-255; and TM7 from Gln-277 to Tyr-297. The first, second, and third extracellular loops are defined using TM boundaries (ECL1 between TM2 and TM3, ECL2 between TM4 and TM5, and ECL3 between TM6 and TM7). ECL2-TM5 and TM6-ECL3 are ECL2 and ECL3 and are expected to adopt a helical structure according to CXCR4 x-ray structure.
b Fold change is compared with WT-CCR5 (KD = 0.69 ± 0.26 nm, lC50 = 1.05 ± 0.61 nm).
c Numbers in parentheses are corrected with the cell-surface expression level of CCR5 mutants compared with that of WT-CCR5 (for the CCR5 mutants Y108A and W190A, binding of radioligands was corrected with their receptor expression level determined using the CTC5 mAb only).
d Data were undetectable.
FIGURE 4.
Effects of CCR5 single mutations on CCL3 (left), gp120 (middle), and MVC (right) binding to the receptor. As in Fig. 3, the 7TM of CCR5 are represented by cylinders, as viewed from the extracellular side of the receptor (top) or in the plane of the plasma membrane (bottom). The side chains of mutated residues are displayed as capped sticks. Atoms are colored as follows: oxygen in red, nitrogen in blue, sulfur in pale yellow, carbon using a white-yellow-red color scale depending on the strength of radioligand binding inhibition (white denotes unchanged binding upon mutation, and red indicates that binding is lost). No accurate measurements could be reported for residues colored in magenta.
The CTC5 and 2D7 mAbs, which recognize epitopes within the N-terminal domain and the second extracellular loop of CCR5, respectively, were used for flow cytometry analysis of cell-surface expression of the CCR5 mutants. These antibodies were chosen because their binding to CCR5 was not affected by MVC (our data not shown). As compared with the wild-type receptor, 10 of the CCR5 mutants show weak (R168A, F189A, Y251F, and E283A), intermediate (L255A and F260A), or drastic (F112A, W248A, Y251A, and Y251I) decrease of cell-surface expression. In contrast to Y251A and Y251I substitutions, the replacement of Tyr-251 by a Phe partly preserved receptor expression, suggesting a role for the aromatic moiety of Tyr-251 in maintaining a proper CCR5 conformation. Similarly, the conservative substitution of Phe-112 by a tyrosine has no effect on receptor expression, although change to Ala substantially altered it. The mutations C178A, W190A, and to a lesser extent Y108A lowered the binding of 2D7 to CCR5, but not the binding of CTC5, in accordance with previous observations (26, 34, 35) indicating conformational rearrangements of the second extracellular loop of the receptor.
The binding to CCR5 of [125I]CCL3 and [35S]gp120, but not MVC, was sensitive to mutations of residues in extracellular loops 1–3 (ECL1, ECL2, and ECL3). In ECL1, the substitution of Trp-94 by an alanine or a methionine abrogated the binding of both radioligands. This residue was conserved among all chemokine receptors except CX3CR1 (26) and was described as critical for CCL5 binding (21) and for infection by some HIV strains (36). In the proximal part of ECL2, the substitution of Arg-168 by an alanine, previously observed to markedly decrease the binding of anti-CCR5 antibodies with antiviral activity (37), only moderately changes the binding of both radioligands. In the central part of ECL2, the mutations C178A and T177A/T177Q compromise the ability of CCR5 to interact with both radioligands. Because it forms a disulfide bridge with Cys-101 in TM3, Cys-178 is a key residue for the proper folding of ECL2, which is actually required for HIV fusion to target cells (19). Changing Ser-180 into a proline, but not into an alanine, abolishes [35S]gp120 binding while it preserves [125I]CCL3 binding. This finding helps explain why murine CCR5 is not suitable for HIV-1 infection (38), because Ser-180 in human CCR5 is equivalent to Pro-182 in murine CCR5. In the distal part of ECL2 (ECL2-TM5 in Table 1), the substitution by an alanine of Tyr-184 or Tyr-187 deteriorated [125I]CCL3 but not [35S]gp120 binding, whereas the substitution of Phe-189 by an alanine deteriorated [35S]gp120 but not [125I]CCL3 binding. Finally, the substitution of Trp-190 by an alanine decreases [125I]CCL3 binding and abrogates [35S]gp120 binding. In ECL3, the F260A mutation significantly decreases [35S]gp120 but not [125I]CCL3 binding. Our data thus suggest that the chemokine and the viral protein bind to distinct yet overlapping regions of extracellular loops in CCR5 and highlight the importance of the correct folding of ECL2 for the binding of both radioligands.
In addition to ECLs, residues in the TMs were found to be involved in the binding of [125I]CCL3 and [35S]gp120 but not of MVC. The substitution in TM1 of Tyr-37 by an alanine, but not a phenylalanine, affects the binding properties of the receptor. This residue is conserved among all CC-chemokine receptors. Our result highlights the functional importance of its aromatic moiety. Of note, Tyr-37 is also required for strong binding of several CCR5 entry inhibitors (TAK-779, AD101, SCH-C, AK317, and AK350, see supplemental Fig. S3). The substitution in TM3 of Leu-104 by a phenylalanine, but not an alanine, also affects [125I]CCL3 and [35S]gp120 binding. The L104F mutation (but not the L104A mutation) was previously reported to impair CCR5 activity in response to chemokines (26). Finally, the replacement in TM6 of the highly conserved Trp-248 by an alanine hinders [125I]CCL3 and [35S]gp120 from binding to the receptor. As mentioned above, the W248A mutation dramatically decreases the receptor expression, suggesting a key structural role for this residue, but preventing any binding measurement for MVC.
Other mutations in the TMs have differential effects on the binding of [125I]CCL3 and [35S]gp120 to CCR5, again emphasizing that both radioligands display different structural requirements for CCR5 recognition. Indeed, mutations of Phe-109 in TM3 (F109H/F109A) reduce the binding of [35S]gp120 only, although substitutions of Asn-252 in TM6 (N252A/N252I) reduce the binding of [125I]CCL3 only. Asn-252 is highly conserved among CC-chemokine receptors (position 6.52 in the Ballesteros-Weinstein numbering system), and its mutations in other receptor systems impact ligand recognition and receptor activation (39–41). A role for Asn-252 in CCR5 activation could explain why N252I and N252A mutations affect CCL3 but not gp120 binding, because the stabilization of the activated form of the receptor by G-proteins is critical for high affinity binding of CCL3 but is dispensable for that of gp120 (16).
The binding to CCR5 of MVC is only sensitive to a few mutations in the receptor 7TM. In detail, the nine mutations that decrease CCR5 affinity for MVC are W86A (TM2), Y108A (TM3), Q194H (TM5), I198A (TM5), Y251A/Y251F/Y251I (TM6) and E283A/E283Q (TM7). Most significant changes were observed for Glu-283. The four other mutations that induce over a 10-fold increase in the IC50 values for half-maximal inhibition of [125I]CCL3 binding either correspond to the loss of the aromatic property (W86A, Y108A, and Y251I) or to the conversion of a bulky aliphatic hydrophobic side chain into a smaller one (I198A). Of note, with the exception of the Q194H mutation, all the mutations in the CCR5 TMs, which affect MVC binding, deteriorate [35S]gp120 binding too. By contrast, only four of them impact [125I]CCL3 binding too: W86A, Y108A, E283A, and E283Q. Moreover, the effect of W86A, Y108A, and E283A mutations on binding is qualitatively much stronger for [35S]gp120 than for [125I]CCL3.
In summary, we found that the structural determinants in CCR5 for the recognition of MVC, [125I]CCL3, and [35S]gp120 have limited overlap, in agreement with the allosteric mode of action of MVC. Our data suggest that binding of radioligands to CCR5 is mainly driven by the recognition of the extracellular surface of the receptor. MVC binding to CCR5 occurs deeper within the 7TM of the receptor.
Modeling of the CCR5 Transmembrane Cavity
We built the three-dimensional structure of the CCR5 7TM by homology to bovine rhodopsin (CCR5_Rho model), bovine ligand-free opsin (OPSD) (CCR5_Ops model), human β2-adrenergic receptor (CCR5_AdrB2 model), human A2A adenosine receptor (AA2A) (CCR5_AA2a model), and human CXC chemokine receptor type 4 (CXCR4) bound either to the small molecule IT1 (CCR5_CXCR4_small model) or to the cyclic peptide CVX15 (CCR5_CXCR4_peptide model). The identities between the target and the template 7TM sequences are 23.3, 25.4, 22.2, and 35.4%, respectively (Table 2 and supplemental Fig. S1). Among the GPCRs of known structure, CXCR4 is obviously the closest homolog of CCR5, and therefore a priori better suits our homology modeling purpose. The amino acid conservation between CCR5 and CXCR4 is, however, not evenly distributed along the sequence of 7TM (supplemental Table S1). Hence, sequences are highly similar in TM2 (53.3% identity) but are much less conserved in TM6 (23.1% identity). The maximal identity between TM6 sequences is actually found by comparing CCR5 to ADBR2 (34.6%). Moreover, the patterns of glycine and proline residues in CCR5 do not perfectly match the sequence of any of the TMs in the templates, except TM2 in CXCR4. Indeed, a TXP motif is found in TM2 of all GPCRs of the angiotensin, opioid, and chemokine families (26, 42). In the CXCR4 crystal structure, this motif induces a tight kink of TM2, with an ∼120° rotation of the helix extracellular end compared with the other GPCR structures. To further investigate the usefulness of using a distant homolog template for CCR5 modeling while taking into account the TXP motif, we created a second model based on the rhodopsin crystal structure, and we replaced TM2 and TM3 by ab initio distorted helices (CCR5_mix model). In total, seven CCR5 models were created (Table 2). General checks for the structure quality indicate that all models have equally good geometry (supplemental Table S2).
TABLE 2.
Statistics about CCR5 three-dimensional models
| CCR5 models |
|||||||
|---|---|---|---|---|---|---|---|
| CCR5_Rho | CCR5_Ops | CCR5_AdrB2 | CCR5_AA2a | CCR5_mix | CCR5_CXCR4_peptide | CCR5_CXCR4_small | |
| Template | Bovine rhodopsin (1u19) | Bovine opsin (3dqb) | Human β2-adrenergic receptor (2rh1) | Human A2A adenosine receptor (3eml) | TM1.4–7 from bovine rhodopsin (1f88), ab initio TM2.3 | Human chemokine receptor type 4 (3oe0) | Human chemokine receptor type 4 (3odu) |
| Modeled domain | 7TMa | 7TMa | 7TMa | 7TMa | 7TMa, N terminus, ECL1, ECL2, and ECL3b | Ser-17 to Ala-298 | A: Pro-19 to Cys-323; B: Cys-20 to Gln-313 |
| Sequence conservation in the 7TM | |||||||
| Identityc | 23.3% | 23.3% | 25.4% | 22.2% | 23.3% | 35.4% | 35.4% |
| Similarityc | 46.0% | 46.0% | 48.1% | 47.1% | 46.0% | 62.4% | 62.4% |
| Geometryd in the 7TM | |||||||
| Main chain | 92.1 | 93.3 | 96.9 | 97.0 | 93.1 | 95.0 | 93.7//92.5 |
| Side chain | 13.3/9.0 | 10.7/8.1 | 11.6/8.3 | 12.1/9.3 | 12.7/12.4 | 10.4/8.7 | 9.7/6.9//11.4/13.9 |
| 7TM cavity volumee | 1083 Å3 | 1092 Å3 | 1089 Å3 | 834 Å3 | 975 Å3 | 942 Å3 | 846//990 Å3 |
a The 7TM is defined as in Table 1.
b The modeled extracellular domains are a tripeptide of the N-terminal domain (N terminus: Val-25 to Gln-27), the first extracellular loop (ECL1, Trp-94 to Gly-97), part of the second extracellular loop (ECL2, Phe-166, Thr-177 to Ser-179, and Trp-109), and part of the third extracellular loop (ECL3, Leu-256 to Gln-261, and Asn-273 to Asp-276).
c Data are between the target and template 7TM sequences. Two residues are noted similar if the score of substitution pair in a blosum62 matrix is positive or null.
d Percent residues in most favored regions of Ramachandran plot (top) and standard deviation from ideal values of χ1 and χ2 angles in degrees (bottom) were computed using Procheck version 3.5 (49).
e The volume of the 7TM cavity (without loops) was measured using the in-house program VolSite.
The overall structure of the seven helix bundle is conserved in the seven models. The r.m.s.d. values computed on the 7TM Cα atoms after their best fit superposition range from 1.2 to 1.5 Å for the models based on the CXCR4 templates, and from 2 to 3.1 Å for models based on the other templates. These values tend to decrease if only the cavity residues are considered, therefore indicating that the geometry of the transmembrane cavity is better preserved that the overall topology of the 7TM. Structural variations between the models reproduce differences existing between the templates (supplemental Fig. S4). Different positioning of the helix extracellular ends directly determines cavity volumes. The smaller volume is observed for the AA2A-based model (∼830 Å3) and the larger volume for OPSD-based and human β2-adrenergic receptor-based models (∼1000 Å3). In the three models with a kinked TM2 (CXCR4-based and CCR5_mix models), the TM cavity has an intermediate size (Table 2).
Evaluation of the CCR5 7TM Models, Agreement with Site-directed Mutagenesis
The present site-directed mutagenesis study tested eight aromatic residues of CCR5 7TM. Data suggest that three of them (Phe-112, Trp-248, and Tyr-251) are essential for receptor folding and expression at the cell surface, and that two of the five remaining ones (Trp-86 and Tyr-108 but not Tyr-37, Trp-94, and Phe-109) contribute to MVC binding either by direct intermolecular interactions or by local constraints on the receptor structure. In all seven CCR5 models, the side chains of Tyr-108, Phe-112, Trp-248, and Tyr-251 point into the receptor cavity and connect TM3 with TM6. Considering Trp-86, only the mixed ab initio/homology model of CCR5 (CCR5_mix) and the two CXCR4-based homology models clearly account for the effects of the Trp to Ala substitution. In these models, Trp-86 positions its side chain into the 7TM cavity with the indole ring being sandwiched between Tyr-37 and Tyr-108. By contrast in the four other models, the Trp-86 side chain faces the lipid bilayer.
This study describes mutants for five aliphatic hydrophobic residues of CCR5. The residues in TM3 (Met-100 and Leu-104), TM4 (Gly-245), and TM6 (Leu-255) do not or hardly affect CCR5 binding properties. These four residues have constant positions in all CCR5 models (Met-100 and Leu-104 are located at the TM2/TM3 interface, Gly-245 at the intracellular end of TM4, and Leu-255 at the TM5/TM6 interface). By contrast, the residue in TM5 (Ile-198) strongly influences the ability of CCR5 to bind gp120 and MVC. In all models except CCR5_AdrB2, the side chain of Ile-198 is directed toward the 7TM cavity center. Because of the vicinity of large hydrophobic or aromatic residues, Ile-198 is constrained but to variable extents in the different models (its side chain is almost fully buried in CCR5_mix and CCR5_AA2a models, whereas it can interact directly with MVC in the other models).
Finally, four polar positions of the CCR5 TMs were tested by site-directed mutagenesis (Gln-194, Asn-252, Gln-280, and Glu-283). Two of them influence the receptor binding properties as follows: Glu-283 in TM7 is required for the binding of CCL3, gp120, and MVC; and Gln-194 in TM5 contributes to MVC binding. In all models, the Glu-283 carboxylate is well accessible into the 7TM cavity. In all the models except CCR5_AA2a, the side chain of Gln-194 is oriented toward TM4 in the vicinity of Pro-162. The availability of Gln-194 terminal amide for the direct binding of MVC, however, varies with models.
In summary, experimental data obtained from mutation of aromatic residues favor the CCR5_mix and CXCR4-based models, because of the Trp-86 side chain orientation. Experimental data obtained for the mutation of polar and hydrophobic residues further point out weaknesses in the CCR5_AdrB2 and CCR5_AA2a models. They also suggest the superiority of CXCR4-based models relative to CCR5_mix, because of Ile-198 solvent accessibility.
Challenging the CCR5 7TM Models by Virtual Screening
The seven CCR5 7TM models were challenged for the prediction of ligand binding by automated docking of the following: (i) members of MVC chemical series with nanomolar affinity for CCR5 (MVC-like set); (ii) chemically diverse potent CCR5 antagonists, such as TAK-779 (DIVERSE-active set); (iii) known CCR5 nonbinders sharing structural similarities with known CCR5 binders (FOCUS-inactive set); and (iv) ligands for other chemokine receptors (CCR1, CCR2, CCR3, CXCR4, and CCR8) with no or weak interaction with CCR5 (ChemoR-decoys set).
Docking results are reported in Table 3. A ligand docking was considered successful if the score of the top-ranked pose exceeds a gentle threshold, which implied a minimal number of inter-molecular interactions; and if, in the corresponding complex, the bound ligand establishes an ionic bond with the Glu-283 residue of the receptor and π-π interactions with at least three aromatic residues of the receptor 7TM.
TABLE 3.
Number of compounds predicted active by the virtual screening of libraries
A compound is tagged active if its docked pose matches the three following criteria: (pki docking score) ≥5 and (distance between compound positive charge and Glu-283 carboxylate) ≤5.5 Å and (number of inter-molecular π-π interaction) ≥3.
| Libraries |
CCR5 models |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Name | Potency | Sizea | CCR5_Rho | CCR5_Ops | CCR5_AdrB2 | CCR5_AA2a | CCR5_mix | CCR5_CXCR4_peptide | CCR5_CXCR4_small |
|
| A chain | B chain | |||||||||
| MVC-like | nm | 9 (11) | 6 (6) | 0 (0) | 0 (0) | 1 (1) | 8 (10) | 6 (6) | 0 (0) | 0 (0) |
| DIVERSE-active | nm | 17 (19) | 3 (3) | 1 (1) | 2 (2) | 1 (1) | 11 (11) | 6 (6) | 0 (0) | 1 (1) |
| FOCUS-inactive | Inactiveb | 87 (178) | 6 (8) | 1 (1) | 5 (7) | 3 (3) | 12 (19) | 0 (0) | 1 (1) | 4 (4) |
| ChemoR-decoys | Unknown | 67 (117) | 12 (16) | 1 (1) | 13 (17) | 5 (6) | 25 (36) | 5 (8) | 2 (2) | 8 (10) |
a Total number of compounds of the library. The total number of isomers is given in parentheses.
b No activity was reported for compounds at 500 μm in competition binding assays as described previously (45).
The receptor models based on OPSD, human β2-adrenergic receptor, and AA2A were not well suited for the identification of CCR5 true binders. None of them yielded acceptable docking of MVC-related ligands or other CCR5 antagonists. The virtual screening of the FOCUS-inactive and ChemoR-decoys libraries using these three CCR5 models picked only a few hits, suggesting somewhat a good capacity of the receptor structures to discard nonbinders. However, considering the overall limited number of docking successes, it is more likely that these three models can hardly accommodate large and positively charged ligands.
The rhodopsin-based models achieved better performances in docking. The CCR5_bRho model identified two-thirds of the MVL-like compounds yet only three other CCR5 antagonists. Figures were improved by using the CCR5_mix model, which retrieved almost all MVC-related compounds and identified 65% of other CCR5 antagonists. Although the fully automated model CCR5_bRho selected only 6.9 and 22.4% of compounds from the FOCUS-inactive and ChemoR-decoys libraries, respectively, the CCR5_mix model was more permissive and selected 13.8% of inactive compounds and 37.3% of compounds known to bind other chemokine receptors. As usually observed for GPCR models, the ligand-based refinement of CCR5 7TM model facilitates docking because it induces a bias in the protein pocket shape (43).
The virtual screening of the four libraries was also achieved using the two CXCR4-based models. According to the data deposited in the Protein Data Bank, CCR5_CXCR4_peptide consists of a single chain and CCR5_CXCR4_small is dimeric. Noteworthy, the two CXCR4 templates were described as protein dimers (23), and thus our two CCR5 models are assumed to represent the dimeric form of the receptor. The results of the retrospective virtual screening obtained for each of the two chains of the CCR5_CXCR4_small model are as bad as those obtained for the CCR5_Ops, CCR5_AdrB2, and CCR5_AA2A models. By contrast, the CCR5_CXCR4_peptide model is better suited to our docking purposes. It identified 67% of the MVL-like compounds and 35% of the other CCR5 antagonists, although it discarded all inactive compounds and 92.5% of compounds known to bind to other chemokine receptors. The 7TM sites are highly similar in the two CXCR4-based models. Considering residues in the 7TM cavity, the r.m.s.d. computed on the Cα positions is lower than 0.8 Å, and no changes in their rotameric state are consistently observed between the single chain of CCR5_CXCR4_peptide and both chains of CCR5_CXCR4_small (supplemental Table S3). However, little adjustment of the Glu-283 carboxylate positioning, as well as tiny variations in the relative positions of TM5 and TM6, can account for the differences in docking. As compared with the CCR5_mix model, the CCR5_CXCR4_peptide model suggested the binding mode for a smaller number of chemically different true binders, but it yielded a significantly lower percent of nonbinders predicted active. This observation is in agreement with a computational analysis of diverse CCR5 antagonist chemotypes that suggest that an ensemble of receptor conformations is required to explain the binding of all CCR5 antagonists (44).
As a conclusion, we can rank CCR5 models and objectively hint at their applicability in computer-aided drug design projects. All tested GPCR templates are suitable for a rough prediction of residue accessibility for ligand binding into the 7TM of the receptor. Models based on a distant homolog can also be useful for the identification of novel ligands by structure-based virtual screening, provided it is sufficiently optimized in its ligand-binding site. An optimized rhodopsin-based CCR5 model indeed allowed the discovery of new ligands of the receptor by high throughput docking of screening collections (45). Finally, only a close homolog, here CXCR4, allows the prediction of the full receptor three-dimensional structure, including loops. High sequence identity between the template and target sequence is obviously necessary for a fine structural interpretation of experimental functional data.
Modeling MVC Binding into CCR5
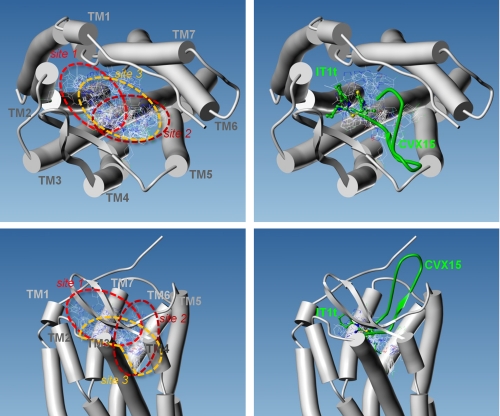
We docked MVC into the most valid model, namely the CCR5_CXCR4_peptide. We predicted docked poses using five different programs (FLEXX, FRED, GOLD, PLANTS, and SURFLEX). These programs were chosen because of their distinctive principles and algorithms, therefore preventing a methodological bias in the docking solutions. Automated docking of MVC into the CCR5 7TM produced a heterogeneous ensemble of 41 poses (Fig. 3). To evaluate the number of different binding modes, we computed the r.m.s.d. of the coordinates of all MVC non-hydrogen atoms. The all-against-all comparison of poses yielded r.m.s.d. values ranging from 0.14 to 15.18 Å. Considering that two poses are similar if the r.m.s.d. is smaller than 2 Å, the conformational ensemble consists of seven clusters with 2–6 members and 20 singletons. The docking data hence represent a total of 27 different binding modes, which roughly delineate three binding sites for MVC in the 7TM cavity of CCR5. The first site (MVC site1) places a few MVC poses in part of the pocket defined by side chains from TM2, TM3, and TM7. MVC site1 is located near the receptor cavity mouth and corresponds to the binding site in CXCR4 for the synthetic molecule IT1t (Fig. 3). In a few other poses, MVC occupies the opposite side of the 7TM cavity and lies on helix V. Here again, MVC is positioned at the entrance of the receptor cavity. This second site (MVC site2) partly overlaps with the CXCR4-binding site for the CVX15 peptide. By contrast to CVX15, docked MVC does not bulge outward to the receptor and deeply buries into a tight channel between the side chains of Tyr-108 in TM3 and Ile-198 in TM5. Finally in the remaining poses, MVC fills most of the volume of the deep part of the pocket. This third site (MVC site3) spans the deeper part of both MVC site1 and MVC site2. It also includes the tight channel between TM3 and TM5, which accommodates either the phenyl group or the cyclohexyl group of MVC. Both MVC site1 and MVC site3 predict MVC-binding modes that are consistent with an ionic bond between the protonated amine of the MVC tropane ring and the carboxylate group of CCR5 Glu-283.
FIGURE 3.
Multiple binding modes for maraviroc into CCR5. All (left) or Glu-283 interacting (right) docked poses of MVC are displayed as thin lines, with CPK color coding of the atomic bonds. The 7TMs of CCR5 are represented by cylinders, as viewed from the extracellular side of the receptor (top) or in the plane of the plasma membrane (bottom). The position of ligands cocrystallized with CXCR4 are indicated in green using ball-and-stick and ribbon representations for the synthetic molecule IT1t and the peptide CVX15, respectively.
DISCUSSION
A collection of 48 CCR5 mutants was designed to probe binding of [125I]CCL3, [35S]gp120, and MVC. The 28 tested positions covered all helices in the receptor cavity and gave emphasis to TM tails and extracellular loops. To date, all site-directed mutagenesis studies published were analyzed in the light of CCR5 three-dimensional models based on the crystal structure of distant GPCR homologs (generally bovine rhodopsin). Here, we have demonstrated that these approximate models have a limited predictive power. We have built and validated a three-dimensional model for CCR5 based on the structure of its close homolog CXCR4. This model includes extracellular loops and gives new insights to ligand recognition by the receptor.
Description of the CCR5 Architecture
The CCR5 model based on CXCR4 in complex with the CVX15 peptide (PDB 3OE0) provides a reasonable framework for discussing our experimental data. It especially suggests a structural and/or functional role for the tested residues. In the following structural description, we have paid special attention to aromatic clusters, intermolecular hydrogen bonding networks, and polar side chains in a hydrophobic environment.
The core of the CCR5 helix bundle is made of the packed side chains of apolar aliphatic residues (Leu, Ile, and Val) and of the stacked rings of aromatic residues (Phe, Tyr, and Trp). Aromatic amino acids are grouped into clusters that bridge the transmembrane helices. If we consider potential π-π interactions, we can arbitrarily delineate five clusters. Starting from the N terminus of the receptor, the first cluster, AR1, consists of Tyr-37 in TM1; Phe-79 and Trp-86 in TM2; and Tyr-108 in TM3. It connects the extracellular tails of TM1, TM2, and TM3. The second cluster, AR2, which consists of Phe-85 and Tyr-89 in TM2 and Trp-94 and Phe-96 in ECL1, determines the folding of ECL1 and indirectly controls the positioning of the central part of ECL2. ECL2 indeed adopts a β-hairpin structure, whose tip is covalently bound to TM3 (disulfide bridge between Cys-178 and Cys-101) and lies on ECL1 (a hydrogen bond is formed between the backbone atoms of Trp-94 and Tyr-176). AR1 and AR2 are indirectly connected via the side chain of Leu-104. The third cluster, AR3, involves TM3, TM6, and TM7. It can be divided into an upper layer, which is accessible to the solvent and includes Tyr-108, Phe-109, Phe-112, and Phe-113 in TM3 and Tyr-251 in TM6, and a lower layer, which is buried into the receptor and includes Tyr-244 and Trp-248 in TM6 and His-289 in TM7. The AR3 cluster possibly ensures cohesion of the receptor and could represent a hinge region for signal transmission. Considering the interaction network between residues, Trp-248 would represent a hub contacting many other residues. The clusters AR3 and AR1 are connected. Indeed, Tyr-108 is common to both clusters and Phe-79, in AR1 is stacked on His-289 in AR3. The fourth cluster, AR4, which consists of Phe-166 and Phe-182 in ECL2, Phe-189 and Trp-190 in TM5-ECL2, and Phe-193 in TM5, plays a role in the proper folding of ECL2. Noteworthy, according to the CXCR4 crystallographic dimer, this cluster could also be involved in the receptor dimerization. The fifth cluster, AR5, consists of four aromatic residues located close in space but not engaged in canonical π-π interactions in our model (Tyr-187 in ECL2 and Phe-260, Phe-263, and Phe-264 in TM6-ECL3). It may be involved in the folding of the N-terminal domain of the receptor and, as mentioned before, in the receptor homodimerization. In the CXCR4 template used for CCR5 modeling, four of the five clusters are also present. The clusters AR1 and AR3 are almost identical in both receptors. The only variations concern TM3 aromatic residues in contact with TM5 aliphatic hydrophobic residues (more precisely, Phe-109, Phe-112, and Phe-113 in CCR5 correspond, respectively, to Thr-117, Leu-120, and Tyr-121 in CXCR4). This high sequence conservation supports the presumed structural role for the two clusters. Clusters AR2 and AR4 are well conserved too, yet three substitutions can account for distinctive structural features in the extracellular region (Tyr-89 to Asp in AR2 and Phe-189 to Leu and Phe-193 to Val in AR4). AR5 is a cluster specific to CCR5.
In the CCR5 model, hydrogen and ionic bonds are established in the solvent-exposed extracellular part of the receptor (including loops and TM tails). In detail, polar interactions are observed between TM5 and TM6-ECL3 (Thr-195/Thr-259/Asn-192/Glu-262/Lys-191), between TM3 and ECL2 (Thr-105/Ser-180/Thr-167), between the N terminus, TM1, and TM7-ECL3 (Lys-22/Asp-276/Asn-24/Gln-280/Gln-277), and between TM7-ECL3 and TM6-ECL3 (Asp-276/Asn-258/Gln-261/Ser-272/Asn-267).
A few polar residues are located in the hydrophobic part of the receptor 7TM, including residues Ser-38, Thr-82, and Ser-160. Thr-82 and Ser-160 are, respectively, in TXP and SXP motifs, which are known to introduce a distortion in a transmembrane helix. Thr-82 was identified as a structural determinant for the chemokine-induced activation of CCR5 (42). The other polar residues buried in a hydrophobic environment are the Gln-194/Lys-197 pair in TM5 and Asn-252 in TM6. The highly conserved Glu-283 in TM7 is located at the 7TM hydrophobic cavity center, yet its polar carboxylic acid group is engaged in a hydrogen bond network together with Tyr-251, Tyr-108 (19), and possibly Tyr-37 via a water molecule. As a conclusion of the structural description, Glu-283 and Trp-248 are key residues for CCR5 structure and dynamics. Both residues occupy a central position in the receptor and establish contacts with many residues in all TMs except TM4. An aromatic core involves TM1, TM2, TM3, TM6, and TM7 and contacts aliphatic residues of TM5 (Leu-203 and Ile-198).
Mapping CCL3 and gp120-binding Sites in CCR5
The single mutations of CCR5, which alter the binding of the chemokine CCL3, and that of the HIV-1 envelope glycoprotein gp120 are represented on the three-dimensional model of the receptor shown in Fig. 4. They first suggest that the apical protrusive part of the ECL2 β-hairpin (from Ser-169 to Thr-177, ECL2(169–177)) is involved in the recognition of both CCL3 and gp120. Three independent observations support this assumption as follows. (i) The C178A mutation abolishes CCL3 and gp120 binding. It disrupts the disulfide bridge between ECL2 and TM3, thereby inducing the bending of ECL2(169–177) toward the outward side of the cavity. (ii) The T177A and T177Q mutations decrease the binding of both CCL3 and gp120. The side chain of Thr-177 is solvent-exposed and in close proximity to the hydroxyl groups of Tyr-89 in TM2 and Ser-179. Thus, the mutations of Thr-177 modify the properties of the binding surface of ECL2(169–177). (iii) Binding of CCL3 and gp120 is hindered by mutations in the aromatic clusters AR1 and AR2. ECL2(169–177) makes extensive contacts with ECL1, which is defined by the configuration of the aromatic cluster AR2. Direct (W94A and W94M) as well as indirect perturbations of AR2 (mutations in AR1 or the L104F mutation) change the positioning of ECL2(169–177).
The spatial distribution of mutations that impair CCL3 and gp120 binding hints at a different penetration of the two proteins within the CCR5 7TM cavity. Mutation at the conserved position 283 in TM7 more dramatically alters the binding of gp120 than that of CCL3. Considering CCL3 binding, the effects of mutations E283A and E283Q are quantitatively equivalent. Two structural interpretations are possible. Either CCL3 does not directly bind the 7TM cavity or the direct recognition of CCL3 by the 7TM cavity is not driven by long distance electrostatic contributions. Gp120 binding is lost with the E283A mutant but is partly preserved with the E283Q mutant. Thus, the negative charge of Glu-283 seems not to be required for gp120 interaction with CCR5, although a direct or a water-mediated hydrogen bond linking gp120 to this residue is likely. Another difference between gp120 and CCL3 is their differential sensitivity to the nine mutations probing five residues in the core aromatic cluster of CCR5 7TM (residues Tyr-108, Phe-109, Phe-112, Trp-248, and Tyr-251 in AR3). Deterioration of CCL3 binding is only observed after mutation of Trp-248, which likely alters the proper folding of the receptor and to a lesser extent after that of Tyr-108. By contrast, gp120 binding is significantly changed by the mutations in AR3. Mutations in the aromatic clusters, which involve the top of TM5 (AR4 and AR5), also have different effects on CCL3 and gp120 binding. The Y187A mutation abrogates the binding of CCL3 but not that of gp120. Interestingly, in our CCR5 model, Tyr-187 takes part in the folding of the receptor N terminus, thus suggesting that proper folding of the CCR5 N terminus is dispensable for interaction of gp120 with CCR5 but not for that of the chemokine. Mutations in AR4 (F189A and W190A) prevents the binding of gp120. In the CCR5 model, Phe-189 and Trp-190 determine the conformation of the C-terminal tail of ECL2 (ECL2(170–190)). A critical role for this region in the tight binding of gp120 is further supported by the data from mutations at position 180. Indeed, the S180P mutation prevented gp120 binding to CCR5, although the S180A mutation had no effects. The effect of the Ser to Pro substitution could result from a distortion of ECL2(170–190) secondary structure elements (indirect effect) and/or be the consequence of the loss of a hydrogen bond between the backbone NH group of Ser-180 and gp120 (direct effect). Such a hydrogen bond exists in the crystal structure of the 1OE0 PDB template between the backbone NH group of CXCR4 Arg-188 and the backbone CO group of Arg-1 in the peptide CVX15. In the gp120-CCR5 complex, the hydrogen bond acceptor could be a backbone atom in the gp120 V3 loop, which would thus form together with ECL2 a four-stranded β-sheet (Fig. 5).
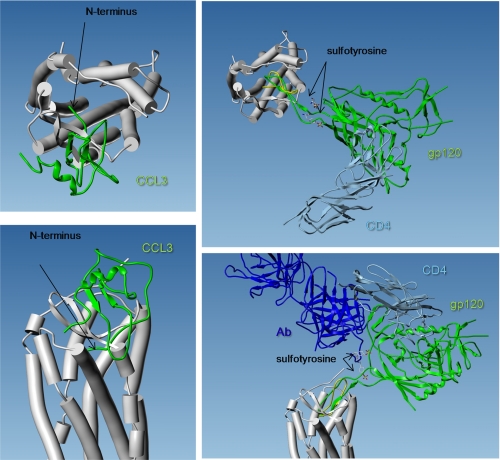
FIGURE 5.
Three-dimensional views of CCR5 model, together with CCL3 crystal structure (left) and gp120 crystal structure (right). The 7TM of CCR5 are represented by cylinders, as viewed from the extracellular side of the receptor (top) or in the plane of the plasma membrane (bottom). The solution structure of CCL3 was extracted from 1B50 PDB entry. It is represented by a green ribbon. Its orientation was chosen based on the electrostatic potentials of the two proteins as follows: the positively charged side of CCL3 faces the negatively charged entry of the membrane receptor. The two proteins were docked to establish H-bonds between the ECL2 hairpin and the CCL3 N-terminal β-strand. Note that the N terminus of the chemokine is truncated (it starts with Asp-5) and that its conformation is not relevant (it bumps into the receptor). The complex between gp120 (green ribbon), CD4 (light blue ribbon), and an antibody (dark blue ribbon) was extracted from 2QAD PDB entry. The antibody includes two sulfotyrosine residues which mimic that of CCR5 N terminus at positions 10 and 14. The orientation was chosen to establish H-bonds between the backbone atoms of CCR5-ECL2 and the tip of gp120V3 loop, according to what was observed between the CXCR4 structure template and the cocrystal peptide CVX15 (yellow ribbon). Note that this orientation is in agreement with an interaction between the gp120 hinge region (between V3 loop and the core protein) and CCR5 sulfotyrosine residues (the CCR5 model here starts at position 17).
Considered together, the data from our site-directed mutagenesis experiments strongly suggest that CCL3 and gp120 show different binding modes, which nevertheless have in common a strong dependence to the ECL2 structure. To schematize, CCL3 would rather act as a lid on the receptor surface, including ECL2(169–177), whereas gp120 would partly fill the receptor cavity while occupying the whole of ECL2.
Mapping MVC-binding Sites in CCR5
According to the quantitative changes in MVC affinity for CCR5 upon mutation, the key receptor residue for MVC recognition is Glu-283. Like all compounds of the chemical series developed as CCR5-mediated HIV entry inhibitors, MVC is positively charged at physiological pH. Its protonated tropane ring is expected to establish an ionic bond with the receptor, and the carboxylate of Glu-283 is the only available negatively charged group with no intramolecular counter-charge (the other acidic residues in the upper cavity mouth, namely Glu-172 in ECL2, Glu-262 in TM6, and Asp-276 in TM7, are close to the positively charged residues Lys-171 in ECL2, Lys-191 in TM5, and Lys-22 in the N terminus, respectively). The CCR5 model shows that the conformation of the Glu-283 side chain is selected by a bulky and mostly apolar environment. More precisely, considering a 4.5-Å inter-atomic distance threshold, the side chain of Glu-283 is surrounded by the side chains of two hydrophobic aliphatic residues (Met-279 and Met-287) and four aromatic residues (Trp-86, Tyr-108, Trp-248, and Tyr-251). The aromatic residues stabilize the carboxylate of Glu-283 in its hydrophobic environment through nonbonded anion-π pairs (46). In addition, the position of the carboxylate is fine-tuned by a hydrogen bond network that includes the hydroxyl groups of the two nearby tyrosines, Tyr-108 and Tyr-251 (19). The mutation of all four aromatic residues in close vicinity of Glu-283 was shown here to impact MVC binding to CCR5. Trp-248 mutation into Ala abrogates MVC binding due to poor receptor expression level. The loss of aromatic characteristics at positions 86, 108, and 251 significantly decreases MVC binding to CCR5 (7.7–75.5-fold raise in the IC50 values for mutation into Ala). The perturbation of the hydrogen bond network (Y251F), which is not expected to largely modify the shape of the receptor pocket, indeed only induces a small increase in the IC50 value for displacement of [125I]CCL3 by MVC (3.3-fold). To summarize, four of the six CCR5 positions found to be important for MVC binding concern Glu-283 and its direct surrounding.
The two other mutations that decrease the apparent affinity of MVC for CCR5 involve Ile-198 and Gln-194, both in TM5. The most dramatic change of the MVC IC50 value is observed for the I198A mutant (131-fold raise). In our model, the side chain of Ile-198 is partly exposed at the cavity surface (the relative solvent-accessible surface computed using nacess is about 30%) and thus could interact directly with MVC. The buried part of the Ile-198 side chain makes contact with the side chains of Phe-109 and Phe-112 in TM3. The replacement of these Phe residues by Ala, His, or Tyr has nevertheless no effect on MVC binding to CCR5, strongly suggesting that the MVC molecular surface does not tightly match the 7TM cavity region defined by the side chains of Ile-198 and its direct neighbors. The buried part of the Ile-198 side chain also contacts the apolar part of the Lys-197 side chain, which in turn interacts with Gln-194. The mutation of Gln-194 into His, but not into Ala, decreases the MVC IC50 value by 5.6-fold. The Q194H mutant is expected to amply disturb the receptor local structure because of the proximity of two basic residues (Lys-197 and His-194). Neither Lys-197 nor Gln-194 is oriented toward the cavity center. Both residues are rather involved in packing TM3, TM4, and TM5. Together with Ile-198, they may control the receptor dynamics so that their mutation may change the receptor conformational equilibrium toward conformations with a lower affinity for MVC. In further support of this assumption, Ile-198 is located in helix V one turn upstream of a hinge region defined by a GXXXP motif, which likely plays a role in receptor conformational flexibility. Indeed, a similar GXXXP motif is located in the central part of the CXCR4 TM5, and stabilization of this motif was reported to be required for the receptor crystallization. This was accomplished by introducing a L125W thermostabilizing mutation in the CXCR4 TM3 in such a way that the indole side chain of the Trp residue stacks on the proline ring of the GXXXP motif (23).
To sum up, among the residues lining the CCR5 TM cavity, Glu-283 is the only residue essential for MVC binding. Five other residues are important for the definition of the MVC-binding site, but the effects of their mutation are most probably not due to the loss of key intermolecular interactions but rather to indirect structural effects. These observations can be accounted for by the fact that there would not be tight complementarity between the shape of drug and the receptor 7TM cavity. Indeed, the modeled cavity of the free receptor is wide and deep, and its volume is about twice larger than that of MVC (∼520 Å3). Its shape and physicochemical properties are consistent with the binding of MVC in different parts of the pocket. Our docking study further supports this assumption and proposes almost 30 different binding modes in three overlapping yet distinct sites (MVC site1, site2, and site3). Twelve of the binding modes suit the establishment of an ionic bond between MVC and Glu-283. They imply a deep penetration of the drug into the 7TM cavity (Fig. 3).
Toward a Clearer Picture for the Allosteric Modulation of CCR5 by MVC
Considering the present mapping of the binding sites in CCR5, the MVC docking poses, and the bulk and shape of the two other ligands (CCL3 from 1B50 PDB entry and gp120 from the 2QAD PDB entry), we can suggest structural foundations for the allosteric modulation of CCR5 by the inhibitor (Fig. 5). MVC has a symmetrical shape that looks like a rough helix. Three hydrophobic wings extend from the charged nitrogen at the molecule center (∼7 Å long each, ∼7 Å wide for the substituted tropane wing, and ∼4 Å wide for the two other wings). In the free CCR5 model, the cavity mouth has an oblong ellipsoidal shape, with an approximate length of 26 Å and a width of 10–12 Å. This likely allows multiple entry routes for MVC.
The most favored routes lead the drug to select one or several receptor conformers that are less suitable to interact with CCL3 and gp120 (16). From a crude geometrical point of view, MVC-bound CCR5 could accommodate CCL3 because, as shown here, the chemokine is expected to anchor to the extracellular top of the receptor (mainly the N terminus and the ECL2 β-hairpin), whereas all the docked poses buried MVC into the 7TM cavity. This strongly suggests that MVC alters CCL3 binding to CCR5 as a result of conformational rearrangements of the extracellular domains rather than by simple steric hindrance. In addition, MVC could also directly act by impeding interaction of the N terminus of the chemokine with CCR5 TM residues, a process that was proposed to be critical for activation of the receptor (11). Indeed, we (16) and others (47) have shown that stabilization of activated, G-protein-coupled CCR5 is required for high affinity binding of agonistic chemokines to the receptor. In this regard, MVC, as an inverse agonist for CCR5, may also act by stabilizing inactive G-protein-uncoupled conformations of the receptor (16). Even more than CCL3, the mutations in the CCR5 7TM cavity that affect the binding of MVC to CCR5 also alter that of gp120 at the receptor surface. Our results also indicate that MVC may also act by hampering key intermolecular interactions for gp120 binding (for example the interaction between Ile-198 and Phe-109). Overall, these results support the notion that MVC binding to CCR5 induces conformational rearrangements that hinder tight association between ECL2 and the V3 loop. It should be noted that our results are also consistent with the V3 loop of the viral glycoprotein partly filling the receptor 7TM cavity. Thus, one can hypothesize that the space left into the 7TM cavity of MVC-bound CCR5 could not be sufficient to accommodate the full V3 loop. However, to what extent this process takes part in the inhibition of gp120 binding by MVC remains elusive, as the degree to which the loop penetrates in the cavity is not known.
Finally, in our recent work (16), we reported that MVC accelerates dissociation of [125I]CCL3 and [35S]gp120 from CCR5 and so acts as an allosteric inhibitor. From a structural point of view, this indicates that MVC can bind to CCR5-CCL3 and CCR5-gp120 binary complexes and that the transient ternary complexes evolve toward CCR5-MVC binary complexes. This implies that the 7TM cavity remains accessible for MVC in CCL3-bound and gp120-bound CCR5. Because the receptor mouth section area (250 Å2) exceeds the gp120 V3 section, a gate exists in the gp120-bound receptor, most probably at the C-terminal part of TM2 (where the top of TM1 and the ECL2 β-hairpin define a cleft in the CCR5 model) or at the opposite side of the entry mouth, i.e. at the N-terminal part of TM5. According to gp120-CCR5 complex model, it is likely that MVC binds close to TM2, in a region of CCR5 corresponding to the above-described MVC site1 (Fig. 3). Rough model of the CCL3-CCR5 complex, which shows that the receptor 7TM cavity is not fully obstructed by the bound chemokine, supports similar conclusions (Fig. 5).
Because the crystal form of CXCR4 modeling template is dimeric (23), our CCR5 model is supposed to represent a receptor dimer. CCR5 indeed exists as a homodimer in intact cells, and negative cooperativity can take place between associated monomers (47, 48). In this context, we cannot rule out that allosteric interactions between MVC and gp120 or CCL3 transmit across dimers. Alternatively, MVC could also interfere with CCR5 dimerization in such a way that gp120 or CCL3 previously bound to receptors are released. We here observed that mutation of residues in the putative CCR5 homodimer interface (Phe-189 and Trp-190 in TM5) is detrimental to gp120 binding and to a lesser extent to CCL3 binding. This suggests that CCR5 dimerization may be a prerequisite for gp120 binding and HIV entry into target cells.
Supplementary Material
Acknowledgments
We acknowledge Mike Westby and Charles Craigh for helpful discussion. We thank Pfizer for providing us with radiolabeled maraviroc.
This work was supported by Instituto de Salud Carlos III, Spanish Ministry of Health Grant FIS PI080752, AIDS Network ISCIII-RETIC Grant RD06/0006, Agence Nationale de Recherche sur le SIDA (ANRS), and FIPSE Grant 36633/07.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4, Tables S1–S3, and additional references.
- MVC
- maraviroc
- ECL
- extracellular loop
- GPCR
- G-protein coupled receptor
- OPSD
- bovine opsin
- PDB
- Protein Data Bank
- r.m.s.d.
- root mean square deviation
- TM
- transmembrane helix
- 7TM
- seven transmembrane domain
- SDM
- site-directed mutagenesis.
REFERENCES
- 1. Blanpain C., Migeotte I., Lee B., Vakili J., Doranz B. J., Govaerts C., Vassart G., Doms R. W., Parmentier M. (1999) Blood 94, 1899–1905 [PubMed] [Google Scholar]
- 2. Alkhatib G., Combadiere C., Broder C. C., Feng Y., Kennedy P. E., Murphy P. M., Berger E. A. (1996) Science 272, 1955–1958 [DOI] [PubMed] [Google Scholar]
- 3. Arenzana-Seisdedos F., Parmentier M. (2006) Semin. Immunol. 18, 387–403 [DOI] [PubMed] [Google Scholar]
- 4. Gaertner H., Cerini F., Escola J. M., Kuenzi G., Melotti A., Offord R., Rossitto-Borlat I., Nedellec R., Salkowitz J., Gorochov G., Mosier D., Hartley O. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 17706–17711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mack M., Luckow B., Nelson P. J., Cihak J., Simmons G., Clapham P. R., Signoret N., Marsh M., Stangassinger M., Borlat F., Wells T. N., Schlöndorff D., Proudfoot A. E. (1998) J. Exp. Med. 187, 1215–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Safarian D., Carnec X., Tsamis F., Kajumo F., Dragic T. (2006) Virology 352, 477–484 [DOI] [PubMed] [Google Scholar]
- 7. Trkola A., Ketas T. J., Nagashima K. A., Zhao L., Cilliers T., Morris L., Moore J. P., Maddon P. J., Olson W. C. (2001) J. Virol. 75, 579–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Maeda K., Nakata H., Koh Y., Miyakawa T., Ogata H., Takaoka Y., Shibayama S., Sagawa K., Fukushima D., Moravek J., Koyanagi Y., Mitsuya H. (2004) J. Virol. 78, 8654–8662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dorr P., Westby M., Dobbs S., Griffin P., Irvine B., Macartney M., Mori J., Rickett G., Smith-Burchnell C., Napier C., Webster R., Armour D., Price D., Stammen B., Wood A., Perros M. (2005) Antimicrob. Agents Chemother. 49, 4721–4732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ray N. (2009) Drug Des. Dev. Ther. 2, 151–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Blanpain C., Doranz B. J., Bondue A., Govaerts C., De Leener A., Vassart G., Doms R. W., Proudfoot A., Parmentier M. (2003) J. Biol. Chem. 278, 5179–5187 [DOI] [PubMed] [Google Scholar]
- 12. Cormier E. G., Dragic T. (2002) J. Virol. 76, 8953–8957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huang C. C., Lam S. N., Acharya P., Tang M., Xiang S. H., Hussan S. S., Stanfield R. L., Robinson J., Sodroski J., Wilson I. A., Wyatt R., Bewley C. A., Kwong P. D. (2007) Science 317, 1930–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Watson C., Jenkinson S., Kazmierski W., Kenakin T. (2005) Mol. Pharmacol. 67, 1268–1282 [DOI] [PubMed] [Google Scholar]
- 15. Muniz-Medina V. M., Jones S., Maglich J. M., Galardi C., Hollingsworth R. E., Kazmierski W. M., Ferris R. G., Edelstein M. P., Chiswell K. E., Kenakin T. P. (2009) Mol. Pharmacol. 75, 490–501 [DOI] [PubMed] [Google Scholar]
- 16. Garcia-Perez J., Rueda P., Staropoli I., Kellenberger E., Alcami J., Arenzana-Seisdedos F., Lagane B. (2011) J. Biol. Chem. 286, 4978–4990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dragic T., Trkola A., Thompson D. A., Cormier E. G., Kajumo F. A., Maxwell E., Lin S. W., Ying W., Smith S. O., Sakmar T. P., Moore J. P. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 5639–5644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Maeda K., Das D., Ogata-Aoki H., Nakata H., Miyakawa T., Tojo Y., Norman R., Takaoka Y., Ding J., Arnold G. F., Arnold E., Mitsuya H. (2006) J. Biol. Chem. 281, 12688–12698 [DOI] [PubMed] [Google Scholar]
- 19. Maeda K., Das D., Yin P. D., Tsuchiya K., Ogata-Aoki H., Nakata H., Norman R. B., Hackney L. A., Takaoka Y., Mitsuya H. (2008) J. Mol. Biol. 381, 956–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tsamis F., Gavrilov S., Kajumo F., Seibert C., Kuhmann S., Ketas T., Trkola A., Palani A., Clader J. W., Tagat J. R., McCombie S., Baroudy B., Moore J. P., Sakmar T. P., Dragic T. (2003) J. Virol. 77, 5201–5208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kondru R., Zhang J., Ji C., Mirzadegan T., Rotstein D., Sankuratri S., Dioszegi M. (2008) Mol. Pharmacol. 73, 789–800 [DOI] [PubMed] [Google Scholar]
- 22. Labrecque J., Metz M., Lau G., Darkes M. C., Wong R. S., Bogucki D., Carpenter B., Chen G., Li T., Nan S., Schols D., Bridger G. J., Fricker S. P., Skerlj R. T. (2011) Virology 413, 231–243 [DOI] [PubMed] [Google Scholar]
- 23. Wu B., Chien E. Y., Mol C. D., Fenalti G., Liu W., Katritch V., Abagyan R., Brooun A., Wells P., Bi F. C., Hamel D. J., Kuhn P., Handel T. M., Cherezov V., Stevens R. C. (2010) Science 330, 1066–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Amara A., Vidy A., Boulla G., Mollier K., Garcia-Perez J., Alcamí J., Blanpain C., Parmentier M., Virelizier J. L., Charneau P., Arenzana-Seisdedos F. (2003) J. Virol. 77, 2550–2558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bissantz C., Logean A., Rognan D. (2004) J. Chem. Inf. Comput. Sci. 44, 1162–1176 [DOI] [PubMed] [Google Scholar]
- 26. Govaerts C., Bondue A., Springael J. Y., Olivella M., Deupi X., Le Poul E., Wodak S. J., Parmentier M., Pardo L., Blanpain C. (2003) J. Biol. Chem. 278, 1892–1903 [DOI] [PubMed] [Google Scholar]
- 27. Wood A., Armour D. (2005) Prog. Med. Chem. 43, 239–271 [DOI] [PubMed] [Google Scholar]
- 28. Verdonk M. L., Cole J. C., Hartshorn M. J., Murray C. W., Taylor R. D. (2003) Proteins 52, 609–623 [DOI] [PubMed] [Google Scholar]
- 29. Jain A. N. (2007) J. Comput. Aided Mol. Des. 21, 281–306 [DOI] [PubMed] [Google Scholar]
- 30. Korb O., Stützle T., Exner T. E. (2009) J. Chem. Inf. Model. 49, 84–96 [DOI] [PubMed] [Google Scholar]
- 31. Rarey M., Kramer B., Lengauer T., Klebe G. (1996) J. Mol. Biol. 261, 470–489 [DOI] [PubMed] [Google Scholar]
- 32. Hawkins P. C., Skillman A. G., Warren G. L., Ellingson B. A., Stahl M. T. (2010) J. Chem. Inf. Model. 50, 572–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Marcou G., Rognan D. (2007) J. Chem. Inf. Model. 47, 195–207 [DOI] [PubMed] [Google Scholar]
- 34. Blanpain C., Lee B., Vakili J., Doranz B. J., Govaerts C., Migeotte I., Sharron M., Dupriez V., Vassart G., Doms R. W., Parmentier M. (1999) J. Biol. Chem. 274, 18902–18908 [DOI] [PubMed] [Google Scholar]
- 35. Zhang J., Rao E., Dioszegi M., Kondru R., DeRosier A., Chan E., Schwoerer S., Cammack N., Brandt M., Sankuratri S., Ji C. (2007) Antimicrob. Agents Chemother. 51, 1386–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Thompson D. A., Cormier E. G., Dragic T. (2002) J. Virol. 76, 3059–3064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Olson W. C., Rabut G. E., Nagashima K. A., Tran D. N., Anselma D. J., Monard S. P., Segal J. P., Thompson D. A., Kajumo F., Guo Y., Moore J. P., Maddon P. J., Dragic T. (1999) J. Virol. 73, 4145–4155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ross T. M., Bieniasz P. D., Cullen B. R. (1998) J. Virol. 72, 1918–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kim J., Wess J., van Rhee A. M., Schöneberg T., Jacobson K. A. (1995) J. Biol. Chem. 270, 13987–13997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kleinau G., Haas A. K., Neumann S., Worth C. L., Hoyer I., Furkert J., Rutz C., Gershengorn M. C., Schülein R., Krause G. (2010) FASEB J. 24, 2347–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mazna P., Berka K., Jelinkova I., Balik A., Svoboda P., Obsilova V., Obsil T., Teisinger J. (2005) Biochem. Biophys. Res. Commun. 332, 726–734 [DOI] [PubMed] [Google Scholar]
- 42. Govaerts C., Blanpain C., Deupi X., Ballet S., Ballesteros J. A., Wodak S. J., Vassart G., Pardo L., Parmentier M. (2001) J. Biol. Chem. 276, 13217–13225 [DOI] [PubMed] [Google Scholar]
- 43. de Graaf C., Rognan D. (2009) Curr. Pharm. Des. 15, 4026–4048 [DOI] [PubMed] [Google Scholar]
- 44. Pérez-Nueno V. I., Ritchie D. W., Borrell J. I., Teixidó J. (2008) J. Chem. Inf. Model. 48, 2146–2165 [DOI] [PubMed] [Google Scholar]
- 45. Kellenberger E., Springael J. Y., Parmentier M., Hachet-Haas M., Galzi J. L., Rognan D. (2007) J. Med. Chem. 50, 1294–1303 [DOI] [PubMed] [Google Scholar]
- 46. Philip V., Harris J., Adams R., Nguyen D., Spiers J., Baudry J., Howell E. E., Hinde R. J. (2011) Biochemistry 50, 2939–2950 [DOI] [PubMed] [Google Scholar]
- 47. Springael J. Y., Le Minh P. N., Urizar E., Costagliola S., Vassart G., Parmentier M. (2006) Mol. Pharmacol. 69, 1652–1661 [DOI] [PubMed] [Google Scholar]
- 48. Issafras H., Angers S., Bulenger S., Blanpain C., Parmentier M., Labbé-Jullié C., Bouvier M., Marullo S. (2002) J. Biol. Chem. 277, 34666–34673 [DOI] [PubMed] [Google Scholar]
- 49. Laskowski R. A., Rullmannn J. A., MacArthur M. W., Kaptein R., Thornton J. M. (1996) J. Biomol. NMR 8, 477–486 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.