Abstract
Voltage-gated sodium (Nav) channels are the molecular targets of β-scorpion toxins, which shift the voltage dependence of activation to more negative membrane potentials by a voltage sensor-trapping mechanism. Molecular determinants of β-scorpion toxin (CssIV) binding and action on rat brain sodium channels are located in the S1-S2 (IIS1-S2) and S3-S4 (IIS3-S4) extracellular linkers of the voltage-sensing module in domain II. In IIS1-S2, mutations of two amino acid residues (Glu779 and Pro782) significantly altered the toxin effect by reducing binding affinity. In IIS3-S4, six positions surrounding the key binding determinant, Gly845, define a hot spot of high-impact residues. Two of these substitutions (A841N and L846A) reduced voltage sensor trapping. The other three substitutions (N842R, V843A, and E844N) increased voltage sensor trapping. These bidirectional effects suggest that the IIS3-S4 loop plays a primary role in determining both toxin affinity and efficacy. A high resolution molecular model constructed with the Rosetta-Membrane modeling system reveals interactions of amino acid residues in sodium channels that are crucial for toxin action with residues in CssIV that are required for its effects. In this model, the wedge-shaped CssIV inserts between the IIS1-S2 and IIS3-S4 loops of the voltage sensor, placing key amino acid residues in position to interact with binding partners in these extracellular loops. These results provide new molecular insights into the voltage sensor-trapping model of toxin action and further define the molecular requirements for the development of antagonists that can prevent or reverse toxicity of scorpion toxins.
Keywords: Mutant, Neurotoxin, Protein structure, Sodium Channels, Sodium Transport, β-Scorpion Toxin, Voltage-gated Sodium Channels, Voltage Sensor Trapping
Introduction
Voltage-gated sodium (Nav)3 channels are responsible for generation and propagation of action potentials in nerves and in skeletal and cardiac muscle cells (1). The pore-forming α subunit of eukaryotic Nav channels has four homologous domains (I-IV), with each domain containing six transmembrane segments, S1-S6 (2). The S1-S4 segments form the voltage-sensing module, whereas the S5 and S6 segments and the P loop between them form the pore module (2). The S4 segments, which contain 4–7 conserved positively charged arginine or lysine residues separated by 2 hydrophobic residues, have been postulated to be the voltage sensors of the channels (2). In response to depolarization, the S4 segments are thought to move outward across the cell membrane, and this movement exerts a force on the S4-S5 linker, pulls S5 segments away from each other, and bends the S6 segment to open the pore (3). Structural modeling suggests that in the resting state, the extracellular S1-S2 and S3-S4 loops are close to each other, whereas in the activated state these two extracellular loops move away from each other (3). These voltage-driven conformational changes in voltage sensors are the molecular targets for modulation of Nav channel function by scorpion toxins (4).
β-Scorpion toxins enhance activation of Nav channels via binding to neurotoxin receptor site 4 (4–7). A β-scorpion toxin, CssIV from the venom of the scorpion Centruroides suffusus suffusus, shifts the voltage dependence of rat brain NaV1.2 channel activation to more negative membrane potentials (8). However, a strong priming depolarizing prepulse is required for the toxin to have its functional effect (8). A voltage sensor-trapping mechanism was proposed to explain the prepulse-dependent enhancement of activation by β-scorpion toxins (8). According to the model, the toxin binds to its receptor site in the extracellular loops connecting transmembrane segments S1-S2 and S3-S4 in homologous domain II (IIS1-S2 and IIS3-S4) in the resting state of the voltage sensor in a concentration-dependent, bimolecular reaction to form the initial toxin-channel complex. Upon strong depolarization, the IIS4 segment moves outward and rotates, and the toxin binds to newly accessible amino acid residues on the IIS1-S2 and IIS3-S4 loops and the extracellular end of the IIS4 segment in a unimolecular, concentration-independent reaction. In this activated position, the voltage sensor is tightly bound to the rigid toxin and is trapped in its activated conformation. Upon repolarization of the cell membrane, the trapped IIS4 voltage sensors remain activated; therefore, subsequent depolarizations activate the channels at more negative membrane potentials because one of the four voltage sensors is already activated (8–10).
Previous studies revealed amino acid residues in IIS1-S2 and IIS3-S4 of rNav1.2a channels whose substitution altered the binding and action of CssIV (8–11). In the present experiments, we have refined the molecular map of the amino acid residues in these two extracellular linkers that are required for CssIV binding and voltage sensor trapping, and we have discovered a hot spot for toxin action where single mutations in adjacent amino acid residues can either enhance or impair toxin action substantially. Our results enable a detailed structural and mechanistic model for CssIV interaction with its receptor site in the voltage sensor of domain II of rNav1.2a channels and provide further support for the voltage sensor-trapping model of scorpion toxin action and the sliding helix model of voltage sensor function.
EXPERIMENTAL PROCEDURES
PCR-directed Mutagenesis
Mutations were introduced by site-directed mutagenesis using a PCR-based strategy as described previously (9). Hydrophobic amino acid residues, serine, cysteine, and threonine were substituted with alanine to prevent toxin-binding interactions with their side chains. Glutamine was substituted for the larger charged residues arginine, lysine, and glutamic acid, whereas asparagine was substituted for aspartic acid residues. Single amino acid chimeras were generated by aligning amino acid sequences of Nav channels subtypes and substituting nonconserved amino acid residues. All the WT and mutant cDNAs were subcloned into pCDM8 plasmid.
cDNA Transfection
The methods of cDNA transfection have been described in detail previously (10). Briefly, cDNAs encoding wild-type (WT) and mutant rNav1.2a channels were transiently expressed in tsA-201 cells by calcium-phosphate transfection. pEBO-pCD8-leu2 was co-transfected into tsA-201 cells, and the cell surface CD8 marker was identified by incubation with polystyrene microspheres pre-coated with anti-CD8 antibody. The expressed CD8 protein was used to identify cells that express rNav1.2a channels. Transfected cells were subcloned 12–18 h after transfection. Electrophysiological recordings were performed 18–72 h after transfection. WT and mutant rNav1.2a cDNAs were always transfected and studied in parallel to ensure that differences from WT were truly representative of the phenotype of the mutation.
Electrophysiological Recording and Analysis
The electrophysiological recording procedures have been described previously (9). Briefly, the whole-cell patch clamp configuration was utilized for Na+ current recording with extracellular recording solution containing (in mm) NaCl (150), Cs-HEPES (10), MgCl2 (1), KCl (2), CaCl2 (1.5), and 0.1% BSA, pH 7.4, and intracellular recording solution containing (in mm) N-methyl-d-glucamine (190), HEPES (10), MgCl2 (10), NaCl (10), and EGTA (5), pH 7.4. Linear leak and residual capacitance currents were subtracted using an online P/−4 subtraction paradigm. To assess the extent of negative shift of the voltage dependence of activation caused by CssIVE15A, the tsA-201 cells were held at −100 mV and test depolarizations were applied to potentials from −100 to +20 mV in 5-mV increments. Current-voltage (I-V) plots were generated from peak currents elicited by each depolarization. Each test depolarization was either applied alone or preceded by a 1-ms prepulse to +50 mV followed by a 60-ms interval at the holding potential. We tested the functional properties of each mutant rNav1.2a construct in the absence of toxin to examine the effect of the mutant residue, followed by recordings in the presence of CssIVE15A. The voltage dependence and kinetics of each mutant channel were initially screened with 500 nm CssIVE15A to detect differences from WT. If a mutant channel did have significant differences in function from WT, further tests were performed to investigate the phenotype of mutant channels in detail. All the data were analyzed with Igor Pro (WaveMetrics, Lake Oswego, OR). Normalized I-V curves were fit with a function including a single Boltzmann component of the form: (V − Vrev) × Gmax/{1 + exp[(V½ − V)/k]}, where V½ is the half-activation voltage in mV, and k is a slope factor in mV. In analyses of the rates of development and reversal of voltage sensor trapping, currents were normalized to the maximal peak current of a preceding I-V plot. All data are presented as mean ± S.E.
Structural Modeling
Homology and de novo modeling of the voltage sensor in domain II of rat Nav1.2a channels was performed using the Rosetta-Membrane method (3, 12). rNav1.2a sequence (residues Leu754-Gly875) was aligned with the Kv1.2-Kv2.1 chimera channel sequence (residues Ser158-Gln315 (13)) using ClustalX software (14). The alignment of the IIS4 segments was manually adjusted to align the first arginine in the IIS4 of rNav1.2 with Gln290 in Kv1.2-Kv2.1 chimera channels, which corresponds to the conserved R0 position in most Kv channels (supplemental Fig. S1). 5,000 models of the voltage sensor were generated, and the lowest scoring model was selected as the best model. Modeling of CssIV toxin was performed using the Rosetta method for modeling of soluble proteins (15). β-Scorpion toxin sequence was aligned with the neurotoxin 2 sequence (X8, Protein Data Bank code 1JZA) using ClustalX software (14) (see supplemental Fig. S1). 5,000 models of the toxin were generated and the lowest scoring model was chosen as the best model. Docking simulations of CssIV binding to the voltage sensor in domain II of the rNav1.2a channel were performed using the Rosetta docking method (16, 17). Backbone flexibility of the extracellular part of the voltage sensor (residues Cys768-Gly800 and Ser832-Arg856) was allowed during simulations and Tyr40 and Tyr42 residues of the β-scorpion CssIV toxin were required to be at the voltage sensor-toxin interface. 10,000 models were generated and the best model was chosen among 20 lowest scoring models as the model that fit the majority of available experimental data on key residues contributing to interaction of the CssIV toxin with voltage-sensing domain II of rat Nav1.2a channels (8, 9, 18).
RESULTS
Modification of Voltage-dependent Activation of NaV1.2 Channels by CssIVE15A Toxin
Our previous work showed that CssIV shifted the voltage dependence of activation of rNav1.2a channels to more negative membrane potentials (8). These functional effects required a strong depolarizing prepulse to activate the channels (8). In the present study we used a high-affinity recombinant derivative of CssIV, CssIVE15A (19). We first verified the requirement of a prepulse to observe the functional effects of CssIVE15A on WT rNav1.2a channels. Without a prepulse, current-voltage (I-V) relationships obtained from rNav1.2a channels using 15-ms depolarizations to potentials from −100 to +20 mV from a holding potential of −100 mV were similar whether or not 500 nm CssIVE15A was present. Under these conditions, no Na+ current was activated at test potentials more negative than −60 mV (Fig. 1). Even in the presence of CssIVE15A, little inward current was activated by a test pulse to −60 mV (Fig. 1B). In contrast, when a 1-ms prepulse to +50 mV was applied 60 ms before each test pulse, marked Na+ current was observed at −60 mV (Fig. 1B), and a substantial component of the Na+ current activated with a negatively shifted voltage dependence (Fig. 1C). These effects were not observed in the absence of CssIVE15A. This negative shift in the voltage dependence of activation reflects voltage sensor trapping by bound CssIV (8). At 500 nm CssIVE15A, a mean of 11.9% of the peak Na+ current activated at this negative membrane potential for WT rNav1.2a channels (Fig. 1 and Table 3). We used this negative shift in the voltage dependence of activation to assay the effects of mutations in IIS1-S2 and IIS3-S4 on CssIV binding and action, and we have termed the additional current observed at a test pulse to −60 mV following a prepulse “IVST,” as an abbreviation for voltage sensor-trapping current. To conserve channel function, site-directed mutations of single amino acid residues were designed to reduce the hydrophobicity or hydrophilicity of amino acid side chains by conversion to Ala, to reduce the charge of amino acid side chains while retaining their hydrophilicity by conversion of Arg, Lys, Glu, and Asp to Asn, Gln, or Cys, or to substitute single amino acid residues from homologous NaV channels having different responses to β-scorpion toxins. The functional properties of several representative mutants are described first, followed by presentation of a molecular map and high-resolution structural model of the toxin-receptor interaction.
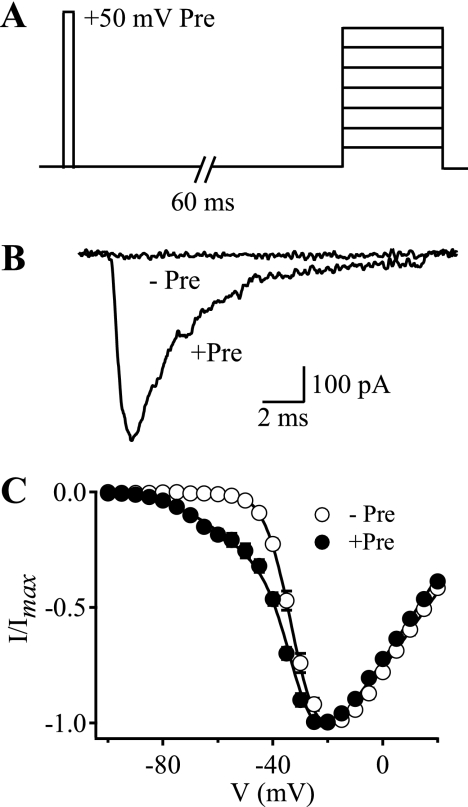
FIGURE 1.
Voltage sensor-trapping activity of CssIVE15A on WT rNav1.2a channels. Cells were stimulated with depolarizations ranging from −100 to +20 mV in 5-mV increments either alone or preceded 60 ms earlier by a 1-ms test pulse to +50 mV in the presence of 500 nm CssIVE15A. A, pulse protocol. B, IVST traces recorded during test pulses to −60 mV obtained without (−Pre) or with (+Pre) the prepulse. C, I-V plots obtained for rNav1.2a channels in the presence of 500 nm CssIVE15A with (filled circles, +Pre) or without (open circles, −Pre) the prepulse. The solid lines are global fits of a function with 2 Boltzmann components to the I-V curves without and with prepulses. The common fit parameters were Va1 = −32.1 mV, Va2 = −53.6 mV. The negative component represented 25.3% of the conductance for the +Pre curve.
TABLE 3.
IVST and KD of WT and mutant channels in the presence of CssIVE15A
The voltage dependence of activation was measured as described under “Experimental Procedures.” IVST is the normalized voltage-sensor trapping current observed at a test pulse to −60 mV following a prepulse. For channels in which saturating effects of CssIVE15A could be recorded, KD values were calculated from fitting the data to the voltage sensor trapping model (supplemental Fig. S2). IVST data are presented as mean ± S.E.
| Channel | Concentration | IVST (−Pre) | IVST (+Pre) | n | KD |
|---|---|---|---|---|---|
| nm | nm | ||||
| WT | 500 | 0 | 11.9 ± 2.3% | 5 | 1130 |
| A841N | 500 | 0 | 2.8 ± 0.7% | 5 | NAa |
| 1000 | 0 | 5.3 ± 1.1% | 4 | ||
| L846A | 500 | 0 | 0.1 ± 0.3% | 4 | NA |
| 1000 | 0 | 0.1 ± 0.3% | 3 | ||
| N842R | 200 | 5.1 ± 0.7% | 26.0 ± 3.2% | 7 | NA |
| 500 | 9.0 ± 1.5% | 53.0 ± 4.5% | 6 | ||
| 1000 | 8.9 ± 0.9% | 55.0 ± 4.0% | 8 | ||
| V843A | 20 | 0 | 4.0 ± 1.0% | 4 | 263 |
| 50 | 0 | 10.4 ± 2.0% | 7 | ||
| 100 | 0 | 19.1 ± 3.5% | 5 | ||
| 200 | 0 | 29.9 ± 2.4% | 7 | ||
| 500 | 0 | 56.0 ± 5.0% | 5 | ||
| 1000 | 0 | 49.2 ± 3.9% | 5 | ||
| E844N | 20 | 0 | 2.9 ± 0.9% | 4 | 425 |
| 50 | 0 | 8.4 ± 1.9% | 3 | ||
| 200 | 0 | 26.3 ± 3.1% | 5 | ||
| 300 | 0 | 38.8 ± 13.7% | 3 | ||
| 400 | 0 | 50.3 ± 7.2% | 4 | ||
| 500 | 0 | 50.3 ± 6.5% | 5 |
a NA, not applicable.
Loss of Voltage Sensor Trapping with Mutants A841N and L846A
In previous studies, we showed that amino acid residues in IIS1-S2 and IIS3-S4 were crucial for the binding and voltage sensor-trapping activity of β-scorpion toxin on rNav1.2a channels (8, 9). In the present work, we used recombinant CssIVE15A to identify individual amino acid residues that are important for toxin action by mutagenesis and characterization of their functional effects by electrophysiological analysis. We found that two mutants (A841N and L846A) in IIS3-S4 greatly reduced the voltage sensor-trapping activity of CssIVE15A.
In the absence of toxin, mutation A841N did not affect the I-V relationship compared with WT (Fig. 2A, Table 1), indicating that this mutation does not alter the voltage-dependent activation of sodium channels. As for WT channels, CssIVE15A (500 nm) did not induce significant Na+ current when cells expressing A841N channels were depolarized to −60 mV without a depolarizing prepulse (Fig. 2B, Table 2). However the I-V relationship of A841N was altered only slightly by a +50-mV, 1-ms depolarizing prepulse in the presence of the toxin (Fig. 2B), and IVST was only 2.8% at 500 nm and increased to 5.3% at 1 μm (Table 3). These results suggest that mutant A841N has reduced affinity for CssIV and possibly also reduced efficacy in voltage sensor trapping.
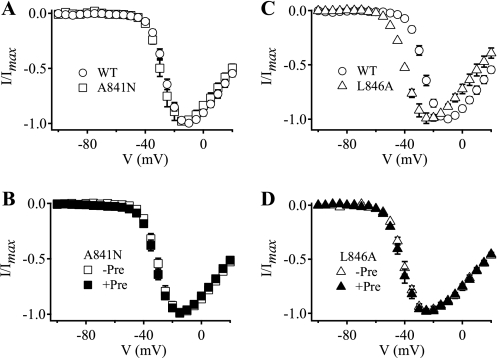
FIGURE 2.
Voltage sensor-trapping activity of CssIVE15A on A841N and L846A mutant channels. A and C, I-V plots in the absence of toxin; B and D, I-V plots in the presence of 500 nm CssIVE15A with or without a prepulse. A, WT rNav1.2a (open circles) and A841N (open squares). B, A841N, in the absence (open squares, −Pre) and presence (filled squares, +Pre) of the prepulse. C, WT rNav1.2a (open circles) and L846A (open triangles). D, L846A in the absence (open triangles, −Pre) and presence (filled triangles, +Pre) of the prepulse.
TABLE 1.
Voltage dependence of activation
The voltage dependence of activation was measured as described under “Experimental Procedures” in the absence of toxin. The voltage of half-activation (V0.5) and the slope factor of each channel were derived from fitting the corresponding voltage-dependent activation curve with a single Boltzmann equation. All data are presented as mean ± S.E.
| Channel | V0.5 | Slope | n |
|---|---|---|---|
| mV | |||
| WT | −24.1 ± 1.0 | −5.5 ± 0.6 | 4 |
| A841N | −26.3 ± 2.4 | −5.2 ± 0.8 | 3 |
| N842R | −26.6 ± 0.6 | −6.0 ± 0.4 | 6 |
| V843A | −31.9 ± 0.8 | −4.4 ± 0.2 | 4 |
| E844N | −21.8 ± 0.3 | −5.8 ± 0.5 | 5 |
| L846A | −40.8 ± 2.7 | −5.6 ± 0.6 | 4 |
TABLE 2.
Voltage dependence of activation with CssIVE15A
The voltage dependence of activation was measured as described under “Experimental Procedures” in the presence of 500 nm CssIVE15A but without the prepulse. The voltage of half-activation (V0.5) and slope factor of each channel in the presence of 500 nm CssIVE15A but without the prepulse are derived from fitting the corresponding voltage-dependent activation curve with a single Boltzmann equation. All data are presented as mean ± S.E.
| Channel | V0.5 | Slope | n |
|---|---|---|---|
| mV | |||
| WT | −29.6 ± 1.1 | −5.9 ± 0.3 | 5 |
| A841N | −27.8 ± 1.2 | −5.5 ± 0.2 | 5 |
| N842R | NAa | NA | NA |
| V843A | −20.3 ± 1.6 | −7.9 ± 0.4 | 5 |
| E844N | −23.0 ± 0.4 | −7.3 ± 0.2 | 5 |
| L846A | −38.5 ± 1.4 | −5.6 ± 0.4 | 4 |
a NA, not applicable because IVST was present without a prepulse.
The nearby mutation L846A also impaired voltage sensor trapping by CssIVE15A, but its effect was even greater. It caused a negative shift in the voltage dependence of activation under control conditions (Fig. 2C, Table 1). However, voltage sensor trapping by CssIVE15A was completely abolished for L846A (Fig. 2D, Table 3). Thus, mutation L846A had two functional effects. First, the mutation itself enhanced activation of the channel, either by stabilizing the IIS4 voltage sensor in the activated conformation, destabilizing the resting conformation, or both. Second, the L846A mutation completely blocked voltage sensor trapping by the toxin, either by complete loss of binding of CssIVE15A to the mutant channel in resting state or by complete loss of voltage sensor trapping efficacy by CssIVE15A in the activated state, or a combination of these effects.
Increased Voltage Sensor Trapping with Mutant N842R
To examine the functional properties of mutant N842R, we first tested its voltage-dependent activation in the absence of toxin. We observed that the I-V relationship of the N842R channel was similar to WT (Fig. 3A, Table 1). However, unlike the WT channel, treatment with 500 nm CssIVE15A induced a significant Na+ current during test pulses to −60 mV without a depolarizing prepulse (Fig. 3, B and C, Table 3), and the I-V relationship revealed a significant component of IVST (Fig. 3C, Table 3). Global fits to the I-V curves indicated that ∼9% of the current activated with the negative voltage dependence without a prepulse. Approximately 55% of the Na+ current activated with negative voltage dependence after a prepulse, about 6-fold more than without the prepulse (Fig. 3C, Table 3). This negatively shifted component of the Na+ current is 4.45-fold greater than that observed for WT (Figs. 1B and 3C, and Table 3). Therefore, the mutation N842R greatly enhanced voltage sensor trapping by CssIVE15A.
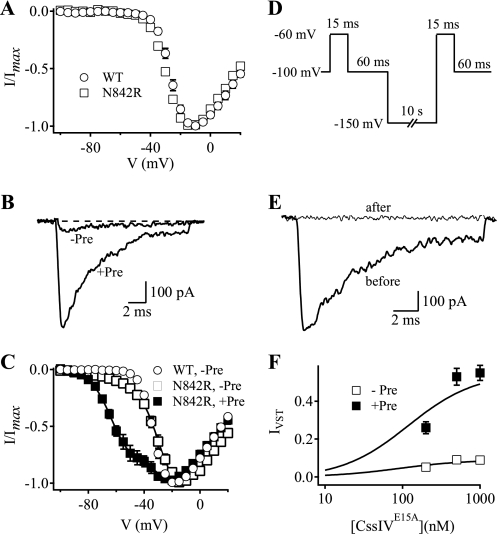
FIGURE 3.
Voltage sensor-trapping activity of CssIVE15A in N842R mutant channels. A, I-V plots obtained in the absence of toxin for WT rNav1.2a channels (circles) and N842R mutant channels (squares). B, IVST traces recorded during a 15-ms test pulse to −60 mV in the absence or presence of a 1-ms prepulse to +50-mV applied 60 ms earlier. C, I-V plots for N842R mutant channels in the presence of CssIVE15A with (filled squares) or without (open squares) the prepulse. The plot for WT channels without prepulse in the presence of toxin is shown for comparison (open circles). The solid lines are global fits to the I-V curves with Va1 = −27.6 mV, k1 = −7.0, Va2 = −61.4 mV, k2 = −7.9 mV. Without the prepulse, 7.5% of the current activated with the negative voltage dependence (Va2). With the prepulse, 55% of the current activated with the negative voltage dependence, Va2. D, voltage protocol used to observe the effects of hyperpolarization upon voltage sensor-trapping activity by CssIVE15A on N842R channels at resting membrane potential. E, IVST traces recorded at −60 mV before or after the hyperpolarization pulse to −150 mV for 10 s. F, concentration-response curves for IVST on N842R mutant channels by CssIVE15A with (filled squares, +Pre) or without (open squares, -Pre) the prepulse. IVST was normalized to the peak of the I-V in the absence of toxin. Concentration-response data were fit with first-order Hill equations (n > = 4).
We hypothesized that the IVST that we observed without a prepulse might be caused by voltage sensor trapping at the holding potential of −100 mV and might be reversed by a stronger hyperpolarization. In fact, the IVST observed with this mutant channel was eliminated by a 10-s hyperpolarization to −150 mV (Fig. 3, D and E). Evidently, CssIVE15A trapped a significant fraction of the IIS4 voltage sensors of N842R channels in an activated state at the resting membrane potential.
We measured the voltage sensor trapping activity of CssIVE15A with N842R channels at three different concentrations of CssIVE15A (Fig. 3F). Values of IVST as a function of concentration were fit with first-order Hill equations (Fig. 3F, Table 3). IVST reached a maximum of 53 ± 4.5% of total Na+ current with an EC50 of 170 nm. Without a prepulse, IVST increased over a similar concentration range to reach a maximum of 9.0 ± 1.5% (Fig. 3F).
Our previous work demonstrated that the rate of voltage sensor trapping by CssIV during the strong depolarizing prepulse is concentration-independent, suggesting that this process does not involve binding of toxin (9). To compare the rate and extent of voltage sensor trapping upon depolarization by CssIVE15A in WT and N842R channels, we applied depolarizations of variable duration (0–5 ms) to +50 mV from a holding potential of −100 mV. Trapping was assayed 60 ms later by a test pulse to −60 mV (Fig. 4A, inset). The peak test pulse current at −60 mV increased as a function of prepulse duration (Fig. 4, A and B). The time courses were fit with single exponential functions to yield the maximum amount of trapping (Fig. 4C, top) and the rate of trapping (Fig. 4C, bottom) as a function of concentration. For WT, higher concentrations of CssIVE15A resulted in greater IVST after a 5-ms depolarization, whereas IVST was maximal for N842R at all three concentrations of CssIVE15A (Fig. 4C, top). These results indicate that voltage sensor trapping by CssIVE15A in N842R channels is saturated at 200 nm. In contrast to our results for the extent of voltage sensor trapping, the time constants for the development of voltage sensor trapping were concentration-independent for both WT and N842R (Fig. 4C, bottom). These results are consistent with the three-step model for voltage sensor trapping: concentration-dependent binding, voltage-dependent channel activation, and finally concentration-independent voltage sensor trapping (8, 9).
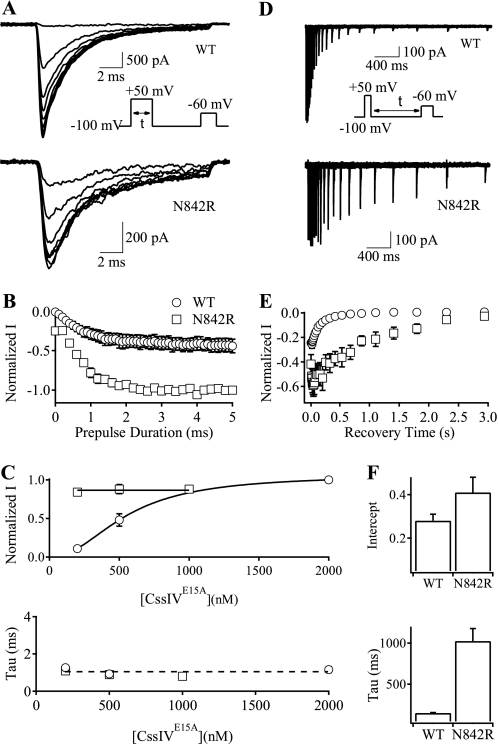
FIGURE 4.
Rates of the development and recovery of voltage sensor trapping by CssIVE15A on N842R mutant channels. To measure the rates of development of voltage sensor trapping (A–C) the cell membrane was depolarized to +50 mV for 0 to 5 ms (0.2-ms increments), followed by repolarization to the resting potential for 60 ms and a test pulse to −60 mV (A, inset). A, IVST traces at −60 mV for WT rNav1.2a (upper panel) and N842R mutant channels (lower panel). Some traces were omitted for clarity. B, plots of normalized IVST versus prepulse duration for WT rNav1.2a (open circles) and N842R mutant channels (open squares). C, magnitude of IVST (upper panel) and time constants (lower panel) of the development of voltage sensor trapping for WT rNav1.2a and N842R mutant channels at a series of CssIVE15A concentrations. To measure the rates of recovery from voltage sensor trapping (D–F), the cell was depolarized to +50 mV for 1 ms followed by a repolarization to resting potential for 20–3000 ms and a test pulse to −60 mV for 15 ms (D, inset). Data were fit with a single exponential to determine the maximal IVST (the intercept) and the rates of recovery from voltage sensor trapping. D, superimposed IVST traces for WT rNav1.2a (upper panel) and N842R mutant channels (lower panel). E, plots of peak currents in D versus recovery time for WT rNav1.2a (open circles) and N842R mutant channels (open squares). F, intercepts (upper panel) and time constants (lower panel) for the recovery of the voltage sensor trapping for WT rNav1.2a and N842R mutant channels.
Repolarization causes reversal of voltage sensor trapping. To measure the rates of reversal, cells were depolarized to +50 mV for 1 ms to activate and trap a population of channels. They were then repolarized to −100 mV for variable times followed by a test depolarization to −60 mV to assay the loss of trapping (Fig. 4D, inset). IVST decayed exponentially with recovery time for both WT and N842R channels (Fig. 4D). However, the rate of decay was ∼6.8-fold slower for N842R compared with WT channels (Fig. 4, D and E), as quantified by the time constants of single exponential fits to these data (Fig. 4F).
Increased Voltage Sensor Trapping with Mutants V843A and E844N
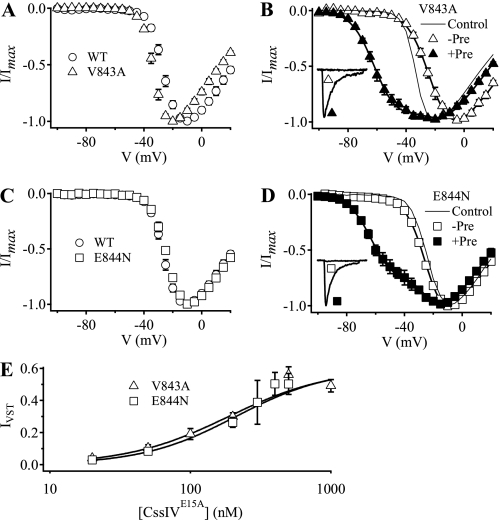
Mutations V843A and E844N also increased voltage sensor trapping following a prepulse (Fig. 5), but the effects were less complex than for mutation N842R. In the absence of CssIVE15A, mutation V843A shifted the voltage dependence of activation ∼7 mV to more negative potentials (Fig. 5A, Table 1), suggesting that the mutation stabilized the activated state of the domain II voltage sensor. In the presence of 500 nm CssIVE15A but without a prepulse, the voltage-dependent activation of V843A was shifted 11.4 mV in the positive direction (Fig. 5B, Tables 1 and 2). This is consistent with the idea that toxin binding stabilizes the voltage sensor in its resting conformation. No IVST was detectable at −60 mV with V843A channels (Fig. 5B, inset). However, following a prepulse to +50 mV in the presence of 500 nm CssIVE15A, IVST was increased to 56 ± 5% of the maximal peak current for V843A (Fig. 5B), which was 4.7-fold greater than WT. In contrast to V843A, the voltage dependence of activation of E844N was similar to WT (Fig. 5C, Table 1). However, following a prepulse to +50 mV in the presence of 500 nm CssIVE15A, IVST was increased to 50.3 ± 6.5% of the maximal peak current for E844N (Fig. 5D), 4.2-fold greater than WT. Thus, both mutations V843A and E844N greatly enhanced voltage sensor trapping by CssIVE15A. Fitting concentration-response curves for voltage sensor trapping gave EC50 values of 187 ± 18 nm for V843A and 210 ± 25 nm for E844N (Fig. 5E).
FIGURE 5.
Voltage sensor trapping activity of CssIVE15A on V843A and E844N mutant channels. A, I-V plots obtained in the absence of toxin for WT rNav1.2a channels (circles) and V843A mutant channels (triangles). B, I-V plots for V843A mutant channels in the presence of 500 nm CssIVE15A with (filled triangles, +Pre) or without (open triangles, −Pre) a +50-mV, 1-ms prepulse applied 60 ms earlier. Inset, IVST traces recorded in the presence of 500 nm CssIVE15A with a 15-ms test pulse to −60 mV in the absence or presence of the prepulse. The solid lines are global fits to I-V curves with and without prepulses with parameters Va1 = −21.1 mV, k1 = −7.9 mV, Va2 = −59.5 mV, k2 = −8.2 mV. Without a prepulse, no current activated with the more negative voltage dependence. With a prepulse, 43.2% of the conductance activated with the negative voltage dependence. The line without symbols is the toxin-free data for V843A from panel A. C, I-V plots obtained in the absence of toxin for WT rNav1.2a channels (circles) and E844N mutant channels (squares). D, I-V plots for E844N mutant channels in the presence of 500 nm CssIVE15A with (filled squares, +Pre) or without (open squares, −Pre) the prepulse. Inset, IVST traces recorded in the presence of 500 nm CssIVE15A with a test pulse depolarizing to −60 mV for 15 ms in the absence or presence of a +50-mV, 1-ms prepulse 60 ms earlier. The solid lines are global fits to I-V curves with and without prepulses with parameters Va1 = 23.3 mV, k1 = −7.0 mV, Va2 = −60.7 mV, k2 = −8.1 mV. Without a prepulse, no current was activated with the more negative voltage dependence. With a prepulse, 43.2% of the conductance activated with the negative voltage dependence. The line without symbols is the toxin-free data for E844N from panel C. E, concentration-response curves for normalized IVST of V843A (open triangles) and E844N (open squares) mutant channels by CssIVE15A with the prepulse. IVST values were normalized to the current at the peak of the I-V curves for the corresponding mutants in the absence of toxin. The concentration-response data were fit with first-order Hill equations (n ≥ 4).
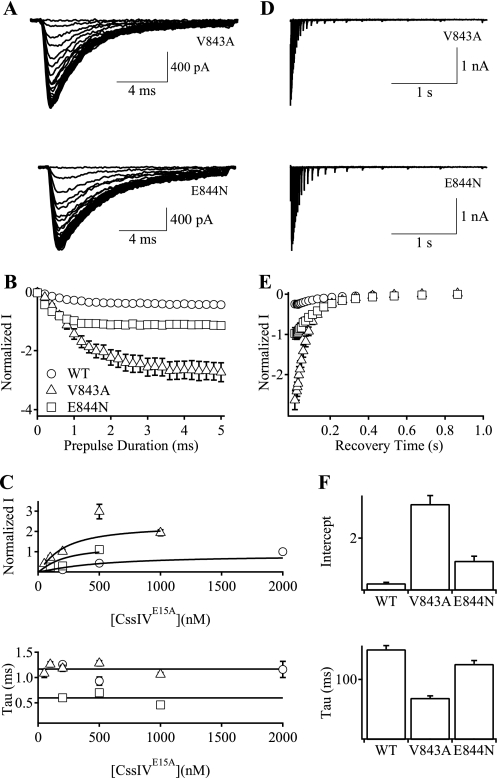
We measured the rates of development of voltage sensor trapping for mutants V843A and E844N (Fig. 6, A and B). The level of IVST increased with the duration of the prepulse and reached maximal effect at ∼1 ms for E844N and 3 ms for V843A (Fig. 6B). The extent of voltage sensor trapping for both mutants was substantially greater than that of WT (Fig. 6B). The extent of voltage sensor trapping also increased with toxin concentration for both mutants (Fig. 6C, top), whereas the rate constants for the development of voltage sensor trapping were concentration-independent for both mutants (Fig. 6C, bottom), consistent with our three-step model for voltage sensor trapping in which the final trapping step is both concentration- and voltage-independent (8, 9).
FIGURE 6.
Rates of the development and recovery of voltage sensor trapping by CssIVE15A on V843A and E844N mutant channels. A–C, the development of voltage sensor trapping was measured using the protocol of Fig. 4A. A, IVST traces at −60 mV for V843A (upper panel) and E844N (lower panel) mutant channels. Some traces were omitted for clarity. B, plots of normalized IVST versus prepulse duration for WT rNav1.2a (circles), V843A (triangles), and E844N (squares) channels. C, maximal magnitude of IVST (upper panel) and the time constants (lower panel) of the development of voltage sensor trapping by CssIVE15A in WT rNav1.2a, V843A, and E844N channels at a series of CssIVE15A concentrations. D–F, the rates of recovery from voltage sensor trapping were measured using the protocol of Fig. 4D. D, IVST traces for V843A (upper panel) and E844N mutant channels (lower panel). E, plots of peak IVST in D versus recovery time for WT rNav1.2a (open circles), V843A (open triangles), and E844N (open squares) channels. F, intercept (upper panel) and time constants (lower panel) of the recovery of the voltage sensor trapping by CssIVE15A in WT rNav1.2a, V843A, and E844N mutant channels.
We also measured the rate of reversal of voltage sensor trapping for mutants V843A and E844N (Fig. 6D). The maximal level of voltage sensor trapping was greater for both V843A and E844N than for WT (Fig. 6, E and F, top). The rate of reversal of voltage sensor trapping at −100 mV for E844N was comparable with that of WT but was accelerated for V843A (Fig. 6F, bottom). The striking differences in the kinetics of the development and reversal of voltage sensor trapping for mutants N842R, V843A, and E844N are considered under “Discussion.”
A Molecular Map of the β-Scorpion Toxin Receptor Site
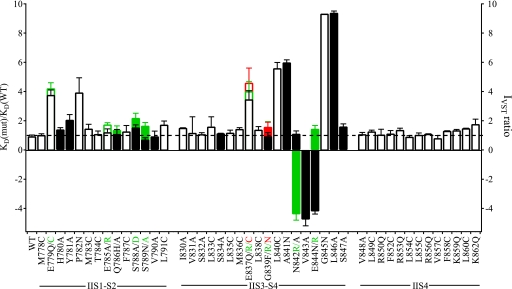
Positively charged and hydrophobic amino acid residues in CssIV are important for its binding (7). Our previous studies of channel chimeras implicated the extracellular IIS1-S2 and IIS3-S4 loops of Nav channels in formation of the receptor site for β-scorpion toxins and provided evidence that the IIS4 segment itself is not directly involved in toxin binding (8–10). To complete the mapping of the receptor site for β-scorpion toxin on Nav channels, we constructed and analyzed 23 mutations in IIS1-S2 and IIS3-S4. A linear map of the functional effects of all of the mutants that have been characterized in this study and previous work is illustrated in Fig. 7 in terms of the Kd ratio (mutant/WT) measured in radioligand binding studies or the IVST ratio (WT/mutant) measured in this work. This complete scan of amino acid residues in the IIS1-S2, IIS3-S4, and IIS4 segments reveals two areas of interest (Fig. 7). Eight residues in the IIS3-S4 loop (Glu837, Leu840, Ala841, Asn842, Val843, Glu844, Gly845, and Leu846) are important in CssIV binding and action on rNav1.2a channels, and seven of them form a hot spot of contiguous amino acid residues for toxin action. Mutations of these amino acid residues can either increase or decrease voltage sensor trapping by CssIV. In addition, two residues in IIS1-S2 (Glu779 and Pro782) also contribute significantly to CssIV binding and action (Fig. 7). These results indicate that the IIS3-S4 loop plays the primary role in binding of CssIV toxin and controls the functional effects of the toxin, whereas two amino acid residues in the IIS1-S2 loop play a secondary role.
FIGURE 7.
Binding affinity and voltage sensor-trapping activity of CssIVE15A in WT and mutant rNav1.2a channels. The ratio of the KD of CssIV binding to mutant channels to that of WT channels (KD(mut)/KD(WT), open bars) studied by Cèstele et al. (8, 9) or the IVST ratio for mutant channels studied functionally here (filled bars). For mutant channels that enhance voltage sensor trapping, the IVST ratio was defined as −1[IVST(WT)/IVST(mut)]. For mutant channels that reduce voltage sensor trapping, the IVST ratio was defined as IVST(WT)/IVST(mut). At some loci multiple mutants were studied and the data corresponding to each mutant and its corresponding label are color coded. For instance, E837Q, E837R, and E837C are plotted in black, green, and red, respectively, in the bar graph. Empty bars represent previously studied amino acid residues (8, 9), whereas the filled bars represent new residues whose function was analyzed in this paper.
Structural Model of β-Scorpion Toxin Binding to the Nav1.2 Channel
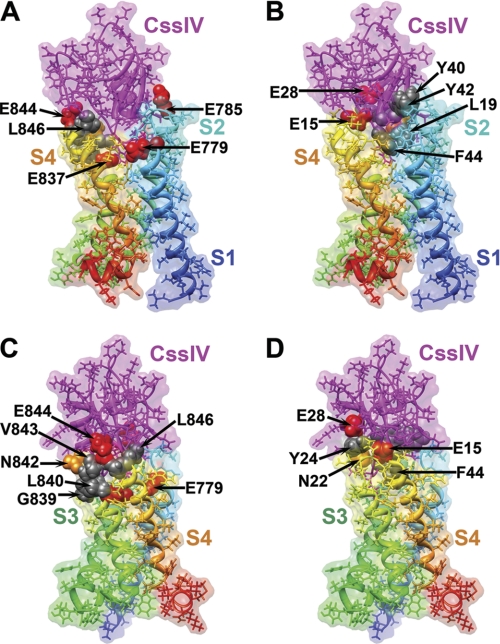
We previously developed a structural model of the CssIV-rNav1.2a complex (9) using the Rosetta rigid-body docking method (16) and the structure of the bacterial KvAP voltage-sensing domain (20) as a template. Here we present a new structural model of the CssIV-rNav1.2a complex that was generated using the Rosetta flexible-backbone docking method (17) and the high-resolution structure of the mammalian Kv1.2-Kv2.1 chimera channel (13) as a template (see “Experimental Procedures”). Our new structural model shows that the CssIV toxin has an extensive surface interacting with the amino acid residues that line the extracellular water-accessible cavity of the rNav1.2a voltage sensor in domain II (Fig. 8, A and B). The overall orientation of the CssIV toxin relative to the voltage sensor in our new model (Fig. 8) is very similar to our original model (9). The majority of residues in the voltage sensor that have significant effects on CssIV toxin binding and voltage sensor trapping are at the interface with the toxin in our model (Fig. 8, A and C). Specifically, Glu779 in S1 interacts with Phe44 of CssIV, Glu837 in S3 interacts with Phe44 of CssIV, and Ala841, Asn842, Glu844, and Leu846 in the IIS3-S4 loop are in direct contact with the CssIV toxin (Fig. 8C), consistent with the significant effects of mutations in these residues on toxin binding and action (Fig. 7). Although Leu840, Val843, and Gly845 seem not to be in direct contact with the CssIV toxin in our model, mutations L840C and V843A may alter the local conformation of the IIS3-S4 loop during channel activation, and the mutation G845N would create a structural conflict for toxin binding according to our model. On the CssIV toxin side of the interaction, Glu15, Asn22, Tyr24, Glu28, and Trp58 are either in direct contact or within close proximity to the IIS3-S4 loop of the channels, which is in agreement with the significant effects of mutations at these positions on toxin binding to the channel (Fig. 8, B and D).
FIGURE 8.
Full atom and molecular surface representation of β-scorpion CssIV binding to the voltage-sensing domain II of rNav1.2a. Models were generated as described under “Experimental Procedures.” Segments IIS1 through IIS4 of the voltage-sensing domain are colored individually and labeled. A and B, side view of the structural model with voltage-sensing domain segments IIS1 and IIS4 on the front. C and D, side view of the structural model with the voltage-sensing domain segments and with IIS3 and IIS4 on the front (rotated 90° counterclockwise when viewed from the extracellular side of the membrane compared with orientation shown in panels A and B). Side chains of key residues for the CssIV-rNav1.2a interaction are shown in space filling representation and all other side chains shown in stick representation. A probe radius of 1.4 Å was used to scan the molecular surface of each structural model. This figure was generated using Chimera (43).
DISCUSSION
The Receptor Site for β-Scorpion Toxins Includes a Hot Spot in the IIS3-S4 Loop
In the experiments described here, we have substantially extended our previous studies of the molecular determinants of voltage sensor trapping (8–10) and further established that both IIS1-S2 and IIS3-S4 loops are required for normal binding and action of β-scorpion toxins, whereas the IIS3-S4 loop plays a dominant role. These data suggest that β-scorpion toxins interact with a short segment of IIS1-S2 and a broader region of IIS3-S4. Evidently, these two distant regions of the primary structure of Nav channels are close to each other in the folded structure of the channels and form a single toxin-binding site. Similarly, the CssIV residues that are crucial for toxin binding to Nav channels come from different regions of the primary structure but are brought together in a wedge-shaped interactive surface in the tertiary structure of the toxin (18).
Seven of the eight influential residues in IIS3-S4 are in consecutive positions in the primary structure of the rNav1.2a channels. This cluster of amino acid residues in the IIS3-S4 loop defines a hot spot for toxin action. The amino acid sequence of this segment is well conserved between rNav1.2a and hNav1.4, consistent with the similar effects of CssIV on these two channels (21). The IIS1-S2 and IIS3-S4 loops may play cooperative roles in toxin binding and voltage sensor trapping. In molecular models of K+ channels, the IIS1-S2 loop is relatively static (22, 23), whereas the IIS3-S4 loop undergoes outward displacement and rotation relative to IIS1-S2 in response to voltage sensor activation, and the cleft between IIS1-S2 and IIS3-S4 widens (3, 24). Molecular models of the voltage sensor of a bacterial Nav channel suggest similar movements during activation (25, 26). As the cleft formed by the IIS1-S2 and IIS3-S4 linkers widens during the transition from the resting state to the activated state, newly accessible residues in both the IIS3-S4 loop and the N-terminal end of the IIS4 segment may interact with the toxin. CssIV may bind in a state-independent manner to the IIS1-S2 loop and then cause voltage sensor trapping by preferential state-dependent interactions with the IIS3-S4 loop in its activated conformation.
Mutations in the IIS3-S4 Loop Can Either Strengthen or Weaken Voltage Sensor Trapping
In the IIS1-S2 and IIS3-S4 extracellular loops, mutations at seven positions greatly weakened or even abolished binding or voltage sensor trapping by CssIV (Fig. 7). This is in agreement with previous findings that mutations in these extracellular loops generally weaken the binding interaction of toxins to a variety of voltage-gated ion channels (8, 9, 27). These results point to the S3-S4 loops of voltage-gated ion channels as the primary molecular target for gating modifier toxins. Surprisingly, and in contrast to these previous studies, we found three mutations that greatly enhanced voltage sensor-trapping activity by a β-scorpion toxin on rNav1.2a channels. The bidirectional effects of mutations in the IIS3-S4 loop on toxin efficacy in voltage sensor trapping suggest that the native residues in these positions make interactions that contribute both positively and negatively to the binding energy of the toxin and that toxin interactions with this extracellular loop determine the efficacy of the voltage sensor-trapping process.
Functional analyses of N842R, V843A, and E844N mutant channels showed that the voltage sensor-trapping action by CssIV was strongly enhanced. Similar enhancement of voltage sensor trapping was observed for mutations that neutralize the two outermost arginine gating charges in the IIS4 segment (10) or the negatively charged residues with which they interact (28). It was proposed that the loss of electrostatic interactions between the negatively charged residues in IIS2 and IIS3 and the arginine gating charges in IIS4 increased the mobility of IIS4 voltage sensor, thereby allowing more rapid and complete voltage sensor trapping (10). This increased mobility might reduce the energy barrier for the voltage sensor to be trapped by β-scorpion toxin in the activated state. N842R, V843A, and E844N differ from those mutants studied previously. First, all of the residues studied previously are localized within the cell membrane electrical field and are thus directly involved in voltage sensing (10, 28). In contrast, Asn842, Val843, and Glu844 are all localized in the IIS3-S4 extracellular loop outside the membrane electrical field and thus cannot contribute directly to voltage sensing. Second, the amino acid residues within the IIS2, IIS3, and IIS4 transmembrane segments do not contribute to the receptor site for β-toxins (9), whereas Asn842, Val843, and Glu844 evidently do play an important role in toxin binding. The gain-of-function effects of the N842R, V843A, and E844N mutations may include both enhanced binding affinity and enhanced efficacy for voltage sensor trapping (see kinetic model below), whereas the gain-of-function effects of mutations in the IIS4 segment must result from specific effects on voltage sensor trapping.
N842R is a single residue chimera between rNav1.2a, on which β-toxins have strong voltage sensor-trapping action, and hNav1.5, on which β-toxins have very weak voltage sensor-trapping action (8). Therefore, it is surprising that N842R strongly enhances the voltage sensor-trapping action of β-toxins and that CssIV can trap the voltage sensor of N842R at the resting membrane potential without a depolarizing prepulse. The adjacent mutations V843A and E844N also increased voltage sensor trapping, but only following a depolarizing prepulse. The striking differences in CssIV action caused by these three adjacent mutations highlight the extreme sensitivity of this hot spot for toxin action in IIS3-S4 to changes in single amino acid residues.
Kinetics of β-Scorpion Toxin Action and Recovery Fit the Voltage Sensor-trapping Model
Previous studies showed that CssIV purified from scorpion venom negatively shifts the voltage dependence of activation following a strong depolarizing prepulse (8, 9). A voltage sensor-trapping mechanism was proposed to explain the prepulse-dependent enhancement of activation by β-scorpion toxins (8, 9). According to the model, before the prepulse, the toxin binds to its receptor site in the resting state of the channels in a bimolecular chemical reaction to form a toxin-channel complex. Upon strong depolarization, the IIS4 segment in the voltage sensor in domain II moves outward, and the toxin binds to newly accessible amino acid residues in the IIS3-S4 loop and the extracellular end of the IIS4 segment. In this activated position, the IIS4 segment is tightly bound to the toxin and trapped in its activated, outward position. Upon repolarization of the cell membrane, the trapped IIS4 voltage sensor remains activated, which reduces the energy required to re-activate the channels because one of the four voltage sensors is already activated. The reduced electrical energy required to activate one fewer voltage sensor causes the negative shift of the voltage dependence of activation. Kinetic analysis showed that voltage sensor trapping by CssIV is well fit by a model that incorporates these three steps in toxin action (9), and kinetic analysis of the effects of a toxin partial agonist (CssIVE15R) further supported a three-step voltage sensor-trapping process. The voltage sensor-trapping mechanism also explains the actions of tarantula huwena toxin-IV (29), β-scorpion toxin Ts1 (30), tarantula protoxin-II (31, 32), α-scorpion toxins (33, 34), and δ-conotoxins (35).
Our kinetic analysis of the effects of gain-of-function mutations in rNav1.2a channels also supports the voltage sensor-trapping mechanism. Below saturating concentrations of toxin, the extent of voltage sensor trapping is increased with increasing toxin concentration, whereas the rates of voltage sensor trapping and the reversal of voltage sensor trapping are both independent of toxin concentration. Our evidence that the effects of both a loss-of-function toxin partial agonist, CssIVE15R (19), and the three gain-of-function rNav1.2a mutants studied here are all well fit by a kinetically similar voltage sensor-trapping mechanism provides additional strong support for this model of toxin action.
In general, our results do not provide sufficient data to separate the effects of the mutations on toxin binding affinity versus efficacy in voltage sensor trapping because we cannot reach saturating effects for mutants with increased EC50 values. However, we were able to use this kinetic model to fit the results and derive Kd values for the mutants with enhanced voltage sensor-trapping activity. The mutations V843A and E844N in the hot spot for CssIV action both reduced the Kd value for binding to the resting state by 2.7–4.3-fold (Table 3). Thus, increased binding affinity makes a substantial contribution to increased voltage sensor trapping by these mutants.
Although voltage sensor trapping by all three gain-of-function mutants in IIS3-S4 of rNav1.2a has generally similar dependence on toxin concentration and voltage, the effects of these mutations on the rate of onset and reversal of voltage sensor trapping are quite different and these factors affect toxin efficacy. N842R greatly slows the reversal of voltage sensor trapping upon repolarization, whereas neither V843A nor E844N have this effect (Figs. 4 and 6). Corrected time courses of the onset of voltage-sensor trapping were generated (supplemental Fig. S2) by taking into account the time course of trapping (Figs. 4B and 6, A and B) and the loss of trapping during the 60-ms repolarization intervals (Figs. 4D and 6D). These corrected time courses show that the amount of trapping for V843A is much greater than for N842R, E844N, or WT, even though the toxin receptor was 100% occupied at the 500 nm test concentrations for each of these mutant channels (Figs. 3F and 5E). These differences likely result from more efficient trapping of the depolarized voltage sensor by the toxin bound to the V843A mutant channel and greater stability of the trapped, activated voltage sensor. Despite these differences in detail, the corrected time courses were well fit with a voltage sensor-trapping model assuming 100% occupancy of the toxin receptor (supplemental Fig. S2D) with the parameters given in supplemental Fig. S2E.
The corrected time course of voltage sensor trapping (supplemental Fig. S2C) for V843A reaches a final value of 3.2 or 320% of the original peak conductance in the absence of toxin. This value indicates that the trapped voltage sensor for this mutant channel causes the peak open probability (PO) for this channel to increase above the maximum level reached in the absence of toxin by at least 3-fold. Based on previous single channel recording studies of NaV channels, this would bring the maximum single channel open probability in the presence of CssIV toxin to nearly the theoretical upper limit of 1.0 for this mutant.
Structural Model for the Toxin-Receptor Complex
The functional surface of β-scorpion toxins is composed of two clusters of discontinuous residues on the two sides of the wedge-shaped toxin molecule (18). One is associated with the α helix of CssIV, which includes a hot spot that is conserved among β-scorpion toxins and contains their primary pharmacophore. The other cluster of important amino acid residues is hydrophobic and is associated mainly with the β2 and β3 strands and the C-terminal tail. This cluster of amino acid residues determines the species selectivity of the β-scorpion toxins (18). Our structural model indicates that residues in IIS1-S2 interact with the species selectivity cluster on the toxin, but not with the pharmacophore cluster, whereas IIS3-S4 residues interact with both clusters on the toxin surface (Fig. 8). Both sets of interactions contribute to toxin binding affinity, but binding to the IIS3-S4 loop appears to be the primary determinant of toxin efficacy in voltage sensor trapping, as illustrated by the large changes in efficacy of voltage-sensor trapping by mutations of the adjacent residues at positions 842, 843, and 844. Thus, the S3-S4 linker emerges not only as the center for high affinity toxin binding but also as the site of interaction that determines whether β-scorpion toxins act as agonists or antagonists of voltage-dependent channel activation.
Voltage Sensor Trapping Supports a Sliding Helix Model of Voltage Sensor Function
β-Scorpion toxins bind to their receptor site in the resting state of Nav channels and then trap the voltage sensor in its activated state (8, 9). Therefore, the receptor site composed of the IIS1-S2 and IIS3-S4 loops must be available for toxin binding at the extracellular surface of the membrane in the resting state of the channels. This characteristic distinguishes the mechanism of β-scorpion toxins from hanatoxin and related cysteine-knot toxins that inhibit potassium channels (36) and channel chimeras (27). The receptor site for these toxins can be transferred from one ion channel to another with significant recovery of toxin action by forming chimeras in which the extracellular half of the S3 and IIS4 segments and the connecting IIS3-S4 loop are inserted (27). These studies support the concept that the IIS3-S4 linker is the primary component of the receptor sites for these gating modifier toxins, as demonstrated in earlier work on α- and β-scorpion toxins (8, 9, 34). However, hanatoxin partitions into the cell membrane and interacts with amino acid residues in the transmembrane part of the S3b-S4 helical hairpin (37, 38), but CssIV does not (39). Our data indicate that β-scorpion toxins can bind to the resting state of Nav channels via the extracellular surface only. Our finding that CssIV bound to the N842R mutant channel can trap its voltage sensor without a depolarizing prepulse further confirms that the IIS3-S4 loop is at the cell surface in a resting state of the channel. These results agree closely with the predictions of the sliding helix model of voltage-dependent gating (40–42), which posits that the IIS4 segments are in a transmembrane position in the resting state of the channel and that, in response to depolarization, the IIS4 segments move outward and rotate. As expected from this gating model, our results confirm that the IIS3-S4 loop is on the extracellular surface of the membrane in the resting state of the channels.
The sliding helix mechanism also predicts that the IIS3-S4 linker undergoes substantial conformational changes during activation. Our results show that the majority of the residues forming the receptor site for β-toxins in sodium channels are in a cluster of consecutive residues in the IIS3-S4 extracellular loop and that the amino acid residues in this loop control the efficacy of voltage sensor trapping by the toxin. The sensitivity of voltage sensor trapping to mutation of these individual residues is also consistent with the movement and conformational changes of the IIS3-S4 loop that are predicted by the sliding helix model of voltage sensor function (3, 42). A major step toward further definition of this mechanism of voltage sensor function, as well as toward understanding the structural basis for β-scorpion toxin action, will eventually come from structural analysis of voltage sensors in defined functional states stabilized by binding of specific toxins and other effectors.
Supplementary Material
Acknowledgments
We thank the late Elizabeth M. Sharp and Ripal Shah for excellent technical assistance with molecular biology.
This work was supported, in whole or in part, by National Institutes of Health Grant U01 NS058039 (to W. A. C. and M. G.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- Nav
- voltage-gated sodium.
REFERENCES
- 1. Hille B. (2001) Ion Channels of Excitable Membranes, 3 Ed., Sinauer, Sunderland, MA [Google Scholar]
- 2. Catterall W. A. (2000) Neuron 26, 13–25 [DOI] [PubMed] [Google Scholar]
- 3. Yarov-Yarovoy V., Baker D., Catterall W. A. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 7292–7297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Catterall W. A., Cestèle S., Yarov-Yarovoy V., Yu F. H., Konoki K., Scheuer T. (2007) Toxicon 49, 124–141 [DOI] [PubMed] [Google Scholar]
- 5. Cahalan M. D. (1975) J. Physiol. 244, 511–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Couraud F., Jover E., Dubois J. M., Rochat H. (1982) Toxicon 20, 9–16 [DOI] [PubMed] [Google Scholar]
- 7. Gurevitz M., Karbat I., Cohen L., Ilan N., Kahn R., Turkov M., Stankiewicz M., Stühmer W., Dong K., Gordon D. (2007) Toxicon 49, 473–489 [DOI] [PubMed] [Google Scholar]
- 8. Cestèle S., Qu Y., Rogers J. C., Rochat H., Scheuer T., Catterall W. A. (1998) Neuron 21, 919–931 [DOI] [PubMed] [Google Scholar]
- 9. Cestèle S., Yarov-Yarovoy V., Qu Y., Sampieri F., Scheuer T., Catterall W. A. (2006) J. Biol. Chem. 281, 21332–21344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cestèle S., Scheuer T., Mantegazza M., Rochat H., Catterall W. A. (2001) J. Gen. Physiol. 118, 291–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Marcotte P., Chen L. Q., Kallen R. G., Chahine M. (1997) Circ. Res. 80, 363–369 [DOI] [PubMed] [Google Scholar]
- 12. Yarov-Yarovoy V., Schonbrun J., Baker D. (2006) Proteins 62, 1010–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Long S. B., Tao X., Campbell E. B., MacKinnon R. (2007) Nature 450, 376–382 [DOI] [PubMed] [Google Scholar]
- 14. Jeanmougin F., Thompson J. D., Gouy M., Higgins D. G., Gibson T. J. (1998) Trends Biochem. Sci. 23, 403–405 [DOI] [PubMed] [Google Scholar]
- 15. Rohl C. A., Strauss C. E., Misura K. M., Baker D. (2004) Methods Enzymol. 383, 66–93 [DOI] [PubMed] [Google Scholar]
- 16. Gray J. J., Moughon S., Wang C., Schueler-Furman O., Kuhlman B., Rohl C. A., Baker D. (2003) J. Mol. Biol. 331, 281–299 [DOI] [PubMed] [Google Scholar]
- 17. Wang C., Bradley P., Baker D. (2007) J. Mol. Biol. 373, 503–519 [DOI] [PubMed] [Google Scholar]
- 18. Cohen L., Karbat I., Gilles N., Ilan N., Benveniste M., Gordon D., Gurevitz M. (2005) J. Biol. Chem. 280, 5045–5053 [DOI] [PubMed] [Google Scholar]
- 19. Karbat I., Ilan N., Zhang J. Z., Cohen L., Kahn R., Benveniste M., Scheuer T., Catterall W. A., Gordon D., Gurevitz M. (2010) J. Biol. Chem. 285, 30531–30538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jiang Y., Lee A., Chen J., Ruta V., Cadene M., Chait B. T., MacKinnon R. (2003) Nature 423, 33–41 [DOI] [PubMed] [Google Scholar]
- 21. Cohen L., Ilan N., Gur M., Stühmer W., Gordon D., Gurevitz M. (2007) J. Biol. Chem. 282, 29424–29430 [DOI] [PubMed] [Google Scholar]
- 22. Ruta V., Chen J., MacKinnon R. (2005) Cell 123, 463–475 [DOI] [PubMed] [Google Scholar]
- 23. Banerjee A., MacKinnon R. (2008) J. Mol. Biol. 381, 569–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pathak M. M., Yarov-Yarovoy V., Agarwal G., Roux B., Barth P., Kohout S., Tombola F., Isacoff E. Y. (2007) Neuron 56, 124–140 [DOI] [PubMed] [Google Scholar]
- 25. DeCaen P. G., Yarov-Yarovoy V., Zhao Y., Scheuer T., Catterall W. A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 15142–15147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. DeCaen P. G., Yarov-Yarovoy V., Sharp E. M., Scheuer T., Catterall W. A. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 22498–22503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bosmans F., Martin-Eauclaire M. F., Swartz K. J. (2008) Nature 456, 202–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mantegazza M., Cestèle S. (2005) J. Physiol. 568, 13–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xiao Y., Bingham J. P., Zhu W., Moczydlowski E., Liang S., Cummins T. R. (2008) J. Biol. Chem. 283, 27300–27313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Campos F. V., Chanda B., Beirão P. S., Bezanilla F. (2007) J. Gen. Physiol. 130, 257–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Smith J. J., Cummins T. R., Alphy S., Blumenthal K. M. (2007) J. Biol. Chem. 282, 12687–12697 [DOI] [PubMed] [Google Scholar]
- 32. Sokolov S., Kraus R. L., Scheuer T., Catterall W. A. (2008) Mol. Pharmacol. 73, 1020–1028 [DOI] [PubMed] [Google Scholar]
- 33. Campos F. V., Chanda B., Beirão P. S., Bezanilla F. (2008) J. Gen. Physiol. 132, 251–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rogers J. C., Qu Y., Tanada T. N., Scheuer T., Catterall W. A. (1996) J. Biol. Chem. 271, 15950–15962 [DOI] [PubMed] [Google Scholar]
- 35. Leipold E., Hansel A., Olivera B. M., Terlau H., Heinemann S. H. (2005) FEBS Lett. 579, 3881–3884 [DOI] [PubMed] [Google Scholar]
- 36. Swartz K. J., MacKinnon R. (1997) Neuron 18, 675–682 [DOI] [PubMed] [Google Scholar]
- 37. Phillips L. R., Milescu M., Li-Smerin Y., Mindell J. A., Kim J. I., Swartz K. J. (2005) Nature 436, 857–860 [DOI] [PubMed] [Google Scholar]
- 38. Milescu M., Vobecky J., Roh S. H., Kim S. H., Jung H. J., Kim J. I., Swartz K. J. (2007) J. Gen. Physiol. 130, 497–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cohen L., Gilles N., Karbat I., Ilan N., Gordon D., Gurevitz M. (2006) J. Biol. Chem. 281, 20673–20679 [DOI] [PubMed] [Google Scholar]
- 40. Catterall W. A. (1986) Annu. Rev. Biochem. 55, 953–985 [DOI] [PubMed] [Google Scholar]
- 41. Guy H. R., Seetharamulu P. (1986) Proc. Natl. Acad. Sci. U.S.A. 83, 508–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Catterall W. A. (2010) Neuron 67, 915–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.