Abstract
Studies implicate an important role for the mixed lineage leukemia (Mll) gene in hematopoiesis, mainly through maintaining Hox gene expression. However, the mechanisms underlying Mll-mediated hematopoiesis during embryogenesis remain largely unclear. Here, we investigate the role of mll during zebrafish embryogenesis, particularly hematopoiesis. Mll depletion caused severe defects in hematopoiesis as indicated by a lack of blood flow and mature blood cells as well as a significant reduction in expression of hematopoietic progenitor and mature blood cell markers. Furthermore, mll depletion prevented the differentiation of hematopoietic progenitors. In addition, we identified the N-terminal mini-peptide of Mll that acted as a dominant negative form to disrupt normal function of mll during embryogenesis. As expected, mll knockdown altered the expression of a subset of Hox genes. However, overexpression of these down-regulated Hox genes only partially rescued the blood deficiency, suggesting that mll may target additional genes to regulate hematopoiesis. In the mll morphants, microarray analysis revealed a dramatic up-regulation of gadd45αa. Multiple assays indicate that mll inhibited gadd45αa expression and that overexpression of gadd45αa mRNA led to a phenotype similar to the one seen in the mll morphants. Taken together, these findings demonstrate that zebrafish mll plays essential roles in hematopoiesis and that gadd45αa may serve as a potential downstream target for mediating the function of mll in hematopoiesis.
Keywords: Development, Gene Regulation, Hematopoiesis, Leukemia, Zebrafish, gadd45aa, hox, mll
Introduction
The mixed lineage leukemia (MLL) gene, a human homolog of Drosophila melanogaster Trithorax (Trx), was originally identified through its involvement in in-frame reciprocal translocations with partner genes primarily associated with leukemias (1–5). To date, over 70 different partner genes have been identified (6–8). The MLL gene encodes a 430-kDa protein containing three AT hook motifs, two speckled nuclear localization domains (SNL1 and SNL2), a CXXC zinc finger motif, multiple plant homology domains (PHDs),2 and a Su/var, Enhancer or zeste, Trithorax (SET), domain. The PHD fingers contribute to protein-protein interactions, and their deletion, as a result of chromosomal translocation, is necessary for MLL fusion proteins to induce hematopoietic cell immortalization (9). The SET domain contributes to modulation of homeobox (HOX) gene expression by facilitating histone H3 Lys-4 methylation at the proximal promoter region (10). Recent studies have also shown that genotoxic stress induces the ATR phosphorylation of MLL, leading to increased histone H3 Lys-4 methylation and subsequent execution of the mammalian S-phase checkpoint (11). The MLL protein normally is cleaved into a specific 320-kDa N-terminal (MLLN) and a 180-kDa C-terminal (MLLC) fragment (12, 13). In vivo, wild-type MLLN and MLLC associate with each other as well as with other factors, such as Menin, WDR5, ASH1/2, HCF2, RBBP5, and MOF (males absent on the first), to form a large nuclear complex involved in chromatin remodeling (14–16).
The function of Mll in vivo has been extensively analyzed in mouse models. Mll heterozygous knock-out mice exhibit growth retardation, hematopoietic defects, and skeletal malformation (17). Insertion of stop codons within exon 3 or exons 12–14 of Mll produce a defect in fetal liver hematopoiesis and are embryonic lethal at E10.5 and E11.5–14.5, respectively (18). In addition, truncation of Mll at exon 5 results in early embryonic death due to failure of preimplantation (19). Chimeric mice expressing lacZ, but not a Myc tag, in exon 8 of Mll develop acute leukemias (20). However, mice that express MLL with a deleted SET domain (MLLΔSET) are viable and fertile (21). Recently, another study demonstrated an essential role for MLL in neurogenesis (22). Thus, the proper formation of an MLL super complex and functions outside of the SET domain are required for early embryonic development and organogenesis.
Although the phenotypes in Mll knock-out mice vary depending on the particular allele knocked out and the tissues in which Mll was deleted, mouse models have conclusively shown that Mll has an essential role in controlling Hox gene expression (17, 19–20, 23, 24). Mll-null mice initiate the expression of stage-specific Hox genes appropriately but do not maintain expression past embryonic day 9.5 (25). Hox genes, as transcription factors, participate in the development of multiple tissues, including the hematopoietic system, by conferring positional identities to cells along the anteroposterior axis (26–29). Moreover, Mll is required for definitive hematopoietic expansion in a Hox-dependent manner and is essential for the maintenance of adult hematopoietic stem cell quiescence and promoting proliferation of the progenitors (28, 31). Mll also plays an important role in fetal and adult hematopoietic stem cell self-renewal (32). Not surprisingly, heterozygous mice (Mll+/−) exhibit hematopoietic abnormalities along with altered Hox gene expression (17). Detailed assessment of yolk sac and fetal liver hematopoiesis demonstrated deficiencies in proliferation and/or survival of Mll null hematopoietic progenitors and delays in onset of differentiation instead of blocking specific lineages (23). Collectively, these findings provide clear evidence indicating that MLL plays a critical role in hematopoiesis, in part by regulating Hox gene expression.
Despite these advances in understanding the role of Mll in hematopoiesis, it remains unclear how Mll-mediated hematopoiesis occurs during embryogenesis due to the early embryonic death of Mll-null mice. In this study, we used morpholinos to study the function of zebrafish mll in hematopoiesis during embryogenesis, and we found that mll plays an essential role in both zebrafish primitive and definitive hematopoiesis.
EXPERIMENTAL PROCEDURES
Maintenance of Fish Stocks and Embryo Collection
For wild-type zebrafish (Danio rerio) (strain AB), flk1-GFP transgenic zebrafish (provided by Jianfang Gui, originally obtained from Shuo Lin) maintenance, breeding, and staging were performed as described previously (33).
Whole Mount in Situ Hybridization and o-Dianisidine Staining
Probes for detecting zebrafish mll, gata1, scl, lmo2, pu.1, runx1, c-myb, mpo, l-plastin, hbbe1, hoxb4a, hoxb5a, hoxb7a, hoxa9a, hoxb6b, hoxc8, hoxd3a, cdx4, and gadd45αa were PCR-amplified from zebrafish cDNA pools and subcloned into the pTA2 vector (Toyobo, Osaka, Japan). Probes for flk (34), pax2a (35), myoD and nkx2.5 (36), eph2a (37), and gadd45αb (38) were described previously. All primers are listed in supplemental Table 1. The procedures for whole mount in situ hybridization and o-dianisidine staining were performed as described previously (39).
Mll Antisense Morpholinos and Their Validations
We designed a morpholino (MO) that targeted the ATG codon of mll to block translation (mll-MO1), TGCGCCATTTTACACTCTGCCTGCC. We also designed an MO that targets the exon1-intron1 splicing junction site of mll (mll-MO2), TTTAGCCAGGGTCCCGCTTACCTCC, and an MO that targets the 5′-UTR region of mll (mll-MO3), ACACTCTGCCTGCCTGCCCTAGGAT. The morpholinos were generated at Gene Tools, LLC (Philomath, OR).
For mll-MO1 validation, the zebrafish mll cDNA fragment that encompassed part of the 5′-UTR (5′-ggcaggcagagttgaaa-3′) and 252-bp fragment (246 bp of exon1 plus 6 bp of exon2) was cloned into pAcGFP-N1 (Clontech) to generate a wild-type mll (exon1)-GFP fusion protein expression vector (WT1). A mutated mll (exon1)-GFP fusion protein expression vector (MT1) was generated by PCR using a forward primer with seven mismatched nucleotides as follows: 5′-cggagccacacagttaa-3′ (seven mismatched nucleotides are underlined).
For mll-MO2 validation, three primers were designed. The sequence of the primer in exon1 (P1) was 5′-ctccttatcagcgcgggaacaaat-3′; the sequence of the primer in exon2 (P3) was 5′-gggctatgtagtcttctgttgtcact-3′; and the sequence of the primer in intron1 (P2) was 5′-agacaacaacaaccaactggcacc-3′.
For mll-MO3 validation, zebrafish mll cDNA fragment that encompassed part of the 5′-UTR (5′-atcctagggcaggcaggcagagtgtaaa-3′) and 252-bp fragment (246 bp of exon1 plus 6 bp of exon2) was cloned into pAcGFP-N1 (Clontech) to generate a wild-type mll (exon1)-GFP fusion protein expression vector (WT2). A mutated mll(exon1)-GFP fusion protein expression vector (MT2) was generated by PCR using a forward primer with 5 mismatched nucleotides: 5′-atccaccgttgcgcaagaaggcagagtgt-3′ (five mismatched nucleotides are underlined).
Semi-quantitative RT-PCR
The primers used for amplifying zebrafish scl, lmo2, gata1, pu.1, runx1, c-myb, flk1, gadd45αa and β-actin are listed in supplemental Table 1. The procedure for semi-quantitative RT-PCR was described previously (40).
Rescue Experiments
The full-length wild-type hoxb4a, hoxb7a, hoxb6b, hoxa9a, and hoxd3a were PCR-amplified from a zebrafish cDNA pool and then subcloned into the Psp64 poly(A) vector (Promega). Synthesized mRNA was injected into embryos at 100 pg/per embryo. The primers are listed in supplemental Table 1.
Luciferase Reporter Assay and mRNA Injection
The 1360-bp promoter of zebrafish gadd45αa (previously named gadd45αl) was PCR-amplified from wild-type zebrafish genomic DNA using the primers listed in supplemental Table 1 and then subcloned into the pGL3-Basic vector (Promega).
For MLL overexpression, 20–30 one-cell stage embryos were injected either with the PCNX-human MLL expression vector (5 pg/per embryo) (provided by Stanley Korsmeyer) combined with the pGL3-gadd45αa promoter vector (0.1 pg/per embryo) and CMV-Renilla (0.1 pg/per embryo, as an internal control) or with empty vector (5 pg/per embryo) combined with the pGL3-gadd45αa-promoter (0.1 pg/per embryo) and CMV-Renilla (0.1 pg/per embryo). For mll knockdown, 20–30 one-cell stage embryos were injected either with mll-MO1 (8 ng/per embryo) combined with pGL3-gadd45αa-promoter vector (0.1 pg/per embryo) and CMV-Renilla (0.1 pg/per embryo) or with control MO (8 ng/per embryo) combined with pGL3-gadd45αa-promoter (0.1 pg/per embryo) and CMV-Renilla (0.1 pg/per embryo). After 12 h, embryos were homogenized, and luciferase activity measurements were taken by a Dual-Luciferase reporter assay system (Promega).
For gadd45αa mRNA injection experiments, the full-length cDNA of zebrafish gadd45αa was amplified and then subcloned into the Psp64 poly(A) vector (Promega). Synthesized mRNA was injected into embryos at 400 pg/per embryo. The primers are listed in supplemental Table 2.
RESULTS
Amino Acid Sequence of Zebrafish mll Is Evolutionarily Conserved with Mammalian MLL
Using the GenBankTM data base, we identified the zebrafish ortholog of the MLL gene (accession number NM_001110279). Mll contains 12,703-bp nucleotides and encodes a protein 4218 amino acids in length, which is longer than that of the human MLL protein (3969 amino acids).
As showed in supplemental Fig. 1, zebrafish mll encodes most all of the functional domains identified in the mammalian MLL gene, including the AT hook motifs, a CXXC zinc-finger domain, a central zinc finger (PHD) region, and a C-terminal SET domain. The functional domains of the human and zebrafish mll gene share a high degree of identity as follows: 63.64% in AT1 and AT2, 77.78% in AT3, 93.88% in CXXC, 78.85% in PHD1, 83.64% in PHD2, 69.35% in PHD3, 71.74% in Bromo, and 85.21% in the SET domain. Compared with the whole protein sequence that shares 51.8% identity, the functional domains of the human and zebrafish proteins show a much higher level of conservation. Zebrafish Mll also contains the two identical proteolytic cleavage sites (Taspase1) found in human and mouse proteins, suggesting that zebrafish Mll also undergoes proteolytic processing into the MLLN and MLLC fragments in vivo.
Zebrafish mll Is Expressed Ubiquitously during Early Embryogenesis
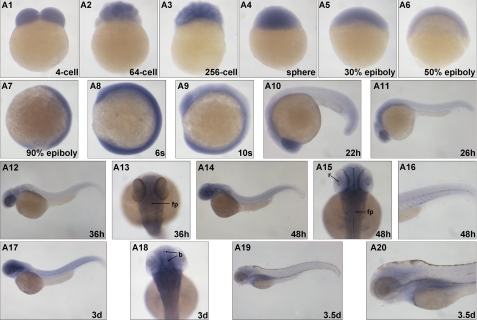
We evaluated the expression patterns of mll during embryo development using whole mount in situ hybridization (Fig. 1). At early developmental stages, mll expression was ubiquitous in all cells, suggesting a maternal expression (Fig. 1, A1–A9). By 22 h post-fertilization (hpf), the mll expression pattern appeared as a gradient, from the anterior head region to the posterior tail (Fig. 1, A10–A13). The anterior head region expressed the highest level of mll with a slight decrease in the anterior trunk region attached to the yolk sac. Other regions of the body expressed lower homogenous mll levels (Fig. 1, A10–A13). By 48 hpf, the anterior head region continued to express the highest levels with a relatively high level of mll expression also found along the blood vessels (Fig. 1, A14–A16). This expression pattern could last up to 3 days post-fertilization (dpf). However, by 3.5 dpf, the highest expression of mll occurred in the kidney region. Although the anterior head region still expressed mll, its expression was not detected in other regions of body (Fig. 1, A19–A20).
FIGURE 1.
Developmental expression of zebrafish mll. Whole mount in situ hybridization analysis of zebrafish mll expression (blue) at the 4-cell (lateral view), 64-cell (lateral view), 256-cell (lateral view), sphere (lateral view), 30% epiboly (lateral view), 50% epiboly (lateral view), 90% epiboly (lateral view), 6-somite (lateral view), 10-somite (lateral view), 22-hpf (lateral view), 26-hpf (lateral view), 36-hpf (lateral view), 36-hpf (dorsal view), 48-hpf (lateral view), 48-hpf (dorsal view), 48-hpf (tail, dorsal view), 3-dpf (lateral view), 3-dpf (dorsal view), and 3.5-dpf (lateral view) stages. s, somite; h, hours post-fertilization; d, days post-fertilization; fp, floor plate; r, retina.
Zebrafish mll Is Essential for Early Embryo Development and Hematopoiesis
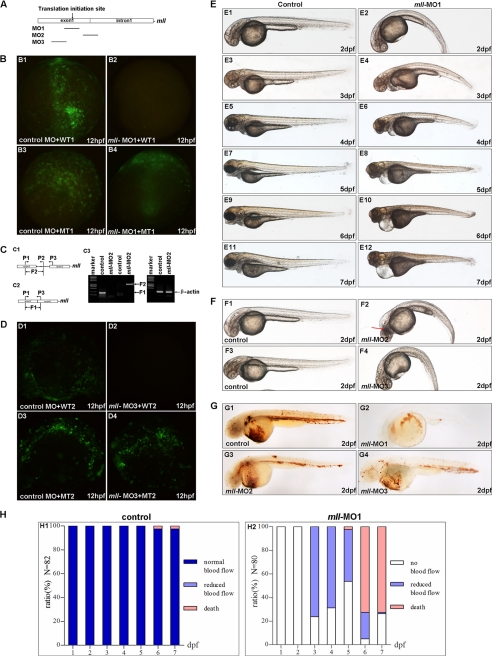
To investigate the roles of mll during zebrafish embryogenesis, we depleted its expression in zebrafish embryos using morpholino (MO)-mediated gene-knockdown. We totally designed three morpholinos based on the cDNA sequence and linkage sequence between exon1 and intron1 of zebrafish mll gene as follows: one blocked mll translation by targeting the ATG site (mll-MO1); another one blocked mll splicing by targeting the linkage sequence between exon1 and intron 1 (mll-MO2), and the third one blocked mll translation by targeting 5′-UTR region (mll-MO3) (Fig. 2A).
FIGURE 2.
Knockdown of mll by morpholinos (mll-MO1, mll-MO2, and mll-MO3) caused defects in erythropoiesis, blood circulation, and morphogenesis. A, schematic diagram depicts the targets of three morpholinos in zebrafish mll genome. B, validation of mll-MO1. Panel B1, embryos were injected with control MO (8 ng/per embryo) and a wild-type mll (exon1)-GFP fusion protein expression vector (WT1) (5 pg/per embryo) and then examined by fluorescence microscopy. Panel B2, embryos were injected with mll-MO1 (8 ng/per embryo) and a wild-type mll (exon1)-GFP fusion protein expression vector (WT1) (5 pg/per embryo) and then examined by fluorescence microscopy. Panel B3, embryos were injected with control-MO (8 ng/per embryo) and a mismatch-mutated mll-GFP fusion protein expression vector (MT1) (5 pg/per embryo) and then examined by fluorescence microscopy. Panel B4, embryos were injected with mll-MO1 (8 ng/per embryo) and a mismatch mutated mll (exon1)-GFP fusion protein expression vector (MT1) (5 pg/per embryo) and then examined by fluorescence microscopy. C, validation of mll-MO2. Panel C1, schematic diagram depicts the genomic fragment of mll exon1, intron1, and exon2. The positions of PCR primers used to detect fragment 2 (F2) are indicated by arrows. Panel C2, schematic diagram depicts the cDNA fragment of mll exon1 and exon2. The positions of PCR primers to detect fragment 1 (F1) are indicated by arrows. Panel C3, gels show that mll-MO2 (8 ng/per embryo) blocked splicing of exon1 and exon2 of mll efficiently. The fragment 1 resulting from normal splicing was not detected in embryos injected with mll-MO2 (8 ng/per embryo) as indicted in 3rd line (from left to right, left panel), but it was detected in embryos without injection (control) as indicated in 2nd line (from left to right, left panel). Fragment 2 resulting from blocking splicing was detected in embryos injected with mll-MO2 (8 ng/per embryo) as indicated in 5th line (from left to right, left panel), but it was not detected in embryos without injection (control) as indicated in 4th line (from left to right, left panel). The cDNA fragment of β-actin can be detected in both control embryos and mll-MO2 morphants as indicted in 2nd and 3rd lines (from left to right, right panel). D, validation of mll-MO3. Panel D1, embryos were injected with control-MO (8 ng/per embryo) and a wild-type mll(exon1)-GFP fusion protein expression vector (WT2) (5 pg/per embryo) and then examined by fluorescence microscopy. Panel D2, embryos were injected with mll-MO3 (8 ng/per embryo) and a wild-type mll (exon1)-GFP fusion protein expression vector (WT2) (5 pg/per embryo) and then examined by fluorescence microscopy. Panes D3, embryos were injected with control-MO (8 ng/per embryo) and a mismatch-mutated mll-GFP fusion protein expression vector (MT2) (5 pg/per embryo) and then examined by fluorescence microscopy. D4, embryos were injected with mll-MO3 (8 ng/per embryo) and a mismatch mutated mll (exon1)-GFP fusion protein expression vector (MT2) (5 pg/per embryo) and then examined by fluorescence microscopy. E, morphological defects in mll-MO1 morphants. Panel E1, 2-dpf embryos without morpholino injection (control) developed normally. Panel E2, mll-MO1 morphants (2-dpf stage) showed a curved body and heart edema. Panel E3, 3-dpf embryos without morpholino injection (control) developed normally. Panel E4, mll-MO1 morphants (3-dpf stage) showed heart edema and a shortened body. Panel E5, 4-dpf embryos without morpholino injection (control) developed normally. Panel E6, mll-MO1 morphants (4-dpf stage) showed severe heart edema, smaller eyes, and a shortened body. Panels E7, E9, and E11, 5–7-dpf embryos without morpholino injection (control) developed normally. Panels E8, E10 and E12, mll-MO1 morphants (5–7-dpf stage) showed severe heart edema, smaller eyes, a shortened body, and an abnormal head. F, morphological defects in mll-MO2 and mll-MO3 morphants. Panel F1, 2-dpf embryos without morpholino injection (control) developed normally. Panel F2, mll-MO2 morphants (2-dpf stage) showed a curved body and head degenerated. Panel F3, 2-dpf embryos without morpholino injection (control) developed normally. Panel F4, mll-MO3 morphants (2-dpf stage) showed a curved body and heart edema. G, defects of erythropoiesis in mll morphants. Panel G1, erythropoiesis in embryos (2-dpf stage) without morpholino injection was normal as revealed by o-dianisidine staining for hemoglobin. Panel G2, erythropoiesis in embryos (2-dpf stages) injected with mll-MO1 (8 ng/per embryo) was blocked as revealed by o-dianisidine staining for hemoglobin. Panel G3, erythropoiesis in embryos (2-dpf stages) injected with mll-MO2 (8 ng/per embryo) was blocked as revealed by o-dianisidine staining for hemoglobin. Panel G4, the erythropoiesis in embryos (2-dpf stages) injected with mll-MO3 (8 ng/per embryo) was blocked as revealed by o-dianisidine staining for hemoglobin. H, quantitative assays for blood circulation in embryos. Panel H1, different stage embryos without morpholino injection showed normal blood flow. Panel H2, different stage mll-MO1 morphants showed no blood flow or reduced blood flow.
For validation of mll-MO1, we used RT-PCR to clone a cDNA fragment that partially encompassed the 5′-UTR (mll-MO1-targeted region) along with exon1 into pAcGFP-N1 (Clontech) to generate a wild-type mll (exon1)-GFP fusion protein expression vector (named as WT1). We also generated a mutated mll (exon1)-GFP fusion protein expression vector (named as MT1, with seven mismatched nucleotides in the mll-MO1-targeting region). We co-injected embryos with mll-MO1 (8 ng/per embryo) and either WT1 or MT1 vector. Mll-MO1 efficiently blocked expression from the WT1 but not MT1 vector (Fig. 2, panels B2 and B4). However, the control MO (control), a standard morpholino described previously (41), could not block expression from either vector (Fig. 2, panels B1 and B3). These observations indicate that mll-MO1 could knock down zebrafish mll expression during embryogenesis efficiently and specifically.
For validation mll-MO2, we designed three primers (P1, P2, and P3) for RT-PCR analysis (Fig. 2, panels C1 and C2). After injecting embryos with control MO (8 ng/per embryo) or mll-MO2 (8 ng/per embryo), we performed RT-PCR. The results showed that mll-MO2 injection could generate a DNA fragment with the size as predicted (976 bp, named F2) (Fig. 2, panel C3), which was further verified by sequencing (data not shown). These results indicated that mll-MO2 could block mll splicing efficiently and specifically.
For validation mll-MO3, we used the same approach as that for validating mll-MO1. As showed in Fig. 3D, mll-MO3 could also knock down mll expression efficiently and specifically.
FIGURE 3.
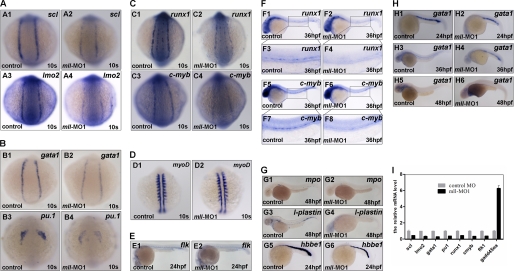
mll is required for embryonic and adult hematopoietic marker gene expression. Whole mount in situ hybridization assays for hematopoiesis markers in control embryos or mll-MO1-injected embryos. A, expression patterns of scl and lmo2 in control embryos or mll-MO1 morphants at the 10-somite stage. Panels A1 and A2, scl; A3 and A4, lmo2. B, expression patterns of gata1 and pu.1 in control embryos or mll-MO1 morphants at 10-somite stage; Panels B1 and B2, gata1; B3 and B4, pu.1. C, expression patterns of runx1 and c-myb in control embryos or mll-MO1 morphants at 10-somite stage, Panels C1 and C2, runx1; C3 and C4, c-myb. D, expression patterns of myoD in control embryos (panel D1) or mll-MO1 morphants (panel D2) at 10-somite stage. E, expression patterns of flk1 in control embryos or mll-MO1 morphants at 24-hpf stage. F, expression patterns of runx1 and c-myb in control embryos or mll-MO1 morphants at 36-hpf stage. Panels F1–F4, runx1; and F5–F8, c-myb. G, expression patterns of mpo, l-plastin, and hbbe1 in control embryos or mll morphants at 48-hpf stage. Panels G1 and G2, mpo; G3 and G4, l-plastin; and G5 and G6, hbbe1. H, Gata1 expression in embryos without mll-MO1 injection at 24-, 36-, and 48-hpf stages. Panels H1, H3 and H5, gata1 expression in embryos without mll-MO1 injection; panels H2, H4, and H6, gata1 expression in mll-MO1 morphants. s, somites; hpf, hours post-fertilization. I, semi-quantitative RT-PCR analysis for the expression regulation of scl, lmo2, gata1, pu.1, runx1, c-myb, flk1, and gadd45αa by mll knockdown using mll-MO1 at 10-somite stage.
Subsequently, we investigated the effect of mll knockdown mediated by morpholino injections during zebrafish embryogenesis. In 10 independent experiments, injection of mll-MO1 (8 ng/per embryo) into zebrafish embryos produced obvious defects in embryogenesis and hematopoiesis (Fig. 1E). Of note, the embryos injected with mll-MO1 showed a similar morphology to that of the embryos injected with the control MO (8 ng/per embryo) up until 26 hpf, the time at which embryonic blood flow normally begins (data not shown). From 2 dpf, we observed significant defects in the mll-MO1 morphants, including curved body axis, heart edema, reduced blood cells, and a lack of blood flow (Fig. 2, panels E1–E10, and data not shown). At 7 dpf, control embryos continued to show normal development (Fig. 2, panel E11), whereas mll-MO1 morphants had shorter bodies, smaller eyes, serious heart edema, and still no blood flow (Fig. 2, panel E12, and data not shown). All mll-MO1 morphants died between 6 and 10 dpf (Fig. 2, panel H2). As predicted, injection of mll-MO2 or mll-MO3 generated similar phenotypes as that of mll-MO1 (Fig. 2F) except that mll-MO2 injection caused more degeneration in the head region (Fig. 2, panel F2, indicated by red arrow). These observations further suggest that all three morpholinos could knock down mll expression efficiently and specifically, and mll-MO2 might have some additional nonspecific effects.
Given that previous studies have shown an important role for MLL in mammalian hematopoiesis (6, 7) and the obvious defects in blood cell number and blood flow we observed in mll morphants, we next focused on investigating the influence of mll on zebrafish hematopoiesis. We first traced blood flow in morphants from 2 to 7 dpf under a dissection microscope. Interestingly, blood flow partially recovered with reduced circulatory blood cells in 78% of morphants injected with 8 ng/per embryo mll-MO1 (n = 80) at 3 dpf (Fig. 2, panel H2, and data not shown), but then gradually decreased after 4 dpf (Fig. 2, panel H2). In 7-dpf morphants, blood flow completely ceased (data not shown). When alive, the mll-MO1 morphants had either no blood cells or only one clump of blood cells located back and in front of beating hearts, whereas control embryos developed normally both in terms of morphology and blood flow. No recovery of blood flow occurred in embryos injected with a higher concentration mll-MO1 (12 ng/per embryo) (data not shown). Moreover, an mll-MO1 concentration of 16 ng/per embryo resulted in embryonic death at 2 dpf (data not shown). These observations not only suggest that mll-MO1 affected zebrafish embryogenesis and hematopoiesis in a dose-dependent manner but also imply that the low concentration of mll-MO1 could completely block definitive hematopoiesis, but only partially block primitive hematopoiesis.
To further determine the role of mll in zebrafish erythropoiesis, o-dianisidine staining for hemoglobin was performed to check whether red cell maturation was affected by mll knockdown. The results indicated that the mature red blood cells were dramatically reduced in mll-depleted embryos mediated by all three morpholino injections compared with control embryos at 2 dpf (Fig. 2G).
In all mll-MO1, mll-MO2, and mll-MO3 morphants, particularly in mll-MO2 morphants, we observed obscured areas in the head and trunk regions, indicating tissue degeneration (Fig. 2, E and F, and data not shown). Further apoptosis staining showed that mll-MO1 and mll-MO2 injection could indeed induce apoptosis in embryos compared with control MO injection, particularly for mll-MO2 injection (supplemental Fig. 2A). This apoptosis induction could be the result from off-target effect of morpholinos (42). To test this possibility, we co-injected embryos with a morpholino directed against p53 (p53-MO) combined with mll-MO1, mll-MO2, or mll-MO3, respectively, and then compared these embryos with those injected with mll-MO1, mll-MO2, or mll-MO3 alone. The embryos with the p53-MO/mll-MO combined injection became much more transparent and had less apoptotic cells than those injected with mll-MO1, mll-MO2, or mll-MO3 alone (data not shown); however, the defects in hematopoiesis remained unchanged (data not shown).
To further figure out whether the defects in hematopoiesis exhibited in mll morphants resulted from the influence of mll on cell proliferation, we performed phosphorate-histone H3 staining (43). As shown in supplemental Fig. 2B, the cell proliferation was not altered obviously in mll morphants as revealed by phospho-H3 immunofluorescent staining. Together, these observations support the specificity of mll morpholinos for mediating mll knockdown and the important roles of mll on hematopoiesis.
Mll Is Required for Maintaining Hematopoietic Progenitors and for Proper Erythroid and Myeloid Formation
To better understand how mll knockdown led to blood cell reduction and blood flow loss, we looked at expression of both hematopoietic cell markers and vascular markers. As shown in Fig. 3A and supplemental Fig. 3B, mll-MO1 morphants at the 5- and 10-somite stage (10 s) dramatically down-regulated expression of the early hemangioblast markers, scl and lmo2 (Fig. 3, panels A1–A4, and supplemental Fig. 3, panels B1–B4), whereas the pronephric duct marker pax2a and precardiac marker nkx2.5 were not altered in the lateral plate mesoderm (supplemental Fig. 3A). gata1 and pu1, markers for erythroid and myeloid progenitors, respectively, were also reduced in morphants at 10-somite stage (Fig. 3, panels B1–B4). In addition, morphants had a decrease in expression of the hematopoietic progenitor marker runx1 as well as c-myb at 10-somite stage (Fig. 3, panels C1–C4). These results suggest that mll depletion might specifically block the hematopoietic program but neither the pronephric cell lineage nor precardiac cell lineage.
To ensure that the alteration of hematopoietic markers in mll-MO1 morphants was not the result of the developmental delay usually observed in some morpholino injections, we used the myogenic marker myoD to monitor developmental stages. The results showed that mll-MO1 injection did not cause developmental delay (Fig. 3D).
Loss of blood flow might result from a defect in vasculature development, a blood deficiency, or both. To explore the real cause, we checked expression of the vascular endothelial cell marker flk1 and aorta marker ephB2a. As shown in Fig. 3E and supplemental Fig. 4A, both flk1 and ephB2a were indeed reduced in mll-depleted embryos. However, they were still expressed in mll-MO1 morphants at the 24- or 36-hpf stage, respectively. To further understand whether vascular development was affected by mll knockdown, we used tracing experiments by taking advantage of flk-GFP transgenic fish (GFP expression driven by flk promoter) and found that the formation of vessels was not defected in morphants by 24 hpf (supplemental Fig. 4, panels B1–B4). Of note, by 36 hpf, the lateral vessels gradually began to recover, from the anterior to the posterior (supplemental Fig. 4, panels B5 and B6), and then by 48 hpf they were almost fully recovered (supplemental Fig. 4, panels B7 and B8). These observations indicate that the loss of blood flow in morphants resulted from hematopoietic deficiency rather than a defect of vasculature development.
To further confirm that definitive hematopoiesis was also affected by mll knockdown, we checked expression of runx1 and c-myb at the 36-hpf stage. As shown in Fig. 3F, both runx1 and c-myb were reduced in mll-MO1 morphants. These observations suggest that mll plays an important role in definitive hematopoiesis as well as that in primitive hematopoiesis.
As expected, mature hematopoietic cell markers, such as mpo (granulocytes), l-plastin (macrophage), and hbbe1 (red blood cells), were all significantly reduced, but not absent, in morphants at 24 or 48 hpf (Fig. 3G). Interestingly, at 24 hpf, the mll-MO1 morphants had a reduced level of erythroid progenitors marked by gata1 (Fig. 3, panels H1 and H2); however, at 36 hpf, the gata1-positive cells were still gathered at the intermediate cell mass of Oellacher region in morphants, whereas in control embryos the gata-1-positive cells disappeared (Fig. 3, panels H3 and H4). Even in 48-hpf mll-MO1 morphants, the gata1-positive cells still could be found around the heart area (Fig. 3, panels H5 and H6). Given that the red blood cells had already matured and entered into the blood circulation at 36 hpf, we hypothesize that mll depletion might also block erythroid progenitor differentiation in addition to preventing erythroid progenitor formation.
To further confirm the similar effect of mll-MO2, we performed all markers staining as well as that for mll-MO1. The result showed that mll-MO2 could cause alteration of gene expression similar to that of mll-MO1 (supplemental Fig. 5 and data not shown). As expected, mll-MO3 caused the alteration of all marker expression that was similar to that of mll-MO1 (data not shown). These observations further confirm the specificity and efficiency of these three mll morpholino-mediated mll knockdowns.
Taken together, these observations suggest that mll is required for maintaining hematopoietic progenitors and for proper erythroid and myeloid formation. Gene expression changes after mll knockdown mediated by mll-MO1 injection were confirmed by semi-quantitative RT-PCR (Fig. 3I).
Mll N-terminal Mini-peptide Disrupts Normal Function of mll
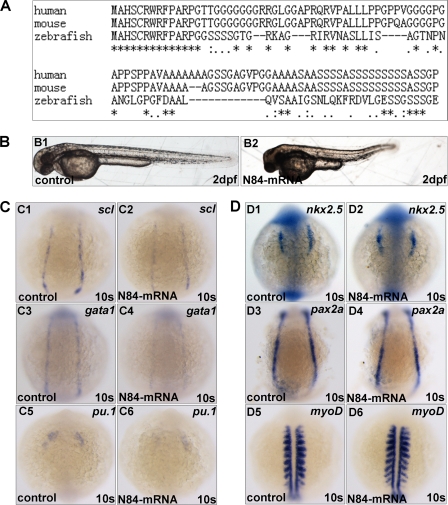
Interestingly, in performing validation experiments for mll-MO1, we noticed that whenever we injected mll (exon1)-GFP vector (named WT1 that contained the 17-bp UTR region plus 246-bp exon1 and 6-bp exon2 of zebrafish mll) into embryos, the embryos exhibited phenotypes quite similar to that of mll morphants if not identical, including shorter body, smaller eyes, heart edema, reduced blood cells, and no blood flow (data not shown). After examining the sequence of WT1, we realized that it encoded an 84-amino acid peptide fused with GFP, which contains 82 amino acids of mll exon1 and -2 amino acids of mll exon2. This phenomenon led us to hypothesize that the 84-amino acid N terminus of zebrafish mll might function as a dominant negative form of mll in affecting hematopoiesis and embryogenesis. The sequence alignment analysis for the 82-amino acid N terminus of mll among human, mouse, and zebrafish indicated that the N terminus of mll was evolutionarily conserved, particularly for the first 14 amino acids (100% identical), which has been identified for Menin binding previously (Fig. 4A) (44). To further test this hypothesis, we synthesized an mRNA encoding 84 amino acids of the mll N terminus and did mRNA injection assays. After this mRNA was injected into embryos at the one cell stage (400 pg/per embryo), the embryos displayed a smaller body size, shortened body axis, degenerated head region, cardiac edema, as well as a lack of blood flow (Fig. 4B and data not shown). These phenotypes were very similar, if not identical, to that of the mll morphants. In addition, we examined the expression of hematopoietic markers scl, gata1, and pu.1 (Fig. 4C), precardiac marker nkx2.5 (Fig. 4, panels D1 and D2), pronephric duct marker pax2a (Fig. 4, panels D3 and D4), and myogenic marker myoD (Fig. 4, panels D5 and D6) after an 84-amino acid mRNA injection. The alteration of the above markers by an 84-amino acid mRNA injection exhibited a similar changing tendency to that of mll morpholino injections (Fig. 4, C and D). These data implicate that the N terminus of mll may function as a dominant negative form of mll, which disrupts normal function of mll during embryogenesis and hematopoiesis.
FIGURE 4.
Overexpression of the zebrafish mll N-terminal 84-amino acid peptide (exon1 plus 2 amino acids of exon2) in embryos caused defects in hematopoiesis similar to that observed in mll morphants. A, alignment of 82 amino acids (exon1) in N terminus of zebrafish mll with its human and mouse orthologs. B, morphology of embryos without injection (control) or injected with mRNA encoding 84 amino acids of mll exon1. C, expression patterns of scl, gata1, and pu.1 in embryos without injection (control) or injected with mRNA of 84-amino acid peptide at 10-somite stage. Panels C1 and C2, scl; panels C3 and C4, gata1; panels C5 and C6, pu.1. D, expression patterns of nkx2.5, pax2a, and myoD. Panels D1 and D2, nkx2.5; and panels D3 and D4, pax2a; and panels D5 and D6, myoD. s, somite.
Mll Depletion Altered Hox Gene Expression and hoxa9a and hoxd3a Partially Rescued the Formation of Early Hematopoietic Progenitors
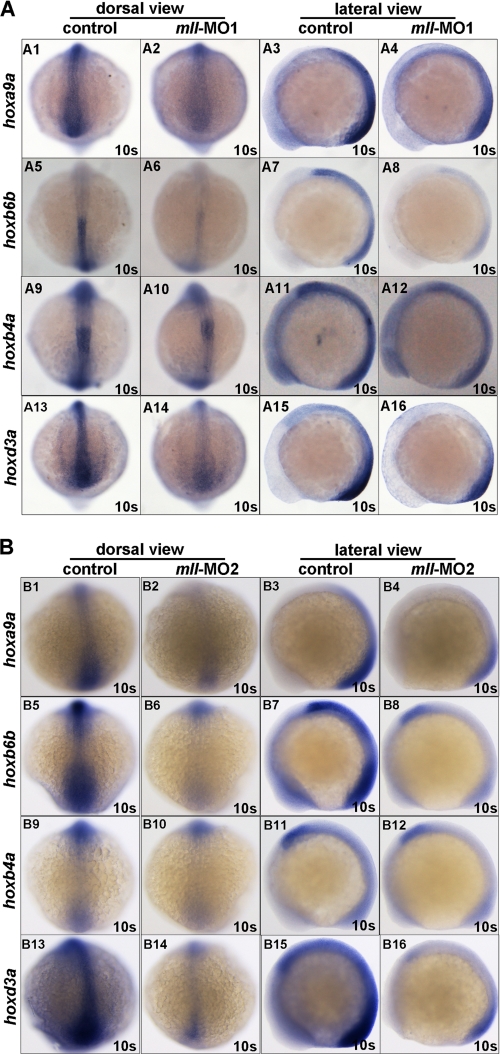
Studies have demonstrated a well defined role for Mll in regulating the expression of a cluster of Hox genes, many of which participate in mammalian hematopoiesis (27, 28, 30, 45–48). Therefore, we next asked whether mll knockdown affected expression of the genes hoxa9a, hoxc8, hoxb7a, hoxb6b, hoxb4, and hoxd3a or expression of their upstream gene cdx4. As shown in Fig. 5, mll-MO1 morphants exhibited a dramatic down-regulation of hoxa9a, hoxb4a, hoxb6b, and hoxd3a, but there was no obvious change in the expression of hoxb5a, hoxb7a, hoxc8, and cdx4 (supplemental Fig. 6). Similar results were obtained by mll-MO2 injection (Fig. 5B and data not shown).
FIGURE 5.
mll is required for Hox gene expression. Whole mount in situ hybridization assays for Hox gene expression in embryos without morpholino injection (control) or injected with mll-MO1 and mll-MO2 at 10-somite stage. A, Hox gene expression in embryos without morpholino injection (control) or injected with mll-MO1 at 10-somite stage. Panels A1–A4, Hoxa9a (dorsal view and lateral view); panels A5–A8, Hoxb6b (dorsal view and lateral view); panels A9–A12, Hoxb4a (dorsal view and lateral view); panels A13–A16, Hoxd3a (dorsal view and lateral view). B, Hox gene expression in embryos without morpholino injection (control) or injected with mll-MO2 at 10-somite stage. Panels B1–B4, Hoxa9a (dorsal view and lateral view); panels B5–B8, Hoxb6b (dorsal view and lateral view); panels B9–B12, Hoxb4a (dorsal view and lateral view); panels B13–B16, Hoxd3a (dorsal view and lateral view). s, somites.
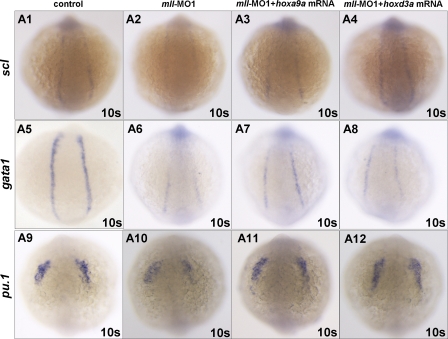
We then performed rescue experiments to determine whether the mll-MO1-mediated effect on hematopoiesis was the result of Hox gene down-regulation. For these experiments, we synthesized hoxa9a, hoxb4a, hoxb7a, hoxb6b, and hoxd3a mRNA and then co-injected embryos with each of the Hox mRNAs (100 pg/per embryo) and mll-MO1 (8 ng/per embryo). Whole mount in situ hybridization showed that only hoxa9a and hoxd3a mRNA could partially rescue expression of the early hematopoietic progenitor markers scl, gata1, and pu.1 (Fig. 6, A3, A7, and A11 and A4, A8, and A12) (for hoxoa9a, n = 32/78; for and rabbit anti, n = 36/87) but not hoxb7a, hoxb6b, and hoxb4a, (supplemental Fig. 6B), which were further confirmed by semi-quantitative RT-PCR assays (data not shown). However, none of the Hox mRNAs rescued blood flow (data not shown). Although the ability of mll to target hoxd3a and hoxa9a expression may contribute to hematopoietic regulation, these results suggest the involvement of other mll targets as well.
FIGURE 6.
Injection of either Hoxa9a mRNA or Hoxd3a mRNA could rescue scl, gata1, and pu.1 gene expression in mll-MO1 morphants. A1, scl expression in control embryos without mll-MO1 injection; A2, scl expression in mll morphants; A3, scl expression in embryos injected with mll-MO1 (8 ng/per embryo) combined with Hoxa9a mRNA (100 pg/per embryo); A4, scl expression in embryos injected with mll-MO1 (8 ng/per embryo) combined with Hoxd3a mRNA (100 pg/per embryo); A5, gata1 expression in control embryos without mll-MO1 injection; A6, gata1 expression in mll morphants; A7, gata1 expression in embryos injected with mll-MO1 (8 ng/per embryo) combined with Hoxa9a mRNA(100 pg/per embryo); A8, gata1 expression in embryos injected with mll-MO1 (8 ng/per embryo) combined with Hoxd3a mRNA (100pg/per embryo); A9, pu.1 expression in control embryos without mll-MO1 injection; A10, pu.1 expression in mll morphants; A11, pu.1 expression in embryos injected with mll-MO1 (8 ng/per embryo) combined with Hoxa9a mRNA (100 pg/per embryo); A12, pu.1 expression in embryos injected with mll-MO1 (8 ng/per embryo) combined with Hoxd3a mRNA (100 pg/per embryo).
To further determine whether hoxa9a or hoxd3a knockdown could recapitulate the phenotypes of mll morphants, we injected hoxa9a-MO (8 ng/per embryo) (CGACAATTTGGTCAGCTTACCTGGA; splicing-blocking) or hoxd3a-MO (8 ng/per embryo) (CATAATAAGTGGCTTTCTGCATTGC; translation-blocking) into one-cell stage embryos, respectively. The results showed that either hoxa9a-knockdown or hoxd3a-knockdown reduced red blood cell number partially (hoxa9a morphants, 35%, n = 108; hoxd3a morphants, 33.7%, n = 83). No other obvious phenotypes were observed (data not shown). These results further suggest that hoxa9a and hoxd3a could only partially account for the role of mll in embryogenesis, even in hematopoiesis.
Gadd45αa Is a Potential mll Target Involved in Zebrafish Hematopoiesis
To better understand the underlying mechanism of how mll regulates hematopoiesis, we performed expression profiling in mll morphants (10-somite stage) using a zebrafish oligonucleotide microarray representing about 44,000 genes (Agilent). To reduce the noise in the microarray data, we performed four parallel experiments using mll-MO1 and mll-MO2. We considered genes whose average fold change in expression was bigger than or equal to 2 in both mll-MO1 and mll-MO2 morphants as mll-regulated targets. As shown in supplemental Table 2, after knockdown of full-length mll we identified 27 known genes that were up-regulated and 45 that were down-regulated. Not surprisingly, most of down-regulated genes were related to hematopoiesis and neurogenesis, for example 4 Hox genes, lmo2, and kdr1, supporting the use of this microarray as an initial screen.
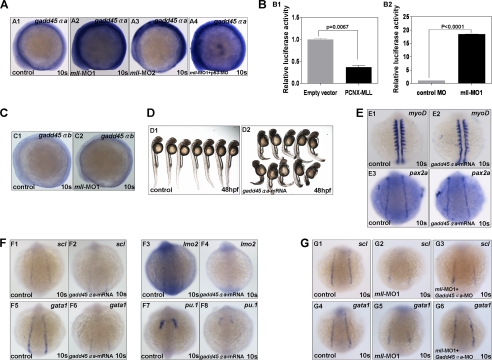
Based on this assay, we identified gadd45αa (previously named as gadd45al), as the gene with the highest fold increase in both mll-MO1 (6.98-fold) and mll-MO2 (224.44-fold) morphants. To confirm the up-regulation of gadd45αa, we performed whole mount in situ hybridization. In both mll-MO1 and mll-MO2 morphants, we observed an increase in gadd45αa expression as compared with the control (Fig. 7A, panels A1–A3). Semi-quantitative RT-PCR assays further confirmed that gadd45αa was up-regulated more than 6-fold after mll knockdown (Fig. 3I).
FIGURE 7.
Zebrafish mll effectively suppressed gadd45αa expression. A, whole mount in situ hybridization assays for gadd45αa expression in embryos at 10-somite stage (lateral view). Panel A1, gadd45αa expression in embryos without mll morpholino injection (control); panel A2, gadd45αa expression in embryos injected with mll-MO1 (8 ng/per embryo); panel A3, gadd45αa expression in embryos injected with mll-MO2 (8 ng/per embryo); panel A4, gadd45αa expression in embryos injected with both mll-MO1 (8 ng/per embryo) and p53-MO(8 ng/per embryo). B, promoter luciferase reporter assays for gadd45αa regulation by either mll overexpression or mll knockdown in embryos. Panel B1, gadd45αa promoter activity was suppressed by human MLL overexpression significantly (p < 0.05); panel B2, gadd45αa promoter activity was enhanced by mll knockdown significantly (p < 0.0001). C, gadd45αb expression in embryos without mll morpholino injection (control) or injected with mll-MO1 (8 ng/per embryos) at 10-somite stage (lateral view). Panel C1, gadd45αb expression in embryos without mll morpholino injection (control); panel C2, gadd45αa expression in embryos injected with mll-MO1; D, morphology of embryos without mRNA injection (control) or injected with gadd45αa mRNA (400 ng/per embryo) at 48-hpf stage. Panel D1, control embryos; panel D2, embryos injected with gadd45αa mRNA. E, expression patterns of myoD and pax2a in embryos without mRNA injection (control) or injected with gadd45αa mRNA (400 pg/per embryo) at 10-somite stage. Panels E1 and E2, myoD; panel E3 and E4, pax2a. F, expression patterns of scl, lmo2, gata1, and pu.1 in embryos without mRNA injection (control) or injected with gadd45αa mRNA (400 ng/per embryo) at 10-somite stage. Panels F1 and F2, scl; panels F3 and F4, lmo2; panels F5 and F6, gata1; panels F7 and F8, pu.1. G, gadd45αa mRNA (400 ng/per embryo) injection could partially rescue scl and gata1 expression in mll-MO1 morphants. Panels G1–G3, scl rescued; panels G4–G6, gata1 rescued.
As shown above, the injection of mll morpholinos had some off-target effects for inducing cell apoptosis. In addition, it was reported previously that gadd45α is a p53-regulated stress protein (49). Thus, it is possible that gadd45αa up-regulation in mll morphants might be the result of p53 activation. To rule out this possibility, we co-injected mll-MO1 and p53-MO into zebrafish embryos and then looked at gadd45αa expression by whole mount in situ hybridization. The result showed that the up-regulation of gadd45αa in embryos with depleted mll and p53 was similar to that with only depleted mll (Fig. 7A, panel A4).
To further validate the suppressive role of mll on gadd45αa, we cloned a 1360-bp promoter of zebrafish gadd45αa into the pGL3-Basic vector to construct a luciferase reporter. As shown in Fig. 7B, mll overexpression significantly inhibited gadd45αa promoter activity (p = 0.0067), although mll knockdown significantly induced promoter activity (p < 0.0001). Furthermore, we checked the expression of gadd45αb (previously named as gadd45α), a homolog of zebrafish gadd45αa, in mll morphants. Unlike gadd45αa, gadd45αb expression was not altered after mll knockdown (Fig. 7C). Taken together, these observations suggest that gadd45αa might serve as a direct target of mll-mediated gene suppression in zebrafish.
We next asked if the targeting of gadd45αa by mll plays a role in hematopoiesis. We synthesized gadd45αa mRNA and then injected embryos (400 pg/per embryo). Embryos with overexpression of gadd45αa mRNA displayed defects in morphogenesis and hematopoiesis almost identical to the ones seen in the mll morphants, including small body size, curved body axis, heart edema, reduced blood cells, and lack of blood flow (Fig. 7D and data not shown). Whole mount in situ hybridization revealed a reduction of hematopoietic markers, including scl, lmo2, gata1, and pu.1 in gadd45αa-overexpressing embryos (Fig. 7F) but not the pronephric marker pax2a (Fig. 7, panels E3 and E4), similar to that observed in mll morphants. MyoD staining showed that gadd45αa mRNA injection also did not cause embryo development delay (Fig. 7, panels E1 and E2). To obtain more evidence for supporting that gadd45αa is a target of mll, we designed and synthesized a morpholino for blocking gadd45αa translation by targeting its ATG code (gadd45αa-MO: CCGTTAAGTTCTTCAAAAGTCATGT) and then performed rescue experiments. As shown in Fig. 7G, gadd45αa-MO could indeed partially rescue hematopoietic marker scl and gata1 expressions in mll-MO1 morphants (Fig. 7G; for scl, n = 10/26; for gata1, n = 12/24), which was further confirmed by semi-quantitative RT-PCR assays (data not shown). However, gadd45αa-MO injection could not rescue both blood flow and morphological deficiency exhibited in mll morphants, suggesting more downstream targets other than Hox genes and gadd45αa mediating the function of mll during embryogenesis.
Given that both Hox genes and gadd45αa-MO regulated by mll, we intended to figure out the relationship between gadd45αa and Hox genes. After embryos were injected with hoxa9a-MO or hoxd3a-MO, the expression of gadd45αa was not altered obviously (supplemental Fig. 7A). After embryos were injected with gadd45αa mRNA, neither hoxa9a nor hoxd3a was changed (supplemental Fig. 7B). Furthermore, gadd45αa-MO injection also did not alter hoxa9a and hoxd3a expression (data not shown). Taken together, these observations suggest that gadd45αa may serve as a mll target in regulating zebrafish hematopoiesis, and the regulation of gadd45αa by mll is parallel to that of Hox genes regulated by mll.
DISCUSSION
Mll Morpholinos Generated a Defective Zebrafish Embryo Phenotype
In this study, we designed three morpholinos targeting zebrafish mll. In comparison with embryos injected with a standard control morpholino, mll morphants had defects in blood cell formation and altered Hox gene expression, consistent with defects seen in MLL null mice (17, 50). Because of the length of the zebrafish mll gene (4218 amino acids), we could not synthesize mll mRNA to perform rescue experiments; however, we observed that the mll-MO1, mll-MO2, and mll-MO3 morphants had very similar phenotypes, particularly in hematopoiesis, suggesting that the MO-mediated knockdown of mll is effective and specific.
In addition, we also showed that the N-terminal mini peptide (84 amino acids) might act as a dominant negative form of mll to disrupt the normal function of mll in hematopoiesis. The N terminus of MLL contains a highly conserved Menin-binding sequence at 5–10 amino acids, essential for MLL fusion protein-mediated leukemogenesis (44). To further determine whether zebrafish menin mediated the effect of N-terminal mini peptide, we designed and synthesized a morpholino to block translation (menin-MO, TCTTCTGAGTTGAACGAATCCCCAT) for zebrafish menin (Ensembl ID ENSDART00000080803). However, the menin-MO injected embryos did not display any detectable phenotypes, even with a very high dosage (24 ng/individual, data not shown). This result implies that menin probably was not responsible for the effect of N-terminal mini peptide. However, we could not rule out that other unidentified copies or homologs of menin played a redundant role. Given that the rearrangement of the MLL gene resulted in the N terminus fused with partner genes causes aggressive acute leukemias, the identification of a dominant negative domain of Mll in N-terminal might provide a useful clue for explaining the cause of leukemogenesis.
Zebrafish Primitive and Definitive Hematopoiesis Required mll
Zebrafish have two waves of hematopoiesis, primitive and definitive hematopoiesis (51). The series of genes that regulate hematopoiesis do so in a strictly controlled, time-dependent and spatially dependent manner. As revealed by the knock-out mouse models, MLL plays a major role in regulating mammalian hematopoiesis; however, the role that it plays in zebrafish hematopoiesis remains unknown, particularly during embryogenesis. In this study, we provide evidence to show that the knockdown of zebrafish mll affected both waves of zebrafish hematopoiesis. In the early embryonic stage, loss of mll resulted in reduced expression of the early hemangioblast markers scl and lmo2, as well as the erythroid progenitor marker gata1 and the myeloid progenitor marker pu1. However, the pronephronic duct markers pax2a and precardiac marker nkx2.5 were not affected. These results suggest that loss of mll specifically modulated hematopoiesis but not other mesoderm-derived tissues or organs. Besides the primitive hematopoietic markers, mll morphants also expressed reduced levels of definitive hematopoietic markers, including runx1 and c-myb. Consequently, mature hematopoietic cell markers, such as mpo, l-plastin, and hbbe1 were all significantly down-regulated. Blood flow tracing and hemoglobin staining demonstrated that mll depletion led to a severe disruption of mature blood cell formation.
Interestingly, 24-hpf mll morphants had a reduction in gata1-staining cells, and at 36 hpf, these gata1-staining cells still could be found in the intermediate cell mass. At this time point, gata1-marked cells should have developed into mature red blood cells. This phenomenon suggests that mll might contribute to controlling hematopoietic progenitor cell differentiation as well as their formation, consistent with its role in the mouse model. Regarding that the apoptosis induction by mll knockdown did not account for the effect of mll on hematopoiesis, that the cell proliferation was not affected by mll knockdown, and that scl and lmo2 were reduced as early as the 5-somite stage in mll morphants, we think that mll might mainly modulate the formation and differentiation of hematopoietic progenitors. Recently, Robinson et al. (52) revealed that zebrafish mll expressed in hematopoietic tissue, also implying its role in hematopoiesis.
In Zebrafish, mll Regulated Hox Gene Expression, but Other mll Targets May Play Larger Roles in Hematopoiesis
As revealed by whole mount in situ hybridization, mll morphants had reduced expression levels of hoxa9a, hoxb6b, hoxb4a, and hoxd3a, whereas microarray analysis showed a down-regulation of hoxa10b, hoxa13a, hoxa2b, and hoxd3a. These results suggest that zebrafish mll plays a critical role in maintaining expression of select Hox genes, similar to what occurs in mammals. However, none of the altered Hox genes could completely rescue the blood and morphology deficiency found in mll morphants, indicating that Hox genes may only be partially responsible for mediating the function of mll in hematopoiesis.
Gadd45αa Is a Potential Downstream Target of mll in Hematopoiesis Regulation
Through microarray analysis, we identified gadd45αa as being dramatically up-regulated in mll morphants. The results from whole mount in situ hybridization staining, promoter assays, and semi-quantitative RT-PCR further implicate gadd45αa as a direct target of mll; however, this will require confirmation through further assays such as chromatin immunoprecipitation and electrophoretic mobility shift assays (EMSA).
The embryos injected with synthesized gadd45αa phenotypically resembled the mll morphants, and co-injection of gadd45αa-MO could partially rescue the hematopoietic marker expression in mll morphants, suggesting a downstream role for gadd45αa in the regulation of hematopoiesis. However, gadd45αa overexpression could not alter Hox gene expression, and knockdown of hoxa9a and hoxd3a could not alter gadd45αa expression, which suggests that the regulation of gadd45αa by mll is parallel to that of Hox genes regulated by mll.
Of note, similar to Hox gene overexpression, gadd45αa knockdown could not rescue morphological phenotypes and blood flow in mll morphants, suggesting that multiple mll targets other than Hox genes and gadd45αa synergistically mediated the function of mll during embryogenesis. To further identify mll's targets during embryogenesis will provide additional clues for understanding mll's function both in morphogenesis and hematopoiesis.
Gadd45α, as a nonenzymatic factor, has been implicated in promoting demethylation (53). Although two studies (54, 55) questioned Gadd45α's function in promoting demethylation, a study using zebrafish embryos provided evidence to show that gadd45 family members, including gadd45α (now named as gadd45αb), gadd45αa, gadd45β, and gadd45γ, participate in promoting 5-meC demethylation, implying an important role of gadd45 in epigenetic regulation. Interestingly, we saw a connection between gadd45αa and mll, another epigenetic regulator. Determining the function of gadd45αa and mll in hematopoiesis will shed light on the underlying mechanisms regulating this important activity and provide a better understanding of how MLL fusion proteins induce leukemogenesis.
Supplementary Material
Acknowledgments
We gratefully acknowledge Drs. Stanley Kormeyers and Jianfang Gui for the generous gift of reagents.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2 and Figs. 1–7.
- PHD
- plant homology domain
- MO
- morpholino
- hpf
- hours post-fertilization
- dpf
- days post-fertilization
- SET
- Su/var, Enhancer or zeste, Trithorax.
REFERENCES
- 1. Ziemin-van der Poel S., McCabe N. R., Gill H. J., Espinosa R., 3rd, Patel Y., Harden A., Rubinelli P., Smith S. D., LeBeau M. M., Rowley J. D., Diaz M. O. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 10735–10739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Djabali M., Selleri L., Parry P., Bower M., Young B. D., Evans G. A. (1992) Nat. Genet. 2, 113–118 [DOI] [PubMed] [Google Scholar]
- 3. McCabe N. R., Burnett R. C., Gill H. J., Thirman M. J., Mbangkollo D., Kipiniak M., van Melle E., Ziemin-van der Poel S., Rowley J. D., Diaz M. O. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 11794–11798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gu Y., Nakamura T., Alder H., Prasad R., Canaani O., Cimino G., Croce C. M., Canaani E. (1992) Cell 71, 701–708 [DOI] [PubMed] [Google Scholar]
- 5. Tkachuk D. C., Kohler S., Cleary M. L. (1992) Cell 71, 691–700 [DOI] [PubMed] [Google Scholar]
- 6. Hess J. L. (2004) Trends Mol. Med. 10, 500–507 [DOI] [PubMed] [Google Scholar]
- 7. Krivtsov A. V., Armstrong S. A. (2007) Nat. Rev. Cancer 7, 823–833 [DOI] [PubMed] [Google Scholar]
- 8. Marschalek R. (2011) Br. J. Haematol. 152, 141–154 [DOI] [PubMed] [Google Scholar]
- 9. Muntean A. G., Giannola D., Udager A. M., Hess J. L. (2008) Blood 112, 4690–4693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Milne T. A., Briggs S. D., Brock H. W., Martin M. E., Gibbs D., Allis C. D., Hess J. L. (2002) Mol. Cell 10, 1107–1117 [DOI] [PubMed] [Google Scholar]
- 11. Liu H., Takeda S., Kumar R., Westergard T. D., Brown E. J., Pandita T. K., Cheng E. H., Hsieh J. J. (2010) Nature 467, 343–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hsieh J. J., Ernst P., Erdjument-Bromage H., Tempst P., Korsmeyer S. J. (2003) Mol. Cell. Biol. 23, 186–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yokoyama A., Kitabayashi I., Ayton P. M., Cleary M. L., Ohki M. (2002) Blood 100, 3710–3718 [DOI] [PubMed] [Google Scholar]
- 14. Yokoyama A., Wang Z., Wysocka J., Sanyal M., Aufiero D. J., Kitabayashi I., Herr W., Cleary M. L. (2004) Mol. Cell. Biol. 24, 5639–5649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wysocka J., Swigut T., Milne T. A., Dou Y., Zhang X., Burlingame A. L., Roeder R. G., Brivanlou A. H., Allis C. D. (2005) Cell 121, 859–872 [DOI] [PubMed] [Google Scholar]
- 16. Dou Y., Milne T. A., Tackett A. J., Smith E. R., Fukuda A., Wysocka J., Allis C. D., Chait B. T., Hess J. L., Roeder R. G. (2005) Cell 121, 873–885 [DOI] [PubMed] [Google Scholar]
- 17. Yu B. D., Hess J. L., Horning S. E., Brown G. A., Korsmeyer S. J. (1995) Nature 378, 505–508 [DOI] [PubMed] [Google Scholar]
- 18. Yagi H., Deguchi K., Aono A., Tani Y., Kishimoto T., Komori T. (1998) Blood 92, 108–117 [PubMed] [Google Scholar]
- 19. Ayton P., Sneddon S. F., Palmer D. B., Rosewell I. R., Owen M. J., Young B., Presley R., Subramanian V. (2001) Genesis 30, 201–212 [DOI] [PubMed] [Google Scholar]
- 20. Dobson C. L., Warren A. J., Pannell R., Forster A., Rabbitts T. H. (2000) EMBO J. 19, 843–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Terranova R., Agherbi H., Boned A., Meresse S., Djabali M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 6629–6634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lim D. A., Huang Y. C., Swigut T., Mirick A. L., Garcia-Verdugo J. M., Wysocka J., Ernst P., Alvarez-Buylla A. (2009) Nature 458, 529–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hess J. L., Yu B. D., Li B., Hanson R., Korsmeyer S. J. (1997) Blood 90, 1799–1806 [PubMed] [Google Scholar]
- 24. Yamashita M., Hirahara K., Shinnakasu R., Hosokawa H., Norikane S., Kimura M. Y., Hasegawa A., Nakayama T. (2006) Immunity 24, 611–622 [DOI] [PubMed] [Google Scholar]
- 25. Yu B. D., Hanson R. D., Hess J. L., Horning S. E., Korsmeyer S. J. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 10632–10636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schilling T. F., Knight R. D. (2001) Philos. Trans. R. Soc. Lond. B Biol. Sci. 356, 1599–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Argiropoulos B., Humphries R. K. (2007) Oncogene 26, 6766–6776 [DOI] [PubMed] [Google Scholar]
- 28. Ernst P., Mabon M., Davidson A. J., Zon L. I., Korsmeyer S. J. (2004) Curr. Biol. 14, 2063–2069 [DOI] [PubMed] [Google Scholar]
- 29. Magli M. C., Barba P., Celetti A., De Vita G., Cillo C., Boncinelli E. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 6348–6352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Krishnaraju K., Hoffman B., Liebermann D. A. (1997) Blood 90, 1840–1849 [PubMed] [Google Scholar]
- 31. Dorshkind K., Witte O. (2004) Mol. Cell 13, 765–766 [DOI] [PubMed] [Google Scholar]
- 32. McMahon K. A., Hiew S. Y., Hadjur S., Veiga-Fernandes H., Menzel U., Price A. J., Kioussis D., Williams O., Brady H. J. (2007) Cell Stem Cell. 1, 338–345 [DOI] [PubMed] [Google Scholar]
- 33. Wan X. Y., Ji W., Mei X., Zhou J. G., Liu J. X., Fang C. C., Xiao W. H. (2010) PLoS One 5, e91118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Thompson M. A., Ransom D. G., Pratt S. J., MacLennan H., Kieran M. W., Detrich H. W., 3rd, Vail B., Huber T. L., Paw B., Brownlie A. J., Oates A. C., Fritz A., Gates M. A., Amores A., Bahary N., Talbot W. S., Her H., Beier D. R., Postlethwait J. H., Zon L. I. (1998) Dev. Biol. 197, 248–269 [DOI] [PubMed] [Google Scholar]
- 35. Krauss S., Johansen T., Korzh V., Fjose A. (1991) Nature 353, 267–270 [DOI] [PubMed] [Google Scholar]
- 36. Tu C. T., Yang T. C., Tsai H. J. (2009) Plos One 4, e4249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhong T. P., Childs S., Leu J. P., Fishman M. C. (2001) Nature 414, 216–220 [DOI] [PubMed] [Google Scholar]
- 38. Rai K., Huggins I. J., James S. R., Karpf A. R., Jones D. A., Cairns B. R. (2008) Cell 135, 1201–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yang Z., Liu N., Lin S. (2001) Dev. Biol. 231, 138–148 [DOI] [PubMed] [Google Scholar]
- 40. Zhou J., Feng X., Ban B., Liu J., Wang Z., Xiao W. (2009) J. Biol. Chem. 284, 19142–19152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lee P., Goishi K., Davidson A. J., Mannix R., Zon L., Klagsbrun M. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 10470–10475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mende M., Christophorou N. A., Streit A. (2008) Mech. Dev. 125, 947–962 [DOI] [PubMed] [Google Scholar]
- 43. Tittle R. K., Sze R., Ng A., Nuckels R. J., Swartz M. E., Anderson R. M., Bosch J., Stainier D. Y., Eberhart J. K., Gross J. M. (2011) Dev. Biol. 350, 50–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yokoyama A., Somervaille T. C., Smith K. S., Rozenblatt-Rosen O., Meyerson M., Cleary M. L. (2005) Cell 123, 207–218 [DOI] [PubMed] [Google Scholar]
- 45. Davidson A. J., Ernst P., Wang Y., Dekens M. P., Kingsley P. D., Palis J., Korsmeyer S. J., Daley G. Q., Zon L. I. (2003) Nature 425, 300–306 [DOI] [PubMed] [Google Scholar]
- 46. Diehl F., Rössig L., Zeiher A. M., Dimmeler S., Urbich C. (2007) Blood 109, 1472–1478 [DOI] [PubMed] [Google Scholar]
- 47. Kyba M., Perlingeiro R. C., Daley G. Q. (2002) Cell 109, 29–37 [DOI] [PubMed] [Google Scholar]
- 48. Antonchuk J., Sauvageau G., Humphries R. K. (2002) Cell 109, 39–45 [DOI] [PubMed] [Google Scholar]
- 49. Rosemary Siafakas A., Richardson D. R. (2009) Int. J. Biochem. Cell Biol. 41, 986–989 [DOI] [PubMed] [Google Scholar]
- 50. Lim D. A., Huang Y. C., Swigut T., Mirick A. L., Garcia-Verdugo J. M., Wysocka J., Ernst P., Alvarez-Buylla A. (2009) Nature 458, 529-U533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. de Jong J. L., Zon L. I. (2005) Annu. Rev. Genet. 39, 481–501 [DOI] [PubMed] [Google Scholar]
- 52. Robinson B. W., Germano G., Song Y., Abrams J., Scott M., Guariento I., Tiso N., Argenton F., Basso G., Rhodes J., Kanki J. P., Look A. T., Balice-Gordon R. J., Felix C. A. (2011) Br. J. Haematol. 152, 307–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Barreto G., Schäfer A., Marhold J., Stach D., Swaminathan S. K., Handa V., Döderlein G., Maltry N., Wu W., Lyko F., Niehrs C. (2007) Nature 445, 671–675 [DOI] [PubMed] [Google Scholar]
- 54. Jin S. G., Guo C., Pfeifer G. P. (2008) PloS Genet. 4, e1000013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Engel N., Tront J. S., Erinle T., Nguyen N., Latham K. E., Sapienza C., Hoffman B., Liebermann D. A. (2009) Epigenetics 4, 98–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.