Abstract
Bone destruction is the major pathological process in many bone metabolic diseases and is a result of increased osteoclast formation and bone resorption. The liver X receptors (α,β), important regulators of cholesterol metabolism and inflammatory signaling, have recently been observed to play a role in both physiological and pathological bone turnover. However, the relationship between liver X receptors (LXR) and osteoclast differentiation/formation remains unknown. Here, we report that the LXR ligand GW3965 is able to clearly and potently inhibit the formation of mature osteoclasts from receptor activator of nuclear factor κB ligand (RANKL)-stimulated human and murine osteoclast precursors. This results in a significant inhibition of bone resorption. We observed that GW3965 significantly inhibited expression of the osteoclast markers tartrate-resistant acid phosphatase, cathepsin K, osteoclast-associated receptor (OSCAR), and calcitonin receptor, appearing to act in an NFATc1/p38/microphthalmia-associated transcription factor (MITF)-dependent mechanism, independently of receptor activator of nuclear factor κB or c-Fos and not directly involving the NFκB pathways. GW3965 was less effective in RAW264.7 monocyte/macrophage cells, which are more committed into the osteoclast lineage. Also, GW3965 seemed to act differently depending on the source of the progenitor cells as it had no effect on calvarial osteoclasts, compared with marrow or blood-derived monocytes. As these effects were abolished in osteoclast precursors derived from LXRβ−/− mice, we suggest that GW3965 acts via an LXRβ-dependent mechanism. Taken together, our results suggest that the LXR can act as an important inhibitor of RANKL-mediated osteoclast differentiation.
Keywords: Bone, Bone Marrow, Differentiation, Mouse, Nuclear Receptors, LXR, Bone Resorption, Osteoclast
Introduction
Bone remodeling occurs continuously throughout life to maintain bone quality in response to mechanical stress and hormonal regulation. To retain a normal bone mass, bone is resorbed by osteoclasts, and new bone matrix is deposited by osteoblasts in a tightly coupled process. Imbalances in this process can lead to excessive resorption by the osteoclasts and increased bone fragility, as observed in many common skeletal diseases (osteoporosis, metastatic bone disease, and Paget disease of bone) (1, 2). Osteoclasts are multinucleated giant cells derived from the pluripotent hematopoietic stem cell lineage. Differentiation of the precursor cells into the osteoclast lineage is principally regulated by macrophage colony-stimulating factor (M-CSF) and receptor activator of NFκB ligand (RANKL)2 (3). Binding of RANKL to its receptor RANK activates different signaling cascades (NFκB, MAPK, and Ca2+ oscillation/calcineurin) to induce nuclear factor of activated T cells c1 (NFATc1), the key transcription factor in osteoclastogenesis (4, 5). NFATc1 in turn induces the transcription of osteoclast-specific genes, including tartrate-resistant acid phosphatase (TRAP/acp5), cathepsin K (ctsk), osteoclast-associated receptor (oscar), and calcitonin receptor (calcr) (6).
The liver X receptors, LXRα and LXRβ, are members of the nuclear receptor superfamily. They form heterodimers with the retinoid X receptor and are activated by naturally occurring ligands, which are specific derivatives of cholesterol called oxysterols, and by the synthetic agonists T0901317 and GW3965 (7, 8). It is the ability of the LXRs to modulate cholesterol metabolism and inhibit inflammation that has led pharmaceutical companies to recognize these receptors as potential drug targets for cardiovascular diseases such as atherosclerosis (9–11).
There are now several studies indicating that LXR may also be important in both physiological and pathological bone homeostasis. In our previous paper (12), we showed that in the absence of LXRα, mice had a significant increase in bone mineral density due to an elevation in cortical density, bone mineral content, cortical thickness, and area. This appeared to be a result of impaired osteoclast activity as suggested by a significant reduction in serum collagen fragments (CTX) and bone TRAP activity. We further showed that LXRβ was important in osteoblast function, with an elevation in osteoblast-associated genes and bone formation markers in the absence of LXRβ (LXRβ−/− mice). Prawitt et al. (13), then recently demonstrated that the LXR agonist (T0901317) significantly decreased bone formation markers in primary calvarial osteoblasts from WT mice. In addition, two studies have demonstrated that LXR treatment is beneficial in suppressing the severe arthritis and joint destruction in a mouse model of collagen-induced arthritis (14, 15). As progressive bone erosions in chronic arthritis are strongly linked to increased osteoclast-mediated bone resorption, it is possible that the LXRs are acting directly on the osteoclasts within this disease state.
However, as yet no studies have investigated if ligand-activated LXR is able to directly affect osteoclastogenesis. Therefore, we aimed to examine the mechanism of LXR action in M-CSF/RANKL-stimulated mouse bone marrow macrophages (BMM), human CD14+ monocytes, mouse calvaria organ cultures, and mouse macrophage/monocyte RAW264.7 cells. Our results showed that the LXR agonist GW3965 acts via LXRβ to potently inhibit osteoclast differentiation, significantly inhibiting the osteoclast differentiation markers (TRAP/acp5, ctsk, oscar, and calcr), via a p-38/MITF/NFATc1-dependent mechanism. This resulted in an absence of mature osteoclasts and cells that were unable to effectively resorb bone.
EXPERIMENTAL PROCEDURES
Animals
LXRα−/− and LXRβ−/− mice were generated by targeted disruption in our laboratory as described previously (16, 17). All mice were backcrossed from a 129/Sv to a C57BL/6 background for 10 generations. Age-matched C57BL/6 mice were used as WT controls. Animals were housed under a 12-h light/dark cycle in the specific pathogen-free facility and were fed a standard mouse chow (R36 Lactamin, Vadstena, Sweden) ad libitum. For the calvarial bone resorption assay, CsA mice from our own inbred colony at Umeå University, Umeå, Sweden, were used. All experiments were approved by the local Animal Experimentation Ethic Committee.
Bone Marrow Macrophages
Female mice (n = 6/group) were obtained from the same colony at 8–12 weeks of age. BMM were obtained as described by Refs. 18, 19. Briefly, the ends of femora, tibiae, and humeri were removed, and the bones were centrifuged at 3,500 rpm for 5 min to flush the bone marrow. The marrow cells were resuspended in 0.16 m NH4Cl, 0.17 m Tris, pH 7.65, in PBS for 5 min to remove red blood cells. Cells from each individual animal were cultured in α-MEM (Invitrogen) containing ribonucleosides and deoxyribonucleosides with 10% FBS (Invitrogen), 0.5% (10 mg/ml) gentamicin (Invitrogen), 2 mm l-glutamine (Invitrogen), and 100 ng/ml M-CSF (R & D Systems) in a 10-cm suspension culture dish (Corning Costar Inc., Corning, NY), to which stromal cells and lymphoid cells cannot adhere, at 37 °C for 3 days. Cells were washed vigorously with PBS to remove any nonadherent cells and then with cold (4 °C) 0.02% EDTA in PBS to dissociate the attached BMM. Cells were seeded at a density of 10,000 cells/cm2 in 24-well plates (Nunc). BMM from WT, LXRα−/−, and LXRβ−/− mice were cultured in α-MEM (as above) + 50 ng/ml M-CSF and 3 ng/ml RANKL (Escherichia coli-derived mouse RANKL Lys158–Asp316, catalog no. 462-TEC, R&D Systems) with 1 μm GW3965 (LXR agonist; GlaxoSmithKline and Sigma) or DMSO (control) for 4 days, with a medium change after 2 days. One μm was chosen as preliminary studies (data not shown) showed that 1–5 μm GW3965 equally inhibited BMM differentiation. As an undifferentiated control, cells were cultured with 100 ng/ml M-CSF. Cells were also treated with 0.5–2 μm T0901317 (Sigma). At the end of the treatment on day 4, cells were stained for TRAP using a leukocyte acid phosphatase kit according to the manufacturer's instructions (Sigma Diagnostics; kit no. 387-A), and total number of TRAP+ cells (cells containing 2 or more nuclei) were counted in three wells/treatment. Number of cells/well with and without GW3965 were expressed as a percentage. To analyze the area of the cells, images were taken at ×10 magnification, and Scion Image was used to quantify the area of 100 cells per treatment group then expressed as a percentage.
Human CD14+ Osteoclast Cultures
Blood was received from healthy blood donors at the Sahlgrenska University Hospital Blood Center. Buffy coats were diluted in 200 ml of PBS and peripheral blood mononuclear cells prepared by Ficoll-Paque PLUS separation according to the manufacturer's instructions (GE Healthcare). CD14+ cells were labeled with CD14 MicroBeads and isolated using a MACS column as recommended by the manufacturer (Miltenyi Biotec). Cells were seeded at a density of 300,000 cells/cm2 in 96-well plates with α-MEM (Invitrogen) supplemented with 10% FBS (Sigma), 2 mm GlutaMAX (Invitrogen), 50 μg/ml gentamicin (Invitrogen), 100 units/ml penicillin, 100 μg/ml streptomycin (PEST, Invitrogen), and 30 ng/ml recombinant human M-CSF (R&D Systems). For osteoclast generation, 2 ng/ml recombinant mouse RANKL was added with 0.1–0.5 μm GW3965. Cells were cultured for 4 days, with a medium change at 3 days. At the end of the treatment, cells were stained for TRAP as above, and total number of TRAP+ cells (cells containing 3 or more nuclei) were counted in 3–4 wells/treatment. For the phagocytosis assay, 5 μl of FITC-conjugated zymosan (1 mg/ml; Invitrogen) was added to each well on day 4. After 1 h, cells were washed three times in PBS, fixed, and stained for TRAP as above.
RAW264.7 Cells
The mouse monocyte/macrophage cell line RAW264.7 was purchased from ATCC. Cells were grown in α-MEM (as described above in BMM) and differentiated into multinucleated cells with 3 ng/ml RANKL and either 1 μm GW3965 or DMSO over 6 days. Medium was collected every 24 h. For TRAP staining, mRNA extraction, and Western blot, 5000 cells/cm2 were plated into 6-well plates; for quantitative analyses, 2500 cells/cm2 were plated into 24-well plates. Cells were stained for TRAP as above, and on day 4 the total number of TRAP+ cells (cells containing two or more nuclei) were counted in three wells/treatment. The area of the cells was analyzed as above. TRAP enzyme activity was measured in media collected from RAW264.7 cells cultured for 0 to 6 days as described previously (12).
Osteoclast Formation on Bone and Resorption Assay
CD14+ cells were seeded at a density of 100,000 cells/bovine bone slice (Immunodiagnostic Systems, Herlev, Denmark) in 96-well plates in α-MEM supplemented as above. Cells were cultured for 10 days with complete change of medium on days 4 and 7. At the end of the experiment, cells were stained for TRAP as above and number of TRAP+ cells (cells containing three or more nuclei) were counted on four bone slices/treatment. Cells were subsequently removed by sonication in 0.5 m ammonium hydroxide and bones were stained with 0.5% toluidine blue to visualize resorption pits. The release of collagen type I fragments into the culture medium during resorption was analyzed by CrossLaps for culture ELISA (IDS Nordic a/s).
RNA Preparation and qPCR
Total RNA was isolated from cells with the E.Z.N.A. kit (Omega Bio-TeK) according to manufacturer's instructions. For qPCR, 1 μg of RNA was reverse-transcribed with 100 ng of random hexamer primer using the Superscript II reverse transcriptase (Invitrogen). Specific primers were designed using the Primer Express software (PE Biosystems, Foster City, CA). For sequences see supplemental Table 1. The following forward and reverse primer pairs were obtained from published studies as follows: TRAP/acp5 (12), ctsk (12), mitf (20), rank (21), c-fos (22), and calcr (23). All genes were analyzed with the SYBR Green detection method using the Applied Biosystems 7500 Real Time PCR machine. Each PCR was run at the annealing temperature of 60 °C. Primers were used at a concentration of 300 nm and tested for specificity using a dissociation curve analysis and efficiency in a standard curve assay with the housekeeping genes. Genes of interest were analyzed from three separate experiments using the comparative CT method. All gene expression data were normalized against both β-actin and 18 S, and the control values expressed as 1 to indicate a precise fold change for each gene of interest.
Western Blot
For the early time points, RAW264.7 cells were grown in 10-cm culture dishes and BMM in 24-well plates as described above. Both were pretreated for 18 h with 1 μm GW3965. Then 3 ng/ml RANKL was added for 5, 15, and 30 min. Cells were scraped into PBS, spun at 3500 rpm for 2 min, and then frozen on dry ice. Cells were lysed in 50 μl of TDS-PBS (PBS with 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 1 mm EDTA with both complete protease inhibitors and PhosSTOP phosphatase inhibitors (Roche Applied Science)) on ice for 30 min. Protein concentration was determined using a BCA protein quantification kit (Pierce) with BSA as a standard, and then 40 μg (RAW264.7 cells) or 15 μg (BMM) of total proteins was loaded. For day 2 and 4 cells, BMM and RAW264.7 cells were grown in 6- or 24-well plates, respectively. SDS lysis buffer containing β-mercaptoethanol (200 or 100 μl) was added to cells and 35 μl of cell lysate was loaded.
Total protein was separated by 10% SDS-polyacrylamide gel and transferred to a nitrocellulose membrane (Whatman). The membrane was blocked with 5% skim milk, and primary antibodies were incubated overnight at 4 °C in 4% BSA, p38, p-p38, and IκB-α (1:1000 Cell Signaling Technology) or 5% milk, TRAP (1:30,000 (24, 25), cathepsin K (1:1000 (24, 25)), OSCAR (1:150 R&D Systems), and NFATc1 (1:200, Pharmingen). Following incubation for 1 h at room temperature with the appropriate horseradish peroxidase-conjugated secondary antibodies (1:10,000, GE Healthcare) in 3% milk, all bands were visualized using the ECL-Plus system (GE Healthcare). The levels of β-actin (1:30,000 for 30 min in 5% milk; Sigma) were analyzed as a control for constant loading. Densitometric values were quantified for each band with ImageJ 1.40. The control density value at time point 0 (for −/+GW3965) is presented as 1 in Figs. 2E and 5F.
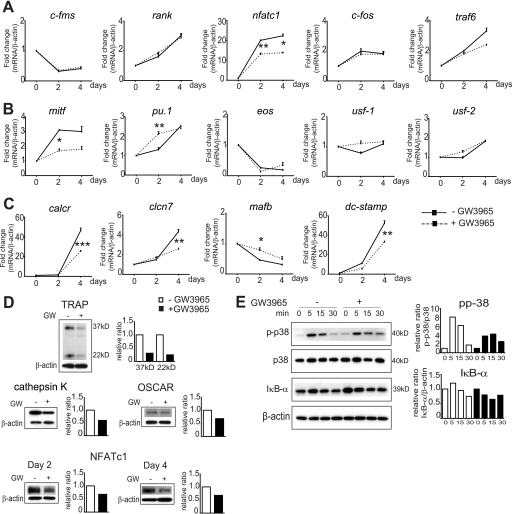
FIGURE 2.
Gene and protein expression of osteoclast differentiation genes from GW3965-stimulated WT BMM. A, gene expression analysis. Treatment over 4 days with GW3965 resulted in no change in the expression of c-fms, rank, c-fos, and traf6 but significantly inhibited nfatc1. B, mitf was also inhibited on day 2; however, this effect was no longer significant on day 4, with no effect on eos or usf-1/2 but an increase in pu.1 on day 2. C, calcr, DC-stamp, and clcn7 were repressed on day 4, whereas the osteoclast differentiation inhibitor mafB was increased on day 2. D, protein expression analysis. GW3965 (GW) reduced TRAP, cathepsin K, and OSCAR protein levels on day 4 BMM from WT mice, also inhibiting NFATc1 at days 2 and 4. The control (−GW3965) density value was expressed as 1. E, Western analysis indicated that GW3965 inhibited p38 phosphorylation, although no effect was observed on the IκB-α subunit. To observe if GW3965 has an effect on phosphorylation each time point 0 (GW− and GW+) was expressed as 1 in the density graph. The gene expression data were expressed as fold change compared with M-CSF only cells (fold value 1) and normalized against β-actin; mean ± S.E. (n = 3) *, p < 0.05; **, p < 0.01; ***, p < 0.001 using a one-way ANOVA with a post hoc Tukey test.
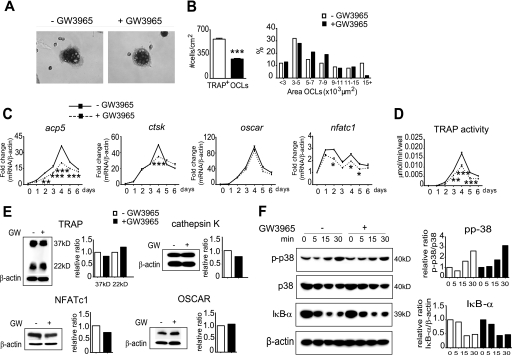
FIGURE 5.
Regulation of osteoclast differentiation from RANKL-stimulated RAW264.7 cells with GW3965. A and B, GW3965 inhibited the formation of TRAP+ OCLs with significantly fewer cells on day 4 of M/R differentiation. However, the cells that do form were of a normal size. C and D, expression of TRAP/acp5 was reduced over 6 days of differentiation (** on day 2; *** on day 3–5), as was TRAP activity in the media (** on day 3; *** on days 4 and 5), and NFATc1, but little or no effect was seen on ctsk or oscar gene expression. E, no effect was observed at the protein level of TRAP, cathepsin K (Cath K), or OSCAR from day 4 cells. F, Western analysis indicated that GW3965 had no effect on either the NFκB pathway, with retained successful degradation of the IκB-α subunit, or on p38 phosphorylation. Each time point 0 (GW− and GW+) was expressed as 1 in the density graph. The gene expression data were expressed as mean ± S.E. (n = 3): *, p < 0.05; **, p < 0.01; ***, p < 0.001 using a one-way ANOVA with a post hoc Tukey test.
Mouse Calvarial Bone Resorption Assay
2-Day-old mice were injected with 1.5 μCi of 45Ca to label the mineral crystals in the skeleton. After 4 days, parietal bones were microdissected and cut into four pieces as described previously (26). The bones were preincubated for 18–24 h in α-MEM containing 0.1% albumin and 1 μmol/liter indomethacin to reduce the basal rate of bone resorption caused by endogenous synthesis of prostaglandins because of dissection trauma (27). Bones were then extensively washed and cultured for 120 h in multiwell culture dishes containing 1.0 ml of indomethacin-free medium with or without test substances. Radioactivity in the culture medium and in demineralized bones was quantified using a β-scintillation counter. Isotope release was expressed as the percent release of the initial amount of isotope (calculated as the sum of radioactivity in medium and bone after culture) (27). The results were expressed as a percentage of the control.
Statistical Analyses
Effects of the LXR agonist GW3965 between treated and untreated groups were analyzed either using a Student's t test or a one-way ANOVA with a post hoc Tukey test. All analyses were performed using the GraphPad Prism 5.0 software. Data were considered statistically significant when a p value less than 0.05 was obtained. All data are expressed as mean ± S.E. All gene experiments were performed three times and normalized against two different housekeeping genes with consistently significant results. Western blots were performed twice with consistent results.
RESULTS
Effect of GW3965 on Osteoclast Differentiation from Mouse Bone Marrow Macrophages
To examine if activating the LXR inhibits the differentiation of murine osteoclast precursors, we treated WT BMM with M-CSF/RANKL (M/R) and added the LXR agonist GW3965 (1 μm) in the culture from days 0–4. Addition of GW3965 significantly inhibited both the formation of TRAP+ osteoclasts (OCLs) at day 4, with a 90% reduction in the number of multinucleated TRAP+ cells, and their size (Fig. 1, A–C). After 4 days, as expected the M/R treatment with no GW3965 increased the expression of TRAP/acp5 by 200-fold when compared with M-CSF (M) only cells at day 0 (fold value 1), also increasing ctsk by 100-fold and oscar by 18-fold. Adding GW3965 significantly inhibited the expression of TRAP/acp5, ctsk, and oscar mRNA on day 4 (Fig. 1D).
FIGURE 1.
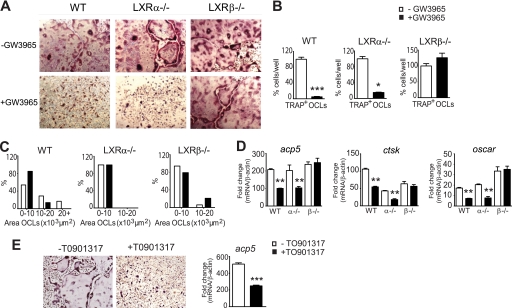
Regulation of osteoclast differentiation from M/R-stimulated mouse BMM with GW3965. A, representative images from BMM isolated from WT, LXRα−/−, and LXRβ−/− mice, cultured for 4 days in the presence of M/R, and stained for TRAP (purple stain), a marker of osteoclasts. TRAP+ multinucleated OCLs were able to form in vitro from both the LXR−/− BMM. After the addition of GW3965, the formation of TRAP+ OCLs from WT and LXRα−/− mice, but not from LXRβ−/− cells, was significantly inhibited. B, cells formed from the LXR−/− mice were smaller than WT, in particular those from the LXRα−/−. GW3965 (1 μm) reduced the size of cells from WT mice but not from LXRβ−/− BMM. C, number of TRAP+ OCLs from WT and LXRα−/− BMM was also significantly reduced but not from LXRβ−/− BMM. D, using qPCR, TRAP/acp5, ctsk, and oscar gene expression was significantly reduced in the WT and LXRα−/− cells on day 4 after GW3965 treatment, with no change in expression in the LXRβ−/− cells. E, LXR agonist TO901317 (0.5 μm) also inhibited the formation of TRAP+ OCLs on day 4 stimulated with M-CSF/RANKL, inhibiting acp5 gene expression. The data in B were expressed as a percentage of total cells; data in C were expressed as a percentage difference to −GW3965 cells; data in D and E were expressed as fold change compared with M-CSF only cells (fold value 1) and normalized against β-actin (n = 4). All data expressed as mean ± S.E. *, p < 0.05; **, p < 0.01; ***, p < 0.001 using a Student's t test.
To determine whether GW3965, a nonsubtype-specific agonist, was acting in an LXRα-, -β-, or -α/β-dependent mechanism, we examined precursor cells derived from the LXRα−/− and β−/− mice. First we observed in the cultures with no GW3965 that BMM from both LXRα−/− and β−/− mice were able to form TRAP+ OCLs in vitro after stimulation for 4 days by M/R, similar to the WT animals (Fig. 1A). However, as the TRAP+ OCLs were smaller than those formed from WT mice, particularly those from LXRα−/− mice, it suggested that both LXR subtypes are important in normal osteoclastogenesis (Fig. 1B). In BMM from the LXRα−/− mice, the addition of GW3965 clearly inhibited the formation of TRAP+ OCLs, with a 70% reduction in the number of cells (Fig. 1C). However, as the cells formed from the BMM of LXRα−/− mice were already small, the effect of the LXR ligand on cell size was more difficult to detect within this group (Fig. 1B). As in the WT cells, GW3965 significantly inhibited the expression of acp5, ctsk, and oscar mRNA by ∼50% in LXRα−/− OCLs (Fig. 1D). However, GW3965 was unable to inhibit both the formation of TRAP+ OCLs and the expression of acp5, ctsk, and oscar in BMM derived from the LXRβ−/− mice (Fig. 1, A–C). This suggested that when both receptors are stimulated, it is the LXRβ that is important in inhibiting osteoclast differentiation.
We also tested another LXR synthetic agonist, TO901317 (0.5 μm), and found it had the same inhibitory effect on multinucleated osteoclast formation in BMM as GW3965 with a 50% decrease in acp5 gene expression at day 4 (Fig. 1E). We also used 1 and 2 μm TO901317 and saw a similar decrease in acp5 expression as 0.5 μm (data not shown).
Effects of GW3965 on Osteoclast Differentiation Genes
To further analyze at which stage of differentiation GW3965 may be having its inhibitory action, we analyzed osteoclast signaling pathway genes using qPCR on days 0, 2, and 4 of differentiation. We found that there was no obvious difference in the M-CSF receptor c-fms, rank, or c-fos after treatment with GW3965; however, a significant reduction in nfatc1 was observed (Fig. 2A). We also saw a significant decrease in mitf at day 2, which collaborates with the ETS family transcription factor PU.1 and NFATc1 to regulate osteoclast-specific genes (28–31), although this effect was no longer significant by day 4 (Fig. 2B). However, no difference was seen in the MITF inhibitor eos or in the upstream regulatory factors usf-1 and -2, which also associate with NFATc1 (Fig. 2B) (32, 33). Both chloride channel 7 (clcn7), regulated by MITF (34), and calcr, regulated by NFATc1 (35), were inhibited by GW3965 (Fig. 2C). Interestingly, both pu.1 and the anti-osteoclastogenic transcription factor mafb were up-regulated on day 2. We also investigated known osteoclast fusion genes. Dendritic cell-specific transmembrane protein (dc-stamp), the major fusion gene and also an NFATc1 target gene (36), was inhibited in the GW3965-treated BMM (Fig. 2C). To confirm the mRNA results in Fig. 1, we showed that GW3965 down-regulated the expression of TRAP, cathepsin K, and OSCAR at the protein level in WT BMM (Fig. 2D). We further showed that GW3965 also down-regulates NFATc1 protein at days 2 and 4 (Fig. 2D).
During the initial stage of differentiation, NFκB is crucial for the induction of NFATc1. Then p38 MAPK signaling was important for the continual up-regulation of NFATc1 via p38 and MITF/PU.1, which in turn regulates the expression of osteoclast-specific genes. Western analysis at early time points following the addition of RANKL showed little effect of GW3965 on the degradation of the NFκB inhibitor IκB-α (Fig. 2E). However, GW3965 did appear to inhibit the levels of RANKL-stimulated phosphorylated p38 (Fig. 2E). These findings indicate that LXR activation inhibits osteoclast differentiation via a p38/NFATc1/MITF pathway.
Effects of GW3965 on Different Stages of Osteoclast Differentiation
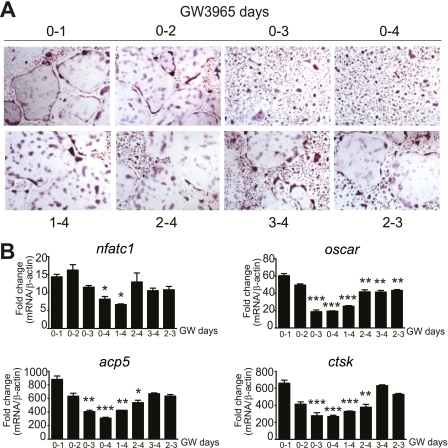
To further investigate at which time point in differentiation GW3965 may be inhibiting osteoclast differentiation, we treated BMM for different periods with the LXR ligand, either by adding GW3965 together with RANKL followed by washout at different days (Fig. 3A, upper panel) or by adding the compound at different time points after RANKL (Fig. 3A, lower panel). Following analysis of specific osteoclast genes at day 4 (Fig. 3B) there appears to be no effect by GW3965 at days 0–2, with only a weak effect at days 3–4. The time period most significantly affected by GW3965 seems to be days 2–3.
FIGURE 3.
Effect of GW3965 on osteoclast formation at different stages of differentiation. A, addition of GW3965 to BMM at early stages (day 0–2) does not affect the formation of multinucleated TRAP-positive osteoclasts but has an inhibitory effect when added from days 2 to 3. B, this effect coincides with the effect on oscar, TRAP/acp5, and ctsk expression, with the most significant inhibition observed when GW3965 is added from days 0 to 3/4. The gene expression data were expressed as fold change compared with M-CSF only cells (fold value 1) and normalized against β-actin; mean ± S.E. (n = 3). Significance was analyzed using a one-way ANOVA with a post hoc Tukey test against day 0–1; *, p < 0.05; **, p < 0.01; ***, p < 0.001.
GW3965 Inhibits Formation of TRAP+ OCLs in Human CD14+ Cells
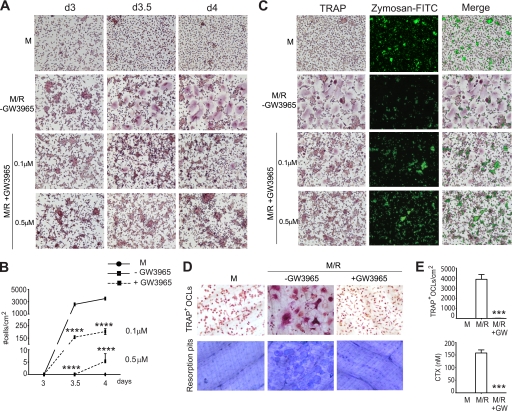
To determine whether the LXR agonist is also able to inhibit differentiation in human cells, CD14+ blood-derived monocytes were differentiated with M/R in the presence of increasing concentrations (0.1–0.5 μm) of GW3965. The formation of TRAP+ OCLs was significantly inhibited in a dose-dependent manner, with 0.5 μm GW3965 reducing the number of TRAP+-multinucleated cells to ∼0.1% of control (Fig. 4, A and B). We also stimulated these cells with a lower concentration of RANKL (0.5 ng/ml) and saw the same inhibitory effect (data not shown). When precursor macrophages differentiate into the osteoclast pathway with M/R, they lose their ability to phagocytose particles, a typical macrophage characteristic. As expected, we observed that M/R stimulation was associated with decreased capability to phagocytose FITC-labeled zymosan particles, compared with cells stimulated with M alone, which were able to phagocytose particles. Interestingly, GW3965 inhibition of osteoclast formation resulted in mononuclear cells capable of phagocytosing zymosan particles, thus apparently displaying a macrophage-like phenotype (Fig. 4C).
FIGURE 4.
Regulation of osteoclast differentiation from M/R-stimulated human CD14+ monocytes with GW3965. A and B, in a dose-response experiment (0.1–0.5 μm LXR ligand), GW3954 inhibited the formation of TRAP+ OCLs stimulated with M-CSF/RANKL, with 1000-fold less TRAP+ OCLs observed with 0.5 μm GW3965. No multinucleated cells were formed in the M group. d, day. C, treatment with M alone resulted in cells able to phagocytose zymosan particles, whereas M/R resulted in multinucleated cells unable to phagocytose. The addition of GW3965 resulted in mononuclear cells capable of phagocytosing zymosan particles and thus appeared to have a macrophage-like phenotype. D and E, GW3965 inhibited the formation of multinucleated OCLs when cultured on bone slices, with almost no OCLs formed. Furthermore, no resorption pits were observed after GW3965 treatment, with no CTX detected in the cell media. No resorption was observed in the M only group. The data are expressed as mean ± S.E. (n = 4) ***, p < 0.001; ****, p < 0.0001 using a one-way ANOVA with a post hoc Tukey test.
GW3965 Inhibits Bone Resorption in Human Osteoclasts
The above experiments were carried out from precursor cells cultured on plastic; therefore, we cultured CD14+ cells on bone slices to confirm these results. GW3965 significantly inhibited the formation of human TRAP+ OCLs on bone with almost no multinucleated osteoclasts formed (Fig. 4, D and E), and as a consequence no resorption pits were observed, and no CTX was detected in the cell media (Fig. 4, D and E). Culturing with M-CSF alone resulted in no multinucleated osteoclasts and no resorption pits.
Effects of GW3965 in RAW264.7 Cells
To determine whether GW3965 is able to inhibit differentiation of a mouse monocyte/macrophage cell line, RAW264.7 cells were cultured for 6 days in the presence of RANKL and GW3965. Although there were 50% fewer TRAP+ OCLs formed, those that did form were of a similar size to the untreated control cells (Fig. 5, A and B). GW3965 also caused a significant decrease in the expression of TRAP/acp5 over the 6 days of culture (Fig. 5C), which was also reflected in the TRAP activity assay (Fig. 5D), but with little effect at the protein levels (Fig. 5E). This discrepancy between assays is likely attributed to the intracellular and secreted pools of TRAP being differentially regulated (37). Furthermore, minor effects of GW3965 were observed in cathepsin K and OSCAR mRNA/protein levels (Fig. 5, C and E). Thus, the LXR agonist appeared to be less effective in inhibiting differentiation in RAW264.7 cells when compared with both murine BMM and human CD14+ cells.
Analysis of signaling pathway genes showed that, similar to BMM, GW3965 caused a decrease in NFATc1 expression and protein (day 4) (Fig. 5, C and E) but had no effect on c-fms, rank, or c-fos (data not shown). Further analysis showed no differences in the expression of traf6, calcr, or mitf-e (data not shown). As RAW264.7 cells are more committed to the osteoclast lineage, we suggest that GW3965 acts early in differentiation and thus may be unable to inhibit as strongly in this cell type. Furthermore, a comparison of lxr mRNA levels between BMM and RAW264.7 cells at day 4 showed that lxrα and -β levels are significantly lower in RAW264.7 cells compared with BMM (lxrα, BMM 1 ± 0.18 versus RAW 0.2 ± 0.04, p value 0.01; lxrβ, BMM 1 ± 0.14 versus RAW 0.4 ± 0.06, p value 0.02; lxrα is expressed as 1-fold). Thus it is possible that RAW264.7 cells may also be less responsive to the agonist.
Western analysis at early time points following the addition of RANKL showed successful degradation of the NFκB inhibitor IκB-α from 0 to 30 min in RAW264.7 cells, with GW3965 having no effect on this response (Fig. 5F). Furthermore, GW3965 did not appear to affect the levels of RANKL-stimulated phosphorylated p38 (Fig. 5F). Thus, GW3965 seems to inhibit osteoclast differentiation in RAW264.7 cells independently of the p38 MAPK or NFκB pathways.
Effects of GW3965 on Calvarial Osteoclasts
To test whether GW3965 had any effects on another model of resorption, we used a stimulated calvarial organ culture. GW3965 had a marginal effect on basal release of 45Ca from cultured neonatal mouse calvarial bones and no effect when 45Ca release was stimulated by either all-trans-retinoic acid (ATRA) or 1,25-dihydroxyvitamin D3 (Table 1). Thus, it appears as if the inhibitory function of GW3965 may differ between periosteal osteoclast progenitors and those derived from the bone marrow.
TABLE 1.
Effect of GW3965 on 45CA release in neonatal mouse calvaria
Data are expressed as % release of 45Ca in 5 days of culture (means ± S.E., n = 5–6).
| Test substance | % release of 45Ca |
|---|---|
| Control | 100 ± 7 |
| GW3965 10−6m | 78 ± 5 |
| ATRA 10−7m | 187 ± 17 |
| ATRA + GW3965 | 161 ± 22 |
| 1,25-Dihydroxyvitamin D3 10−8m | 161 ± 19 |
| 1,25-Dihydroxyvitamin D3 + GW3965 | 165 ± 18 |
DISCUSSION
It is widely accepted that the majority of bone metabolic diseases are due to excessive osteoclastic activity, resulting in elevated bone resorption and destruction of bone. Studies indicate that activating the LXR appears to be beneficial in suppressing the bone destruction associated with severe arthritis (14, 15); however, the effects of an LXR agonist directly on osteoclast formation/function have not been studied. As LXR agonists are currently being investigated as potential therapeutic agents in a number of lipid and inflammatory diseases, it is important to understand their potential impact on skeletal biology. In this respect, our findings presented here that activation of LXRβ leads to potent suppression of osteoclast differentiation and bone resorption are of significant clinical relevance.
Differentiation of osteoclasts from monocytic precursors occurs via a well organized series of events beginning with M-CSF and followed by RANKL stimulation (38). The LXR agonist GW3965 inhibits differentiation of osteoclasts; however, it does not appear to act proximally to RANK signaling because it does not decrease the rank receptor or traf6 expression. Furthermore, downstream of RANK signaling, the NFκB pathway is activated normally in the presence of GW3965, with no difference in degradation of the IκB-α subunit.
RANK also cooperates with immunoreceptor tyrosine-based activation motifs that stimulate osteoclast differentiation via calcium signals (39). One of the receptors that associates with this complex is OSCAR (40). Although GW3965 inhibits the expression of OSCAR mRNA and protein in day 4 differentiated TRAP+ OCLs, this effect occurs late in differentiation (data not shown), so it is likely to be an indirect downstream effect. Finally, as we see no effect of GW3965 on the expression of c-fos mRNA, we suggest that LXR inhibits differentiation independently of c-Fos expression.
NFATc1, the key transcription factor required for the differentiation of osteoclasts (6), is significantly reduced after GW3965 treatment both at the level of RNA and protein, suggesting that the observed inhibitory effects on differentiation are mediated via this transcription factor. Further evidence for this contention was provided by the reduced expression of NFATc1-regulated osteoclast-specific genes as follows: TRAP/acp5, ctsk, oscar, and calcr (4, 35, 41–43). NFATc1 has been demonstrated to not only be a transcriptional partner of AP-1 (c-Fos) (4) but also of MITF and PU.1, with MITF:PU.1 regulating specific differentiation genes such as acp5, ctsk, oscar, and clcn7 (28, 30, 31, 34, 41, 42, 44). As mitf is also repressed after GW3965 treatment at day 2, as is its target gene clcn7, this transcription factor may also be involved in LXR-mediated inhibition of osteoclast differentiation. We further show that GW3965 acts independently of the MITF/PU.1 inhibitory protein Eos (45) and of USF-1 and -2 (32, 33).
The p38 MAPK pathway is particularly important in mediating osteoclast differentiation as it phosphorylates and activates transcription factors involved in differentiation such as NFATc1 (42) and MITF (46). As we observe an inhibition of phosphorylated p38 in BMM after activation of LXR, we suggest that the observed effects of GW3965 may involve the p38 MAPK pathway.
To attempt to further dissect at which stage of differentiation LXR is acting, we added GW3965 at different stages throughout the 4 days. It appears as if the time period most affected by GW3965 are days 2–3. This is likely to be the stage at which cells are fusing; however, most probably all cells in the culture are not in a homogeneous stage of differentiation. There was no effect at days 0–2 and only a weak effect at days 3–4. As we also observed a significant reduction in the expression of dc-stamp, the key player in osteoclast fusion (36, 47) and an NFATc1 target gene (48), it further suggests that GW3965 may be acting at the level of fusion.
We observed a similar inhibitory effect of GW3965 on osteoclast differentiation in human CD14+ cells with few mature multinucleated osteoclasts formed and, furthermore, a significant reduction in bone resorption. Interestingly, we also found that activation of LXR appears to retain a population of mononuclear cells that are able to phagocytose particles. However, it is unknown if these macrophage-like cells form as a direct result of GW3965 treatment or indirectly because of a reduced population of TRAP+-multinucleated cells. In mouse BMM and human CD14+ cells, the LXR agonist has a potent inhibitory effect on the differentiation of osteoclasts from monocyte/macrophage precursors, resulting in the formation of few mature osteoclasts and complete inhibition of bone resorption.
In RAW264.7 mouse macrophage cells, we observed that activation of LXR results in a significant decrease in the number of TRAP+ OCLs similar to BMM and CD14+ cells, with a significant decrease in NFATc1 and TRAP mRNA and protein; however, the overall effects were less significant. As lxrα and -β mRNA levels are both significantly lower in RAW264.7 cells compared with BMM, it may indicate that these cells are less responsive to the agonist at the dose given. In addition, as RAW264.7 cells are more committed to the osteoclast lineage, it is possible that these monocyte/macrophage cells are more resistant to the inhibitory effects of GW3965.
Furthermore, the action of the LXR agonist appears to act differently depending on the source of the precursor cells. For instance, we observed no effect of GW3965 on resorption in a stimulated (ATRA or dihydroxyvitamin D3) calvarial organ culture model. This assay measures only differentiation and activity of periosteal progenitor cells, as any mature osteoclasts that have remained in the 6–7-day-old calvariae are lost during the pre-culture period after exposure to basic medium and indomethacin (49). These progenitors are likely to represent a distinct pool from bone marrow progenitors and may either be derived from circulating committed progenitors or represent a pool of tissue macrophages (50). Heterogeneity among osteoclasts and osteoblasts in different parts of the skeleton has been recognized during recent years (51) consistent with our own findings that ATRA is an inhibitor of RANKL-induced osteoclastogenesis in BMM cultures (52), whereas ATRA stimulates osteoclastic genes and bone resorption in mouse calvariae (59).
Although we show that GW3965 likely inhibits the differentiation of osteoclasts by repressing NFATc1 in a mechanism that is independent of RANK and independent of NF-κB and c-Fos inhibition, but is likely to involve the p-38/MITF pathway, the exact mechanism is unclear. Chromatin immunoprecipitation (ChIP) analysis has turned out to be very useful in clearly outlining the early differentiation events regarding the induction of NFATc1, showing that within 1 h of RANKL stimulation, NFκB (p50/p65) and NFATc2 are recruited to the NFATc1 promoter (53). The function of NFκB in initiating differentiation is particularly interesting with respect to known LXR action. LXRs are able to inhibit NFκB-induced inflammatory responses in a SUMOylation-dependent mechanism (54, 55). After activation of LXR by GW3965, small ubiquitin-like modifier 2/3 is conjugated to the LXR ligand-binding domain. This targets LXR to NCoR repressor complexes and thus prevents the clearance of these complexes from the promoter of inflammatory genes, which in turn prevents NF-κB from activating transcription (56). Thus, it is possible that GW3965 could inhibit NFATc1 induction in osteoclasts by preventing NF-κB action in the nucleus.
Furthermore, we observed in our study that GW3965 acts specifically through LXRβ to inhibit osteoclast differentiation, with no effect of the agonist in BMM derived from LXRβ−/− mice. Such a subtype selectivity of GW3965 has been shown in a recent paper by Venteclef et al. (57), where the anti-inflammatory actions of GW3965 in the hepatic acute phase response are selectively mediated through LXRβ. Although we show in this study that the LXR agonist acts via LXRβ, both subtypes seem to be important in normal osteoclastogenesis with smaller osteoclasts formed in BMM derived from the LXRα−/− and -β−/− mice. We also showed previously using the LXR−/− mice that a nonfunctional LXRα and LXRβ both have a negative effect on osteoclasts, with a significant reduction in serum CTX detected in these mice (12). Osteoclast differentiation and activity are tightly regulated by osteoblasts/stromal cells through RANKL and other systemic factors. As these are global knockouts and both LXRα and -β are not only detectable in osteoclasts but also in osteoblasts (12) and numerous other cell types, the negative effect in vivo on the osteoclasts is likely to be influenced by the local and/or systemic environment.
In summary, this study reveals an important role of the LXR in inhibiting osteoclast differentiation and bone resorption in vitro. We observed that the LXR agonist GW3965 acts through LXRβ to profoundly inhibit osteoclast differentiation from mouse and human monocyte/macrophage precursors present in either bone marrow or blood circulation. This inhibition is likely to occur via the p38/MITF/NFATc1 pathway (see Fig. 6 for summary diagram), significantly inhibiting the formation of large multinucleated osteoclasts and impairing bone resorption. In this respect, use of an LXR agonist would likely have a beneficial therapeutic use in a clinical setting, for instance in various metabolic conditions also known to negatively affect bone turnover such as diabetes. However, as pharmacological activation of LXR also inhibits osteoblastogenesis (13), we propose that long term clinical use of an LXR agonist would not only affect reverse cholesterol transport and inflammatory pathways but could significantly and negatively affect bone remodeling. A long term negative effect on bone remodeling can also be observed with commonly used anti-resorptives, i.e. bisphosphonates (58). Although these drugs are clearly beneficial in inhibiting increased bone remodeling over several years, the long term treatment (10 years of more) is associated with atypical fractures because of the impaired repair of fatigued bone.
FIGURE 6.
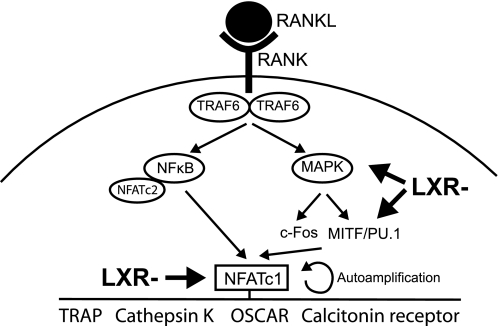
A schematic diagram of RANKL signaling, proposing where activated LXR may act to inhibit (LXR−) osteoclast differentiation.
Supplementary Material
Acknowledgments
We thank Maria Norgård for invaluable discussions and technical help and Ingrid Boström for technical assistance with the calvarial organ cultures.
This work was supported by the Swedish Research Council Grants K209-52X-10363-17-3 and 2006-17820-42007-12, the Robert A. Welch Foundation, the Karolinska Institute Foundation for Geriatric Diseases, the Karolinska Institute Research Foundation, the Apotekare Hedbergs Fund for Medical Research, the Swedish Rheumatism Association, and the Royal 80 Year Fund of King Gustav V.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
- RANKL
- receptor activator of nuclear factor κB ligand
- LXR
- liver X receptor
- RANK
- receptor activator of nuclear factor κB
- TRAP
- tartrate-resistant acid phosphatase
- calcr
- calcitonin receptor
- ANOVA
- analysis of variance
- BMM
- bone marrow macrophage
- OCL
- osteoclast
- α-MEM
- α-minimal essential medium
- M/R
- M-CSF/RANKL
- M
- M-CSF
- qPCR
- quantitative PCR
- ATRA
- all-trans-retinoic acid
- MITF
- microphthalmia-associated transcription factor
- CTX
- carboxyl-terminal cross-linking telopeptide of bone collagen.
REFERENCES
- 1. Boyle W. J., Simonet W. S., Lacey D. L. (2003) Nature 423, 337–342 [DOI] [PubMed] [Google Scholar]
- 2. Teitelbaum S. L. (2000) Science 289, 1504–1508 [DOI] [PubMed] [Google Scholar]
- 3. Asagiri M., Takayanagi H. (2007) Bone 40, 251–264 [DOI] [PubMed] [Google Scholar]
- 4. Takayanagi H., Kim S., Koga T., Nishina H., Isshiki M., Yoshida H., Saiura A., Isobe M., Yokochi T., Inoue J., Wagner E. F., Mak T. W., Kodama T., Taniguchi T. (2002) Dev. Cell 3, 889–901 [DOI] [PubMed] [Google Scholar]
- 5. Gohda J., Akiyama T., Koga T., Takayanagi H., Tanaka S., Inoue J. (2005) EMBO J. 24, 790–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Takayanagi H. (2007) Ann. N.Y. Acad. Sci. 1116, 227–237 [DOI] [PubMed] [Google Scholar]
- 7. Janowski B. A., Willy P. J., Devi T. R., Falck J. R., Mangelsdorf D. J. (1996) Nature 383, 728–731 [DOI] [PubMed] [Google Scholar]
- 8. Lehmann J. M., Kliewer S. A., Moore L. B., Smith-Oliver T. A., Oliver B. B., Su J. L., Sundseth S. S., Winegar D. A., Blanchard D. E., Spencer T. A., Willson T. M. (1997) J. Biol. Chem. 272, 3137–3140 [DOI] [PubMed] [Google Scholar]
- 9. Joseph S. B., McKilligin E., Pei L., Watson M. A., Collins A. R., Laffitte B. A., Chen M., Noh G., Goodman J., Hagger G. N., Tran J., Tippin T. K., Wang X., Lusis A. J., Hsueh W. A., Law R. E., Collins J. L., Willson T. M., Tontonoz P. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 7604–7609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tangirala R. K., Bischoff E. D., Joseph S. B., Wagner B. L., Walczak R., Laffitte B. A., Daige C. L., Thomas D., Heyman R. A., Mangelsdorf D. J., Wang X., Lusis A. J., Tontonoz P., Schulman I. G. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 11896–11901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Levin N., Bischoff E. D., Daige C. L., Thomas D., Vu C. T., Heyman R. A., Tangirala R. K., Schulman I. G. (2005) Arterioscler. Thromb. Vasc. Biol. 25, 135–142 [DOI] [PubMed] [Google Scholar]
- 12. Robertson K. M., Norgård M., Windahl S. H., Hultenby K., Ohlsson C., Andersson G., Gustafsson J. A. (2006) J. Bone Miner. Res. 21, 1276–1287 [DOI] [PubMed] [Google Scholar]
- 13. Prawitt J., Beil F. T., Marshall R. P., Bartelt A., Ruether W., Heeren J., Amling M., Staels B., Niemeier A. (2011) Bone 48, 339–346 [DOI] [PubMed] [Google Scholar]
- 14. Park M. C., Kwon Y. J., Chung S. J., Park Y. B., Lee S. K. (2010) Rheumatology 49, 882–890 [DOI] [PubMed] [Google Scholar]
- 15. Chintalacharuvu S. R., Sandusky G. E., Burris T. P., Burmer G. C., Nagpal S. (2007) Arthritis Rheum. 56, 1365–1367 [DOI] [PubMed] [Google Scholar]
- 16. Alberti S., Schuster G., Parini P., Feltkamp D., Diczfalusy U., Rudling M., Angelin B., Björkhem I., Pettersson S., Gustafsson J. A. (2001) J. Clin. Invest. 107, 565–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schuster G. U., Parini P., Wang L., Alberti S., Steffensen K. R., Hansson G. K., Angelin B., Gustafsson J. A. (2002) Circulation 106, 1147–1153 [DOI] [PubMed] [Google Scholar]
- 18. Granholm S., Lundberg P., Lerner U. H. (2007) J. Endocrinol. 195, 415–427 [DOI] [PubMed] [Google Scholar]
- 19. Takeshita S., Kaji K., Kudo A. (2000) J. Bone Miner. Res. 15, 1477–1488 [DOI] [PubMed] [Google Scholar]
- 20. Lu S. Y., Li M., Lin Y. L. (2010) Mol. Biol. Cell 21, 1763–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kawase Y., Hoshino T., Yokota K., Kuzuhara A., Nakamura M., Maeda Y., Nishiwaki E., Zenmyo M., Hiraoka K., Aizawa H., Yoshino K. (2003) J. Bone Miner. Res. 18, 975–983 [DOI] [PubMed] [Google Scholar]
- 22. Kinser S., Li M., Jia Q., Pestka J. J. (2005) J. Nutr. Biochem. 16, 88–95 [DOI] [PubMed] [Google Scholar]
- 23. Bandyopadhyay S., Lion J. M., Mentaverri R., Ricupero D. A., Kamel S., Romero J. R., Chattopadhyay N. (2006) Biochem. Pharmacol. 72, 184–197 [DOI] [PubMed] [Google Scholar]
- 24. Yamaza T., Goto T., Kamiya T., Kobayashi Y., Sakai H., Tanaka T. (1998) Bone 23, 499–509 [DOI] [PubMed] [Google Scholar]
- 25. Ek-Rylander B., Barkhem T., Ljusberg J., Ohman L., Andersson K. K., Andersson G. (1997) Biochem. J. 321, 305–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ljunggren O., Ransjö M., Lerner U. H. (1991) J. Bone Miner. Res. 6, 543–550 [DOI] [PubMed] [Google Scholar]
- 27. Lerner U. H. (1987) J. Bone Miner. Res. 2, 375–383 [DOI] [PubMed] [Google Scholar]
- 28. Luchin A., Purdom G., Murphy K., Clark M. Y., Angel N., Cassady A. I., Hume D. A., Ostrowski M. C. (2000) J. Bone Miner. Res. 15, 451–460 [DOI] [PubMed] [Google Scholar]
- 29. Mansky K. C., Sulzbacher S., Purdom G., Nelsen L., Hume D. A., Rehli M., Ostrowski M. C. (2002) J. Leukocyte Biol. 71, 304–310 [PubMed] [Google Scholar]
- 30. Motyckova G., Weilbaecher K. N., Horstmann M., Rieman D. J., Fisher D. Z., Fisher D. E. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 5798–5803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. So H., Rho J., Jeong D., Park R., Fisher D. E., Ostrowski M. C., Choi Y., Kim N. (2003) J. Biol. Chem. 278, 24209–24216 [DOI] [PubMed] [Google Scholar]
- 32. Liu Y., Shi Z., Silveira A., Liu J., Sawadogo M., Yang H., Feng X. (2003) J. Biol. Chem. 278, 20603–20611 [DOI] [PubMed] [Google Scholar]
- 33. Kim J. H., Kim K., Jin H. M., Youn B. U., Song I., Choi H. S., Kim N. (2008) J. Mol. Biol. 383, 502–511 [DOI] [PubMed] [Google Scholar]
- 34. Meadows N. A., Sharma S. M., Faulkner G. J., Ostrowski M. C., Hume D. A., Cassady A. I. (2007) J. Biol. Chem. 282, 1891–1904 [DOI] [PubMed] [Google Scholar]
- 35. Matsuo K., Galson D. L., Zhao C., Peng L., Laplace C., Wang K. Z., Bachler M. A., Amano H., Aburatani H., Ishikawa H., Wagner E. F. (2004) J. Biol. Chem. 279, 26475–26480 [DOI] [PubMed] [Google Scholar]
- 36. Kukita T., Wada N., Kukita A., Kakimoto T., Sandra F., Toh K., Nagata K., Iijima T., Horiuchi M., Matsusaki H., Hieshima K., Yoshie O., Nomiyama H. (2004) J. Exp. Med. 200, 941–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zenger S., Ek-Rylander B., Andersson G. (2010) Biochem. Biophys. Res. Commun. 394, 743–749 [DOI] [PubMed] [Google Scholar]
- 38. Shinohara M., Takayanagi H. (2007) Curr. Osteoporos. Rep. 5, 67–72 [DOI] [PubMed] [Google Scholar]
- 39. Koga T., Inui M., Inoue K., Kim S., Suematsu A., Kobayashi E., Iwata T., Ohnishi H., Matozaki T., Kodama T., Taniguchi T., Takayanagi H., Takai T. (2004) Nature 428, 758–763 [DOI] [PubMed] [Google Scholar]
- 40. Kim N., Takami M., Rho J., Josien R., Choi Y. (2002) J. Exp. Med. 195, 201–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kim Y., Sato K., Asagiri M., Morita I., Soma K., Takayanagi H. (2005) J. Biol. Chem. 280, 32905–32913 [DOI] [PubMed] [Google Scholar]
- 42. Matsumoto M., Kogawa M., Wada S., Takayanagi H., Tsujimoto M., Katayama S., Hisatake K., Nogi Y. (2004) J. Biol. Chem. 279, 45969–45979 [DOI] [PubMed] [Google Scholar]
- 43. Kim K., Kim J. H., Lee J., Jin H. M., Lee S. H., Fisher D. E., Kook H., Kim K. K., Choi Y., Kim N. (2005) J. Biol. Chem. 280, 35209–35216 [DOI] [PubMed] [Google Scholar]
- 44. Luchin A., Suchting S., Merson T., Rosol T. J., Hume D. A., Cassady A. I., Ostrowski M. C. (2001) J. Biol. Chem. 276, 36703–36710 [DOI] [PubMed] [Google Scholar]
- 45. Hu R., Sharma S. M., Bronisz A., Srinivasan R., Sankar U., Ostrowski M. C. (2007) Mol. Cell. Biol. 27, 4018–4027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mansky K. C., Sankar U., Han J., Ostrowski M. C. (2002) J. Biol. Chem. 277, 11077–11083 [DOI] [PubMed] [Google Scholar]
- 47. Yagi M., Miyamoto T., Sawatani Y., Iwamoto K., Hosogane N., Fujita N., Morita K., Ninomiya K., Suzuki T., Miyamoto K., Oike Y., Takeya M., Toyama Y., Suda T. (2005) J. Exp. Med. 202, 345–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kim K., Lee S. H., Ha Kim J., Choi Y., Kim N. (2008) Mol. Endocrinol. 22, 176–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lerner U. H., Johansson L., Ranjsö M., Rosenquist J. B., Reinholt F. P., Grubb A. (1997) Acta Physiol. Scand. 161, 81–92 [DOI] [PubMed] [Google Scholar]
- 50. Pettit A. R., Chang M. K., Hume D. A., Raggatt L. J. (2008) Bone 43, 976–982 [DOI] [PubMed] [Google Scholar]
- 51. Everts V., de Vries T. J., Helfrich M. H. (2009) Biochim. Biophys. Acta 1792, 757–765 [DOI] [PubMed] [Google Scholar]
- 52. Conaway H. H., Persson E., Halén M., Granholm S., Svensson O., Pettersson U., Lie A., Lerner U. H. (2009) FASEB J. 23, 3526–3538 [DOI] [PubMed] [Google Scholar]
- 53. Asagiri M., Sato K., Usami T., Ochi S., Nishina H., Yoshida H., Morita I., Wagner E. F., Mak T. W., Serfling E., Takayanagi H. (2005) J. Exp. Med. 202, 1261–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ogawa S., Lozach J., Benner C., Pascual G., Tangirala R. K., Westin S., Hoffmann A., Subramaniam S., David M., Rosenfeld M. G., Glass C. K. (2005) Cell 122, 707–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Treuter E., Gustafsson J. A. (2007) Mol. Cell 25, 178–180 [DOI] [PubMed] [Google Scholar]
- 56. Ghisletti S., Huang W., Ogawa S., Pascual G., Lin M. E., Willson T. M., Rosenfeld M. G., Glass C. K. (2007) Mol. Cell 25, 57–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Venteclef N., Jakobsson T., Ehrlund A., Damdimopoulos A., Mikkonen L., Ellis E., Nilsson L. M., Parini P., Jänne O. A., Gustafsson J. A., Steffensen K. R., Treuter E. (2010) Genes Dev. 24, 381–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Schilcher J., Michaëlsson K., Aspenberg P. (2011) N. Engl. J. Med. 364, 1728–1737 [DOI] [PubMed] [Google Scholar]
- 59. Conaway H. H., Pirhayati A., Persson E., Pettersson U., Svensson O., Lindholm C., Henning P., Tuckermann J., Lerner U. H. (June 29, 2011) J. Biol. Chem. 10.1074/jbc.M111247734 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.