Abstract
The second messenger cAMP is proapoptotic for numerous cell types, but the mechanism for this proapoptotic action is not defined. Here, we use murine CD4+/CD8+ S49 lymphoma cells and isolated thymocytes to assess this mechanism. In WT S49 cells, cAMP acts via protein kinase A (PKA) to induce G1 phase cell cycle arrest and apoptosis. Treatment of WT and cAMP-Deathless (D-) S49 cells, which lack cAMP-promoted apoptosis, with the PKA agonist 8-(4-chlorophenylthio)-cAMP (CPT-cAMP) differentially regulates transcripts for numerous proapoptotic and antiapoptotic proteins. In contrast, kin-S49 cells (which lack PKA) show no cAMP-promoted changes in transcript expression. In this study, we use knockdown and overexpression approaches to define the role in cAMP/PKA-promoted apoptosis of the proapoptotic factor BIM (Bcl-2 interacting mediator of cell death), whose expression prominently increases in response to CPT-cAMP treatment of WT but not D- or kin- S49 cells. Conditional expression of BimL, one of the three major forms of Bim, increases apoptosis of WT, D-, and kin-S49 cells, whereas inhibition of cAMP-mediated induction of Bim isoforms by shRNAi attenuates CPT-cAMP-mediated apoptosis of WT S49 cells. Bim protein levels increase in subpopulations of CPT-cAMP-treated cells that undergo apoptosis. Thymic CD4+/CD8+ cells isolated from Bim−/− mice corroborated the requirement of Bim expression for cAMP-promoted apoptosis. Thus, up-regulation of Bim appears to be an important determinant of cAMP/PKA-mediated apoptosis in immature T cells and may be a mechanism for such apoptosis in other cell types as well.
Keywords: Apoptosis, Cyclic AMP (cAMP), Lymphocyte, Lymphoma, Protein Kinase A (PKA), shRNA, T Cell, Bim−/−, Bim Isoform
Introduction
Apoptosis, programmed cell death, contributes to a variety of cellular processes that include embryonic development, tissue homeostasis, and immune responses. Apoptosis can be stimulated by a variety of “death stimuli,” including DNA damage, oxidative stress, hormones, cytokines, and drugs and is often targeted in cancer therapy. The second messenger cAMP has cell type-dependent effects on apoptosis (as recently reviewed (1)), being anti-apoptotic in certain cell types (e.g. neutrophils (2), eosinophils (3), hepatocytes (4), gastrointestinal epithelial cells (5), and several others) whereas having proapoptotic actions in other types of cells (e.g. cardiac myocytes (6) and certain lymphoid cells (7), in particular poorly differentiated lymphoblastic cells (8, 9)). Hematological malignancies, including large B cell lymphoma and chronic lymphocytic leukemia, are associated with a deficiency in apoptosis (10, 11). We and others have implicated the cAMP/PKA pathway as a promising one to enhance killing of lymphoma and leukemia cells (7, 8, 12–14). Despite the proapoptotic ability of cAMP/PKA, the mechanisms for this action are poorly defined.
Using murine S49 lymphoma cells, CD4+/CD8+ T cells that undergo growth arrest in the G1 phase of the cell cycle and apoptosis in response to cAMP-promoted activation of PKA2 (15), we identified mRNAs that differ between WT and kin- S49 cells, which lack PKA (16). In other studies, we showed that expression of certain apoptotic pathway members are differentially regulated in WT and cAMP-deathless (D-) S49 cells (7, 12), a clonal isolate that undergoes G1 arrest but is resistant to cAMP/PKA-promoted apoptosis (17).
The proapoptotic protein Bim, a BH3-only Bcl family member protein, shows a pronounced difference in expression between WT and D- cells (7, 12), with Bim expression being higher and longer in the WT cells treated with a cAMP analog or with a β-adrenergic agonist; moreover, increase in cAMP does not increase Bim expression in kin- cells (7, 12). Bim triggers apoptosis by promoting the release of cytochrome C from mitochondria. However, Bim may not be sufficient for inducing apoptosis, which can involve other BH3-only proteins (18). Multiple Bim isoforms exist (19), certain of which have been implicated in promoting T cell and B cell apoptosis (20). In this study, we used S49 cells (WT, D-, and kin- mutants) and CD4+/CD8+ thymocytes isolated from WT and Bim−/− mice to test whether Bim mediates cAMP/PKA-promoted apoptotic cell death in CD4+/CD8+ cells.
EXPERIMENTAL PROCEDURES
Materials
WT, kin-, and D- S49 cells were obtained from the University of San Francisco cell culture facility. 8-(4-chlorophenylthio)-adenosine-3′,5′-cyclic monophosphate (CPT-cAMP) was obtained from Sigma-Aldrich. BimL cDNA was purchased from Addgene (21). Protease inhibitor mixture was obtained from Sigma. NuPage 4–12% Bis-Tris gel, pENTR/H1/TO, pENTR-D TOPO, pENTR5′ TOPO, HEK293FT cells, ViraPower packaging, LR Clonase II enzyme mix, TRIzol, and the SuperScript III cDNA synthesis kit were obtained from Invitrogen. The annexin V apoptosis detection kit was obtained from BD Pharmingen, the pan T cell isolation kit from Miltenyi Biotec, Bim polyclonal antibody from BD Pharmingen, the subcellular proteome extraction kit from Calbiochem, and cleaved caspase 3 monoclonal antibody from Cell Signaling Technology, Inc. β-Tubulin polyclonal, Calnexin polyclonal, and goat anti-rabbit-HRP secondary antibodies were obtained from Abcam. The rtTA cDNA and tetO and EF1a promoters were kindly provided by Dr. Bruce Conklin (J. David Gladstone Institute, University of San Francisco). The 2K7Bsd lentiviral vector was kindly provided by Dr. David Suter, University of Geneva (22). Bim−/− mice were kindly provided by Dr. Gregg Silverman (University of California San Diego).
S49 Cell Culture
WT and D- S49 cells were seeded in suspension cultures at a density of 2.5 × 105 cells/ml with Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated horse serum in a humidified atmosphere containing 10% CO2 at 37 °C. Cells were incubated with 100 μm CPT-cAMP or 100 μm isoproterenol for the indicated times.
Generation of Bim shRNAi and Tetracycline-regulatable Constructs
Two Bim short hairpins RNAis were used for experiments that involved knockdown of Bim. Short hairpin sequences were designed with Invitrogen BLOCK-iTtm RNAi Designer. The following double stranded shRNAi oligonucleotides were used: shBim215, 5′-ggagacgagttcaacgaaactcgaaagtttcgttgaactcgtctcctttttt-3′; shBim272, 5′-gctgaagaccaccctcaaatgcgaacatttgagggtggtcttcagctttttt-3; and shBimScramble, 5′-ggttcagaggcagcacgtgcctcgagccacgtgctgcctctgaacctttttt-3′. The shBimScramble was designed with the GeneScript scramble software. The sequences were ligated into the pENTR/H1/TO plasmid according to the manufacturer's instructions. A three-way Gateway recombination (23) reaction was then carried out into the 2K7 lentiviral vector backbone (22) to generate the GFP/shRNAi-expressing vector depicted in Fig. 1B. To conditionally express BimL we employed a similar three-way Gateway strategy using the 2K7 vector backbone. The EF1a and tetO promoter elements were PCR-cloned into the pENTR5′ plasmids, and the rtTA transactivator (24) and the BimL cDNAs were PCR-cloned into the pENTR-D-TOPO vectors according to the manufacturer's instructions. These “entry” vectors were then used in three-way Gateway recombination reactions in the 2K7 vector backbones (22) to produce two lentiviral vectors, one with the EF1a promoter driving rtTA in the 2K7Bsd vector (rendering transduced cells resistant to blasticidin) and the other with the tetO response element upstream of the BimL cDNA in the 2K7Neo vector (rendering transduced cells resistant to neomycin).
FIGURE 1.
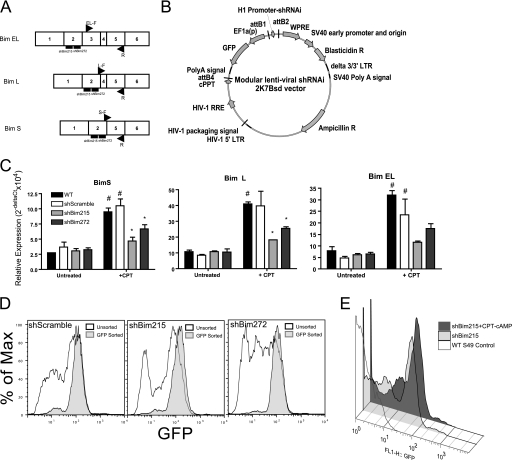
Bim isoform expression and shRNAi-mediated knockdown in WT S49 lymphoma cells. A, schematic of the Bim isoforms and isoform-specific real-time PCR primer and shRNAi target regions. B, graphic of modular 2K7Bsd (22) shRNAi plasmid that bicistronically expresses GFP. cPPT, central polypurine tract; WPRE, woodchuck hepatitis virus posttranscriptional regulator element; HIV-1 RRE, HIV-1 Rev response element; LTR, long terminal repeat. C, relative mRNA expression of Bim isoforms in WT, shBimScramble (control), shBim215, and shBim272 S49 cells, assessed by real-time PCR after 24-h treatment with 100 μm CPT-cAMP. #, p < 0.05 versus untreated; *, p < 0.05 versus +CPT shScramble (n = 3). D, histograms of GFP expression in shRNAi lines (shScramble, shBim215, and shBim272) before and after sorting cells for their GFP expression. E, overlay of histograms of GFP-positive cells (GFP reporter is bicistronically expressed in the shRNAi plasmid) in shBim215-expressing S49 cells treated for 48 h with 100 μm CPT-cAMP.
Transduction and Generation of Stable S49 Cells
Lentiviral particles were generated by cotransfection of HEK293FT cells with the lentivectors and ViraPower packaging mix in Optimem media according to the manufacturer's instructions. Two days after transfection, the extracellular medium was collected and filtered through a 0.45-μ filter. The filtrates were then spun at 25,000 rpm at 4 °C for 1.5 h. Lentiviral-containing pellets were resuspended in 150 μl of PBS with 6 μg/ml polybrene and mixed with 5 × 105 S49 cells. Cells and virus were spun (900 relative centrifugal force for 20 min), transferred to 1 ml of complete medium, and incubated overnight. Forty-eight hours after infection, cells were placed under selective conditions (10 μg/ml blasticidin, 500 μg/ml G418, or both) for approximately 2 weeks, a time when there was complete selection of mock-infected cells.
Isolation and Culture of Thymocytes
Thymuses were isolated from adult female C57BL/6 WT and Bim−/− mice and mechanically dissociated in 10 ml Hanks' Balanced Salt Solution (with 10 mm HEPES). Cells were pelleted (900 relative centrifugal force for 5 min) and washed once with MACS buffer (1× PBS, 1% FBS, 10 mm HEPES), repelleted, resuspended, and filtered (0.45-μ pore size). T cells were isolated from 107 total thymocytes with the pan T cell isolation kit (Miltenyi Biotec) according to the manufacturer's instructions. Isolated T cells were plated in 24-well plates at a concentration of 4 × 105 cells/ml in 1640 RPMI medium containing 30 ng/ml IL7; 0.1 mm nonessential amino acids; 1 mm sodium pyruvate; 10 mm HEPES; 0.1% β-mercaptoethanol; and 15% heat-inactivated, charcoal-stripped FBS. Cells were then cultured in a humidified incubator with 10% CO2 at 37 °C. All procedures were approved by the Institutional Animal Care and Use Committee and conform to the National Institutes of Health guidelines for animal research.
Real-time Quantitative RT-PCR
Total RNA was isolated from cells with TRIzol and converted to cDNA with a Super Script III cDNA synthesis kit according to the manufacturer's instructions. cDNA amplicons were quantified by incorporation of SYBR Green (Anaspec) into double-stranded DNA. Three Bim isoforms (EL, L, and S) were differentiated by real-time PCR using an identical reverse primer (R, 5′-TAA GTT TCG TTG AAC TCG TCT CC-3′) and unique forward primers (EL-F, 5′-GGT CCT CCA GTG GGT ATT TCT C-3′; L-F, 5′-GAA CCG CAA GAC AGG AGC CC-3′; and S-F, 5′-AGA ACC GCA AGC TTC CAT ACG AC-3′). We compared samples by using the relative (comparative) Ct method. Fold induction or fold repression was measured relative to controls and calculated after adjusting for 18 S RNA using 2−ΔΔCt, where ΔCt = Ct of the gene of interest - Ct 18 S and ΔΔ Ct = Δ Ct treatment - Δ Ct control.
Flow Cytometry and Assessment of Annexin V, CD4, and CD8 Labeling
Control and treated cells were grown in triplicate in 2 ml at 3 × 105/ml in 6-well plates for 24, 48, 72, or 96 h. Cells were then prepared for CD4-FITC, annexin V-PE, and CD8-APC staining according to the manufacturer's instructions as described (7). Briefly, 2 × 105 cells were stained in 100 ml of 1× staining buffer with 5 ml of annexin V-PE, 1 ml of CD4-FITC, and 1 ml of CD8-APC for 15 min at room temperature. Stained cells were assessed using a BD FacScan, and files were analyzed with FlowJo software.
Western Blotting
Cells were harvested by centrifugation, and proteins from whole cell lysates were solubilized with 200 ml of a radioimmune precipitation assay buffer (PBS, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1%SDS, 200 mm phenylmethylsulfonyl fluoride, 200 mm sodium orthovanadate, 1 mm DTT). Lysed cells were sonicated twice for 15 s. Subcellular fractions (cytosolic and mitochondrial/membrane fractions) were harvested using a subcellular proteome extraction kit. Lithium dodecyl sulfate loading buffer containing β-mercaptoethanol was added, and samples were heated (70 °C for 10 min). Protein lysates (20 μg) were electrophoresed in lanes on precast NuPage 4–12% Bis-Tris gels. Proteins separated from whole cell lysates were transferred onto PVDF membranes and then incubated with antibodies as indicated.
Statistical Analysis
Data are presented as mean ± S.E. and derived from an average of three independent experiments unless stated otherwise. The significance of differences between controls and experimental groups was determined by analysis of variance or Student's t tests. A value of p < 0.05 was considered statistically significant. Statistical analyses were performed with GraphPad Prism 4.0c (GraphPad Software, Inc.).
RESULTS
S49 lymphoma cells are CD4+/CD8+ T cells that undergo G1 cell cycle arrest and mitochondria-dependent apoptosis in response to activation of PKA by increases in endogenous cAMP or exogenous cAMP analogs (12, 15, 16). DNA microarray studies revealed that numerous transcripts change in response to increases in cAMP in WT S49 cells but not in kin- S49 cells that lack PKA. Certain transcripts are differentially expressed in WT S49 cells compared with D- S49 cells, which fail to undergo cAMP-promoted apoptosis. The expression of the proapoptotic protein BIM prominently increases in WT but not kin- or D- S49 cells (7, 16). We thus set out to test the hypothesis that BIM is a critical determinant of cAMP/PKA-promoted apoptosis in S49 cells and other CD4+/CD8+ cells. Accordingly, we sought to generate cells in which BIM expression was overexpressed or knocked down and then assessed the impact of these changes in BIM expression on apoptosis of the cells.
Bim Contributes to cAMP-mediated Apoptosis
Three isoforms of Bim (BimEL, BimL, and BimS) are generated via alternative splicing of exons 3 and 4 of the Bim transcript (25) (Fig. 1A). To assess the three isoforms by knocking down their expression, we created two Bim shRNAi lentiviral constructs (referred to hereafter as shBim215 and shBim272) that target exon 2 of Bim mRNA, which is common to all three isoforms. We also generated isoform-specific RT-PCR primers by flanking unique intron/exon junctions to each isoform (Fig. 1A).
We used the 2K7 lentiviral expression vector (22) to generate stable shBim-expressing WT S49 cells. This vector contains a central polypurine tract sequence element derived from the HIV-1 pol gene that increases transduction in T lymphocytes (26). We designed the vector to express Bim shRNAi and a GFP marker gene under control of the EF1a promoter, thus enabling us to track transduction efficiency and shRNAi expression in stably selected cells (Fig. 1B).
Stable expression of the shBim hairpins inhibited CPT-cAMP-induced up-regulation of all three Bim mRNA isoforms in S49 cells (Fig. 1C). BimL and BimEL are the predominant mRNA isoforms that are expressed. Bim mRNA expression was attenuated to a greater extent in the shBim215 line than the sh272 line. Basal levels of Bim mRNA were similar in the WT and shRNAi lines (Fig. 1C). We hypothesized that this might be attributable to heterogeneity in the knockdown of Bim in the shRNAi lines (because pools of selected cells were used) as a consequence of variability in lentiviral integration sites or potential silencing efficacy of shRNAi constructs. Because the shBim-expressing cell lines also expressed GFP, heterogeneity in GFP expression provided a means to indirectly assess shRNAi expression in the stably selected cells. We detected a moderately high GFP-negative population (40% ± 2.6) in the shScramble and shBim215 cell lines (Fig. 1D) and an even higher GFP-negative population in the shBim272 cell line (57% ± 0.65). The differences in the percentage of GFP+ cells in the shBim215 and shBim272 lines correlated with differences in extent of Bim knockdown in the two lines, likely explaining why greater knockdown of Bim mRNA occurs in the shBim215 line (Fig. 1C). Treatment of the shBim215 cells with CPT-cAMP decreased the GFP-negative population and enriched for GFP+ cells (Fig. 1E). We obtained similar results with the shBim272 line (data not shown). To focus on cells expressing the construct, we isolated the GFP+ population from each line by cell sorting (Fig. 1D). GFP-sorted cells retained enrichment for GFP+ over several passages. Thus, to use more homogeneous cell populations, we characterized the effect of repression of Bim (by shRNAi) in the GFP+-sorted lines.
The enriched population of GFP-positive shRNAi lines showed significantly (p < 0.05) lower levels of basal Bim expression in the shBim215 and shBim272 lines compared with the shScramble controls (Fig. 2A). Immunoblotting of membrane-enriched fractions of shScramble, shBim215, and shBim272 cells treated with 100 μm CPT-cAMP for 24 h revealed that the BimEL and BimL isoforms were repressed in shBim215 and shBim272 cells (Fig. 2B).
FIGURE 2.
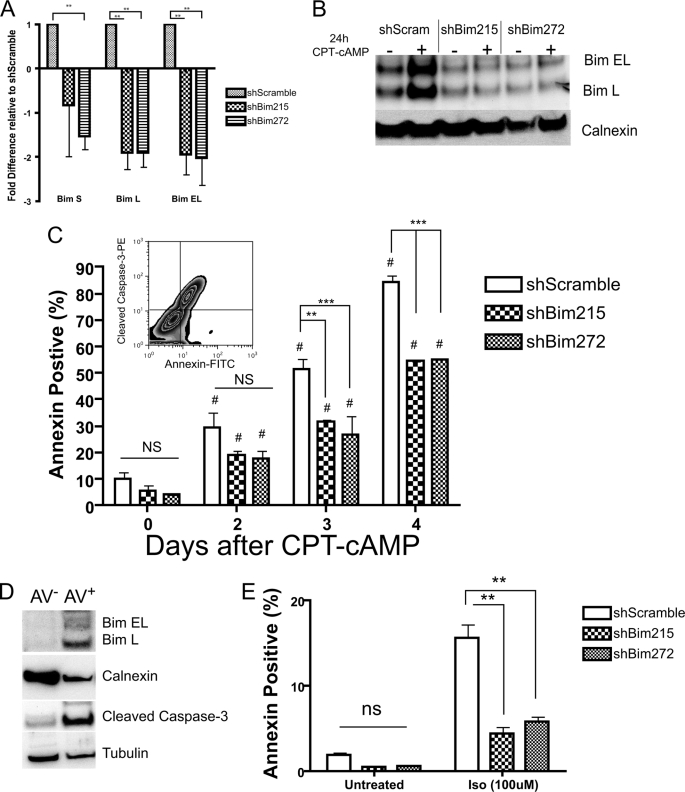
Up-regulation of Bim is important for cAMP/PKA-promoted apoptosis of S49 cells. A, freshly sorted GFP-positive shBim215 and 272 cells have lower basal levels of Bim mRNA than do shScramble (control) cells. **, p < 0.01. B, representative immunoblot analysis of Bim protein levels in membrane fractions of shBim RNAi lines 24 h after CPT-cAMP treatment. Calnexin was used as a loading control. C, quantification of annexin V-PE staining in shBim215, shBim272, and shScramble (control) S49 cells after treatment with 100 μm CPT-cAMP for the indicated times (n = 3). #, p < 0.05 versus untreated; **, p < 0.01; ***, p < 0.005; ns, NS, not significant versus +CPT shScramble (n = 3). The inset shows annexin V-positive cells containing cleaved caspase 3 expression. D, selective increase in Bim protein and cleaved caspase 3 in annexin V+ sorted cells. Cells were stimulated with 100 μm CPT-cAMP for 3 days and sorted for annexin V-positive (AV+) and -negative (AV-) populations. Membrane and soluble fractions were prepared and immunoblotted for Bim and cleaved caspase 3, respectively. Calnexin and tubulin were used as loading controls for membrane and soluble fractions, respectively. E, Bim knockdown protects against isoproterenol (Iso)-induced apoptosis. Cells were treated for 72 h with 100 μm Iso each 24 h. **, p < 0.005; ns, not significant.
We assessed for the induction of apoptosis by annexin V labeling and flow cytometry (Fig. 2C). Annexin V labeling detects plasma membrane alterations that occur during the early stages of apoptosis (27). Annexin V-positive S49 cells also contain cleaved caspase 3 (Fig. 2C, inset), a hallmark of apoptotic cells (28). Compared with shScramble control cells, both shBim215- and shBim272-expressing lines were significantly (p < 0.05) resistant to CPT-cAMP-promoted apoptosis (Fig. 2C).
We assessed the two different populations (annexin V-positive and annexin V-negative) that occur after CPT-cAMP treatment, hypothesizing that the annexin V-positive population would have higher levels of Bim protein and cleaved caspase 3 compared with annexin V-negative cells. We used cell sorting to purify each population of shScramble cells treated with CPT-cAMP for 72 h and found that the Annexin-V-positive cells had increased levels of Bim and cleaved caspase 3 protein (Fig. 2D).
We also examined the effect of increasing endogenous cAMP levels on cell viability by stimulating β-adrenergic receptors with the agonist isoproterenol (100 μm). Both shBim215- and shBim272-expressing S49 cells were protected from apoptosis induced by isoproterenol compared with what occurred in response to similar treatment of WT cells (Fig. 2E).
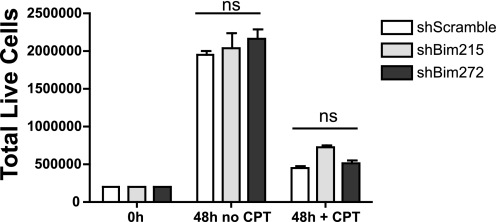
Knockdown of BIM Does Not Alter Cell Growth Kinetics of S49 Cells
Treatment of WT S49 cells induces growth arrest in the G1 phase of the cell cycle. G1 growth arrest occurs within 24 h of PKA activation, thus preceding apoptosis (12, 29). Because hematopoietic progenitor cells from Bim knockout mice have a delayed transition from G1 to S phase (30), we assessed whether Bim knockdown by shRNAi alters the growth kinetics of WT S49 cells. We found that Bim shRNAi-expressing and shScramble control cells have similar cell growth kinetics (Fig. 3).
FIGURE 3.
Bim knockdown blunts CPT-cAMP-promoted apoptosis but does not alter cAMP-mediated growth arrest. S49 cell lines were plated at a density of 2.5 × 105 cells/ml and grown in the presence or absence of 100 μm CPT-cAMP for 48 h. Total live cells were assessed at 48 h. ns, not significant.
These results are consistent with evidence that the induction of G1 arrest is independent of cAMP/PKA-promoted apoptosis in S49 cells and thus occurs by mechanisms with different kinetics, G1 arrest occurring within 24h in the majority of cells and apoptosis occurring more gradually. Rapid growth arrest that is followed by gradual increases in apoptosis also occurs in human epidermoid carcinoma (A431) (31), murine splenocytes (32) and myeloid leukemia cells (33).
Up-regulation of Bim Induces Apoptosis in S49 Cells and Correlates with Increases in Bim Protein Levels
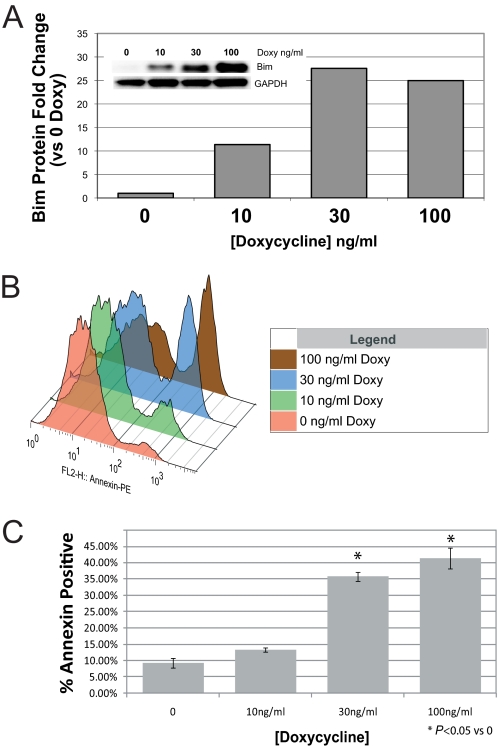
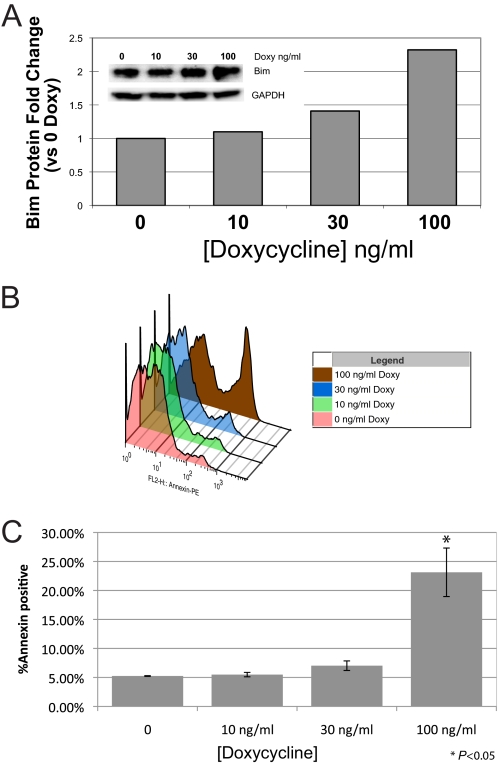
To determine whether up-regulation of BimL induces apoptosis of S49 cells, we generated double-transgenic S49 cells that stably express the reverse tetracycline transactivator (rtTA) under control of the Elongation Factor 1 promoter (EF1a) and the tetO response element upstream of the BimL cDNA so as to regulate BimL expression with doxycycline (Tet-On) (24).
Double-transgenic WT (Fig. 4A) and D- (Fig. 5A) S49 cells displayed a doxycycline-dependent increase in BimL protein levels. Alterations in the doxycycline response observed between WT and D- cells likely results from variations in viral titer and multiplicity of infection, as evidenced by the longer recovery time of D- cells during selection after viral transduction. The dose-dependent increase in BimL protein levels correlated closely with the number of WT (Fig. 4) and D- (Fig. 5) cells undergoing apoptosis. Similar results were observed in double transgenic kin- cells (data not shown). As a control, we treated single transgenic EF1a-rtTA-expressing WT cells with a similar range of doxycycline (up to 1 μg/ml) and found no increase in annexin V labeling (data not shown). These results indicate that increased expression of BimL induces apoptosis of WT, D-, and kin- S49 cells and imply that an inability of D- and kin- cells to increase Bim expression in response to cAMP accounts for their lack of cAMP/PKA-promoted apoptosis.
FIGURE 4.
Up-regulation of BimL induces apoptosis in WT S49 cells. WT S49 cells were stably transduced with lentiviral vectors containing the reverse tetracycline transactivator rtTA driven by the EF1a promoter (EF1a-rtTA) and BimL under control of the tetracycline-responsive element (TetO-BimL). Double-transgenic cells were stimulated for 24 h with the indicated concentrations of doxycycline (Doxy) and then assessed for BimL protein expression on the basis of data from the immunoblot shown in the inset and with quantitation by densitometry (A), representative data of annexin V staining with increasing concentrations of doxycycline after 24 h (B), and quantification of annexin V staining (n = 3) (C). The inset represents an overlay of representative histograms of annexin V-stained cells treated with increasing doses of doxycycline. *, p < 0.05 compared with time 0.
FIGURE 5.
Up-regulation of BimL induces apoptosis in D- S49 cells. D- S49 cells were stably transduced with lentiviral vectors and stimulated for 24 h with the indicated concentrations of doxycycline (Doxy) and then assessed for BimL protein expression on the basis of data from the immunoblot analysis shown in the inset and with quantitation by densitometry (A), representative data of annexin V staining with increasing concentrations of doxycycline after 24 h (B), and quantification of annexin V staining (n = 3) (C). The inset represents an overlay of representative histograms of annexin V-stained cells treated with increasing doses of doxycycline. *, p < 0.05 compared with time 0.
Bim Expression Is Required for cAMP-mediated Apoptosis of CD4+/CD8+ Thymocytes
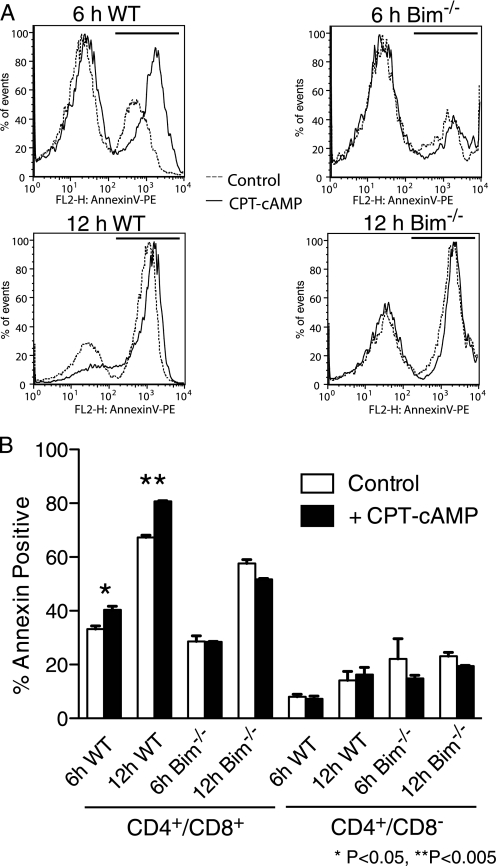
We isolated T cells from WT and Bim−/− knockout mice (34) to determine whether the cAMP pathway mediates apoptosis in primary isolates of different populations of T cells. We found that CPT-cAMP significantly increases apoptosis of WT CD4+/CD8+ thymocytes but has no effect on Bim−/− CD4+/CD8+ thymocytes. Moreover, the apoptotic sensitivity to CPT-cAMP is specific to CD4+/CD8+ double-positive because WT and Bim−/− CD4+/CD8− thymocytes showed no response to CPT-cAMP (Fig. 6). Thus, PKA activation selectively increases the apoptosis of double-positive, i.e. immature, thymocytes.
FIGURE 6.
Bim is required for CPT-cAMP-mediated apoptosis of CD4+/CD8+ thymocytes. T cells were isolated from Bim+/+ (WT) and Bim−/− mice, incubated with CPT-cAMP, and then assessed for apoptosis, as determined by the expression of annexin V. A, representative annexin V histograms of CD4+/CD8+-gated cell populations isolated from WT C57Bl6 and Bim−/− mice incubated for 6 h and 12 h ± 100 μm CPT-cAMP. The solid lines indicate cells treated with 100 μm CPT-cAMP, and the dashed lines represent vehicle-treated cells. Annexin V-positive gates are represented as solid lines above the histograms. B, quantification of annexin V staining in CD4+/CD8+ double-positive and CD4+/CD8− single-positive thymocytes shows enhanced CPT-cAMP-promoted apoptosis of the double-positive cells. n = 3 replicates of a representative experiment. *, p < 0.05; **, p < 0.005.
DISCUSSION
WT S49 cells, but not kin- cells that lack PKA activity, show numerous changes in expression of transcripts in response to treatment with CPT-cAMP, thus indicating that such changes in S49 cells are mediated by PKA and not other cAMP-regulated signaling pathways (e.g. Epac activation) (16). We also identified transcripts of pro- and antiapoptotic genes that are differentially regulated by CPT-cAMP treatment of WT and D- S49 cells (7, 12). On the basis of their resistance to apoptosis but not other effects of cAMP (17), the D- cells enabled us to focus on transcripts that mediate proapoptotic effects of cAMP/PKA. WT, but not D-, cells show up-regulation of the proapoptotic gene Bim 24 h after CPT-cAMP treatment, suggesting that Bim may mediate cAMP killing (7). The current data show that the transcriptional regulation of Bim by PKA is likely to be an important mechanism for cAMP-promoted apoptosis of S49 cells.
CD4+/CD8+ thymocytes (the phenotype as WT S49 cells (35)) are more susceptible to a variety of death stimuli compared with CD4 or CD8 single-positive cells (36). The current findings support this idea because CD4+/CD8+ thymocytes display higher levels of basal apoptosis than do CD4+/CD8− thymocytes. In vitro culture conditions likely induce the high levels of basal apoptosis in CD4+/CD8+ thymocytes (Fig. 6, 37, 38). In light of this, we modified culture conditions in an attempt to increase thymocyte viability (i.e. by increasing IL7 and lowering glucose levels), but these maneuvers did not improve viability (data not shown). CD4+/CD8+ thymocytes express high levels of the death receptor Fas (37, 39, 40). Activating the Fas pathway can induce apoptosis of CD4+/CD8+ thymocytes independent of activation or induction of Bim (37).
It has been postulated that double-positive thymocytes are “primed for death” and contain mitochondria that are enriched with activators of mitochondrial outer membrane permeabilization (e.g. BAX, BAK) (41). This priming renders the mitochondria more sensitive to exposure to BH3-only domain proteins (such as Bim). The failure of Bim−/− cells to prime their mitochondria is postulated to result in the loss of negative selection during T cell maturation and autoimmunity (36, 42). Our results suggest that the cAMP/PKA pathway, which is controlled by numerous extracellular signaling molecules through G protein-coupled receptor activation, may contribute to the negative selection of thymocytes.
Consistent with this idea, the innervation of the thymus by the sympathetic nervous system increases postnatally (43). Little is known, however, about the physiological role of this sympathetic innervation. Studies in S49 cells suggest that sympathetic nerve termini may trigger apoptosis of CD4+/CD8+ thymocytes by releasing neurotransmitters that increase intracellular cAMP levels (e.g. norepinephrine), in turn activating PKA and up-regulating Bim. Our results with thymocytes from Bim knockout mice support this possibility (Fig. 6). cAMP/PKA induces apoptosis in CD4+/CD8+ cells but not single-positive cells. Such results suggest that increases in cAMP act via PKA to promote the negative selection of CD4+/CD8+ cells in vivo.
Bim is regulated by transcriptional and posttranscriptional events, including phosphorylation and ubiquitination (18). For example, Bim can be phosphorylated by ERK and then targeted for ubiquitin-mediated degradation, thereby protecting cells from apoptosis (44). Results in S49 cells imply that cAMP/PKA regulate Bim at the transcriptional level because apoptosis commences 24 h after treatment with CPT-cAMP (Fig. 3A), a time that correlates with accumulation of Bim protein (12). The PKA-regulated transcription factors ATF1 and cAMP response element-binding protein may regulate Bim. Using the RVista analysis tool (45), we identify an evolutionarily conserved cAMP response element approximately 1.5 kb upstream from the transcriptional start site of Bim, suggesting that it may be targeted by cAMP/PKA-regulated transcription factors.
The data shown here raise the possibility that Bim may contribute to cAMP/PKA-mediated cell death in other settings where activation of this signaling pathway induces apoptosis (e.g. cardiac myocytes (6) and chronic lymphocytic leukemia (8, 9)). However, further studies are required to test this idea. Glucocorticoids also can induce Bim expression in lymphoid cells, including S49 cells (12, 46–48) and can promote apoptosis of certain lymphoid cells. Such effects can be additive or synergistic with cAMP/PKA (49, 50).
The current results thus demonstrate that the transcriptional regulation of Bim is a key step in PKA-regulated apoptosis of S49 cells and, by inference, other cell types in which cAMP/PKA are proapoptotic. Altered transcriptional kinetics of Bim also define the basis of the cAMP-deathless phenotype in D- S49 cells. As such, these cells, as well as kin- cells, should be useful in further studies of how cAMP/PKA control the transcription of Bim as a forerunner of apoptosis initiated by cAMP/PKA in S49 cells and potentially other cell types.
Acknowledgment
We thank Angela Baja for technical assistance.
This work was supported, in whole or in part, by IRACDA National Institutes of Health Grant GM068524 and research grants. This work was also supported by American Heart Association Scientist Development Grant 10SDG2630130 and research grants from the Lymphoma and Leukemia Society.
- PKA
- protein kinase A
- D-
- deathless
- CPT-cAMP
- 8-(4-chlorophenylthio)-cAMP
- rtTA
- reverse tetracycline transactivate.
REFERENCES
- 1. Insel P. A., Zhang L., Murray F., Yokouchi H., Zambon A. C. (2011) Acta Physiol. doi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Martin M. C., Dransfield I., Haslett C., Rossi A. G. (2001) J. Biol. Chem. 276, 45041–45050 [DOI] [PubMed] [Google Scholar]
- 3. Chang H. S., Jeon K. W., Kim Y. H., Chung I. Y., Park C. S. (2000) Cell. Immunol. 203, 29–38 [DOI] [PubMed] [Google Scholar]
- 4. Wang Y., Kim P. K., Peng X., Loughran P., Vodovotz Y., Zhang B., Billiar T. R. (2006) Apoptosis 11, 441–451 [DOI] [PubMed] [Google Scholar]
- 5. Nishihara H., Kizaka-Kondoh S., Insel P. A., Eckmann L. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 8921–8926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Singh K., Xiao L., Remondino A., Sawyer D. B., Colucci W. S. (2001) J. Cell. Physiol. 189, 257–265 [DOI] [PubMed] [Google Scholar]
- 7. Zhang L., Zambon A. C., Vranizan K., Pothula K., Conklin B. R., Insel P. A. (2008) J. Biol. Chem. 283, 4304–4313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lerner A., Epstein P. M. (2006) Biochem. J. 393, 21–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang L., Murray F., Zahno A., Kanter J. R., Chou D., Suda R., Fenlon M., Rassenti L., Cottam H., Kipps T. J., Insel P. A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 19532–19537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cillessen S. A., Meijer C. J., Notoya M., Ossenkoppele G. J., Oudejans J. J. (2010) J. Pathol. 220, 509–520 [DOI] [PubMed] [Google Scholar]
- 11. Danilov A. V., Danilova O. V., Klein A. K., Huber B. T. (2006) Curr. Mol. Med. 6, 665–675 [DOI] [PubMed] [Google Scholar]
- 12. Zhang L., Insel P. A. (2004) J. Biol. Chem. 279, 20858–20865 [DOI] [PubMed] [Google Scholar]
- 13. Moon E. Y., Lerner A. (2003) Blood 101, 4122–4130 [DOI] [PubMed] [Google Scholar]
- 14. Naviglio S., Caraglia M., Abbruzzese A., Chiosi E., Di Gesto D., Marra M., Romano M., Sorrentino A., Sorvillo L., Spina A., Illiano G. (2009) Expert Opin. Ther. Targets 13, 83–92 [DOI] [PubMed] [Google Scholar]
- 15. Coffino P., Bourne H. R., Tomkins G. M. (1975) Am J. Pathol. 81, 199–204 [PMC free article] [PubMed] [Google Scholar]
- 16. Zambon A. C., Zhang L., Minovitsky S., Kanter J. R., Prabhakar S., Salomonis N., Vranizan K., Dubchak I., Conklin B. R., Insel P. A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 8561–8566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lemaire I., Coffino P. (1977) Cell 11, 149–155 [DOI] [PubMed] [Google Scholar]
- 18. Gillings A. S., Balmanno K., Wiggins C. M., Johnson M., Cook S. J. (2009) FEBS J. 276, 6050–6062 [DOI] [PubMed] [Google Scholar]
- 19. Miao J., Chen G. G., Yun J. P., Chun S. Y., Zheng Z. Z., Ho R. L., Chak E. C., Xia N. S., Lai P. B. (2007) Apoptosis 12, 1691–1701 [DOI] [PubMed] [Google Scholar]
- 20. Akiyama T., Dass C. R., Choong P. F. (2009) Mol. Cancer Ther. 8, 3173–3180 [DOI] [PubMed] [Google Scholar]
- 21. Harada H., Quearry B., Ruiz-Vela A., Korsmeyer S. J. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 15313–15317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Suter D. M., Cartier L., Bettiol E., Tirefort D., Jaconi M. E., Dubois-Dauphin M., Krause K. H. (2006) Stem Cells 24, 615–623 [DOI] [PubMed] [Google Scholar]
- 23. Magnani E., Bartling L., Hake S. (2006) BMC Mol. Biol. 7, 46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gossen M., Freundlieb S., Bender G., Müller G., Hillen W., Bujard H. (1995) Science 268, 1766–1769 [DOI] [PubMed] [Google Scholar]
- 25. Bouillet P., Zhang L. C., Huang D. C., Webb G. C., Bottema C. D., Shore P., Eyre H. J., Sutherland G. R., Adams J. M. (2001) Mamm. Genome 12, 163–168 [DOI] [PubMed] [Google Scholar]
- 26. Manganini M., Serafini M., Bambacioni F., Casati C., Erba E., Follenzi A., Naldini L., Bernasconi S., Gaipa G., Rambaldi A., Biondi A., Golay J., Introna M. (2002) Hum. Gene Ther. 13, 1793–1807 [DOI] [PubMed] [Google Scholar]
- 27. Vermes I., Haanen C., Steffens-Nakken H., Reutelingsperger C. (1995) J. Immunol. Methods 184, 39–51 [DOI] [PubMed] [Google Scholar]
- 28. Jänicke R. U., Sprengart M. L., Wati M. R., Porter A. G. (1998) J. Biol. Chem. 273, 9357–9360 [DOI] [PubMed] [Google Scholar]
- 29. Coffino P., Gray J. W. (1978) Cancer Res. 38, 4285–4288 [PubMed] [Google Scholar]
- 30. Kozuma Y., Ninomiya H., Murata S., Kono T., Mukai H. Y., Kojima H. (2010) J. Thromb. Haemost. 8, 1088–1097 [DOI] [PubMed] [Google Scholar]
- 31. Mantena S. K., Sharma S. D., Katiyar S. K. (2006) Carcinogenesis 27, 2018–2027 [DOI] [PubMed] [Google Scholar]
- 32. Heinrichs S., Deppert W. (2003) Oncogene 22, 555–571 [DOI] [PubMed] [Google Scholar]
- 33. Gunji H., Hass R., Kufe D. (1992) J. Clin. Invest. 89, 954–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bouillet P., Metcalf D., Huang D. C., Tarlinton D. M., Kay T. W., Köntgen F., Adams J. M., Strasser A. (1999) Science 286, 1735–1738 [DOI] [PubMed] [Google Scholar]
- 35. Yan L., Herrmann V., Hofer J. K., Insel P. A. (2000) Am. J. Physiol. Cell Physiol. 279, C1665–1674 [DOI] [PubMed] [Google Scholar]
- 36. Ryan J. A., Brunelle J. K., Letai A. (2010) Proc. Natl. Acad. Sci. U.S.A. doi: 10.1073/pnas.0914878107 [DOI] [Google Scholar]
- 37. Xing Y., Wang X., Igarashi H., Kawamoto H., Sakaguchi N. (2008) Mol. Immunol. 45, 2028–2037 [DOI] [PubMed] [Google Scholar]
- 38. Ivanov V. N., Nikolić-Zugić J. (1997) J. Biol. Chem. 272, 8558–8566 [PubMed] [Google Scholar]
- 39. Drappa J., Brot N., Elkon K. B. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 10340–10344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ogasawara J., Suda T., Nagata S. (1995) J. Exp. Med. 181, 485–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Häcker G., Weber A. (2007) Arch. Biochem. Biophys. 462, 150–155 [DOI] [PubMed] [Google Scholar]
- 42. Bouillet P., Purton J. F., Godfrey D. I., Zhang L. C., Coultas L., Puthalakath H., Pellegrini M., Cory S., Adams J. M., Strasser A. (2002) Nature 415, 922–926 [DOI] [PubMed] [Google Scholar]
- 43. Leposavić G., Mićić M., Ugresić N., Bogojević M., Isaković K. (1992) Thymus. 19, 77–87 [PubMed] [Google Scholar]
- 44. Ley R., Balmanno K., Hadfield K., Weston C., Cook S. J. (2003) J. Biol. Chem. 278, 18811–18816 [DOI] [PubMed] [Google Scholar]
- 45. Loots G. G., Ovcharenko I., Pachter L., Dubchak I., Rubin E. M. (2002) Genome Res. 12, 832–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kfir-Erenfeld S., Sionov R. V., Spokoini R., Cohen O., Yefenof E. (2010) Leuk. Lymphoma 51, 1968–2005 [DOI] [PubMed] [Google Scholar]
- 47. Molitoris J. K., McColl K. S., Distelhorst C. W. (2011) Mol. Endocrinol. 25, 409–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang Z., Malone M. H., He H., McColl K. S., Distelhorst C. W. (2003) J. Biol. Chem. 278, 23861–23867 [DOI] [PubMed] [Google Scholar]
- 49. Ji Z., Mei F. C., Miller A. L., Thompson E. B., Cheng X. (2008) J. Biol. Chem. 283, 21920–21925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Medh R. D., Saeed M. F., Johnson B. H., Thompson E. B. (1998) Cancer Res. 58, 3684–3693 [PubMed] [Google Scholar]