Background: Proteolytic cleavage of MerTK leads to inhibition of thrombosis and efferocytosis.
Results: In macrophages, lipopolysaccharide required reactive oxygen species to activate protein kinase Cdelta and then p38 MAPK, culminating in ADAM17-mediated proteolysis of MerTK at proline 485.
Conclusion: ADAM17 is a key protease required during pattern recognition receptor-induced MerTK cleavage.
Significance: These findings uncover targets to test the consequences of MerTK cleavage in vivo.
Keywords: ADAM ADAMTS, Lipopolysaccharide (LPS), Macrophages, Phagocytosis, Shedding, MerTK, PKCdelta, ROS, TACE, Efferocytosis
Abstract
Mer tyrosine kinase (MerTK) is an integral membrane protein that is preferentially expressed by phagocytic cells, where it promotes efferocytosis and inhibits inflammatory signaling. Proteolytic cleavage of MerTK at an unidentified site leads to shedding of its soluble ectodomain (soluble MER; sMER), which can inhibit thrombosis in mice and efferocytosis in vitro. Herein, we show that MerTK is cleaved at proline 485 in murine macrophages. Site-directed deletion of 6 amino acids spanning proline 485 rendered MerTK resistant to proteolysis and suppression of efferocytosis by cleavage-inducing stimuli. LPS is a known inducer of MerTK cleavage, and the intracellular signaling pathways required for this action are unknown. LPS/TLR4-mediated generation of sMER required disintegrin and metalloproteinase ADAM17 and was independent of Myd88, instead requiring TRIF adaptor signaling. LPS-induced cleavage was suppressed by deficiency of NADPH oxidase 2 (Nox2) and PKCδ. The addition of the antioxidant N-acetyl cysteine inhibited PKCδ, and silencing of PKCδ inhibited MAPK p38, which was also required. In a mouse model of endotoxemia, we discovered that LPS induced plasma sMER, and this was suppressed by Adam17 deficiency. Thus, a TRIF-mediated pattern recognition receptor signaling cascade requires NADPH oxidase to activate PKCδ and then p38, culminating in ADAM17-mediated proteolysis of MerTK. These findings link innate pattern recognition receptor signaling to proteolytic inactivation of MerTK and generation of sMER and uncover targets to test how MerTK cleavage affects efferocytosis efficiency and inflammation resolution in vivo.
Introduction
MerTK (also known as c-Eyk, Nyk, and Tyro12) is a tyrosine kinase receptor for the growth arrest-specific protein GAS6 and anticoagulant Protein S (1, 2). Engagement of MerTK with either GAS6 or Protein S has been linked to numerous functions, including cell survival, thrombosis, and the phagocytosis of apoptotic cells (efferocytosis) (3–5). In the case of efferocytosis, both GAS6 and Protein S serve as bridging molecules that link MerTK to phosphatidylserine on dying cells (6). This leads to activation of intracellular signaling pathways that culminate in actin-driven apoptotic cell engulfment (7). MerTK is expressed predominantly in monocytic, epithelial, and reproductive tissue (8). In epithelial cells of the eye, naturally occurring mutations in Mertk are associated with onset of autosomal recessive retinitis pigmentosa (9). This is due to a defect of retinal pigment epithelial cells to promote clearance of adjacent light-sensing photoreceptor outer segments (10). Defects in MerTK are linked to other disease phenotypes. For example, in rodents, apoptotic thymocyte removal is defective in mice carrying a kinase-dead Mertk (MertkKD) (3). MerTK deficiency in turn promotes autoantibody production and can stimulate lupus-like autoimmunity (11). Our group has shown that Mertk deficiency promotes defective efferocytosis that is associated with increased vascular wall necrosis in advanced atherosclerotic plaque (12). Thus, MerTK has a critical anti-inflammatory role in a number of clinically relevant disease states.
At the structural level, MerTK is a type I transmembrane (TM)3 protein that encodes four extracellular domains: two fibronectin type-III domains and two extracellular immunoglobulin-like domains (13). Its cytoplasmic tail encodes a tyrosine kinase and controls distinct and separable effects that promote efferocytosis and inflammation resolution (8, 14). This domain homology is shared by two other molecules, Axl and Tyro3, to make up the TAM receptor family of tyrosine kinases (15). In some cases, a truncated TM-less isoform of MerTK can be generated by alternative mRNA splicing. A recent report by Sather et al. (16) indicates that a soluble form of MER (sMER) can also be induced through proteolytic cleavage of its ectodomain, leaving behind a carboxyl-terminal portion of the cleaved MER that remains cell-associated. In addition, sMER that is shed can act as a competitive inhibitor of MerTK during efferocytosis and platelet aggregation by acting as a decoy for its ligand GAS6. In our own hands (17) and others (18), sMER has been identified in inflammatory cardiovascular lesions, a disease linked to defective efferocytosis.
The identification of sMER seats MerTK in a growing list of TM-anchored protein receptors that are regulated by proteolytic shedding (19). Shedding of these cell surface proteins is often catalyzed by metalloproteinases. In the case of TAM receptor tyrosine kinases, mass spectrometric analysis indicates that closely related AXL is cleaved by the metalloproteinase ADAM17 (20). ADAM17 is a disintegrin and metalloproteinase, otherwise known as tumor necrosis factor (TNF)-α-converting enzyme (TACE), for its role in cleaving and releasing active TNFα (21, 22). Although the degradome of ADAM17 indicates a wide range of susceptible substrate proteins, our mechanistic understanding of ADAM17 activation remains incomplete (23, 24). Furthermore, the majority of studies that investigate ADAM17 activity utilize non-physiological inducers, such as the phorbol ester phorbol 12-myristate 13-acetate (PMA). Therefore, an important objective is to determine how physiologic stimuli may differentially signal to activate ADAM17 in health and disease.
Our interest in ADAM17 and mechanisms of MerTK shedding originate from our studies of MerTK-mediated efferocytosis and macrophage responses to pathogen-associated molecular patterns (25). Herein we report the identification of the MerTK proteolytic cleavage site and conclusively show that ADAM17 is the key protease required for sMER shedding induced by the pattern recognition receptor ligand lipopolysaccharide (LPS). We also for the first time elucidate the key signaling intermediates between Toll-like receptor 4 (TLR4) and ADAM17-mediated MerTK cleavage and discuss the in vivo implications of these findings in the context of inflammatory disease.
MATERIALS AND METHODS
Reagents
Antibodies
Polyclonal goat anti-mouse MERTK was from R&D (catalogue no. AF591). For immunoblots, anti-goat IgG-HRP was also from R&D (catalogue no. HAF109). Rabbit polyclonal antibody to ADAM17 was from ABCAM (ab2051). Rabbit antibody to ADAM17 (phospho-Thr735) at 1 mg/ml stock was from ABCAM (ab60996). Adam10 antibody was ab1997. Total PKCδ antibody was SC-937 (C-20). Rabbit anti-phospho-PKCδ (Thr505) 9374S was from Cell signaling. Phospho-p38 MAPK (Thr180/Tyr182) 12F8 Rabbit was from Cell Signaling. Phospho-MKK3 (Ser189)/MKK6 (Ser207) (22A8) Rabbit monoclonal antibody was from Cell Signaling (catalogue no. 9236).
Cleavage Inducers
Lipopolysaccharide (LPS) purified by gel filtration chromatography was from Sigma (product number L4391) from Escherichia coli 0111:B4. Lipoteichoic acid and poly(I:C) were from InvivoGen. PMA and 4α-phorbol 12-myristate 13-acetate were from Sigma.
Chemical Inhibitors
Gö 6976 and Gö 6983 were from EMD Biosciencies. p38 inhibitor, SB 202190, was from Sigma (catalogue no. S7067) and used at 10 μm. TAPI-0 was from Calbiochem (catalogue no. 579050). N-acetyl cysteine was prepared fresh before use and was from Sigma.
Bryostatin 1 (catalogue no. B 7431) was from Sigma. 1,10-Phenanthroline monohydrate, reagent grade, in methanol was from Sigma (catalogue no. P9375).
Detection Reagents
5-(and -6)-chloromethyl-2′,7′-dichloro dihydrofluorescein diacetate, acetyl ester (CM-H2DCFDA) was from Invitrogen. Human Mer sandwich ELISA from R&D DuoSet IC (DYC891-2).
siRNA
TLR4 siRNA (mouse) was from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA) (catalogue no. sc-40261). PKCδ siRNA1 is Mm_Prkcd_2 (SI01388730) (target sequence CCG GGT GGA CAC ACC ACA CTA), and PKCδ siRNA2 is Mm_Prkcd_3 (SI01388737) (target sequence TTG AAT GTA GTT ATT GAA ATA) (Qiagen). Adam17 siRNA was from Qiagen (Mm_Adam17_6 SI02689190 target sequence TTG AAG AAT ACT TGT AAA TTA and Mm_Adam17_5 SI02669261 target sequence CCC GGG TAT TAT GTA CCT GAA). Adam10 siRNA was from Qiagen (Mm_Adam10_5 SI02666062 target sequence CAC AGT GTG CAT TCA AGT CAA and Mm_Adam10_1 SI00165760 target sequence CCA GCA GAG AGA TAC ATT AAA). siRNAs were added to primary macrophages and J774 cells with Lipofectamine 2000 from Invitrogen.
Mice
Wild-type macrophages were obtained from 8–10-week-old female C57Bl6/J mice (Jackson Laboratories). For Adam17-deficient studies, macrophages were from 8–10-week-old female Adam17fl/flLysmcre mice (Adam17ΔMACROPHAGE), which have deficient ADAM17 expression, or from control littermate Adam17fl/fl mice, which have normal ADAM17 expression (26). 8–10-week-old female Nox2 mice were from Jackson, Strain B6.129S6-Cybb, stock number 002364. B6 Myd88−/− (009088) and Tfir−/− mice were also from Jackson.
Isolation of sMER and Synthesis of MER Ectodomain Peptide for Mass Spectrometry
sMER was isolated from serum-free medium over 80% confluent J774 cells after a 1-h treatment with 50 nm PMA. Cell supernatant was clarified by centrifugation to remove cellular and membrane debris. Clarified supernatant was immunoprecipitated with polyclonal anti-MER (AF591) and protein A/G plus-agarose (sc-2003), and the sample was resolved by reducing SDS-PAGE. Confirmation of capture was performed by treating glycosylated MER extracellular domain with glycanase PNGase F. Post-PNGase treatment, sMER resolved at a molecular mass of 65 kDa, similar to the predicted size of the MerTK ectodomain. For the synthetic peptide, a peptide matching the sequence of the semispecific Arg-C proteolytic fragment of MERTK (as described below) was custom synthesized by NeoBioscience (Cambridge, MA). The sequence is as follows: IAA ITK GGI GPF SEP VNI IIP EHS KVD YAP. Its identity was confirmed by accurate mass and tandem mass spectrum.
Mass Spectrometry
In-gel sMER was subjected to proteolytic digestion. The gel was rinsed, reduced with DTT, and alkylated with iodoacetamide and digested with trypsin, chymotrypsin, or Arg-C overnight at 37 °C. Supernatant from the gel was collected, and the gel pieces were further extracted with 50% acetonitrile, 0.1% formic acid, and followed by 10% acetonitrile and 0.1% formic acid. The two washes were combined with the original supernatant, dried down, and suspended in 15 μl of 5% acetonitrile, 0.1% formic acid.
LC-ESI-MS/MS
Extracted in-gel digests were injected onto a C18 trap column (Magic AQ C18 200A, 5 μm, 0.1 × 20 mm, Michrom Bioresources, Inc.), desalted for 15 min with water, 0.1% formic acid (4 μl/min), eluted onto an analytical column (Magic AQ C18 90A, 5 μm, 0.1 × 200 mm, Michrom Bioresources, Inc.), and separated at a flow rate of 0.4 μl/min over 90 min, using a linear gradient of 5–35% acetonitrile, 0.1% formic acid in 0.1% formic acid on a NanoAquity HPLC (Waters, Milford, MA). Positive ion mass spectra were acquired with electrospray ionization in a hybrid linear ion trap-Orbitrap mass spectrometer (LTQ Orbitrap XL, Thermo Fisher, San Jose, CA) with data-dependent acquisition of MS/MS scans (linear ion trap) on the eight most abundant ions in the survey scan (Orbitrap, resolution 30,000). An exclusion window of 45 s was used after two repeated acquisitions of the same precursor ion. Extracted in-gel digests and the synthetic peptide were further analyzed by targeted LC-ESI-MS/MS on the Arg-C semispecific proteolytic fragment at m/z 1045.5 (3+) and 784.4 (4+). A high resolution full scan MS in the Orbitrap (resolution 30,000) was altered with two targeted MS/MS scans with precursor selection window 2.5 Da in the linear trap and two high resolution MS/MS scans in the Orbitrap (resolution 10,000).
Protein Identification
For identification of MERTK, MS/MS spectra were matched against the mouse Uniprot/SwissProt database (mouse version 3.54, April 2010), using the SEQUEST (version 2.7) search engine with fixed Cys carbamidomethylation and variable Met oxidation modifications and no enzyme specificity (semispecific restriction was applied to the results of the data base search). The mass tolerance for precursor ions was 50 ppm (LTQ-Orbitrap data); SEQUEST default tolerance was accepted for product ions. SEQUEST results were further validated with PeptideProphet and ProteinProphet, using an adjusted probability of ≥0.90 for peptides and ≥0.95 for proteins. Each charge state of a peptide was considered a unique identification. Identity of the semispecific Arg-C proteolytic fragment was further confirmed by a Mascot data base search (version 2.1, mouse SwissProt data base, v.XX, Matrix Science) on the MS/MS spectrum of the m/z 1045.5 (semitryptic specificity, mass tolerance 50 ppm precursor, 0.4-Da fragments, modifications: fixed Cys+57.021, variable Met+15.99).
Site-directed Mutagenesis and Analysis of Mutant MerTK Post-transfection
Mutant MerTKs were generated from pIRES2-EGFP Mer from Addgene. Site-directed mutagenesis was performed using the QuikChange Lightning site-directed mutagenesis kit from Stratagene and in cooperation with Genewiz. Successful deletion and sequence integrity were confirmed by sequencing analysis at the Columbia University Core Sequencing Facility. For transfection assays, pIRES2-EGFP was used as a control. DNAs were transfected with Lipofectamine 2000 reagent from Invitrogen into HEK-293A cells (Invitrogen). Cellular MerTK and sMER were assessed post-transfection as described below. For efferocytosis analysis, apoptotic cells were labeled with Cell Tracker Orange from Invitrogen to measure internalization of apoptotic cells by fluorescent microscopy.
Primary Tissue Culture and Harvest of sMER
Peritoneal macrophages were obtained from 12-week-old female C57BL6/J mice from Jackson Laboratories unless indicated otherwise. Macrophages were harvested from these mice by peritoneal lavage after an immunization protocol of intradermal and intraperitoneal methyl-BSA injection or intraperitoneal concanavalin A injection (27). Peritoneal cells were cultured in 20% L-cell-conditioned DMEM for a minimum of 48 h before experimental treatments. Experiments on adherent macrophages were conducted at a typical confluence of 80%. To induce and harvest sMER, cultures were typically from 12-well tissue culture-treated plates that were overlaid with 50 ng/ml LPS in serum-free medium for the indicated times. Cell supernatants were concentrated 10-fold with Amicon Ultra centrifugal filters (10,000 molecular weight cut-off). Cell surface MerTK was measured using the Thermos Scientific Pierce cell surface protein isolation kit (catalogue no. 89881) with sulfo-N-hydroxysulfosuccinimide-SS-biotin (Sulfo-NHS-SS-Biotin).
Fluorescent Analysis of Peroxide Accumulation
Macrophages were loaded with 5-(and 6)-chloromethyl-2′,7′-dihydrodichlorofluorescein (DCF) diacetate acetyl ester (Invitrogen). After 30 min, the cells were washed and viewed immediately at room temperature with an inverted fluorescent microscope (IX-70) equipped with filters appropriate for fluorescein, and images were obtained with a charge-coupled device camera (Cool Snap) equipped with imaging software. Three fields of 700 cells/field were photographed for each condition, and the number of DCF-positive cells in each field was counted and expressed as a percentage of the total number of cells.
Subcellular Fractionation
To measure membrane translocation of PKCδ, primary macrophages were resuspended in 20 mm Tris-HCl (pH 7.5), 0.25 m sucrose, 2 mm EGTA, 2 mm EDTA, and protease and phosphatase inhibitor mixture. Cells were subjected for sonication at 4 °C for 5 s. Cell lysates were subjected to centrifugation at 600 × g to remove nuclei and cellular debris. Supernatant was next spun at 100,000 × g for 1 h. Supernatant was soluble fraction, and pellet was membrane fraction. Proteins were resolved via reducing SDS-PAGE.
Immunoblots
Cell extracts were electrophoresed on 4–20% gradient SDS-polyacrylamide gels and transferred to 0.45-μm nitrocellulose membranes. The membrane was blocked in Tris-buffered saline, 0.1% Tween 20 (TBST) containing 5% (w/v) nonfat milk at room temperature for 1 h and then incubated with the primary antibody in TBST containing 5% (w/v) nonfat milk or 5% bovine serum albumin at 4 °C overnight, followed by incubation with the appropriate secondary antibody coupled to horseradish peroxidase. Proteins were detected by ECL Supersignal West Pico chemiluminescence (Pierce).
Plasma Analysis
Plasma was collected from the left ventricle of the heart after intraperitoneal injection of LPS, and ELISA was performed for sMER. Capture antibody (mouse MER affinity-purified polyclonal antibody, goat IgG, catalogue no. AF591, R&D) was overlaid onto ELISA plates at 0.2 μg/ml. Detection antibody (0.2 μg/ml) was mouse MER affinity-purified polyclonal antibody goat IgG (catalogue no. BAF591, R&D). Signal from MertkKD mice was not above background. For TNFα, measurements were performed at the University of Maryland Cytokine Core Laboratory (Baltimore, MD). All experiments were performed in triplicate, and results were extrapolated from a standard curve.
Statistical Analysis
Results are presented as means ± S.E. Differences between multiple groups were compared by analysis of variance (one- or two-way), and differences between two groups were compared by paired or unpaired Student's t test. p < 0.05 was considered significant. Stated n values are biological replicates.
RESULTS
Identification of the MerTK Proteolysis Site by Mass Spectrometry
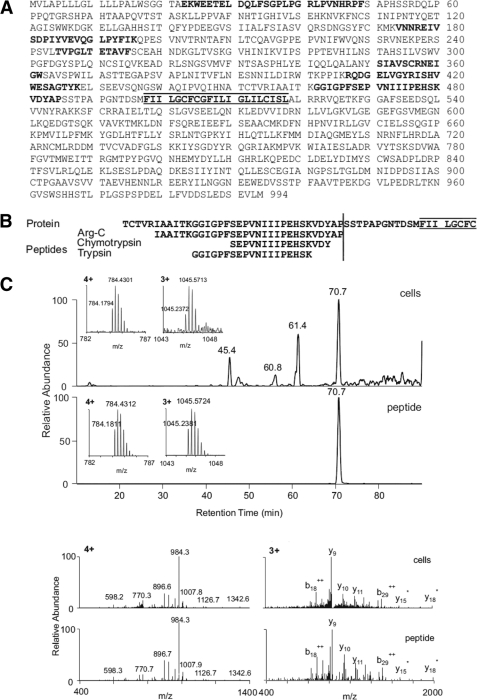
To identify the site at which MerTK is susceptible to proteolysis, we induced cleavage and then immunopurified sMER from murine macrophage cell supernatants. Under these conditions, cleavage resulted in both generation of sMER and a reduction of cell surface MERTK as determined by flow cytometry (data not shown). SDS-PAGE-purified sMER was subjected in parallel to trypsin, chymotrypsin, and endoproteinase-Arg-C (clostripain) digestion and LC-MS/MS to identify MerTK. As an initial test of purity of the immunoprecipitated material, the combined results from the three separate proteolytic digests identified only peptides originating from the MerTK ectodomain. All three protease digests identified peptides in close proximity of the putative transmembrane domain (Fig. 1A). Significantly, the trypsin (K↓GGIGPFSEPVNIIIPEHSK↓V) and chymotrypsin (F↓SEPVNIIIPEHSKVDY↓A) proteolytic peptides were cleaved at enzyme-specific sites at both termini, whereas the Arg-C proteolytic peptide C terminus (R↓IAAITKGGIGPFSEPVNIIIPEHSKVDYAP↓S) was after a proline and not after the usual site of Arg-C cleavage, arginine (Fig. 1B). Two independent MS search engines (Sequest and Mascot) identified the peptides with high confidence. We further synthesized the peptide IAAIT KGGIG PFSEP VNIII PEHSK VDYAP and subjected it to LC-MS/MS under the same conditions as above. Both synthetic and cell-derived Arg-C semispecific proteolytic peptides showed identical retention time, accurate mass at both 4+ and 3+ charge states (mass accuracy of <3 ppm), and MS/MS spectra of both 4+ and 3+ ions (Fig. 1C). Collectively, these data identify Pro485-Ser486 as the induced MerTK proteolytic site.
FIGURE 1.
Identification of the proteolytic cleavage site of MerTK. Soluble MER was isolated by immunoprecipitation and SDS-PAGE. Gel-purified bands were subjected to mass spectrometric analysis. LC-MS/MS analysis of three separate proteolytic digests (trypsin, chymotrypsin, and Arg-C) identified only peptides originating from the ectodomain of MERTK (A). The trypsin and chymotrypsin digests identified specific peptides in the near proximity of the putative transmembrane domain (sequence highlighted by bars above and below). In contrast, the Arg-C digest identified a peptide with a nonspecific site Pro485-Ser486 at the C terminus (B). The Arg-C peptide from the in-gel digest and the synthetic peptide IAA ITK GGI GPF SEP VNI IIP EHS KVD YAP were analyzed side-by-side by targeted LC-MS/MS on a high resolution instrument (LTQ-Orbitrap). The identical retention time, accurate mass at both 4+ and 3+ charge states (mass accuracy of <3 ppm), and MS/MS spectra of both 4+ and 3+ ions demonstrate correct identification (C).
Deletion of Six Amino Acids Spanning Proline 485 Renders MerTK Resistant to Induced Proteolysis and Efferocytosis Suppression by Cleavage Stimuli
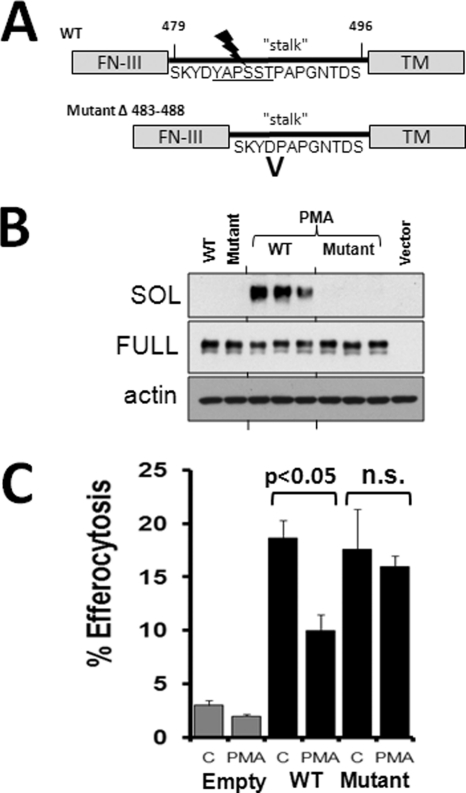
If amino acids including and proximal to proline 485 encode susceptibility to proteolysis, then targeted deletion of these residues could confer MerTK resistance to cleavage inducers. By site-directed mutagenesis, we engineered a six-amino acid deletion mutant of MerTK lacking amino acids 483–488 (Fig. 2A). Mutant MERTKΔ483–488 was transfected into HEK-293 cells, and shedding was induced by the addition of PMA, a known inducer of MerTK proteolysis (16). As indicated in Fig. 2B, expression of MerTKΔ483–488 was equal to WT expression by immunoblot, whereas generation of sMER was nearly completely abrogated in the mutant MerTK post-PMA treatment. A principle function of MERTK is to promote efferocytosis (3). To determine if MerTKΔ483–488 was functional, we transfected cleavage-resistant MertkΔ483–488 into HEK 293 cells, which do not express MerTK and do not engulf apoptotic cells (Fig. 2C, nontransfected cells). As indicated in Fig. 2C, and consistent with previous findings (7), transfection of WT Mertk induced the capacity of HEK cells to promote efferocytosis of UV-irradiated apoptotic Jurkat cells. The cleavage-resistant MerTK promoted efferocytosis in HEK cells to a comparable extent. We next measured efferocytosis after adding cleavage inducer PMA. Efferocytosis was significantly reduced in cells transfected with WT cDNA post-PMA; however, the mutant was resistant to PMA-induced efferocytosis suppression (p < 0.05). Thus, deletion of MerTK amino acids 483–488 confers resistance to induced MerTK cleavage and to suppression of efferocytosis by cleavage stimuli.
FIGURE 2.
Deletion of six amino acids including proline 485 renders MerTK resistant to induced proteolysis and efferocytosis suppression by cleavage stimuli. A, mutation scheme indicating the region of site-directed deletion of residues 483–488 from the MERTK stalk, between the fibronectin-III ectodomain and the TM domain. B, Western blot for cellular MERTK and supernatant sMER after overnight transfection of WT or MERTKΔ483–488 into HEK-293 cells. Post-transfection, cells were treated with or without PMA (50 nm) for 1 h, and cell supernatants and cell extracts were harvested for analysis. C, efferocytosis of apoptotic cells by HEK293 cells was measured by fluorescent microscopy after transfecting with wild type or mutant MerTKΔ483–488 cDNA with or without PMA. p < 0.05, as indicated. n.s., not significant. Error bars, S.E.
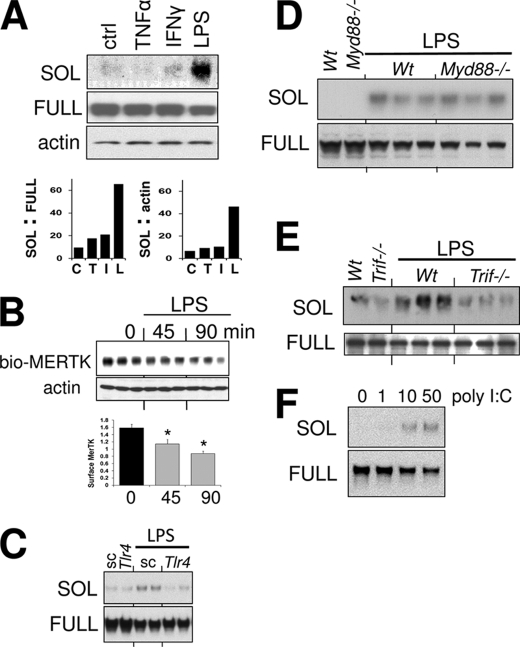
LPS-induced MerTK Cleavage Requires TLR4-TRIF Signaling Independent of Myd88 and Is Inhibited by NADPH Deficiency
We next sought to elucidate the signaling pathway that leads to generation of sMER. The two known inducers of MerTK cleavage are PMA and LPS. PMA (50 nm)-induced cleavage of MerTK from macrophages can be detected by immunoblot as early as 15 min and 1–2 h after 50 ng/ml LPS (16). Besides LPS, we asked if other prototypic inflammatory stimuli could acutely induce MerTK cleavage; however, sMER was not detected in cell supernatants after treating primary macrophages with TNFα or IFNγ for 1 h (Fig. 3A). Generation of sMER was concomitant with reductions in cell surface MerTK as determined by surface biotinylation (Fig. 3B). As expected, silencing of TLR4 with siRNA significantly inhibited LPS-induced sMER generation (Fig. 3C). Interestingly, LPS-mediated cleavage of sMER was not affected by Myd88 deficiency as indicated both by generation of sMER and reductions in full-length cell-associated MerTK (Fig. 3D). Similarly, inhibition of LPS-mediated NF-κB activation, which is downstream of MYD88 signaling, also failed to inhibit formation of sMER (data not shown). Instead, cleavage was suppressed by deficiency of the TLR4 adaptor Trif (Fig. 3E). Consistent with a TRIF-mediated signaling pathway, the TLR3 ligand poly(I:C) (28) also was capable of inducing MerTK shedding (Fig. 3F).
FIGURE 3.
LPS-induced MerTK cleavage requires TLR4-TRIF signaling independent of Myd88. Primary macrophages were incubated with 10 ng/ml TNFα, 10 ng/ml IFNγ, or 50 ng/ml of LPS for 1 h (A), and cell supernatants were harvested for sMER (SOL) immunoblot. Cell lysates were run for full-length cellular MERTK in parallel, and densitometric analysis (bar graph) of the ratio of sMER/full-length MERTK and sMER/actin-loading control were measured. B, cell surface MerTK was measured by immunoblot after surface biotinylation and capture. Macrophages were treated with LPS for the indicated times followed by biotinylation. Densitometric analysis below was normalized to actin loading control. *, p < 0.05. C, sMER generation was measured by immunoblot post-LPS after silencing TLR4 with RNAi versus scrambled (sc) control. D, sMER generation post-LPS treatment in WT and Myd88-deficient primary macrophages. E, sMER generation requires Trif post-LPS treatment, as determined in Trif−/− macrophages. F, poly(I:C) (shown in μg/ml) induces solMER. Primary macrophages were treated with the indicated doses of poly(I:C) for 2 h, and cell supernatants and cell extracts were subjected to immunoblot for soluble and full-length MER, respectively. Error bars, S.E.
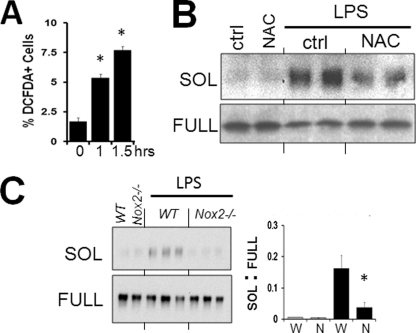
Phagocytes produce reactive oxygen species (ROS) during phagocytosis or after stimulation with a wide variety of agents, including LPS (29). DCF staining was used as a measure of intracellular peroxide/ROS accumulation. We found that LPS caused a substantial increase in the percentage of DCF-positive cells in macrophages at times when sMER can be detected (Fig. 4A). The ROS scavenger, N-actetylcysteine, inhibited sMER shedding (Fig. 4B) and DCF-staining (data not shown). Generation of ROS can occur through plasma membrane assembly and activation of nicotinamide adenine dinucleotide phosphate reduced (NADPH) oxidase (NOX) (30). Shedding was significantly inhibited by Nox2 deficiency in LPS-treated macrophages (Fig. 4C).
FIGURE 4.
LPS-mediated MerTK cleavage requires NADPH. A, monolayers of elicited primary peritoneal macrophages were treated with 50 ng/ml LPS on tissue culture plates, and intracellular peroxide accumulation was assayed by DCF fluorescence as described under “Materials and Methods.” Three fields for each sample were quantified and expressed as a percentage of DCF-positive cells. B, Western blot of solMER from primary murine macrophage supernatants after treating cells with 1 mm NAC. NAC (freshly prepared) was preincubated with macrophages for 60 min prior to directly adding LPS. Corresponding cell extracts of full-length MERTK are shown below. C, sMER generation by LPS was measured from Nox2-deficient cells by Western blot. Densitometric analysis of the ratio of sMER (SOL) to full-length MERTK (FULL) is to the right. *, p < 0.05. Error bars, S.E.
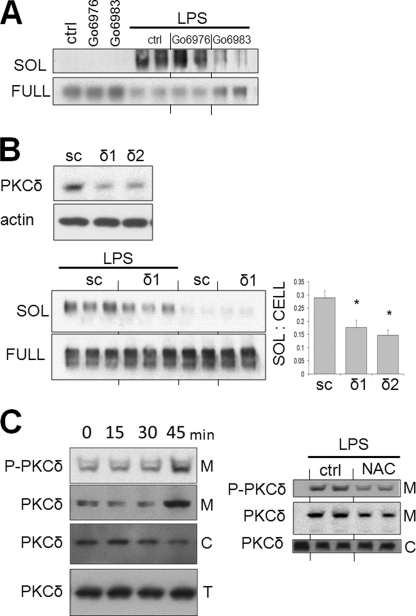
PKCδ Is Required for LPS-induced MerTK Shedding
PMA, a PKC activator, induces robust MerTK cleavage (16). In our own hands, sMER was robustly induced by PMA; however, the PKC-inactive analog of PMA, 4α-PMA, failed to induce MerTK cleavage (data not shown). PKCs are a family of serine-threonine kinases, which are classified into three major groups based on homology and cofactor requirements: “conventional” PKCs, “novel” PKCs, and “atypical” PKCs (31). LPS-induced cleavage was inhibited by the pan-PKC inhibitor Go6983 (32) but not by classical PKC inhibitor Go6976 (Fig. 5A). In addition, co-cultivation of the atypical PKCζ pseudopeptide failed to suppress MerTK cleavage (data not shown). These data suggested that PKC activation did not involve members of the classical or atypical PKCs during MerTK cleavage. We next considered the novel PKCs, particularly PKCδ. Knockdown of PKCδ by two separate siRNAs each yielded greater than 78% reduction of basal PKCδ levels (Fig. 5B). LPS-mediated cleavage, after knockdown with each siRNA, was reduced in both instances (p < 0.05 in each instance). Consistent with a role for PKCδ after LPS activation in macrophages, both membrane-bound and phospho-PKCδ (at Thr505) were elevated 45 min after adding LPS (Fig. 5C). In addition, both PKCδ phosphorylation and membrane translocation were reduced after adding the antioxidant NAC, implicating PKCδ action downstream of NADPH activation during signaling, leading to generation of sMER (Fig. 5C, right).
FIGURE 5.
PKCδ is required for MerTK cleavage. A, primary macrophages were pretreated for 30 min with 250 nm Go6976 or 250 nm Go6983 in complete medium and subsequently treated with 50 ng/ml LPS. Subsequently, levels of sMER from cell supernatants and levels of full-length (FULL) MERTK from cell extracts were measured by Western blot. B, primary macrophages were incubated with PKCδ siRNA for 48 h, and the top panel exhibits representative knockdown efficiency of two PKCδ siRNAs (δ1 and δ2) by Western blot. In parallel, macrophages were cultured with LPS in the presence of PKCδ siRNA and scrambled (sc) control and sMER measured from supernatants and full-length MERTK from cell extracts by immunoblot. Densitometric analysis is shown to the right after knockdown with both PKCδ siRNAs. C, membrane translocation of PKCδ and phospho-PKCδ (PKCδ-P) post-LPS was determined by immunoblot after isolation of membrane pellets as described under “Materials and Methods.” Membrane translocation was also measured after treatment with NAC (right). M, membranous fraction; C, cytosolic fraction; T, total cellular lysate. Error bars, S.E.
MerTK Proteolytic Cleavage Requires ADAM17 and MAPK p38
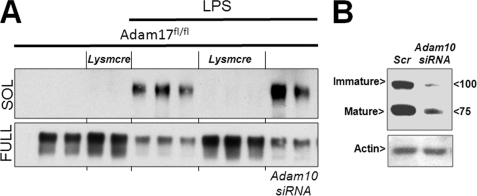
Shedding of MerTK is inhibitable by TAPI-0 (16), a hydroxymate-based inhibitor of collagenase, gelatinase, and the membrane-associated protease ADAM17/TACE (33). To determine if germ line Adam17 is required for proteolytic cleavage of MERTK, we measured sMER production in Adam17fl/flLysmcre macrophages (26). As shown in Fig. 6A, gene inactivation of Adam17 completely inhibited LPS-mediated sMER generation. Similar findings were seen after acute knockdown of Adam17 with siRNA in both primary macrophages and J774 macrophages (data not shown). In some cases, for example, when ADAM17 is inactivated or, alternatively, if cells are activated by ionomycin, the structurally similar ADAM10 can also shed ADAM17 substrates (34). However, siRNA-mediated reduction in ADAM10 (73% knockdown efficiency as indicated in Fig. 6B) did not reduce MerTK cleavage in WT macrophages. Thus, ADAM17 is the primary and non-redundant sheddase of MerTK.
FIGURE 6.
Generation of solMER by LPS requires ADAM17. A, immunoblots of sMER (SOL) and full-length MerTK (full) from Adam17fl/fl and Adam17fl/flLysmcre peritoneal macrophages with or without LPS. Macrophages from the indicated genotype were elicited and purified by adherence to tissue culture-treated plates in the presence of L-cell conditioned medium for 2 days. Subsequently, 50 ng/ml LPS in serum-free medium was added where indicated for 2 h, and supernatant was collected and concentrated from all samples for immunoblot of sMER. Parallel immunoblot of cellular lysates for full-length MerTK is indicated below. Where indicated, macrophages were pretreated with Adam10 siRNA. B, representative immunoblot of ADAM10 protein after either scrambled (sc) or siRNA (si) knockdown of ADAM10 immature (I) and mature (M) forms and indicated molecular weights. A, actin loading control.
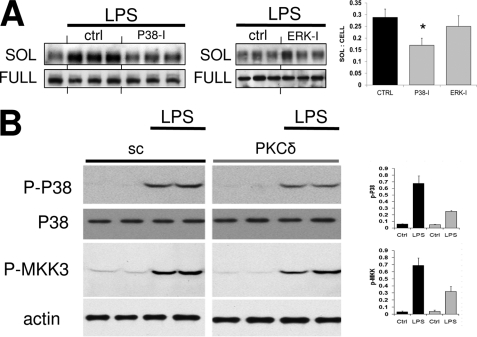
We next considered how LPS might activate ADAM17. Previous reports suggest a role for MAPKs, including ERK1/2 and p38 during the phosphorylation or activation of ADAM17 (35–38). Although the ERK inhibitor PD98059 failed to reduce sMER levels, the p38 inhibitor SB 202190 (SB) partially reduced sMER generation (Fig. 7A). p38 phosphorylation was measured 45 min post-LPS addition and was partially suppressed after silencing PKCδ with siRNA. The upstream MAPK for p38 is MKK3/6 (39). Similarly, MKK3/6 was also activated after LPS and partially suppressed by PKCδ silencing (Fig. 7B).
FIGURE 7.
LPS-induced MerTK cleavage requires p38. A, the effects of p38 inhibition and ERK inhibition were determined on LPS-induced sMER generation. Primary peritoneal macrophages were preincubated with p38 (SB 202190, 10 μm) and ERK (PD98059, 10 μm) MAPK inhibitors for 30 min. Subsequently, 50 ng/ml LPS was added where indicated, and cell supernatant and cell extracts were probed by Western blot for sMER and cellular full-length MerTK, respectively. Densitometric measurement is to the right. *, p < 0.05. B, phosphorylation of p38 (P-38; Thr180/Tyr182) and MKK3 (P-MKK3; MKK3 Ser189/MKK6 Ser207) after LPS treatment of primary macrophages was assessed by immunoblot of cell extracts. Analysis was also performed in the presence of PKCδ siRNA versus scrambled (sc) control. Densitometric analysis is shown to the right and normalized to actin loading control. Error bars, S.E.
sMER Is Induced in Vivo post-LPS Injection
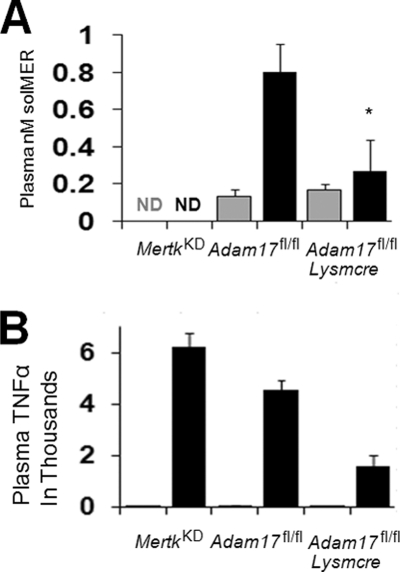
We set out to determine the in vivo significance of our findings. sMER has been identified in human and murine plasma (16), and we asked if LPS could stimulate sMER in a model of endotoxemia. LPS was injected into the peritoneum, and plasma was harvested 3 h later. Plasma sMER levels were then measured by ELISA. As shown in Fig. 8A, sMER levels were significantly increased after LPS injection in control mice. sMER was not detected above background in MerTK-deficient mice before or after LPS injection. In addition, sMER generation was dependent on ADAM17, because Adam17fl/flLysmcre mice failed to induce sMER post-LPS injection. Consistent with previous results, LPS induced robust TNFα production in WT mice, and this was elevated in Mertk-deficient mice and suppressed in Adam17-deficient mice (Fig. 8B). Thus, sMER is induced by LPS treatment in vivo, and this requires ADAM17.
FIGURE 8.
LPS induces sMER generation in vivo. Levels of sMER in murine plasma were determined by ELISA after injection of LPS (black bars) or control saline (gray bars). 8–10-week-old MertkKD, Adam17fl/fl, and Adam17fl/flLysmcre mice were injected with 100 μg of LPS or saline into the peritoneum, and plasma was harvested 3 h later from the left ventricle of the heart. Systemic plasma sMER (A) and plasma TNFα (B) were measured by ELISA as described under “Materials and Methods.” Each bar represents the mean of at least four animals per strain. *, p < 0.05. ND, not detected. Error bars, S.E.
DISCUSSION
As expected, the cleavage site of MerTK does not conform to any previously documented motif for ADAM17 substrates. In fact, mutational analysis of ADAM17 substrates, such as the IL-6 receptor, suggests relaxed sequence specificity proximal to the ADAM17 cleavage site (40). Instead, the length of the membrane-proximal stalk has been implicated as a factor that controls susceptibility to cleavage (41, 42). Based primarily on its structure, MerTK is grouped into the TAM receptor family of tyrosine kinases, which include Tyro3, Axl, and Mertk. Mass spectrometric analysis indicates that TAM family member AXL is cleaved by ADAM17 (20), and shed AXL has been identified in both human and murine serum. The cleavage site of human AXL has been mapped to a 14-amino acid region proximal to the predicted TM domain (43, 44). Cleavage of TYRO3 has not been reported. Based on our own sequence analysis, the stalk regions of murine TAMs fail to exhibit a consensus motif for cleavage. However, murine MerTK and human MerTK do share a significant number of proline residues within their stalk region, leading us to speculate that human MerTK could also be cleaved after a proline.
The degradome of ADAM17 indicates a wide range of susceptible substrates. Therefore, how ADAM17 activation is finely regulated or, alternatively, a preference for specific cleavage substrates may be key to understanding substrate specificity under disparate homeostatic and pathophysiological contexts. Previous reports indicate that Gram-positive bacteria can stimulate the transcription of ADAM17 (45, 46). However, LPS-mediated cleavage was not inhibited by actinomycin D treatment (data not shown) or cylcoheximide (16), implicating a post-translational mechanism. Furthermore, generation of sMER was specific to the TLR4 agonist, Gram-negative endotoxin. The Gram-positive cell wall component lipoteichoic acid and TLR2 agonist (47) was unable to induce sMER shedding (data not shown). Although the link between LPS and ADAM17/TACE is well established, surprisingly little is known about the intermediary signaling molecules required. A previous study showed that endotoxin-induced MYD88 was upstream of ADAM17 processing during generation of EGF receptor ligands in nonhematopoietic cells (48). Although MerTK cleavage required TLR4, it was independent of MYD88 and instead signaled through TRIF. Cleavage could also be activated by the TLR3 agonist poly(I:C). Indeed, in epithelial cells, multiple Toll-like receptors, including TLR3, have been implicated in ADAM17-mediated shedding (49).
Mitochondrial ROS have been implicated in GPCR-induced TACE-dependent TGFα shedding (50). In the case of MerTK shedding post-LPS, the ROS scavenger NAC and NADPH deficiency both blocked cleavage (Fig. 4). Although there are examples of TLR4 activation of NADPH through MYD88 (51), TRIF-medicated activation of NADPH is lacking. Interestingly, Park et al. (52) reported that NADPH oxidase subunits can directly interact with TLR4 to promote ROS generation. Furthermore, redox agents have been shown to regulate mature ADAM17 during neutrophil-mediated shedding of L-selectin (53). ROS has been suggested to activate ADAM17 (54, 55), in part through activation of PKCδ, and in some cell types, PKCδ is a redox-sensitive kinase (56, 57). A role for PKCδ in ADAM17 activation was previously implicated only based on data using the nonspecific PKC inhibitor Rottlerin. Once activated by ROS, PKCδ may in turn promote additional ROS activation (58).
Besides LPS, numerous other stimulators of ADAMs have been implicated. These include activators of protein kinase C, such as 12-O-tetradecanoylphorbol-13-acetate and PMA. Previously, a link between PKC signaling and LPS/TLR4 was shown for the PKC isozyme ϵ. PKCϵ was found to phosphorylate Trif-related adapter molecule downstream of LPS (59). In addition, PKCδ has been found to bind the TLR4/TLR2 adaptor protein TIRAP/Mal (60). In vitro activation by LPS has been shown to induce rapid release of soluble FMS-like tyrosine kinase-1 receptor (sFlt-1), concomitant with phosphorylation of PKCδ (61). Furthermore, a PKCδ-p38 MAPK cascade has been identified in Caenorhabditis elegans (62). PKCδ-mediated activation of p38 may lead to p38 interaction with ADAM17 (37), and both p38 and ERK can phosphorylate ADAM17 at threonine 735 (35). However, activation of ADAM17 by PMA does not depend on its cytoplasmic domain, arguing against inside-out regulation via cytoplasmic phosphorylation as an underlying mechanism (24, 63). One possible explanation is that the ADAM17 cytoplasmic tail contains an inhibitory residue that must be phosphorylated for activation of ADAM17. Another possibility may be explained by differences in cell types utilized for the aforementioned studies. Finally, a still unidentified molecule may act to link PKCδ and p38 to ADAM17.
Proteolytic cleavage is known to regulate the activity of many transmembrane-anchored proteins. In the case of growth factors and cytokines, such as EGF, TFGα, and TNFα, proteolysis can lead to the biological activation of inactive precursors and their autocrine or paracrine release into the extracellular milieu. In the case of transmembrane receptors, such as TNFα receptor-I, TNF-α receptor-II, and L-selectin, proteolytic cleavage leading to ectodomain shedding can often lead to antagonist functions. Besides the loss of a cell surface signaling conduit, the shed ectodomains of cell surface receptors can also function as competitive decoys to bind receptor ligands. However, concentration is a critical factor. Low levels of soluble TNF receptor enhance TNFα action, whereas high concentrations are inhibitory (64). In the case of MerTK, recombinant sMER has been shown to be inhibitory by two accounts: first through suppression of efferocytosis in vitro and, second, through inhibition of thrombus formation in vivo (16). Interestingly, in the case of efferocytosis, LPS has been reported to inhibit the clearance of neutrophils in vitro, in part through induction of the ADAM17 target TNFα and suppression of macrophage-derived GAS6, the ligand for MerTK (65). These data suggest a coordinated response by macrophages to suppress MerTK function upon recognition of LPS. MerTK inactivation by cleavage would also suppress its anti-inflammatory function, thereby permitting the phagocyte to become fully activated. Future studies that seek to determine the in vivo/physiological relevance of MerTK cleavage, both in the context of bacterial challenge and during diseases of chronic inflammation and defective efferocytosis, will benefit from the identification of the cleavage site and signaling pathways revealed herein.
Acknowledgment
We sincerely thank Dr. Mary Reyland for help and consultation regarding PKC-related experiments.
This work was supported, in whole or in part, by National Institutes of Health (NIH), NHLBI, Program of Excellence in Nanotechnology (PEN) Award Contract HHSN268201000045C and NIH Grants HL54591 and HL75662 (to I. T.), GM64750 (to C. B.), and UL1 RR 024156 from the National Center for Research Resources (NCRR), a component of the NIH, and 1K99HL097021 (to E. T.).
- TM
- transmembrane
- KD
- kinase-dead
- NAC
- N-acteylcysteine
- PMA
- phorbol 12-myristate 13-acetate
- sMER
- soluble MER
- TACE
- TNF-α-converting enzyme
- DCF
- dihydrodichlorofluorescein
- ESI
- electrospray ionization
- ROS
- reactive oxygen species
- TAM
- Tyro3, Axl, and Mer.
REFERENCES
- 1. Nagata K., Ohashi K., Nakano T., Arita H., Zong C., Hanafusa H., Mizuno K. (1996) J. Biol. Chem. 271, 30022–30027 [DOI] [PubMed] [Google Scholar]
- 2. Uehara H., Shacter E. (2008) J. Immunol. 180, 2522–2530 [DOI] [PubMed] [Google Scholar]
- 3. Scott R. S., McMahon E. J., Pop S. M., Reap E. A., Caricchio R., Cohen P. L., Earp H. S., Matsushima G. K. (2001) Nature 411, 207–211 [DOI] [PubMed] [Google Scholar]
- 4. Chen C., Li Q., Darrow A. L., Wang Y., Derian C. K., Yang J., de Garavilla L., Andrade-Gordon P., Damiano B. P. (2004) Arterioscler. Thromb. Vasc. Biol. 24, 1118–1123 [DOI] [PubMed] [Google Scholar]
- 5. Anwar A., Keating A. K., Joung D., Sather S., Kim G. K., Sawczyn K. K., Brandão L., Henson P. M., Graham D. K. (2009) J. Leukoc. Biol. 86, 73–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ishimoto Y., Ohashi K., Mizuno K., Nakano T. (2000) J. Biochem. 127, 411–417 [DOI] [PubMed] [Google Scholar]
- 7. Wu Y., Singh S., Georgescu M. M., Birge R. B. (2005) J. Cell Sci. 118, 539–553 [DOI] [PubMed] [Google Scholar]
- 8. Graham D. K., Dawson T. L., Mullaney D. L., Snodgrass H. R., Earp H. S. (1994) Cell Growth Differ. 5, 647–657 [PubMed] [Google Scholar]
- 9. Gal A., Li Y., Thompson D. A., Weir J., Orth U., Jacobson S. G., Apfelstedt-Sylla E., Vollrath D. (2000) Nat. Genet. 26, 270–271 [DOI] [PubMed] [Google Scholar]
- 10. Vollrath D., Feng W., Duncan J. L., Yasumura D., D'Cruz P. M., Chappelow A., Matthes M. T., Kay M. A., LaVail M. M. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 12584–12589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cohen P. L., Caricchio R., Abraham V., Camenisch T. D., Jennette J. C., Roubey R. A., Earp H. S., Matsushima G., Reap E. A. (2002) J. Exp. Med. 196, 135–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Thorp E., Cui D., Schrijvers D. M., Kuriakose G., Tabas I. (2008) Arterioscler. Thromb. Vasc. Biol. 28, 1421–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Graham D. K., Bowman G. W., Dawson T. L., Stanford W. L., Earp H. S., Snodgrass H. R. (1995) Oncogene 10, 2349–2359 [PubMed] [Google Scholar]
- 14. Tibrewal N., Wu Y., D'mello V., Akakura R., George T. C., Varnum B., Birge R. B. (2008) J. Biol. Chem. 283, 3618–3627 [DOI] [PubMed] [Google Scholar]
- 15. Lemke G., Rothlin C. V. (2008) Nat. Rev. Immunol. 8, 327–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sather S., Kenyon K. D., Lefkowitz J. B., Liang X., Varnum B. C., Henson P. M., Graham D. K. (2007) Blood 109, 1026–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Thorp E., Schrijvers D., Tabas I. (2010) Abstracts: Arteriosclerosis, Thrombosis, and Vascular Biology 2010 Scientific Session, Abstract 15, Lippincott Williams & Wilkins, Philadelphia [Google Scholar]
- 18. Hurtado B., Muñoz X., Recarte-Pelz P., García N., Luque A., Krupinski J., Sala N., García de Frutos P. (2011) Thromb. Haemost. 105, 873–882 [DOI] [PubMed] [Google Scholar]
- 19. Blobel C. P. (2005) Nat. Rev. Mol. Cell Biol. 6, 32–43 [DOI] [PubMed] [Google Scholar]
- 20. Müllberg J., Rauch C. T., Wolfson M. F., Castner B., Fitzner J. N., Otten-Evans C., Mohler K. M., Cosman D., Black R. A. (1997) FEBS Lett. 401, 235–238 [DOI] [PubMed] [Google Scholar]
- 21. Black R. A., Rauch C. T., Kozlosky C. J., Peschon J. J., Slack J. L., Wolfson M. F., Castner B. J., Stocking K. L., Reddy P., Srinivasan S., Nelson N., Boiani N., Schooley K. A., Gerhart M., Davis R., Fitzner J. N., Johnson R. S., Paxton R. J., March C. J., Cerretti D. P. (1997) Nature 385, 729–733 [DOI] [PubMed] [Google Scholar]
- 22. Moss M. L., Jin S. L., Milla M. E., Bickett D. M., Burkhart W., Carter H. L., Chen W. J., Clay W. C., Didsbury J. R., Hassler D., Hoffman C. R., Kost T. A., Lambert M. H., Leesnitzer M. A., McCauley P., McGeehan G., Mitchell J., Moyer M., Pahel G., Rocque W., Overton L. K., Schoenen F., Seaton T., Su J. L., Becherer J. D. (1997) Nature 385, 733–736 [DOI] [PubMed] [Google Scholar]
- 23. Murphy G. (2008) Nat. Rev. Cancer 8, 929–941 [DOI] [PubMed] [Google Scholar]
- 24. Le Gall S. M., Maretzky T., Issuree P. D., Niu X. D., Reiss K., Saftig P., Khokha R., Lundell D., Blobel C. P. (2010) J. Cell Sci. 123, 3913–3922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tabas I. (2010) Nat. Rev. Immunol. 10, 36–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Horiuchi K., Kimura T., Miyamoto T., Takaishi H., Okada Y., Toyama Y., Blobel C. P. (2007) J. Immunol. 179, 2686–2689 [DOI] [PubMed] [Google Scholar]
- 27. Li Y., Tabas I. (2006) J. Leukoc. Biol. 81, 483–491 [DOI] [PubMed] [Google Scholar]
- 28. Glenn G., van der Geer P. (2008) FEBS Lett. 582, 911–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pabst M. J., Johnston R. B., Jr. (1980) J. Exp. Med. 151, 101–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ushio-Fukai M. (2006) Sci. STKE 2006, re8. [DOI] [PubMed] [Google Scholar]
- 31. Reyland M. E. (2009) Front. Biosci. 14, 2386–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Young L. H., Balin B. J., Weis M. T. (2005) Cardiovasc. Drug Rev. 23, 255–272 [DOI] [PubMed] [Google Scholar]
- 33. Crowe P. D., Walter B. N., Mohler K. M., Otten-Evans C., Black R. A., Ware C. F. (1995) J. Exp. Med. 181, 1205–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Le Gall S. M., Bobé P., Reiss K., Horiuchi K., Niu X. D., Lundell D., Gibb D. R., Conrad D., Saftig P., Blobel C. P. (2009) Mol. Biol. Cell 20, 1785–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Soond S. M., Everson B., Riches D. W., Murphy G. (2005) J. Cell Sci. 118, 2371–2380 [DOI] [PubMed] [Google Scholar]
- 36. Díaz-Rodríguez E., Montero J. C., Esparís-Ogando A., Yuste L., Pandiella A. (2002) Mol. Biol. Cell 13, 2031–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xu P., Derynck R. (2010) Mol. Cell 37, 551–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rousseau S., Papoutsopoulou M., Symons A., Cook D., Lucocq J. M., Prescott A. R., O'Garra A., Ley S. C., Cohen P. (2008) J. Cell Sci. 121, 149–154 [DOI] [PubMed] [Google Scholar]
- 39. Raingeaud J., Whitmarsh A. J., Barrett T., Dérijard B., Davis R. J. (1996) Mol. Cell Biol. 16, 1247–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Althoff K., Reddy P., Voltz N., Rose-John S., Müllberg J. (2000) Eur. J. Biochem. 267, 2624–2631 [DOI] [PubMed] [Google Scholar]
- 41. Becherer J. D., Blobel C. P. (2003) Curr. Top. Dev. Biol. 54, 101–123 [DOI] [PubMed] [Google Scholar]
- 42. Caescu C. I., Jeschke G. R., Turk B. E. (2009) Biochem. J. 424, 79–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. O'Bryan J. P., Fridell Y. W., Koski R., Varnum B., Liu E. T. (1995) J. Biol. Chem. 270, 551–557 [DOI] [PubMed] [Google Scholar]
- 44. Costa M., Bellosta P., Basilico C. (1996) J. Cell Physiol. 168, 737–744 [DOI] [PubMed] [Google Scholar]
- 45. Gómez M. I., Sokol S. H., Muir A. B., Soong G., Bastien J., Prince A. S. (2005) J. Immunol. 175, 1930–1936 [DOI] [PubMed] [Google Scholar]
- 46. Bostanci N., Reddi D., Rangarajan M., Curtis M. A., Belibasakis G. N. (2009) Oral Microbiol. Immunol. 24, 146–151 [DOI] [PubMed] [Google Scholar]
- 47. Takeuchi O., Hoshino K., Kawai T., Sanjo H., Takada H., Ogawa T., Takeda K., Akira S. (1999) Immunity 11, 443–451 [DOI] [PubMed] [Google Scholar]
- 48. Brandl K., Sun L., Neppl C., Siggs O. M., Le Gall S. M., Tomisato W., Li X., Du X., Maennel D. N., Blobel C. P., Beutler B. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 19967–19972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Koff J. L., Shao M. X., Ueki I. F., Nadel J. A. (2008) Am. J. Physiol. Lung Cell Mol. Physiol. 294, L1068–L1075 [DOI] [PubMed] [Google Scholar]
- 50. Myers T. J., Brennaman L. H., Stevenson M., Higashiyama S., Russell W. E., Lee D. C., Sunnarborg S. W. (2009) Mol. Biol. Cell 20, 5236–5249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fan J., Frey R. S., Malik A. B. (2003) J. Clin. Invest. 112, 1234–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Park H. S., Jung H. Y., Park E. Y., Kim J., Lee W. J., Bae Y. S. (2004) J. Immunol. 173, 3589–3593 [DOI] [PubMed] [Google Scholar]
- 53. Wang Y., Herrera A. H., Li Y., Belani K. K., Walcheck B. (2009) J. Immunol. 182, 2449–2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhang Z., Oliver P., Lancaster J. R., Jr., Schwarzenberger P. O., Joshi M. S., Cork J., Kolls J. K. (2001) FASEB J. 15, 303–305 [DOI] [PubMed] [Google Scholar]
- 55. Shao M. X., Nadel J. A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 767–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kanthasamy A. G., Kitazawa M., Kanthasamy A., Anantharam V. (2003) Antioxid. Redox Signal. 5, 609–620 [DOI] [PubMed] [Google Scholar]
- 57. Konishi H., Tanaka M., Takemura Y., Matsuzaki H., Ono Y., Kikkawa U., Nishizuka Y. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 11233–11237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chen C. L., Chan P. C., Wang S. H., Pan Y. R., Chen H. C. (2010) J. Cell Sci. 123, 2901–2913 [DOI] [PubMed] [Google Scholar]
- 59. McGettrick A. F., Brint E. K., Palsson-McDermott E. M., Rowe D. C., Golenbock D. T., Gay N. J., Fitzgerald K. A., O'Neill L. A. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 9196–9201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kubo-Murai M., Hazeki K., Sukenobu N., Yoshikawa K., Nigorikawa K., Inoue K., Yamamoto T., Matsumoto M., Seya T., Inoue N., Hazeki O. (2007) Mol. Immunol. 44, 2257–2264 [DOI] [PubMed] [Google Scholar]
- 61. Lee M. C., Wei S. C., Tsai-Wu J. J., Wu C. H., Tsao P. N. (2008) J. Leukoc. Biol. 84, 835–841 [DOI] [PubMed] [Google Scholar]
- 62. Ziegler K., Kurz C. L., Cypowyj S., Couillault C., Pophillat M., Pujol N., Ewbank J. J. (2009) Cell Host Microbe 5, 341–352 [DOI] [PubMed] [Google Scholar]
- 63. Reddy P., Slack J. L., Davis R., Cerretti D. P., Kozlosky C. J., Blanton R. A., Shows D., Peschon J. J., Black R. A. (2000) J. Biol. Chem. 275, 14608–14614 [DOI] [PubMed] [Google Scholar]
- 64. Aderka D., Engelmann H., Maor Y., Brakebusch C., Wallach D. (1992) J. Exp. Med. 175, 323–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Feng X., Deng T., Zhang Y., Su S., Wei C., Han D. (2011) Immunology 132, 287–295 [DOI] [PMC free article] [PubMed] [Google Scholar]