Abstract
The urokinase-type plasminogen activator receptor (uPAR) is a glycolipid-anchored membrane protein with an established role in focalizing uPA-mediated plasminogen activation on cell surfaces. Distinct from this function, uPAR also modulates cell adhesion and migration on vitronectin-rich matrices. Although uPA and vitronectin engage structurally distinct binding sites on uPAR, they nonetheless cooperate functionally, as uPA binding potentiates uPAR-dependent induction of lamellipodia on vitronectin matrices. We now present data advancing the possibility that it is the burial of the β-hairpin in uPA per se into the hydrophobic ligand binding cavity of uPAR that modulates the function of this receptor. Based on these data, we now propose a model in which the inherent interdomain mobility in uPAR plays a major role in modulating its function. Particularly one uPAR conformation, which is stabilized by engagement of the β-hairpin in uPA, favors the proper assembly of an active, compact receptor structure that stimulates lamellipodia induction on vitronectin. This molecular model has wide implications for drug development targeting uPAR function.
Keywords: Cell Adhesion, Epitope Mapping, Extracellular Matrix, Protein Conformation, Protein-Protein Interactions, Surface Plasmon Resonance (SPR), CD87, Lamellipodia, uPAR, Vitronectin
Introduction
Timely controlled cell migration is a decisive factor for a plethora of important biological processes that occur during development and adulthood. Controlled cell migration is thus intimately involved in both maintenance and dynamic remodeling of tissue architectures during, e.g. wound healing and mammary gland development (1). These processes are executed and tightly regulated via a complicated cross-talk between specific cell surface receptors (e.g. integrins) and insoluble protein components deposited in the extracellular matrix. The extracellular matrix is nonetheless thought to play a dual role in regulating cell migration, as it provides both the focal adhesion sites required for cellular traction and opposes migration by generating physical barriers (2, 3). Cell migration in vivo, therefore, requires a coordinated regulation of extracellular matrix proteolysis, adhesion, and signaling (4). The urokinase-type plasminogen activator receptor (uPAR)2 may allegedly assist a rendezvous between these functions, as it has the potential to exert control at all three levels. Besides being responsible for focalizing uPA-mediated plasminogen activation on cell surfaces (5–7), uPAR also facilitates adherence to vitronectin embedded in the extracellular matrix (8–10), and as a consequence, it promotes intracellular signaling (4, 11).
The glycolipid-anchored uPAR is a modular glycoprotein composed of three homologous Ly6/uPAR-type (LU) protein domain repeats (5, 12). The far majority of proteins belonging to this domain family contain only a single copy of the LU module, as exemplified by the glycolipid-anchored CD59, the extracellular ligand binding domain in the TGF-β receptors, and the diverse group of secreted snake venom α-neurotoxins (13). In the human genome, five genes are recognized so far to encode proteins with multiple LU domains, and these are all confined to a small gene cluster located on chromosome 19q13 (14). Among these modular proteins, the structure of only uPAR has presently been solved (7, 15–17), and it reveals that all three LU domains in this receptor adopt the archetypical three-finger fold that is found in the snake venom α-neurotoxins. More importantly, all three LU domains in uPAR cooperate to form a large hydrophobic ligand binding cavity for uPA. A notable flexibility in the modular assembly of uPAR is nevertheless displayed by these structures dependent on which ligand is actually buried in this ligand binding cavity (5, 7, 15, 16). The functional binding epitope for vitronectin, another established uPAR ligand, is also confined to a composite receptor interface, which in this case is located between uPAR domain I (DI) and domain II (DII) as determined by alanine-scanning mutagenesis (18). This functional epitope aligns excellently with the receptor surface buried at the interface between the small N-terminal somatomedin B domain (SMB) of vitronectin and uPAR as determined by x-ray crystallography (17). Consequently, the interaction with vitronectin could possibly be sensitive to conformational rearrangements of the individual uPAR modules, which may be elicited by receptor binding to other ligands, e.g. uPA.
With a view to this proposition, it is noteworthy that ample evidence exists in the literature from different laboratories to suggest that uPA binding modulates the interaction between uPAR and vitronectin both at the biochemical (8, 18, 19) and the cellular level (9, 10, 20–25). The molecular basis underlying this dependence is, however, still controversial, and several models, including uPAR dimerization (9, 19, 26), direct interactions with integrins (27–29), or other adaptor proteins (30, 31) have been advocated. In the present study, we have revisited this molecular interplay guided by the structural data obtained recently on this ternary complex (17, 18), and we now present independent functional data pointing to a crucial role of the molecular flexibility in uPAR.
MATERIALS AND METHODS
Chemicals and Reagents
Linear peptides were synthesized and HPLC-purified as described (32). Thiols of cysteine-containing peptides were air-oxidized in 10% (v/v) DMSO to yield cyclic disulfide-linked peptides. Proper oxidation was verified after HPLC purification by the monoisotopic masses resolved by MALDI-MS (Autoflex TOF/TOF, Bruker Daltonics, Bremen, Germany). An inventory of peptides applied in this study is provided in Table 1.
TABLE 1.
Properties of synthetic peptides and uPA-derived protein ligands for uPAR
| Codea | Sequenceb | Molecular massc | IC50d | References |
|---|---|---|---|---|
| Da | m | |||
| AE234 | cNKYFSNICW | 1274.97 (0.45) | 0.15 × 10−6 | 41,42 |
| AE235 | CIYKNWFNcS | 1274.48 (0.05) | >10−6 | |
| AE105 | DXFsrYLWS | 1225.50 (0.11) | 11 × 10−9 | 32 |
| AE120 | (DXFsrYLWSG)2-βAK | 2745.46 (0.05) | 2.7 × 10−9 | 32 |
| AE151 | (rWDXSLsFYG)2-βAK | 2745.51 (0.09) | >10−6 | 32 |
| GFD | Residues 1–48 of human uPA | 5362.30 (0.04) | 0.56 × 10−9 | 48 |
a AE234 is equivalent to cyclo[21,29][d-Cys19-Cys29]-uPA21–30 (42) and AE235 is a scrambled control.
b The sequences are shown in the single-letter code, where capital letters represent l-amino acids and lower case letters represent d-amino acids. Underlined cysteines (C) indicate that these thiols are oxidized to create an intramolecular disulfide bond (cyclic peptide). X is β-cyclohexyl-l-alanine, and βA is β-l-alanine.
c Molecular masses were determined on a Bruker Autoflex TOF/TOF mass spectrometer using α-cyano-4-hydroxycinnamic acid as desorption matrix except for GFD, which was analyzed as described (48). The determined monoisotopic masses are shown along with the deviations from the theoretical masses in parentheses.
d These IC50 values were determined by a surface plasmon resonance-based assay, where their impact on the association rate (vobs) for 1 nm uPAR to a high density of immobilized pro-uPA (>0.1 pmol/mm2) was measured relative to a serial 2-fold dilution curve for uPAR (Fig. 3). Because of the high affinity of AE120 and GFD, only 0.5 nm uPAR was used in these competition assays.
Purified Protein Preparations
Soluble forms of recombinant human uPAR (residues 1–283) were expressed by stably transfected Drosophila melanogaster S2 cells (33), and a library of >300 purified uPAR mutants carrying single-site substitutions was prepared and characterized as described (34). Recombinant human pro-uPAS356A (residues 1–411) without catalytic activity due to the active-site mutation, pro-uPAΔGFD (residues 45–411), and the N-terminal fragment (ATF) of uPA (residues 1–143) were all expressed by D. melanogaster S2 cells and affinity-purified using the immobilized anti-uPA monoclonal antibody, clone-6 (34). The growth factor-like domain of human uPA (GFD1–48) was a kind gift from S. Rosenberg (35). Purified, native human vitronectin was purchased from Molecular Innovations (Novi, MI) or Invitrogen. The somatomedin B (SMB1–47) domain of human vitronectin was expressed in Pichia pastoris and purified as described (36). Monoclonal anti-uPAR antibodies R2, R3, R4, R5, R8, R9, and R24 were produced in-house as outlined (37), but R3, R4, and R5 are also commercially available from BioPorto (Gentofte, Denmark). R20 and R21 were generated by immunizing uPAR-deficient mice with purified human uPAR, whereas purified murine uPAR was used for the generation of mR1 (38). VIM-5 was from Acris Antibodies (Herford, Cambridge, UK).
HEK293 Cells Expressing uPARwt and Selected Mutants
Stable clones of HEK293 cells expressing comparable levels of human uPARwt, uPARW32A, and uPARY57A were established and characterized by FACS analyses as described (21). These cells were cultured in minimum Eagle's medium (Invitrogen) supplemented with Glutamax I, non-essential amino acids, 10% (v/v) FCS, 100 units/ml penicillin, 100 μg/ml streptomycin, and 400 μg/ml Geneticin at 37 °C in a 5% CO2-humidified incubator.
Induction and Scoring of Lamellipodia Formation in HEK293 Cells Plated on Vitronectin
Cells were seeded in 24-well culture plates onto vitronectin-coated glass coverslips. Coating was accomplished by incubating each coverslip with 70 μl of vitronectin (5 μg/ml in PBS) for 1 h before blocking with 2% (w/v) BSA for 1 h in the same buffer. Subsequently, HEK293 cells (0.5 ml medium containing 5 × 104 cells) were added to each well and allowed to attach for 24 h under the same culture conditions as listed in the previous section. The culture medium was carefully replaced by new medium containing specified additives, e.g. pro-uPA, GFD, peptide surrogates, or monoclonal antibodies. After cultivation for another 24 h, the cells were finally evaluated for lamellipodia formation. Seeding cell densities was optimized for the individual HEK293 clones to reach a semiconfluent culture for the nontreated cells at the end of each experiment.
To facilitate the examination of lamellipodia formation, the adherent cells were fixed on the coverslips, permeabilized, and stained with Alexa 488-conjugated phalloidin to visualize actin filaments (Invitrogen). To this end, coverslips were washed gently with PBS at 37 °C, and the adherent cells were subsequently fixed for 10 min at room temperature in 4% (w/v) paraformaldehyde in PBS before they were permeabilized in 0.2% (v/v) Triton X-100 in PBS for 5 min at room temperature. Finally, the coverslips were recovered from the culture wells and incubated with Alexa 488-labeled phalloidin (0.4 units/ml in PBS with 1% (w/v) BSA) by placing them upside-down in 80-μl droplets on Parafilm sheets for 30 min at room temperature. Extensive washing in PBS was performed after each of the above incubations. To preserve fluorescence, the coverslips were washed once in H2O and mounted in ProLong® Gold antifade reagent (Invitrogen).
An unbiased evaluation of the lamellipodia-positive cells was achieved by recording micrographs of six randomly selected fields for each experimental condition using a Leica DM4000B fluorescence microscope equipped with a Leica DFC-480 camera. These fields were subsequently randomized and examined by four skilled researchers who were unaware of the sample identities. Each field was then assigned the number 0 (no lamellipodia-positive cells present) or 1 (the field contains at least one lamellipodia-positive cell) by the individual investigators. The protrusion index is then defined as the cumulative score (range 0–24) obtained from all investigators for all fields of the sample in question as outlined previously (21).
Evaluation of Untransfected Monocytic Cell Lines
The human monocytic cell lines U937, THP-1, and HL-60 were obtained from ATCC (codes CRL-1593, TIB-202, and CCL-240, respectively) and propagated in RPMI 1640 medium with Glutamax I, 10% (v/v) FCS, 100 units/ml penicillin, and 100 μg/ml streptomycin. These cell lines were seeded on vitronectin-coated coverslips for 24 h as described for HEK293 cells, except that the medium was supplemented with 100 nm phorbol 12-myristate 13-acetate (PMA) with or without 10 nm pro-uPAS356A just before seeding. After this initial stimulation, the culture medium was carefully replaced with freshly prepared medium containing 100 nm PMA and various combinations of 10 nm pro-uPAS356A and 100 nm anti-uPAR mAbs. After an additional incubation for 24 h, cells were processed for microscopic evaluation as described for HEK293 cells.
Rate Constants for the mAb-uPAR interactions Measured by Surface Plasmon Resonance
All interaction studies were carried out on a Biacore 3000TM (Biacore, Uppsala, Sweden) using 10 mm HEPES, 150 mm NaCl, 3 mm EDTA, and 0.005% (v/v) surfactant P-20 at pH 7.4 as running buffer. Purified mouse anti-uPAR mAbs (1 μg/ml) were covalently immobilized on a carboxymethylated dextran matrix (CM5 sensor chip) using N-hydroxysuccinimide/N-ethyl-N′-[3-(diethylamino)propyl]carbodiimide, yielding levels of 400–1000 RU-immobilized mAbs (39). To establish the kinetics for the mAb-uPAR interactions, serial 2-fold dilutions of purified human uPAR (0.4–200 nm in running buffer) were analyzed at 20 °C with 50 μl/min flow rates. To certify reproducibility of the analyses, 50 nm uPAR was re-tested at the end of each experiment. A typical outcome for such an interaction analysis is shown in supplemental Fig. S1. The kinetic rate constants, kon and koff, were subsequently derived from these real-time interaction analyses by fitting the association and dissociation phases to a bimolecular interaction model using the BIAevaluation 4.1 software (Biacore) as described in detail previously (34). For low affinity interactions, KD was also estimated by fitting binding isotherms at equilibrium. After each run, the sensor chip was regenerated by two consecutive injections of 0.1 m acetic acid in 0.5 m NaCl.
Epitope Mapping of Anti-uPAR mAbs
To identify the functional epitopes for various uPAR-specific mAbs, we analyzed their binding kinetics toward a large number of purified uPAR mutants carrying defined single-site alanine substitutions (34). In some cases we analyzed the interaction at 5 °C to minimize the risk of irreversibly denaturing the less stable mAbs during the repeat cycles of regeneration. To narrow the number of uPAR mutants included in this library screen, the overall domain reactivity of the various mAbs was initially established by determining their reactivity toward the following isolated domain constructs of uPAR using Western blotting and/or surface plasmon resonance: uPAR DI1–92, uPAR DII+DIII88–283, and uPAR DIII182–283 (supplemental Table SI).
Determination of IC50 Values for Various Antagonists of the uPA-uPAR Interaction
To determine the IC50 values for small synthetic peptide inhibitors of the uPA-uPAR interaction, we initially immobilized high levels of pro-uPAS356A on a CM5 sensor chip (>5,000 RU ∼ 0.1 pmol/mm2). At such high uPA-coating densities, the observed rate for uPAR association (vobs) is directly proportional to the concentration of the analyte when only low concentrations of uPAR are tested (<2 nm). The IC50 values of the peptide antagonist under study were subsequently determined by measuring vobs for a fixed uPAR concentration (e.g. 0.5 nm) preincubated with a 3-fold dilution series of that compound. The residual uPAR activity was calculated from the dose-response curve analyzed in parallel in the absence of inhibitor and then fitted to a four-parameter logistic model. Running and regeneration conditions for the Biacore3000 instrument were as outlined in the previous paragraphs.
The IC50 values for competition of the uPA-uPAR interaction by various anti-uPAR mAbs were determined by a time-resolved immunofluorescence assay. In brief, White Maxisorp fluoroplates (Nunc, Roskilde, Denmark) were coated with 2 μg/ml catalytically inactive pro-uPA (pro-uPAS356A) in 0.5 m carbonate buffer, pH 9.6. Excess binding sites were blocked by incubation with SuperBlockTM (Pierce) diluted with 1 volume of 40 mm phosphate buffer containing 0.3 m NaCl, pH 7.4. The interaction between immobilized pro-uPAS356A and mixtures containing 2.5 nm uPAR preincubated with a 3-fold dilution series of the relevant mAbs (range 0.1 nm to 8 μm) was allowed to proceed for 1 h at room temperature on an orbital shaker. The uPA-bound uPAR was detected by incubation for 1 h with 0.6 μg/ml Eu3+-labeled R2 (a noninhibitory anti-uPAR mAb). The residual uPAR binding activity in each sample is quantified relative to a standard curve acquired from a parallel incubation of a 2-fold dilution of uPAR in buffer (0.04 nm to 5 nm uPAR). All incubations and dilutions were performed in DELFIATM assay buffer (50 mm Tris-buffered saline, pH 7.8, containing 0.5% (w/v) BSA, bovine immunoglobulin, 0.04% (v/v) Tween 40, and 20 μm diethylenetriaminepentaacetic acid) except for dissociation of Eu3+ from its chelator, which was performed in the final step by incubation in DELFIATM enhancement solution. Fluorescence of the free Eu3+ was measured by time-resolved fluorescence using a Fluostar Optima fluorometer (PerkinElmer Life Sciences) with excitation set at 340 nm and reading emission at 615 nm with a 400-μs delay and a 400-μs acquisition window. All graphic artwork on protein structures were generated by PyMol (Schrödinger, Portland, OR).
RESULTS AND DISCUSSION
Receptor Occupancy by uPA Promotes uPAR-mediated Lamellipodia Formation on Vitronectin
Several independent studies have shown previously that high expression levels of uPAR confer increased adherence, motility, and induction of lamellipodia to cells in a process dependent on their interaction with matrix-embedded vitronectin (8, 9, 11, 21). In addition, in vitro evidence suggests that complex formation with uPA, its bona fide high affinity ligand, further potentiates this uPAR-dependent interaction on vitronectin in various cell types (22–25). To explore this molecular interplay between uPA, membrane-bound uPAR, and immobilized vitronectin in more detail, we adopted an assay system in which uPAR-induced morphology changes in stably transfected HEK293 cell lines expressing comparable levels of selected uPAR mutants are evaluated by a semiquantitative scoring method (21). In accordance with previous studies (9, 11, 21), we found that stably transfected HEK293 cells expressing uPARwt exhibited a pronounced interaction with vitronectin-coated coverslips, causing flattening of the cell body and extensive formation of lamellipodia (Fig. 1). We could also repeat previous observations (9, 21) showing that cell lines expressing the single-site mutants uPARW32A and uPARY57A were unable to elicit these vitronectin-induced morphology changes (Fig. 1). Despite their similar impact on cell morphology, these mutations nonetheless target completely different structural epitopes on uPAR. In the crystal structure solved for the SMB-uPAR-ATF complex (17), Trp32 is thus buried in the vitronectin binding interface, and its mutation to alanine obviously impairs this interaction significantly, whereas the affinity for uPA is unaffected (18). In contrast, Tyr57 is located at the bottom of the hydrophobic uPA binding cavity and is completely shielded from solvent in the corresponding uPAR-ATF complex (16, 40). The affinity of uPARY57A for uPA is accordingly decreased by ∼7-fold compared with both uPARwt and uPARW32A (34, 39), whereas vitronectin binding is unaffected.
FIGURE 1.
Regulation of uPAR-dependent lamellipodia formation on vitronectin by uPA ligation. Panel A shows representative micrographs of transfected HEK293 cells expressing high levels of either uPARwt, uPARW32A, or uPARY57A that are plated on a reconstituted vitronectin matrix for 24 h in the presence or absence of 100 nm concentrations of the stated uPA-derivatives. To enhance detection of lamellipodia, cells were fixed, permeabilized, and stained with Alexa 488-labeled phalloidin before micrographs were taken in a fluorescence microscope. Note the absence of lamellipodia in HEK293 cells expressing either uPARW32A or uPARY57A and the reappearance of these structures after the addition of uPA or GFD. Panel B shows an unbiased semiquantitative assessment of the induction of lamellipodia in these cells using the protocol outlined under “Materials and Methods.”
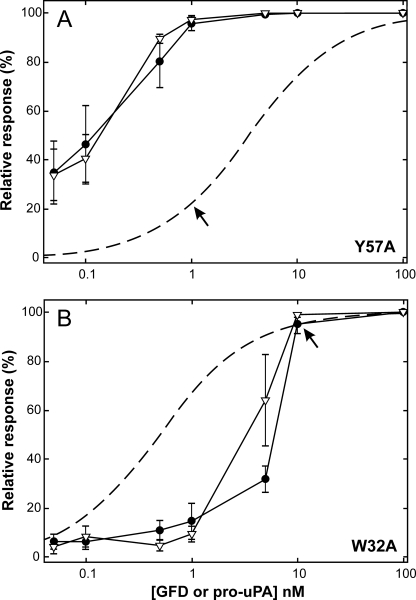
When we add the catalytically inactive pro-uPAS356A or its receptor-binding protein modules ATF and GFD to transfected HEK293 cells seeded on vitronectin-coated coverslips, they all within 24 h induced lamellipodia irrespective of which of the two uPAR mutants the cell lines express (Fig. 1). The amount of uPA derivatives added was in this case sufficiently high (100 nm) to ensure a complete receptor saturation, even for the uPARY57A mutant (18). High concentrations of uPA lacking the receptor binding module (ΔGFD) did not induce lamellipodia in either uPARW32A- or uPARY57A-expressing cell lines (Fig. 1). Intriguingly, dose-response experiments revealed, however, that HEK293 cells expressing uPARY57A required ∼10-fold less pro-uPA or GFD to induce a manifest lamellipodia formation compared with cells expressing uPARW32A (1 versus 10 nm, Fig. 2). This may at first sight appear counterintuitive, bearing in mind that the KD for the interaction between pro-uPA and uPARY57A is 3.5 nm compared with only 0.5 nm for uPARW32A (34). This paradox is, however, easily reconciled by taking into account that the uPA-uPAR Y57A complexes exhibit a vitronectin binding similar to that of uPARwt complexes, whereas uPA-uPAR W32A complexes display an impaired vitronectin binding due to the mutation of a hotspot residue (Trp32) for this interaction (18). A much lower “threshold density” of uPA-uPAR Y57A complexes was, hence, needed on the cell surface to elicit a manifest lamellipodia induction compared with uPA-uPARW32A (Fig. 2).
FIGURE 2.
Dose-response curve for GFD and pro-uPAS356A-induced lamellipodia in HEK293 cells transfected with uPARY57A or uPARW32A. Two stably transfected HEK293 cell lines were stimulated with different levels of GFD (▿) or pro-uPAS356A (●), ranging from 0.05 to 100 nm for 24 h, and protrusion indices were subsequently evaluated as outlined under “Material and Methods.” Data shown represent the mean of 6–8 independent experiments for each concentration of added ligand (bars indicate S.E.). To allow direct comparison to ligand occupancy, the protrusion index is depicted as relative scores. The theoretical saturation curves for uPARW32A and uPARY57A (dashed curves) are calculated based on the KD values previously determined by surface plasmon resonance, 0.5 and 3.5 nm, respectively (34). These graphs reveal that HEK293 cells expressing uPARY57A require only ∼20% ligand saturation to accomplish a complete protrusion score (i.e. at 1 nm ligand), whereas cells expressing uPARW32A require 90–95% ligand saturation (i.e. at 10 nm ligand) as indicated by arrows. It is also clear from these experiments that GFD is just as efficient as pro-uPA in stimulating uPAR-dependent protrusions.
When added at 1 nm, human pro-uPA, but not murine pro-uPA, induced lamellipodia in the uPARY57A-transfected HEK293 cell lines (supplemental Fig. S2), which is concordant with the well established species selectivity of this interaction (7). Importantly, this scenario can be reversed by mutating only two residues in the receptor binding β-hairpin of the GFD module in uPA (supplemental Fig. S2), which causes a switch in the species selectivity of the uPA-uPAR interaction (7). Combined, these data thus clearly demonstrate that the capacity of uPA to induce morphological changes in HEK293 cells expressing two different uPAR mutants, which cannot per se elicit such changes in their unoccupied states, is strictly dependent on ligand binding.
The Small β-Hairpin of GFD Is Sufficient for Efficient Stimulation of uPAR
To explore the minimal structural requirements for this promoting activity of uPA in more detail, we subsequently investigated the impact of two small antagonizing synthetic peptides, both of which engage the central hydrophobic uPA binding cavity of uPAR.
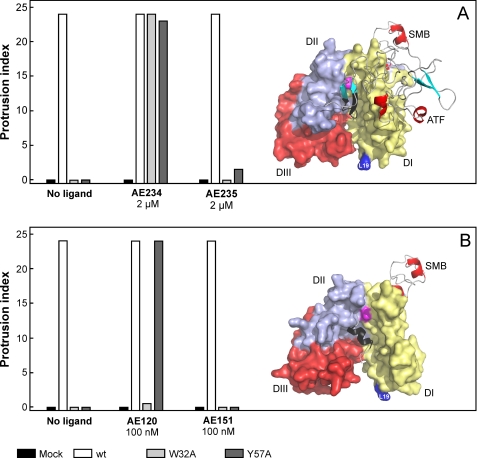
One peptide antagonist (AE234) is designed to mimic the β-hairpin region of GFD, which is responsible for the tight receptor binding properties of uPA (41, 42). This compound is a small 10-mer cyclic peptide, and it inhibited the uPA-uPAR interaction with an IC50 value of 150 nm (Table 1 and Fig. 3). A scrambled version of this peptide (AE235) was also synthesized as a negative control, and it was accordingly shown to have negligible inhibitory effect on the uPA-uPAR interaction (Fig. 3). When these small synthetic peptides were tested on the transfected HEK293 cell lines, AE234 did induce lamellipodia in all uPAR-expressing cell lines. This induction was specific, as the scrambled control peptide (AE235) was unable to provoke a similar response, and the control cells transfected with an empty vector remained devoid of lamellipodia in all experiments (Fig. 4A). This in vitro efficacy of AE234 is truly impressive considering we used only 2 μm, which is ∼10-fold above the determined IC50 value (Table 1). Importantly, this experiment clearly demonstrates that burial of AE234 into the hydrophobic ligand binding cavity of uPAR is enough to restore its lamellipodia-inducing activity in the otherwise defective uPARW32A and uPARY57A cell lines. As this engagement leaves only a very limited surface of AE234 exposed to the solvent and the bound AE234 is located topographically distinct from the SMB binding site (Fig. 4A), this experiment clearly argues for an indirect “allosteric” effect of this peptide antagonist.
FIGURE 3.
Estimation of IC50 values for low molecular weight antagonists of the uPA-uPAR interaction. Panel A shows the linear relationship of the association rate of uPAR (vobs; 10−18 mol/s per mm2) to a high density of immobilized pro-uPAS356A (5800 RU ∼ 0.13 × 10−12 mol/mm2) as a function of the analyzed uPAR concentration (0.5–0.0008 nm). The SDS-PAGE analysis to the left shows the quality of the purified proteins after reduction and alkylation, whereas the raw sensorgrams recorded by surface plasmon resonance for the interaction of a serial 2-fold dilution of uPAR starting at 1 nm are shown to the right. The reproducibility of this data set is illustrated by the repeat analysis of 0.5 nm uPAR at the end of the experiment (shown in blue). vobs was calculated as ΔRU/s from 280 to 415 s and was converted to mol/s assuming that 1 RU ∼ 1 pg/mm2. Panel B shows the competition profile for 0.5 nm uPAR binding to immobilized pro-uPAS356A by a serial 3-fold dilution of the linear peptide antagonist AE120 (300–0.005 nm). The efficacy of the obtained inhibition is visualized by the recorded sensorgrams, and the quality of the data is demonstrated by the repeat analysis of 0.5 nm uPAR without competitor (blue curve). The green curves in A and B are buffer runs. Panel C, the residual levels of unoccupied uPAR in the presence of various concentrations of peptide antagonists were subsequently calculated from recorded vobs (as exemplified for AE120 in B) and the corresponding standard curves (A). The resulting inhibition profiles are shown along with their four-parameter logistic fits for GFD, a cyclic decapeptide derived from the β-hairpin of GFD (AE234) and its scrambled control (AE235), a linear nonapeptide antagonist (AE105), as well as a pseudosymmetrical analog (AE120) and its scrambled control (AE151). The derived IC50 values are shown in Table 1. suPAR, soluble uPAR.
FIGURE 4.
Two different peptide antagonists behave differently as surrogates for stimulation of uPAR-dependent lamellipodia formation. Panel A shows that 2 μm AE234 is capable of inducing robust lamellipodia formation in HEK293 cell lines transfected with uPARW32A or uPARY57A. A scrambled, non-binding version of this peptide (AE235) does not induce this phenomenon. AE234 is a cyclic peptide surrogate of the receptor binding β-hairpin in uPA, where it mimics the region comprising residues 21–30 (shown in black in the model). This region of uPA is almost completely buried in the ATF-uPAR-SMB complex (17). In this schematic, uPAR is shown in a surface representation (DI, yellow; DII, light blue; DIII, red), whereas ATF and SMB are shown as ribbon representations. The position of Leu19 in uPAR is highlighted (blue) to facilitate comparison with data shown in Figs. 5 and 6. Panel B shows that AE120 only is capable of inducing lamellipodia in HEK293 cells transfected with uPARY57A, whereas HEK293 cells expressing uPARW32A are refractory to this stimulation. Its scrambled non-binding version (AE151) is inactive in both transfectants. AE120 is a linear peptide antagonist of the uPA-uPAR interaction, and the crystal structure of uPAR in complex with a truncated analog of AE120 (15) reveals that this antagonist engages the same region as the receptor binding β-hairpin in uPA (see the model). Nevertheless, it stabilizes a notably more open conformation of uPAR relative to that stabilized by ATF. The SMB is added in this schematic merely to illustrate the position of the vitronectin-binding epitope on uPAR. The position of Tyr57 in the central ligand binding cavity at the DI-DII interface is shown in magenta in both figures.
To explore this proposition further, we subsequently tested an unrelated, linear peptide antagonist of uPA binding for its potential to induce lamellipodia in the uPARW32A and uPARY57A mutant cell lines. This peptide (AE120) is a pseudosymmetrical covalent dimer of a 9-mer core peptide (AE105) discovered by phage-display technology and combinatorial chemistry (32). As shown in Fig. 3 and Table 1, AE120 inhibited the uPA-uPAR interaction, with an IC50 value of only 2.7 nm. Being composed of a combination of natural and non-natural amino acids, it is particularly resilient to proteolytic degradation (32). We have previously solved the three-dimensional structure of uPAR in complex with a derivative of AE105 (15), and this AE147-uPAR complex revealed that the linear peptide is indeed buried in the central uPA binding cavity, where its receptor binding core region adopts an α-helical structure (Fig. 4B). Intriguingly, uPAR adopted slightly different three-dimensional structures dependent on which ligand occupies the central ligand binding cavity (compare structures in Figs. 4, A and B). In complex with AE147, uPAR forms the more open structure with a relatively wide breach between DI and DIII. These structural differences become particularly interesting when the efficacies of AE234 and AE120 in inducing lamellipodia are compared. As opposed to uPA, ATF, GFD, and their minimal peptide surrogate AE234, the linear antagonist AE120 only induced lamellipodia in uPARY57A-expressing cells, whereas cells expressing uPARW32A remained completely devoid of such membrane protrusions (Fig. 4B). The corresponding scrambled version (AE151) was negative for lamellipodia induction in all cell lines. This remarkable difference could consequently have a bearing on the distinct uPAR conformations that are stabilized by AE234 and AE120, respectively (Fig. 4). Reassuringly, the smaller core peptides AE105 and AE147, the latter of which was used to assist crystallization of uPAR (15), behaved similarly to AE120 in promoting lamellipodia formation albeit with slightly lower potencies (supplemental Fig. S3). These results strongly argue for a model in which the “closed state” of uPAR, which is stabilized by AE234 and uPA, provides an optimal conformation for induction of vitronectin-dependent changes in cell morphology, whereas the “intermediate state” stabilized by AE120 is less efficient.
Mapping Epitopes and Inhibitory Properties of Monoclonal Anti-uPAR Antibodies
To provide additional independent research tools enabling further interrogation of this alleged conformational plasticity of uPAR, we commenced characterizing the domain reactivity and epitope localization of several monoclonal anti-uPAR antibodies as well as testing their impact on uPA and vitronectin binding.
First, the rate constants for the interactions between the individual mAbs and human uPAR were measured by surface plasmon resonance (Table 2). This revealed that all mAbs tested were of relatively high affinity, displaying KD values between 20 pm and 10 nm (Table 2). To document the quality of these data, the recorded sensorgrams and corresponding fittings of the kinetic parameters for one of these interactions (R21) are shown in supplemental Fig. S1. The overall domain reactivity of these mAbs was subsequently assigned by surface plasmon resonance using defined purified domain constructs of uPAR (supplemental Table SI). In Table 2, mAbs with mutually exclusive binding sites are grouped as defined by pairwise epitope binning (data not shown). The functional epitopes for each of these mAbs were finally confined to single-residue resolution by measuring rate constants for a complete alanine-scanning mutant library of purified human uPAR (34) by surface plasmon resonance. As shown in Table 2, we find that the epitopes for all seven mAbs reactive with the uPAR DI cluster within only two immunogenic hotspot bins located at opposite poles of this LU domain.
TABLE 2.
Epitope-mapping and kinetic rate constants for selected anti-uPAR mAbs
| mAb | Domain reactivitya | Hotspot residuesb | konc | koffc | KDc | IC50d |
|---|---|---|---|---|---|---|
| 105m−1s−1 | 10−4s−1 | 10−9m | 10−9m | |||
| R3 | DI, site 1 | Glu33, Leu61, Lys62 | 1.4 ± 0.6 | 0.52 ± 0.22 | 0.37 | 2.4 |
| R21 | DI, site 1 | Thr59, Gly60, Leu61, Lys62 | 4.1 ± 1.0 | 3.2 ± 0.3 | 0.78 | 1.2 |
| VIM-5 | DI, site 1 | Leu61, Lys62, Ile63 | 1.9 ± 0.5 | 16.3 ± 0.8 | 8.6 | 0.20 × 103 |
| R5 | DI, site 2 | Arg2, Glu16, Leu19, Gly20 | 3.6 ± 0.6 | 10.8 ± 0.04 | 3.1 | 0.22 × 103 |
| R9 | DI, site 2 | Arg2, Glu16, Asp74 | 1.4 ± 0.9 | 9.5 ± 0.03 | 7.7 | 0.20 × 103 |
| R20 | DI, site 2 | Glu16, Leu19, Asp22, Asp74 | 2.9 ± 1.5 | 10.7 ± 0.03 | 4.6 | >10 × 103 |
| mR1 | DI, site 2 | Leu19, Asp22 | 2.1 ± 0.2 | 1.1 ± 0.07 | 0.54 | 0.11 × 103 |
| R4 | DII(DIII) | Arg192, Asp214, Gly217, Ser269 | 4.5 ± 1.3 | 2.8 ± 0.4 | 0.62 | >10 × 103 |
| R8 | DII(DIII) | Arg192, Asp214, Gly217, Ser269 | 6.5 ± 2.1 | 2.4 ± 0.3 | 0.43 | >10 × 103 |
| R2 | DIII | Asp275, Leu276 | 3.2 ± 0.6 | 0.06 ± 0.03 | 0.02 | NAe |
| R24 | DIII | Asp275 | 6.7 ± 1.9 | 6.1 ± 0.08 | 1.0 | NAe |
a The overall domain reactivity for these mAbs was established by surface plasmon resonance using intact uPAR (1–283), uPAR DI (1–92), uPAR DII+DIII (88–283), and uPAR DIII (182–283); see supplemental Table SI. In Table 2, mAbs are clustered in four different bins displaying non-overlapping epitopes between groups but not within groups, as revealed by pairwise binding.
b Functional hotspot residues in epitopes for these mAbs are defined as the 1–4 residues, where alanine substitutions have the greatest impact on the dissociation rate constants (koff) as measured by surface plasmon resonance, as illustrated in Figs. 5 and 6 and supplemental Figs. 4–7.
c The kinetic constants for the interaction between intact soluble uPAR (residues 1–283) and the immobilized mAbs (∼7 fmol/mm2) were measured at 20 °C at a flow rate of 50 μl/min as specified under “Materials and Methods.” Data for R3 were, however, collected at 5 °C because of the lower inherent protein stability of this mAb. Supplemental Fig S1 demonstrates the data quality typically acquired for these analyses, as illustrated for the interaction between immobilized R21 and uPARwt and uPARK62A.
d The IC50 values of the uPA-uPAR interaction were determined by a time-resolved immunofluorescence assay, where the binding of 2.5 nm uPAR to immobilized pro-uPA was detected by Eu3+-labeled R2 as a function of preincubation with a 3-fold dilution series of the respective mAbs.
e NA, not applicable, as these mAbs interfere with the detecting Eu3+-labeled R2.
The first antigenic epitope bin (site 1) is shared by mAbs R3, R21, and VIM-5 (Table 2). It is confined to the solvent-exposed loop 3 in uPAR DI (Leu61 and Lys62), which joins the β-strands βIE and βIF. This shared epitope localization is illustrated for mAbs R21 (Fig. 5) and VIM-5 (supplemental Fig. S4). A common functional denominator for these site 1-reactive mAbs is that their interaction with uPAR precludes subsequent binding of either uPA or vitronectin (Fig. 5, B and C). As judged from the location of their functional epitopes, this inhibitory effect is most likely exerted by a direct steric hindrance of the encounter complex between uPA or SMB and their respective binding sites on uPAR by the bound mAbs.
FIGURE 5.
Defining the site 1 epitope bin in uPAR DI. The data shown in panel A for mAb R21 highlight the location of one of two dominating immunogenic hotspots on DI in intact uPAR (site 1). The interaction between immobilized mAb R21 and a serial 2-fold dilution series of purified human recombinant uPAR mutants (range 6–200 nm) was measured by surface plasmon resonance (Biacore 3000TM). The determined dissociation rate constants (koff) are shown (n = 6) as a function of the positions in the primary sequence of human uPAR DI1–87 that were mutated individually to alanine (omitting positions occupied by cysteines). Secondary structure elements of uPAR DI are shown in the upper section following a previously established nomenclature (15), whereas the primary sequence of DI is shown at the bottom along with the disulfide connectivity and the sequence conservation relative to mouse uPAR (asterisks represent identical residues). The molecular model shown to the left visualizes the location of the hotspot binding site for R21 (i.e. Thr59, Gly60, Leu61, and Lys62, colored blue) relative to the binding sites of the two bona fide ligands using the PDB accession code 3BT1. Bound uPA (represented by ATF) and vitronectin (represented by SMB) are both shown as ribbon diagrams, whereas uPAR is shown in a surface representation with DI, DII, and DIII colored yellow, light blue, and red. The sensorgrams in panel B show that uPAR bound to immobilized R21 is unable to bind 200 nm concentrations of either ATF or GFD (red curves), whereas uPAR bound to immobilized R2, analyzed in parallel and shown for comparison, displays an uncompromised GFD binding (blue curve). The black curves represent binary uPAR-R21 and uPAR-R2 complexes. Panel C shows that uPAR bound to immobilized R21 binds neither 200 nm ATF nor the SMB domain of vitronectin (tested in 2-fold dilution series ranging from 0.1 to 9 μm). As a positive control, the corresponding interactions with uPAR immobilized on mAb R2 were measured in parallel in another flow cell (red curve R2-uPAR and blue curve R2-uPAR-ATF) yielding a KD of 1.8 ± 0.2 μm for the interaction between SMB and R2-uPAR (inset).
Antibodies recognizing the second antigenic epitope bin on uPAR DI (site 2) display, however, much more complex inhibitory profiles. This site, which is shared by mAbs R5, R9, R20, and mR1 (Table 2), is primarily centered on the small loop connecting β-strands βIB and βIC in the disulfide-rich core of this LU domain (i.e. Glu16, Leu19, and Asp22), but contributions from proximal elements, which are located at the N- and C-terminal regions of uPAR DI (i.e. Arg2 and Asp74), are in some cases also evident (Fig. 6 and supplemental Figs. S5 and S6). Despite that site 2 residues in DI do not overlap with the ligand binding site for uPA in intact uPAR (16, 34), three of the four mAbs recognizing this epitope (mR1, R5, and R9) do nonetheless inhibit uPA binding, although with unexpectedly high IC50 values compared to their respective KD values for the interaction with intact uPAR (Table 2). A closer examination of the inhibitory profiles for these particular mAbs at non-equilibrium conditions recorded in real time by surface plasmon resonance reveals, however, that a ternary mAb-uPAR-ATF complex is in fact transiently formed with both mR1 (Fig. 6B), R5 (supplemental Fig. S5), and R9 (38, 43). Interestingly, concomitant binding of either ATF1–143 or GFD4–43 increased the apparent koff for these mAb-uPAR complexes by 10–20-fold, as illustrated in Figs. 6 and supplemental Fig. S5. A similar displacing effect was also observed for the minimal 10-mer peptide mimetic of the β-hairpin in GFD (i.e. AE234), which destabilized these interactions, causing a 3–5-fold increase in their apparent koff value (supplemental Fig. S5C). Importantly, the site 2 epitope bin recognized by these mAbs is non-overlapping with the binding site for the small receptor binding uPA derivatives. These data are in complete accordance with a model in which the engagement of the central, hydrophobic cavity in such a ligand binding elicits a slow rearrangement within uPAR. This rearrangement, in turn, partially distorts the site 2 epitope, thus destabilizing the mAb-uPAR interaction and increasing its dissociation rate constant.
FIGURE 6.
Defining the site 2 epitope bin in uPAR DI. Data shown for mAb mR1, recognizing the second dominating immunogenic epitope (site 2), are shown in A. This panel is organized as in Fig. 5 except that the molecular model for ATF-uPAR-SMB is rotated 180° horizontally as illustrated. The hotspot for mR1 binding is shown in blue, and that for the DIII-reactive mAb R2 is shown in cyan for comparison. The sensorgrams in panel B show that uPAR bound to immobilized mR1 does indeed bind the uPA derivatives ATF (red curve) and GFD (blue curve) when these are injected at 200 nm. The formed ternary complexes are, however, relatively more unstable than the binary uPAR-mR1 complex (black curve), leading to a roughly 15-fold increase in the apparent koff for ATF-uPAR-mR1, 25-fold for GFD-uPAR-mR1, and 3-fold for AE234-uPAR-mR1 (data not shown) during injection of the respective ligands at saturating conditions (B). The sensorgrams in panel C show that SMB does bind uPAR-mR1complexes, but this occurs with a moderately decreased affinity (KD is 4.4 ± 0.5 μm) as compared to that measured for uPAR-R2 complexes measured in parallel (KD is 1.8 ± 0.2 μm), as derived from the equilibrium binding isotherms shown in the inset.
Disruption of uPAR-sustained Lamellipodia by Epitope-mapped mAbs
To test the impact of these monoclonal antibodies on uPAR-dependent lamellipodia formation, they were added to uPARwt-expressing HEK293 cells, which already tightly adhered to vitronectin-coated coverslips. After a subsequent incubation for 24 h with 15 μg/ml (∼100 nm) of the individual mAbs, the protrusion indices were evaluated as described under “Materials and Methods.” As illustrated in Fig. 7A, mAbs recognizing site 1 in human uPAR DI (i.e. R3, R21, and VIM-5) efficiently disrupt already formed lamellipodia. This effect may be anticipated, as all these mAbs efficiently block the vitronectin binding site in uPAR by sterically hindering the accessibility of this site. Therefore, each dissociation event among the multiple uPAR-vitronectin complexes will be followed by blocking uPAR with mAbs rather than renewed complex formation with vitronectin. With time, this will lead to a critical decrease in the number of cell-matrix contacts, thus leading to disruption of lamellipodia. Unexpectedly, all mAbs recognizing site 2 in uPAR DI (i.e. R5, R9, R20, and mR1) exert a similar disruptive effect despite that their epitopes are located remotely from the vitronectin binding site on uPAR. As these particular epitopes are sensitive to rearrangements induced by uPA and AE234 ligation (Fig. 6 and supplemental Fig. S5), it is conceivable that the corresponding mAbs recognize and trap a more “open” conformation of uPAR, which is suboptimal for vitronectin binding. This scenario would thus resemble the one outlined previously for the linear peptide antagonist AE120. As a negative control, a number of mAbs recognizing epitopes on uPAR DIII that do not involve mobile domain interfaces (i.e. R2, R4, R8, and R24) were tested in parallel, and they did not affect the uPAR-induced lamellipodia (Fig. 7A and supplemental Fig. S7).
FIGURE 7.
Effect of various epitope-mapped anti-uPAR monoclonal antibodies on lamellipodia formation. The inhibitory effect of various monoclonal antibodies on uPAR-induced lamellipodia is shown as bar diagrams. In panel A these mAbs were added at a final concentration of 15 μg/ml (∼100 nm) to uPARwt-transfected HEK293 cells already firmly adherent to the vitronectin-coated coverslips. Protrusions were scored 24 h after the addition of the respective mAbs. Identical results were obtained if these mAbs were added during cell seeding (not shown). In panel B, uPARW32A-transfected HEK293 cells were primed to adhere to the vitronectin-coated coverslips by seeding in the presence of 20 nm GFD for 24 h. mAbs representing the different epitope bins (R21, mR1, and R2) were added at a final concentration of 15 μg/ml (∼100 nm) in the presence of 20 nm GFD, and incubation proceeded for an additional 24 h before protrusions were scored. Each bar represents the mean value of four independent measurements. The micrographs show representative appearances of the evaluated HEK293 cells.
To extend these studies to include lamellipodia induction by the ligand-mediated closure of uPAR per se, we tested the inhibitory potential of representative mAbs from the different epitope bins on GFD-induced lamellipodia in HEK293 cells expressing uPARW32A. When these cells were seeded for 24 h on vitronectin-coated coverslips in the presence of 20 nm GFD, they mounted a significant induction of lamellipodia compared to those seeded in the absence of GFD (Fig. 7B). Subsequent incubation for an additional 24 h in the presence of 100 nm mAb and 20 nm GFD shows that mAbs belonging to epitope bins 1 or 2 (R21 and mR1, respectively) efficiently abrogate the GFD-induced lamellipodia in uPARW32A-expressing HEK293 cells, whereas mAb R2 is without effect.
Turning to non-transfected cell lines, it has previously been reported that the anchorage-independent, monocytic cell lines U937, HL60, and THP-1 differentiate into adherent, macrophage-like cells upon cytokine stimulation. Importantly, the acquisition of the adherent phenotype is allegedly dependent on up-regulation of both uPA and uPAR, leading to increased surface densities of the corresponding uPA-uPAR complexes (44, 45). Accordingly, we chose to stimulate these three monocytic cell lines with 100 nm PMA before plating them on vitronectin-coated coverslips in the presence or absence of 10 nm pro-uPAS356A and 100 nm anti-uPAR mAbs representing the various epitope bins. Unfortunately, we failed to induce a high frequency of adherent cells displaying distinct lamellipodia by the combined PMA and uPA treatment, which is opposite to the scenario we observed for unstimulated uPARwt-expressing HEK293 cells. This self-evidently precludes subsequent quantitative studies on the inhibitory potential of the various anti-uPAR mAbs. Nevertheless, even in the presence of 100 nm R21, R5 or mR1, we did occasionally observe a few scattered cells retaining lamellipodia-like structures (supplemental Fig. S8). This circumstantial finding thus tends to suggest that these few cells may have acquired an integrin repertoire (αvβ3 or αvβ5) proficient in enabling adherence to vitronectin and inducing lamellipodia independent of an auxiliary contribution from uPA-uPAR complexes. Further studies are, therefore, clearly required to define the functional significance of the uPA-uPAR axis, including the present molecular model, for the migration of non-transfected cells on vitronectin-rich matrices in vitro and in vivo.
Conclusions
Based on the experimental data discussed above, we now propose a model for the molecular interplay of the uPA-uPAR-vitronectin complexes that addresses the regulatory function of uPAR in the induction of lamellipodia on vitronectin-rich matrices. According to this model, the multidomain uPAR reversibly populates discrete conformational states that differ in their capacity to induce lamellipodia on vitronectin-coated surfaces (Fig. 8). In the absence of uPA, we postulate that a sizable fraction of the glycolipid-anchored uPAR adopts an open conformation on the cell surface, which is unable to induce lamellipodia. The robust induction we and others nevertheless consistently observe in the absence of uPA by cells expressing large amounts of uPARwt (9, 11, 21) is according to this model mediated by a small residual fraction of unoccupied uPAR adopting either an intermediate or closed conformation (see Fig. 8). One obvious corollary to our proposition is that any perturbation of this equilibrium, shifting it toward the open uPAR conformation, should in principle attenuate lamellipodia induction and vice versa. In the present study we challenged this scenario by two different modes of action. In the first, an open conformation of uPAR was allegedly induced and stabilized by mAbs recognizing site 2 on uPAR DI, and reassuringly, this binding abrogated the manifest lamellipodia formation in uPARwt HEK293 cells (Fig. 7). Introducing mutations that destabilize the interdomain interfaces in the modular uPAR structure is our second modus operandi for driving this equilibrium toward a more open uPAR conformation. Accordingly, we show that uPARY57A is indeed unable to induce lamellipodia, but importantly, this activity can be recovered by merely shifting the equilibrium back toward the intermediate or closed uPAR conformation by ligation with AE120 or AE234/GFD, respectively. This model aligns well with data from a previous comprehensive mutagenesis study on glycosylphosphatidylinositol-anchored uPAR expressed in HEK293 cells (9), which showed that individual alanine substitution of several residues located at interdomain interfaces or in the hydrophobic ligand binding cavity of uPAR actually impairs lamellipodia induction (e.g. Leu31, Arg53, Leu55, Tyr57, Leu66, Leu113, Glu120, Leu123, Arg145, and Gly146). None of these residues is located at the vitronectin binding interface in uPAR (16, 18). In our view these mutations accordingly act by destabilizing the interdomain assembly of the glycosylphosphatidylinositol-anchored uPAR, which decreases the surface density of active receptors displaying a proper compact conformation for induction of lamellipodia.
FIGURE 8.
Conformational switch in uPAR regulating lamellipodia formation. This figure schematically depicts the model we propose for ligand-induced regulation of uPAR-mediated lamellipodia formation on vitronectin-rich matrices. We postulate that there is a considerable inherent conformational flexibility in the multidomain assembly of the three homologous LU domains in uPAR, enabling the receptor to explore different conformational states. The equilibrium between these states is sensitive to mutations as well as engagement of different mAbs and ligands as indicated in the figure. The open conformation is inferred from our functional biochemical data, whereas the intermediate and closed conformations are confirmed by x-ray crystal structures of AE147-uPAR (PDB accession code 1YWH) and ATF-uPAR (PDB accession code 2FD6), respectively. The location of Tyr57 at the DI-DII interface is indicated in magenta. Vn, vitronectin.
To dissect the impact of ligand binding on lamellipodia induction, we needed to attenuate the “activity” of unoccupied uPAR to avoid the robust base-line stimulation inherent to the high receptor levels provided by the transfected HEK293 cells. This was accomplished by the uPARW32A mutant, which exhibits a severely compromised vitronectin binding even in the uPA-uPARW32A complex (18). Studies using this particular uPAR mutant accordingly revealed an obligate requirement for the closed conformation to achieve a manifest induction of lamellipodia as ligation with AE120 proved unproductive (intermediate conformation), whereas pro-uPA, ATF, GFD, and their minimal peptide surrogate AE234 endowed uPARW32A with a lamellipodia-inducing activity (stabilizing the closed conformation).
One obvious ramification of this model is the decisive role that adequate surface densities of uPAR have on the outcome for induction of lamellipodia via this pathway. Concordant with this notion, circumstantial evidence in the literature reports that up-regulation of uPAR expression by cytokine treatments induces adherence on vitronectin in certain cell lines (8, 22, 44). This dependence could also be recapitulated using clones of a stably transfected murine myeloid 32D cell line expressing different uPAR levels (25). Along the same lines, these studies showed that uPA binding would lower the apparent threshold of uPAR expression that is required for this induction. Our titration studies on HEK293 cells expressing uPARY57A demonstrate that 20% ligand occupancy is required and sufficient for the manifest induction of lamellipodia in this system (Fig. 2).
Another very important ramification of our molecular model pertains to drug discovery focusing on small inhibitors of the uPA-uPAR interaction. Our model raises the major concern that some small binding antagonists may in fact possess undesirable agonist effects on uPAR-mediated adhesion and migration on vitronectin-rich matrices. This ambiguity of action is illustrated by the inductive effects we observe for AE234, which actually was considered a potential lead compound for pharmaceutical intervention of uPA binding (42, 46). Similar concerns are, however, less relevant for compounds belonging to the AE105-derived class of inhibitors. Such considerations clearly have to be scrutinized when designing new uPAR-targeted intervention modalities (47).
Finally, our model predicts that any hypothetical mutation or mAb stabilizing the intermediate or closed conformation of uPAR will increase its efficacy in promoting lamellipodia formation in its unoccupied state. We are currently pursuing this intriguing possibility by a rational protein design aiming at stabilizing the closed conformation of uPAR in the absence of bound ligand.
Supplementary Material
Acknowledgments
We thank Gitte Juhl Funch, Haldis Egholm, and John Post for excellent technical assistance.
This work was supported by grants from The Danish National Research Foundation (Centre for Proteases and Cancer) and The Lundbeck Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table SI and Figs. S1–S8.
- uPAR
- urokinase-type plasminogen activator (uPA) receptor
- uPAS356A
- catalytically inactive pro-uPA
- ATF
- N-terminal fragment of uPA
- GFD
- growth factor-like domain
- LU domain
- Ly6/uPAR-type like domain
- SMB
- somatomedin B-like domain of vitronectin
- RU
- resonance units
- DI and DII
- domains I and II, respectively.
REFERENCES
- 1. Friedl P., Gilmour D. (2009) Nat. Rev. Mol. Cell Biol. 10, 445–457 [DOI] [PubMed] [Google Scholar]
- 2. Friedl P., Wolf K. (2009) Cancer Metastasis Rev. 28, 129–135 [DOI] [PubMed] [Google Scholar]
- 3. Rozario T., DeSimone D. W. (2010) Dev. Biol. 341, 126–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Smith H. W., Marshall C. J. (2010) Nat. Rev. Mol. Cell Biol. 11, 23–36 [DOI] [PubMed] [Google Scholar]
- 5. Kjaergaard M., Hansen L. V., Jacobsen B., Gårdsvoll H., Ploug M. (2008) Front. Biosci. 13, 5441–5461 [DOI] [PubMed] [Google Scholar]
- 6. Ellis V., Scully M. F., Kakkar V. V. (1989) J. Biol. Chem. 264, 2185–2188 [PubMed] [Google Scholar]
- 7. Lin L., Gårdsvoll H., Huai Q., Huang M., Ploug M. (2010) J. Biol. Chem. 285, 10982–10992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wei Y., Waltz D. A., Rao N., Drummond R. J., Rosenberg S., Chapman H. A. (1994) J. Biol. Chem. 269, 32380–32388 [PubMed] [Google Scholar]
- 9. Madsen C. D., Ferraris G. M., Andolfo A., Cunningham O., Sidenius N. (2007) J. Cell Biol. 177, 927–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Salasznyk R. M., Zappala M., Zheng M., Yu L., Wilkins-Port C., McKeown-Longo P. J. (2007) Matrix Biol. 26, 359–370 [DOI] [PubMed] [Google Scholar]
- 11. Kjøller L., Hall A. (2001) J. Cell Biol. 152, 1145–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ploug M., Rønne E., Behrendt N., Jensen A. L., Blasi F., Danø K. (1991) J. Biol. Chem. 266, 1926–1933 [PubMed] [Google Scholar]
- 13. Galat A. (2008) Cell. Mol. Life Sci. 65, 3481–3493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jacobsen B., Ploug M. (2008) Curr. Med. Chem. 15, 2559–2573 [DOI] [PubMed] [Google Scholar]
- 15. Llinas P., Le Du M. H., Gårdsvoll H., Danø K., Ploug M., Gilquin B., Stura E. A., Ménez A. (2005) EMBO J. 24, 1655–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huai Q., Mazar A. P., Kuo A., Parry G. C., Shaw D. E., Callahan J., Li Y., Yuan C., Bian C., Chen L., Furie B., Furie B. C., Cines D. B., Huang M. (2006) Science 311, 656–659 [DOI] [PubMed] [Google Scholar]
- 17. Huai Q., Zhou A., Lin L., Mazar A. P., Parry G. C., Callahan J., Shaw D. E., Furie B., Furie B. C., Huang M. (2008) Nat. Struct. Mol. Biol. 15, 422–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gårdsvoll H., Ploug M. (2007) J. Biol. Chem. 282, 13561–13572 [DOI] [PubMed] [Google Scholar]
- 19. Sidenius N., Andolfo A., Fesce R., Blasi F. (2002) J. Biol. Chem. 277, 27982–27990 [DOI] [PubMed] [Google Scholar]
- 20. Kanse S. M., Kost C., Wilhelm O. G., Andreasen P. A., Preissner K. T. (1996) Exp. Cell Res. 224, 344–353 [DOI] [PubMed] [Google Scholar]
- 21. Hillig T., Engelholm L. H., Ingvarsen S., Madsen D. H., Gårdsvoll H., Larsen J. K., Ploug M., Danø K., Kjøller L., Behrendt N. (2008) J. Biol. Chem. 283, 15217–15223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Waltz D. A., Chapman H. A. (1994) J. Biol. Chem. 269, 14746–14750 [PubMed] [Google Scholar]
- 23. Stahl A., Mueller B. M. (1997) Int. J. Cancer 71, 116–122 [DOI] [PubMed] [Google Scholar]
- 24. Chang A. W., Kuo A., Barnathan E. S., Okada S. S. (1998) Arterioscler. Thromb. Vasc. Biol. 18, 1855–1860 [DOI] [PubMed] [Google Scholar]
- 25. Sidenius N., Blasi F. (2000) FEBS Lett. 470, 40–46 [DOI] [PubMed] [Google Scholar]
- 26. Cunningham O., Andolfo A., Santovito M. L., Iuzzolino L., Blasi F., Sidenius N. (2003) EMBO J. 22, 5994–6003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pluskota E., Soloviev D. A., Plow E. F. (2003) Blood 101, 1582–1590 [DOI] [PubMed] [Google Scholar]
- 28. Wei Y., Czekay R. P., Robillard L., Kugler M. C., Zhang F., Kim K. K., Xiong J. P., Humphries M. J., Chapman H. A. (2005) J. Cell Biol. 168, 501–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wei Y., Lukashev M., Simon D. I., Bodary S. C., Rosenberg S., Doyle M. V., Chapman H. A. (1996) Science 273, 1551–1555 [DOI] [PubMed] [Google Scholar]
- 30. Fazioli F., Resnati M., Sidenius N., Higashimoto Y., Appella E., Blasi F. (1997) EMBO J. 16, 7279–7286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Resnati M., Pallavicini I., Wang J. M., Oppenheim J., Serhan C. N., Romano M., Blasi F. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 1359–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ploug M., Østergaard S., Gårdsvoll H., Kovalski K., Holst-Hansen C., Holm A., Ossowski L., Danø K. (2001) Biochemistry 40, 12157–12168 [DOI] [PubMed] [Google Scholar]
- 33. Gårdsvoll H., Werner F., Søndergaard L., Danø K., Ploug M. (2004) Protein Expr. Purif. 34, 284–295 [DOI] [PubMed] [Google Scholar]
- 34. Gårdsvoll H., Gilquin B., Le Du M. H., Ménèz A., Jørgensen T. J., Ploug M. (2006) J. Biol. Chem. 281, 19260–19272 [DOI] [PubMed] [Google Scholar]
- 35. Stratton-Thomas J. R., Min H. Y., Kaufman S. E., Chiu C. Y., Mullenbach G. T., Rosenberg S. (1995) Protein Eng. 8, 463–470 [DOI] [PubMed] [Google Scholar]
- 36. Kjaergaard M., Gårdsvoll H., Hirschberg D., Nielbo S., Mayasundari A., Peterson C. B., Jansson A., Jørgensen T. J., Poulsen F. M., Ploug M. (2007) Protein Sci. 16, 1934–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rønne E., Behrendt N., Ellis V., Ploug M., Danø K., Høyer-Hansen G. (1991) FEBS Lett. 288, 233–236 [DOI] [PubMed] [Google Scholar]
- 38. Pass J., Jögi A., Lund I. K., Rønø B., Rasch M. G., Gårdsvoll H., Lund L. R., Ploug M., Rømer J., Danø K., Høyer-Hansen G. (2007) Thromb. Haemost. 97, 1013–1022 [PubMed] [Google Scholar]
- 39. Gårdsvoll H., Danø K., Ploug M. (1999) J. Biol. Chem. 274, 37995–38003 [DOI] [PubMed] [Google Scholar]
- 40. Ploug M., Rahbek-Nielsen H., Ellis V., Roepstorff P., Danø K. (1995) Biochemistry 34, 12524–12534 [DOI] [PubMed] [Google Scholar]
- 41. Magdolen V., Bürgle M., de Prada N. A., Schmiedeberg N., Riemer C., Schroeck F., Kellermann J., Degitz K., Wilhelm O. G., Schmitt M., Kessler H. (2001) Biol. Chem. 382, 1197–1205 [DOI] [PubMed] [Google Scholar]
- 42. Schmiedeberg N., Schmitt M., Rölz C., Truffault V., Sukopp M., Bürgle M., Wilhelm O. G., Schmalix W., Magdolen V., Kessler H. (2002) J. Med. Chem. 45, 4984–4994 [DOI] [PubMed] [Google Scholar]
- 43. List K., Høyer-Hansen G., Rønne E., Danø K., Behrendt N. (1999) J. Immunol. Methods 222, 125–133 [DOI] [PubMed] [Google Scholar]
- 44. Waltz D. A., Sailor L. Z., Chapman H. A. (1993) J. Clin. Invest. 91, 1541–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nusrat A. R., Chapman H. A., Jr. (1991) J. Clin. Invest. 87, 1091–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sato S., Kopitz C., Schmalix W. A., Muehlenweg B., Kessler H., Schmitt M., Krüger A., Magdolen V. (2002) FEBS Lett. 528, 212–216 [DOI] [PubMed] [Google Scholar]
- 47. Kriegbaum M., Persson M., Haldager L., Alpizar-Alpizar W., Jacobsen B., Gårdsvoll H., Kjær A., Ploug M. (2011) Current Drug Targets 12, in press [DOI] [PubMed] [Google Scholar]
- 48. Jørgensen T. J., Gårdsvoll H., Danø K., Roepstorff P., Ploug M. (2004) Biochemistry 43, 15044–15057 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.