Abstract
Matrix metalloproteinases (MMPs) have been shown to be key players in both extracellular matrix remodeling and cell migration during cancer metastasis. MMP-14, a membrane-anchored MMP, in particular, is closely associated with these processes. The hemopexin (PEX) domain of MMP-14 has been proposed as the modulating region involved in the molecular cross-talk that initiates cell migration through homodimerization of MMP-14 as well as heterodimerization with the cell surface adhesion molecule CD44. In this study, minimal regions required for function within the PEX domain were investigated through a series of substitution mutations. Blades I and IV were found to be involved in cell migration. We found that blade IV is necessary for MMP-14 homodimerization and that blade I is required for CD44 MMP-14 heterodimerization. Cross-talk between MMP-14 and CD44 results in phosphorylation of EGF receptor and downstream activation of the MAPK and PI3K signaling pathways involved in cell migration. Based on these mutagenesis analyses, peptides mimicking the essential outermost strand motifs within the PEX domain of MMP-14 were designed. These synthetic peptides inhibit MMP-14-enhanced cell migration in a dose-dependent manner but have no effect on the function of other MMPs. Furthermore, these peptides interfere with cancer metastasis without affecting primary tumor growth. Thus, targeting the MMP-14 hemopexin domain represents a novel approach to inhibit MMP-14-mediated cancer dissemination.
Keywords: Cancer Therapy, Cell Migration, Matrix Metalloproteinase (MMP), Peptides, Signal Transduction, Hemopexin Domain, MMP-14, Invasion, Metastasis
Introduction
Matrix metalloproteinases (MMPs)4 are a class of zinc-dependent endopeptidases that collectively are responsible for the degradation of nearly all extracellular matrix and basement membrane components at times of protease-mediated tissue remodeling (1). MMPs play roles within various areas of cancer pathology, including tumor growth, metastasis, and angiogenesis, and MMP activation is increased in nearly all human cancers when compared with normal tissue (2). The catalytic domains of all MMPs share high amino acid similarity, and their active sites are extensively conserved. As a consequence, distinguishing between MMPs with small molecule substrates or inhibitors is extraordinarily difficult. Lack of specificity, off-target effects, and troubling side effects led to difficulty in the clinic with active-site directed MMP inhibitors (3), thus reinforcing the current need for a more in-depth analysis of unique molecular features of individual MMPs.
With the exception of six membrane-anchored MMPs, the remaining 17 human MMPs are destined for secretion into the extracellular milieu (4). MMP-14, a membrane-type MMP (MT1-MMP), is a critical protein in cancer invasion and metastasis. Invasion through collagen networks and subsequent collagenolysis relies principally on MMP-14 and not on secreted MMPs (5). After MMP-14 trafficking from the trans-Golgi network to the cell surface, proMMP-2 and proMMP-13 are activated, which further enhances the proteolytic repertoire characteristic of invasive cancer cells (6, 7). In an effort to minimize the extracellular matrix remodeling necessary for tissue invasion, MMP-14 is localized at the invadopodia or the leading edge of the cell (8). This migratory front not only focuses extracellular matrix degradation, but it also enables cells to migrate through the tissue in the direction led by MMP-14. This inclination is accomplished through interaction of MMP-14 with a membrane-associated glycoprotein, CD44 (9).
The domain organization of most MMP family members is conserved, including that of MMP-14. Starting at the N terminus, a signal peptide, a propeptide, a catalytic domain, a hinge region, and a hemopexin-like (PEX) domain comprise the conserved domains. In addition to these five typical domains, MMP-14, MMP-15, MMP-16, and MMP-24 also contain a transmembrane region and a short cytoplasmic tail (10). The PEX domain of MMP-14 is the region responsible for association with CD44 that leads to intracellular cytoskeleton rearrangements and the processing of migration and invasion machinery, including proteolysis of proMMP-2 by MMP-14 (9, 11). The MMP hemopexin domain is a four-blade β-propeller (12, 13). On the cell surface, MMP-14 homodimerizes through its PEX domain, and dimer formation is a component of proMMP-2 activation (14).
In this study, a detailed analysis of the MMP-14 hemopexin domain, including homo- and heterodimerizations, was undertaken to dissect the mechanism of MMP-14-mediated cell migration. The data presented provide detailed insight into how the individual blades of the MMP-14 PEX domain promote both cellular migration as well as invasion. Furthermore, synthetic peptides comprising the outer-strand amino acid sequences of the individual PEX domain blades were prepared and found to interfere with MMP-14-induced cell migration in vitro and to prevent cancer cell metastasis in vivo. The physiological impact of these small peptides serves as proof of principle for development of PEX domain-specific MMP inhibitory drugs.
EXPERIMENTAL PROCEDURES
Reagents
Oligo primers were purchased from Operon (Huntsville, AL). The pcDNA3.1myc expression vectors were purchased from Invitrogen. Anti-Myc and anti-HA antibodies were purchased from Roche. Anti-tubulin, anti-AKT, anti-pAKT, anti-ERK, anti-pERK, anti-pEGFR, and anti-EGFR antibodies were purchased from Cell Signaling Technology, Inc. (Danvers, MA). The MMP-16 and MMP-25 plasmids were generously provided by Dr. Steven Weiss (University of Michigan). The CD44H plasmid was generously provided by Dr. Bryan Toole (University of South Carolina).
Cell Culture and Transfection
The COS-1 African green monkey epithelial, human fibrosarcoma HT-1080, breast cancer MCF-7 and MDA-MB-231, prostate cancer LNCaP, and human cancer MDA-MB-435 cell lines were purchased from the ATCC and were maintained in Dulbecco's modified Eagle's medium (Invitrogen) with 10% fetal calf serum. Transfection of plasmid DNA (human) into cells was achieved using polyethylenimine (Polysciences), and the transfected cells were incubated for 48 h at 37 °C followed by biochemical and biological assays.
DNA Construction
The construction of MMP-14-green fluorescent protein (MMP14-GFP) chimeric cDNA was reported previously (15). A construct to express MMP-14 with a carboxy-terminal Myc-tag (MMP14Myc) was engineered by inserting the MMP-14 open reading frame into the pcDNA3.1Myc vector (Invitrogen) to generate the MMP14Myc chimeric cDNA. Employing a two-step PCR approach (15), cDNA encoding a MMP-14 with an HA (human influenza hemagglutinin) tag (MMP14HA) between the propeptide and catalytic domains was generated. Briefly, the first step PCR was performed to amplify fragments A and B containing an HA tag with the complementary sequence at both the 3′ end of fragment A and the 5′ end of fragment B using the primer set listed in supplemental Table 1. The second PCR fused fragments A and B using forward primer 1563 and reverse primer 1564, and was followed by subcloning the entire cDNA into the pcDNA3 vector (Invitrogen). This approach was employed to generate all the substitution mutants in which blade strand sequences in the MMP-14 PEX domain were replaced with the corresponding sequences from MMP-1 to generate MMP14HA-MMP1IS4, MMP14HA-MMP1IIS4, MMP14HA-MMP1IIIS4, andMMP14HA-MMP1IVS4, respectively. The substitution mutant amino acid sequences are listed in supplemental Table 2.
To generate GFP-tagged MMP14-MMP1 mutants, MMP14-GFP chimeric cDNA was first cloned into retroviral vector pQXCIP (Clontech) to generate MMP14-GFP/pQXCIP. PCR was then performed to amplify MMP14HA-MMP1 chimeras using primers 1563 (forward primer, see supplemental Table 1) and 1486 (reverse primer, 5′ GAAGATCTGA CCTTGTCCAGCAGGGAACG 3′). The PCR fragments were digested by XhoI and BglII, and the resultant XhoI-BglII fragments were cloned into the MMP14-GFP/pQXCIP vector (XhoI and BamHI) to generate MMP14-MMP1IS4, MMP14-MMP1IIS4, MMP14-MMP1IIIS4, andMMP14-MMP1IVS4-GFP/pQXCIP. The accuracy of all the constructs was confirmed by DNA sequencing.
Peptide Synthesis
The peptides were synthesized by EZBiolab (Carmel, IN), and purity was determined to be greater than 98% by HPLC. Peptides were capped at the N and C termini with acetyl and amide groups, respectively. Peptide sequences are listed in supplemental Table 3.
Data Mining Analysis of MMP-14
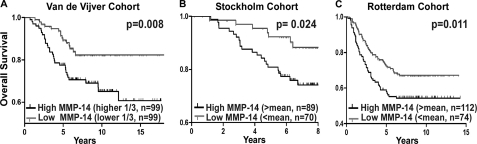
Three independent cohorts (16–18) were analyzed for the correlation of MMP-14 with the survival probability of patients with breast cancer. Stockholm (GSE 1456) and Rotterdam (GSE 2034) cohorts were obtained through the NCBI/GenBanktm GEO database. In the Stockholm dataset, there were 159 patients with mean age of 58 at diagnosis and a mean tumor diameter of 22 mm. There were 286 patients with ages ranging from 26–83 years and without any adjuvant therapy in the Rotterdam cohort. Dichotomization of the patients was done by using the average expression value of MMP-14 for both cohorts. Then, patients were placed into two groups: high MMP-14 (> mean) and low MMP-14(< mean). A Kaplan-Meier analysis was performed to correlate MMP-14 expression with the patient survival rate. A van de Vijver cohort was retrieved from Rosetta Inpharmatics. The age at diagnosis was 52 years or younger. High MMP-14 and low MMP-14 dichotomization was done by using the upper one-third and lower one-third of the patients. A Kaplan-Meier analysis was performed to correlate MMP-14 expression with the patient survival rate. Log Rank (Mantel Cox) was used to determine the statistical significance for the survival curve analysis of each cohort.
The correlation of MMP-14 expression with probability of metastasis was also evaluated in a van de Vijver cohort (16). MMP-14 expression in the metastasis and metastasis-free groups was analyzed by using a two-tailed t test with 95% confidence intervals.
Transwell Chamber Migration Assay
Polycarbonate membranes with 8-μm pore size were assembled in blind well chemotactic chambers (Neuro Probe, MD). The lower chamber was filled with DMEM containing 10% FCS. The upper chamber was filled with 20,000 cells suspended in the same FCS-containing medium. Chambers were incubated for 18 h at 37 °C. The cells remaining on the top surface of the membrane were removed with application of a cotton swab followed by three PBS washes. The cells on the bottom surface of the membrane were fixed, stained (0.1% crystal violet), and quantified by counting 10 fields on the membrane under a ×20 objective. For migration assays examining peptide inhibition, cells were incubated with peptides for 30 min prior to application to migration chambers.
Coimmunoprecipitation
Cells were lysed in 1% CHAPS lysis buffer containing a protease inhibitor mixture (Sigma). Cell lysates were incubated with either anti-Myc or anti-CD44 antibodies overnight. Antigen-antibody complexes were then precipitated by addition of protein A-agarose beads (Invitrogen) for 1 h at 4 °C. Immunoblotting was carried out with anti-HA antibody.
Three Dimensional (3D) Invasion Assay
Cells mixed with an equal volume of type I collagen (3 mg/ml) were dotted onto a 96-well plate followed by covering with additional layer of type I collagen gel (1.5 mg/ml). Media containing peptides were added and incubated for 24 h, followed by the counting of invaded cells.
Gelatin Zymography
Gelatin zymography was carried out using 10% SDS-polyacrylamide gels containing 0.1% gelatin. After electrophoresis, SDS was removed by incubation of the gel with 2.5% Triton X-100, and gelatinase activity was recovered by incubation in a Tris-based buffer containing 10 mm calcium for 24 h. Gels were stained with Coomassie Brilliant Blue, and enzymatic activity was detected by observing the lack of gelatin protein in sample lanes, represented by a clear band.
Retrovirus Infection
A retroviral supernatant was obtained by cotransfection of a vector encoding the envelope gene (pAmphoteric) and a retroviral expression vector containing shRNA against CD44 or wild-type and mutant MMP14-GFP constructs into human embryonic kidney GP2–293 packaging cells (Clontech) according to the manufacturer's protocol. Cells were infected with the supernatant containing the retrovirus in the presence of 4 μg/ml polybrene and selected with 4 μg/ml of puromycin.
Chorioallantoic Membrane (CAM) Angiogenesis Assay
This assay was performed as described previously (19). Fertilized white chicken eggs (Charles River Laboratory, Wilmington, MA) were incubated at 37 °C in 75% humidity for 3 days. The chicken embryo was then removed from the shell and placed in a sterile Petri dish for an additional 7 days in the incubator. Implantation of a tumor cell-soaked sponge was carried out on day 10 as described previously (19, 20). Peptides were added to the sponge every day. On day 4 after implantation, new blood vessels were examined, photographed, and counted.
In Vivo Animal Studies
MDA-MB-435 cells expressing GFP (2 × 106) were inoculated subcutaneously into mammary tissue of 4- to 5-week-old female NCR-Nu mice with six mice per group (Taconic). Once palpable, tumors were measured twice/week, and the volume was calculated using the following formula: length × height × 0.5236. Mice were treated with generic scrambled control peptide (supplemental Table 3), IS4, or IVS4 peptide (20 mg/kg). Peptides were administered through intraperitoneal and intratumoral injection on alternate days, 6 days per week. At the end of the experiment, mice were sacrificed, and tumor and lungs were dissected. Fresh lung sections were cut (approximately 5 mm thick) and examined for the presence of GFP-expressing tumor foci. Lung metastases were visualized under the microscope using a FITC filter to detect metastatic MDA-MB-435/GFP cells.
Statistical Analysis
Data are expressed as the mean ± S.E. of triplicates. Each experiment was repeated at least three times. Student's t test and analysis of variance were used to assess differences (*, p < 0.05; **, p < 0.01; ***, p < 0.001.
RESULTS
Correlation between Cancer Patient Survival Rate and MMP-14 Expression
Mounting evidence has demonstrated that MMP-14 plays an important role in cancer dissemination (21). To gain knowledge of the clinical significance of MMP-14 in patients with cancer, we employed a data mining approach to correlate MMP-14 expression with patient outcome. In the van de Vijver cohort, which contains 295 patients with different stages of breast cancers (16), MMP-14 was up-regulated in patients with metastatic breast cancer as compared with metastasis-free patients (p = 0.003).
To determine the correlation between MMP-14 expression with patient survival rates, a Kaplan-Meier survival analysis of publicly available microarray datasets was carried out. High expression of MMP-14 inversely correlated with patient survival time in all datasets analyzed (Fig. 1, A–C) (16–18). These analyses emphasize that MMP-14 can be used as a prognostic marker for patients with cancer and provides clinical evidence for the role of MMP-14 in cancer dissemination. Furthermore, these analyses suggest that targeting MMP-14 is a viable approach to prevent cancer dissemination and to improve survival rates for patients with cancer.
FIGURE 1.
Clinical relevance of MMP-14 in patients with breast cancer. DNA microarray data mining of van de Vijver, Stockholm, and Rotterdam cohorts reveal a correlation between MMP-14 expression and overall survival rate using Kaplan-Meier survival analysis. Levels of MMP-14 RNA were dichotomized at upper versus lower third (A) and mean (B and C). n = cases. Log Rank (Mantel Cox) was used to determine the statistical significance for the survival curve analysis of each group.
Identification of Motifs within the PEX Domain of MMP-14 Required for Enhanced Cell Migration
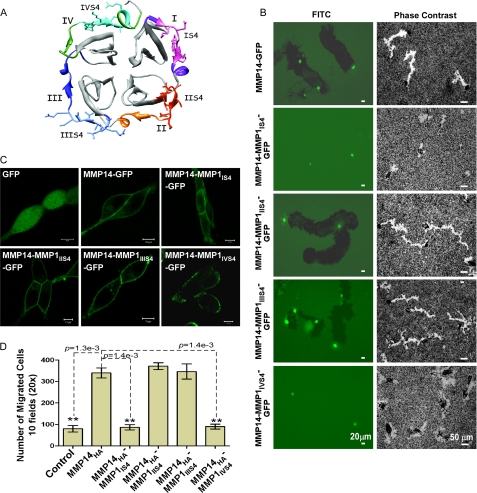
The PEX domains of all MMPs exhibit a highly conserved structure composed of a disc-like shape, with the chain folded into a β-propeller structure that has a pseudo-four-fold symmetry (22). Each propeller contains a sheet of four antiparallel strands with peptide loops linking one sheet to the next, as illustrated in Fig. 2A. Only inner strands 1–3 within each blade are topologically conserved and can be superimposed (12, 13). The fourth outer strand (S4) of each blade deviates considerably between different MMP PEX domains (12). This suggests that the inner three strands constitute the structural framework of the β-propeller architecture, whereas the outer strand of each blade mediates contacts with other specific protein components.
FIGURE 2.
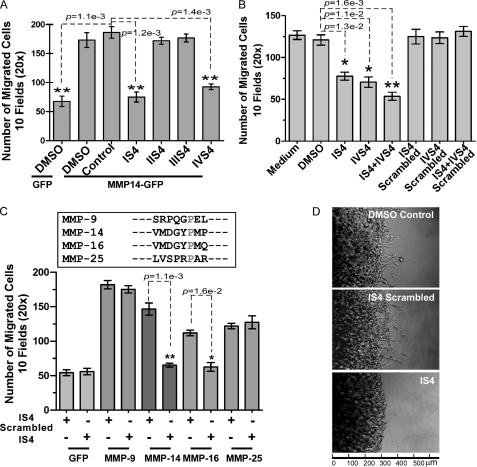
Motifs within the PEX domain are required for cell migration. A, ribbon diagram of the PEX domain of MMP-14. The hemopexin domain is composed of four blades (I to IV). The outermost strands are labeled IS4 to IVS4 and colored magenta, orange, blue, and green, respectively. The amino acid residues corresponding to the IS4, IIS4, IIIS4, and IVS4 peptides synthesized and tested are shown in stick form in pink, deep orange, light blue, and cyan. The schematic was created from 3C7X.pdb (13) using UCSF Chimera. B, assays for cell migration. Left panel, FITC substrate (fibronectin) degradation and cell migration assay employing wild-type and chimeric mutations of MMP-14-GFP expressed in COS-1 cells. Right panel, phagokinetic assay monitoring migration tracks of LNCaP cells expressing wild-type MMP14-GFP and chimeric constructs MMP14-MMP1IS4-IVS4-GFP. C, microscopic examination of wild-type and mutant MMP-14 distribution in LNCaP cells. LNCaP cells stably expressing GFP, MMP14-GFP, and mutant MMP14-GFPs were microscopically examined under a confocal fluorescent microscope. D, Transwell chamber migration assay. COS-1 cells transfected with cDNAs as indicated were subjected to a Transwell chamber migration assay. Each construct was assayed in triplicate, and the experiments were repeated three times. **, p < 0.01, two-tailed Student's t test. Scale bars = 10 μm.
Given the PEX domain topology, the four outermost strands within the PEX domain of MMP-14 were individually replaced with corresponding regions of MMP-1 to generate MMP14HA-MMP1 chimeras. The PEX domain of MMP-1 has a low similarity to the PEX domain of MMP-14 (36% amino acid identity and 51% similarity). To visualize protein expression and trafficking, GFP cDNA was fused to the C terminus of the wild-type and mutant MMP-14 by a PCR approach. The resultant PCR products were cloned into a retroviral vector (see “Experimental Procedures”). These chimeras were examined for their ability to induce cell migration. COS-1 cells infected with retroviruses encoding each of the MMP14-MMP1-GFP chimeras were plated onto FITC-labeled fibronectin-coated coverslips to simultaneously evaluate substrate degradation and cell migration (23). In agreement with our previous observation (24), the wild-type MMP-14-infected cells digested fibronectin substrate, as observed by a loss of fluorescence-labeled substrate, and the cells migrated as shown by the substrate degradation tracks (Fig. 2B, left panels). Similar patterns were observed in the cells expressing the MMP14-MMP1IIS4-GFP and MMP14-MMP1IIIS4-GFP chimeras, whereas both MMP14-MMP1IS4-GFP and MMP14-MMP1IVS4-GFP chimeras failed to cleave the substrate. This defect in the ability to degrade the substrate was not due to diminished enzyme biosynthesis and plasma membrane expression of the chimeras, as detected by GFP expression in the cells (Fig. 2C).
To corroborate that mutation of blades I and IV results in defective cell migration, a second approach employing a phagokinetic assay was undertaken. In accordance with the FITC-substrate degradation assay shown in Fig. 2B (left panels), no migratory tracks were observed in LNCaP cells expressing blade I and IV chimeras as compared with cells expressing wild-type and blade II and III chimeras (Fig. 2B, right panels). This observation was further validated using a Transwell chamber migration assay (Fig. 2D). These data substantiate that the outermost β-strands of blades I and IV are required for MMP-14-enhanced cell migration.
Requirement of the Outermost Strands of Blades I and IV in proMMP-2 Activation by MMP-14
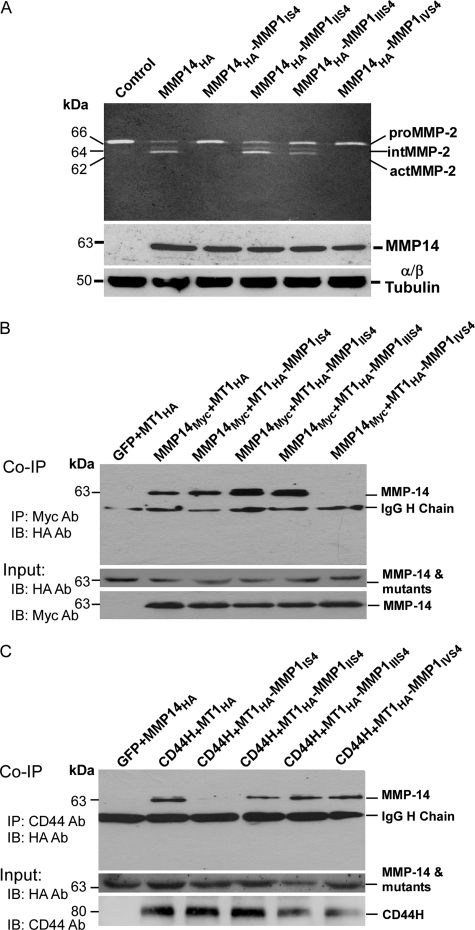
It has been reported that the PEX domain of MMP-14 is required for proMMP-2 activation (14, 25). To determine whether the four outermost strands are involved in proMMP-2 activation, gelatin zymography was carried out. COS-1 cells expressing MMP-14 cDNA as well as the MMP14HA-MMP1IIS4 chimera resulted in the conversion of proMMP-2 from its 72-kDa latent form to its 62-kDa fully active form via a 66-kDa intermediate form (Fig. 3A). Although the activation process of proMMP-2 occurred in COS-1 cells expressing MMP14HA-MMP1IIIS4, the degree of proMMP-2 activation was somewhat reduced. In contrast, mutation to blades IS4 and IVS4 completely disrupted the MMP-14-regulated activation of MMP-2. The failure of proMMP-2 activation was not due to loss of protein synthesis, as detected by comparable expression of wild-type and mutant MMP14-MMP1 proteins in the transfected cells (Fig. 3A, center panel).
FIGURE 3.
Requirement of dimerization of MMP-14 for proMMP-2 activation. A, gelatin zymography and Western blotting. The conditioned media from COS-1 cells transfected with wild-type and mutant MMP-14 cDNAs were examined by gelatin zymography. The total cell lysates were examined for protein biosynthesis using anti-HA antibody. α- and β- tubulin antibodies were employed as a loading control. B, coimmunoprecipitation (Co-IP). COS-1 cells transfected with the MMP14Myc and MMP14HA or MMP14HA-MMP1 mutation chimeras were pulled down with anti-Myc antibody followed by Western blotting (IB) with anti-HA antibody. Aliquots of total cell lysates served as input controls and were examined by Western blotting using anti-HA and anti-Myc antibodies, respectively. IP, immunoprecipitation. C, coimmunoprecipitation. COS-1 cells transfected with CD44H and MMP14HA or MMP14HA-MMP1 chimeras were pulled down with anti-CD44 antibody followed by Western blotting with anti-HA antibody. Aliquots of total cell lysates served as input controls and were examined by Western blotting using anti-HA, and anti-CD44 antibodies, respectively.
Previous reports have indicated that MMP-14 forms homodimers as well as heterodimers with adjacent CD44 via the PEX domain (9, 14). To determine whether the loss of proMMP-2 activation and cell migration by mutant MMP-14 is due to defective dimer formation of MMP-14, a coimmunoprecipitation approach was employed. COS-1 cells were cotransfected with HA-tagged wild-type and mutant MMP-14 cDNAs along with Myc-tagged wild-type MMP-14 cDNA (MMP14Myc), followed by a combination of coimmunoprecipitation and Western blotting (26). Using this approach, we observed that MMP14HA-MMP1IVS4 failed to coprecipitate with wild-type MMP-14 (Fig. 3B), suggesting that the outermost strand of blade IV is required for homodimer formation.
Employing analogous immunoprecipitation experiments, motifs within the PEX domain of MMP-14 critical for interacting with CD44 were examined. Coimmunoprecipitation with anti-CD44 antibody was carried out, followed by Western blotting with anti-HA antibody in COS-1 cells cotransfected with CD44H cDNA along with the MMP14HA or MMP14HA-MMP1 chimeras. CD44H coprecipitated with MMP14HA, as well as with MMP14HA-MMP1IIS4, MMP14HA-MMP1IIIS4, and MMP14HA-MMP1IVS4 (Fig. 3C). However, the blade I mutant (MMP14HA-MMP1IS4) failed to coprecipitate with CD44H, suggesting that blade I is required for interacting with CD44H at the cell surface.
Identification of an MMP14-CD44-EGFR Signaling Cascade for Cell Migration
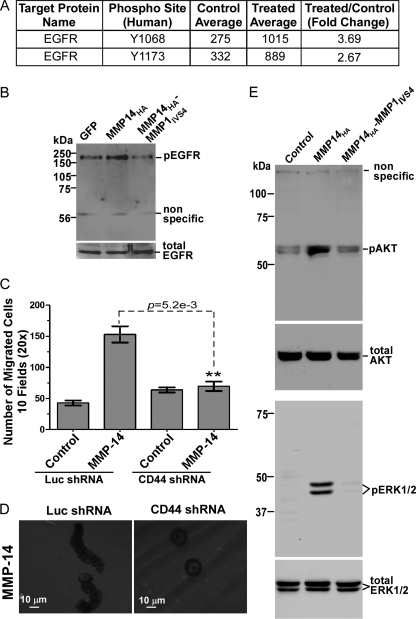
Utilizing an antibody array analysis, the amount of phosphorylated epidermal growth factor receptor (EGFR) was found to be increased in MMP-14-expressing COS-1 cells compared with mock-transfected COS-1 cells (Fig. 4A). Based on the established interactions between EGFR and MMPs, this avenue was pursued further. To validate the active EGFR status in MMP-14-expressing cells, a Western blotting analysis using anti-phospho-EGFR antibody (pT1068) was employed. An increase of phospho-EGFR was confirmed in MMP-14 cells as compared with vector- and MMP14-MMP1IVS4-transfected cells (Fig. 4B).
FIGURE 4.
MMP-14 mediates cell migration through CD44 and EGFR activation. A, Kinex antibody microarray analysis. MCF-7 cells stably expressing MMP-14 as well as a control were analyzed using the Kinex antibody assay service. EGFR was significantly activated in MMP-14-expressing cells. B, Western blotting. COS-1 cells transfected with GFP control, MMP14HA, and MMP14HA-MMP1IVS4 cDNAs were examined by Western blotting using anti-pEGFR and anti-EGFR antibodies. C, Transwell chamber migration assay. COS-1 cells stably expressing luciferase shRNA or CD44 shRNA were transfected with GFP control and MMP-14 cDNAs followed by a Transwell chamber migration assay. Each construct was assayed in triplicate, and the experiments were repeated three times. **, p < 0.01, two-tailed Student's t test. Luc, luciferase. D, a FITC-substrate (fibronectin) degradation and cell migration assay was also performed in CD44 or luciferase shRNA-silenced COS-1 cells transfected with MMP-14 cDNA. E, activation of AKT and ERK1/2 examined by Western blotting. The phosphorylation status of AKT (upper panel) and ERK1/2 (lower panel) in MCF-7 cells transfected with vector control, MMP14HA and MMP14HA-MMP1IVS4 mutant cDNAs was examined.
Given the substantial evidence for MMP-14/CD44 interaction, a cascade whereby MMP-14 cross-talks with CD44 and signals through EGFR to the MAPK and PI3K pathways for cell migration was postulated. To test this hypothesis, endogenous CD44 in COS-1 cells was silenced by introducing a shRNA specific for endogenous CD44 and CD44 variants, as we reported previously (26). Silencing CD44 in COS-1 cells resulted in failure to induce cell migration by MMP-14, examined by a Transwell chamber migration assay (Fig. 4C). This observation was further validated by using the fibronectin substrate degradation/cell migration assay (Fig. 4D). Silencing CD44 in COS-1 cells interfered with MMP-14-mediated cell migration without affecting proteolysis of the underlying matrix.
To validate the requirement for CD44 in MMP-14-induced EGFR activation, phospho-EGFR status was examined in CD44-silenced COS-1 cells. Upon silencing of CD44, MMP-14 failed to activate EGFR, consistent with a role for CD44 in the MMP-14 signaling pathway (data not shown).
Activation of the MAPK and PI3K pathways is believed to be mediated through CD44 and EGFR cross-talk (26). Phosphorylation of two common molecules in these signaling pathways, ERK1/2 and AKT, have been used to determine the activation status of the MAPK and PI3K pathways (27). MCF-7 cells, a weakly invasive breast cancer cell line, were transfected with vector control, MMP14HA, and MMP14HA-MMP1IVS4 cDNAs. Phosphorylated ERK1/2 and AKT were detected in immunoblots. A substantial decrease in pERK1/2 and pAKT was observed in cells expressing mutant MMP14HA-MMP1IVS4 as compared with wild-type MMP-14-expressing cells (Fig. 4E). This reduction is presumably due to an inability to initiate cross-talk between CD44 and downstream signaling pathways.
Design of Inhibitory Peptides Mimicking MMP-14 Critical Motifs Required for Function
Because dimerization of MMP-14 is required for function, we hypothesized that interference with MMP-14 dimerization will prevent MMP-14-signaling of cell migration. On the basis of the critical regions identified by our mutagenesis studies, peptides were synthesized to interfere with MMP-14 homodimer and CD44-MMP-14 heterodimer formation. These eight-amino acid peptides IS4, IIS4, IIIS4, and IVS4 comprise the amino acid sequence of the outermost strand of each PEX domain blade (supplemental Table 3). MCF-7 cells transfected with MMP14-GFP cDNAs were preincubated with these peptides, and the effect of the peptides on cell migratory ability was assessed by a Transwell chamber migration assay. Significant inhibition of MMP-14-mediated cell migration was observed in MCF-7 cells stably expressing the MMP14-GFP chimera treated with IS4 and IVS4 peptides (Fig. 5A). IIS4, IIIS4, or a scrambled peptide had no effect on cell migration.
FIGURE 5.
Inhibitory effect of designed peptides on MMP-14-mediated cell migration and invasion. A, Transwell chamber migration assay in cells expressing exogenous MMP-14. MCF-7 cells stably expressing GFP control or MMP14-GFP were pretreated with scrambled (Control) or blade-specific peptides (100 μm) followed by a Transwell chamber migration assay. **, p < 0.01, two-tailed Student's t test. B, Transwell chamber migration assay in cells expressing endogenous MMP-14. HT-1080 cells expressing endogenous MMP-14 were examined by a Transwell chamber migration assay in the presence of scrambled or blade-specific peptides (100 μm). *, p < 0.05; **, p < 0.01; two-tailed Student's t test. DMSO, dimethyl sulfoxide. C, Transwell chamber migration assay in cells expressing different MMPs. Upper panel, comparison of the amino acid sequences between MMP-14 IS4 and that of other MMPs. Lower panel, COS-1 cells transfected with MMP cDNAs were examined by a Transwell chamber migration assay in the presence of IS4 or IS4-scrambled peptides (100 μm). *, p < 0.05; **, p < 0.01; two-tailed Student's t test. D, 3D invasion assay. MDA-MB-231 cells (1 × 104) were examined by a 3D invasion assay in the presence of IS4 or scrambled peptides (100 μm). Invading cells at the cell-collagen interface were microscopically counted after an 18-hour incubation. Each construct was assayed in triplicate, and the experiments were repeated three times.
The peptide inhibitors were validated in additional cell lines, including COS-1 cells overexpressing MMP-14 (data not shown). Moreover, the peptides displayed dose-dependent inhibition in MMP-14-mediated cell migration (supplemental Fig. 1). To determine whether the peptides also affect cancer cell migration in cells producing endogenous MMP-14, human HT-1080 fibrosarcoma cells expressing high endogenous levels of MMP-14 were employed. HT-1080 cells were preincubated with IS4, IVS4, a combination of both peptides, or scrambled peptides, followed by a Transwell chamber migration assay. Only the IS4 and IVS4 peptides inhibited the migration of HT-1080 cells (Fig. 5B). A mixture of both peptides demonstrated an additive inhibitory effect on cell migration. A similar result was observed in human MDA-MB-435 cancer cells (data not shown).
To determine the specificity of the developed peptides, we tested inhibition by IS4 peptides on cells producing different secretory and membrane-bound MMPs. IS4 peptide did not interfere with MMP-9- and MMP-25 (MT6-MMP)-mediated COS-1 cell migration but did have an inhibitory effect on MMP-14 (MT1-MMP)- and MMP-16 (MT3-MMP)-mediated COS-1 cell migration (Fig. 5C, lower panel). The IS4 peptide sequence of MMP-14 is nearly identical to the corresponding sequence of MMP-16 blade I. In contrast, no significant conservation between the MMP-14 IS4 strand and other MMPs was found (Fig. 5C, upper panel).
Because cell migration is a critical determinate of cancer invasion, the IS4 peptide was assessed for its ability to inhibit cancer cell invasion. MDA-MB-231 cells express endogenous MMP-14 and are naturally invasive (supplemental Fig. 2). MDA-MB-231 cells were examined in a 3D type I collagen invasion assay in the presence of IS4 peptide and IS4 scrambled control peptides. Cells treated with control peptide invaded into the surrounding collagen, whereas cells incubated with IS4 peptide displayed a decreased invasive ability (Fig. 5D).
In Vivo Evaluation of the Specific Peptides in Tumor Growth and Metastasis
To examine the potential efficacy of the peptides in vivo, the peptides were assessed for their stability in mouse serum. The peptides were incubated with undiluted mouse serum for various times, followed by assessment of peptide function in a cell migration assay. There was no notable change in ability of the peptides to inhibit cell migration over 72 h (data not shown).
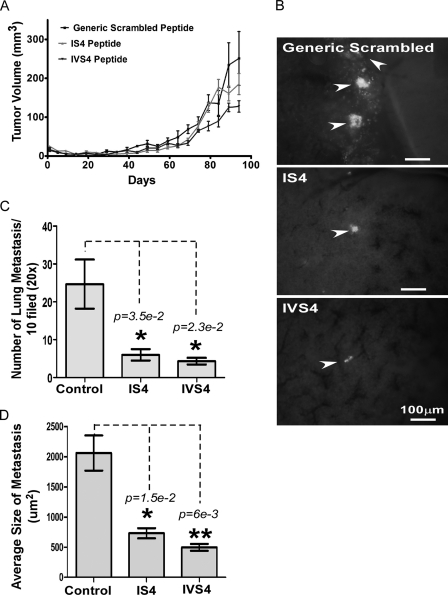
The in vivo effects of the peptides were then assessed using MDA-MB-435 cancer cells stably expressing GFP cDNA, which facilitated visualization of metastatic lesions as well as tumor growth. This cell line was chosen for two reasons: 1) MDA-MB-435 cells express a high levels of endogenous MMP-14, and 2) MDA-MB-435 cells possess a high metastatic potential (28–30). Peptides were alternatively administered intratumorally and intraperitoneally. Although no statistically significant difference in tumor volume was observed in different groups, there was a trend toward more inhibition in the mice treated with IVS4 peptides (Fig. 6A). To analyze the effects of the peptides on tumor metastasis, lung sections of the tumor-bearing mice were harvested, and 5-mm lung sections were examined under fluorescent microscopy for a GFP signal (Fig. 6B). Scrambled control peptide-treated mice exhibited multiple large nodules, whereas the degree of lung metastasis was significantly reduced in IS4 and IVS4 peptide-treated mice (Fig. 6C). The tumor area in the lung was subsequently quantified. Metastatic foci in mice treated with IS4 and IVS4 peptides were significantly smaller than mice treated with scrambled peptide (Fig. 6D). Throughout the 13-week period, no significant changes in body weight or signs of peptide toxicity were observed.
FIGURE 6.
Interference with tumor metastasis by IS4 and IVS4 peptides. A, xenographic mouse tumor model. Mice bearing MDA-MB-435/GFP tumors were treated with the peptides (20 mg/kg) 6 days per week. Tumor growth was monitored twice per week. B, imaging of lung sections (approximately 5 mm) at the end of experiment to identify metastasis. Arrowheads indicate metastatic lesions in the lung, as identified with green fluorescent signal. C and D, lung metastases were microscopically counted, and the size of metastatic nodules was determined using ImageJ software analysis based on GFP expression. The area of metastatic foci per field of examination was quantified from 10 random sites of three different slides for each mouse. *, p < 0.05; **, p < 0.01.
Role of MMP-14 Hemopexin Domain in Angiogenesis
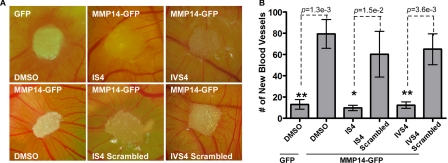
Because angiogenesis and the development of metastases are intrinsically connected (31), we examined whether inhibition of cancer metastasis by the MMP-14 specific peptides is due to interference with MMP-14-induced angiogenesis. To this end, a CAM angiogenesis assay was employed. MCF-7 cells stably expressing MMP14-GFP were loaded onto the CAM and treated with inhibitory peptides as well as scrambled controls on a daily basis. After 4 days, we observed angiogenesis as represented by the fine neovessels contacting and radiating from the sponge absorbed with MMP-14-expressing MCF-7 cells in a spoke-like pattern. Treatment of MMP14-GFP transfected MCF-7 cells with inhibitory peptides reduced MMP-14-mediated angiogenesis, as made evident by a lack of developed vessels (thin and narrow vessels) and a decreased number of new blood vessels (Fig. 7).
FIGURE 7.
Decrease of new blood vessel formation by the peptides. A CAM assay was performed to measure new blood vessel formation. Sponges absorbed with tumor cells were loaded onto the CAM. The peptides (100 μm) were added every day. New blood vessels were photographed (A) and counted under a stereo microscope (B). *, p < 0.05; **, p < 0.01; two-tailed Student's t test.
DISCUSSION
In this study, we employed a mutagenesis approach to identify minimal motifs within the outer strands of the PEX domain of MMP-14 required for cell migration. MMP-14 homodimerization and heterodimerization, dependent on blades IV and I of the PEX domain, respectively, initiate a signaling cascade leading to cell migration. Synthetic peptide-based inhibitors of these PEX domain motifs were shown to abrogate in vitro and in vivo MMP-14-mediated cell migration, invasion, and metastasis.
Upon trafficking of MMP-14 to the plasma membrane and subsequent docking, Itoh et al. (32) reported that homodimer formation of MMP-14 through the PEX domain facilitates proMMP-2 activation on the cell surface and promotes tumor cell invasion. It has also been established that cross-talk between MMP-14 and CD44 exists whereby heterodimerization of the surface proteins regulates the invasive front of the cell (33). Our micro-dissection of the hemopexin domain reveals, for the first time, the precise nature of the interaction between MMP-14 homodimers and MMP-14-CD44 heterodimers by a biochemical approach.
One of the important roles of MMP-14 is to activate proMMP-2 (10). This activation is reported to require MMP-14 dimerization via the PEX domain (14). However, the requirement of the MMP-14 PEX domain on proMMP-2 activation has been challenged on the basis of the observation that deletion of the PEX domain did not affect MMP-14-mediated proMMP-2 activation (34–36). These contrary findings raise the question of whether MMP-14 homodimerization via the PEX domain is required for proMMP-2 activation. Our data indicate that dimerization of MMP-14 via the PEX domain is required for proMMP-2 activation because of the unique structures within the PEX domain.
The structure of isolated MMP-14 PEX domain protein was recently characterized by x-ray crystallography, and as expected, the propeller tertiary structure characteristic of a hemopexin domain is formed (13). In addition, bulges in the outermost β strands of blades I and IV, which are unique to MMP-14, were observed (13). Interestingly, the PEX domain is presented as having two modes of potential dimerization: symmetrical (PEX domain blades II/III and III/II) and asymmetrical (PEX domain blades I/II and I/II/III/IV) dimers.
In contrast to a previous report by Itoh et al. (14), which showed that interruption of homodimer formation abrogates proMMP-2 activation and cell invasion, Tochowicz et al. (13) recently demonstrated that interference of MMP-14 homodimer formation only slightly reduces proMMP-2 activation without affecting MMP-14-enhanced cell migration. Our data are in agreement with the published reports stating that homodimerization is a prerequisite for MMP-14-mediated proMMP-2 activation. Our data further agree with the observation that mutations of key amino acids within the outermost strands of blade III of the PEX domain have only a slight effect on activation of proMMP-2. However, there is a disparity between our report with Tochowicz et al. (13) in terms of mode of homodimerization and the role homodimers play in proMMP-2 activation. In this study, we found that mutation at blades II and III did not impair MMP-14-mediated proMMP-2 activation and cell migration (Figs. 2 and 3). In contrast, substitution of the bulged blade I and blade IV β-strands disrupts dimer formation in full-length, membrane-bound MMP-14 with failure to enhance cell migration and induce proMMP-2 activation. Our data suggest that in the native structure, the unique bulges of blades I and IV of MMP-14 play a critical role in dimerization and functionality. The discrepancy between our work, which employed full-length native MMP-14, as does the earlier work of Itoh et al. (14), and the more recent crystallography paper of Tochowicz et al. (13) most likely arises from the fact that Tochowicz employed isolated PEX protein purified from Escherichia coli as opposed to the native protein. Further studies are required to clarify this issue.
The paradigm by which MMP-14 homodimerizes through blade IV and interacts with CD44 through blade I of the PEX domain leads us to believe that these interactions are key in mediating downstream signaling that promotes cell invasion. Expression of MMP-14 in cells results in activation of downstream signaling molecules, including ERK1/2 and AKT, both of which are required for MMP-14-mediated cell migration (37, 38). Given the fact that deletion of the cytoplasmic domain of MMP-14 does not affect MMP-14-enhanced cell migration and invasion (36, 39), it is likely that intracellular signaling relies on interactions with cell surface proteins (40), particularly CD44, which colocalizes with MMP-14 and has been reported previously to be necessary for the migration-promoting activity (9, 33, 41). Activation of EGFR requires the presence of both MMP-14 and CD44, and activated EGFR results in downstream phosphorylation of ERK1/2 and AKT to enable cell migration (Fig. 4). It has been demonstrated that cleavage of CD44 by MMP-14 is a prerequisite for MMP-14-enhanced cell migration (9, 33, 42, 43), although MMP-14 is reported to be unnecessary for CD44-shedding in melanoma cells (44). Using a functional assay, we observed that coexpression of MMP-14 but not catalytic inactive MMP-14 mutant (E240→A) in COS-1 cells significantly reduced cell adhesion ability to hyaluronic acid (a CD44 ligand)-coated culture plates (supplemental Fig. 3), which is consistent with cleavage of CD44 by MMP-14. It still remains to be understood how cleaved CD44 subsequently activates receptor tyrosine kinases (i.e. EGFR) for additional signaling transduction. Nevertheless, our data outline a cascade of MMP-14-CD44-EGFR-MAPK/PI3K signaling resulting in cell migration. Of interest, we recently reported that expression of latent MMP-9 in transfected COS-1 cells induces cell migration through an interaction with CD44, resulting in a similar activation of the EGFR pathway (26).
We reported previously that the different domains of MMP-14 play distinct roles in substrate degradation and cell migration (24). We further demonstrated that targeting either the catalytic domain or PEX domain efficiently inhibited MMP-14-mediated cell scattering/invasion using a 3D cell scattering assay (24). Because the overall topology of the enzyme active site of MMP-14 is highly conserved with other MMPs (45), we hypothesized that developing an inhibitory reagent targeting the PEX domain can achieve inhibitor specificity (targeting only MMP-14) and tissue selectivity (targeting only MMP-14-expressing cancer cells).
Highly active antifunctional antibodies against the catalytic domain of MMP-14 have been demonstrated recently to be effective in blocking MMP-14-mediated tumor progression and metastasis in a preclinical model, suggesting that specific targeting of MMP-14 is a viable therapeutic approach (46, 47). Antibodies directed against the PEX domain (39) and purified recombinant PEX domain alone significantly inhibited MMP-14-mediated cell migration (supplemental Fig. 4). Although larger-size proteins such as monoclonal antibodies have shown clinical potential as tumor targeting agents, they are limited by their large molecular size and resulting poor tumor penetration as well as by their immunogenicity. These limitations can be overcome by using peptide ligands, which are smaller, less immunogenic, and easier to produce and manipulate. On the basis of our mutagenesis studies, we designed and characterized competitive peptides that interfere with MMP-14-mediated cancer cell migration and invasion. These peptides are highly specific for the PEX domain of MMP-14 and do not react with less conserved MMPs.
MMP-14 expression has been correlated with primary tumor growth and metastasis, as well as in vitro and in vivo angiogenesis (48). Detailed analysis of MMP-14-promoted tumor growth has suggested that the cytoplasmic domain, rather than the catalytic domain, is required for MMP-14 enhanced tumor growth (49, 50). Consistent with these observations, inhibition of cell migratory ability by the administration of IS4 and IVS4 peptides to mice did not significantly reduce MDA-MB-435 primary tumor growth (Fig. 6). However, these peptides inhibited metastasis to the lungs of tumor-bearing mice, presumably through interference with MMP-14-mediated cell migration, invasion, and angiogenesis. Reduction of metastasis without affecting primary tumor growth has been previously reported with anti-metastatic genes and drugs targeting cancer cell migration and invasion (30, 51, 52).
In summary, we have identified a novel axis of signaling between MMP-14 homodimers and CD44 heterodimers which initiates downstream effectors for cell migration. The critical motifs within the PEX domain of MMP-14 that are required for such interactions were identified, allowing for the development of highly specific inhibitory agents targeting MMP-14-mediated cell migration and invasion. These data, in addition to our previous observations regarding the PEX domain of MMP-9 (26, 53), suggest that a common mechanism is required for cell function. Furthermore, we have achieved efficient control of spontaneous cancer metastasis in a preclinical model by targeting the PEX domain of MMP-14. In light of the fact that metastatic human cancers often express high level of MMP-14, our PEX-domain strategy provides proof of concept that targeting non-catalytic domains of MMPs is a viable approach to prevent cancer dissemination.
Supplementary Material
This study was supported, in whole or in part, by National Institutes of Health Grant 5R01CA113553 (to J. C. and N. S. S.). This work was also supported by a Carol Baldwin Breast Cancer Foundation grant (to J. C.), by New York State Foundation for Science, Technology, and Innovation Faculty Development Program Grant C040076 (to N. S.), by a Veterans Affairs Merit Review grant, and by a Carol Baldwin Breast Cancer Foundation grant (to S. Z.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4 and supplemental Tables 1–3.
- MMP
- matrix metalloproteinase
- MMP-14
- membrane type 1-MMP
- PEX
- hemopexin-like
- 3D
- three dimensional
- CAM
- chorioallantoic membrane
- EGFR
- EGF receptor.
REFERENCES
- 1. Roy R., Yang J., Moses M. A. (2009) J. Clin. Oncol. 27, 5287–5297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Egeblad M., Werb Z. (2002) Nat. Rev. Cancer 2, 161–174 [DOI] [PubMed] [Google Scholar]
- 3. Lauer-Fields J. L., Chalmers M. J., Busby S. A., Minond D., Griffin P. R., Fields G. B. (2009) J. Biol. Chem. 284, 24017–24024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Poincloux R., Lizárraga F., Chavrier P. (2009) J. Cell Sci. 122, 3015–3024 [DOI] [PubMed] [Google Scholar]
- 5. Sabeh F., Li X. Y., Saunders T. L., Rowe R. G., Weiss S. J. (2009) J. Biol. Chem. 284, 23001–23011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Itoh Y., Seiki M. (2006) J. Cell Physiol. 206, 1–8 [DOI] [PubMed] [Google Scholar]
- 7. Stetler-Stevenson W. G., Aznavoorian S., Liotta L. A. (1993) Annu. Rev. Cell Biol. 9, 541–573 [DOI] [PubMed] [Google Scholar]
- 8. Artym V. V., Zhang Y., Seillier-Moiseiwitsch F., Yamada K. M., Mueller S. C. (2006) Cancer Res. 66, 3034–3043 [DOI] [PubMed] [Google Scholar]
- 9. Mori H., Tomari T., Koshikawa N., Kajita M., Itoh Y., Sato H., Tojo H., Yana I., Seiki M. (2002) EMBO J. 21, 3949–3959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sato H., Takino T., Okada Y., Cao J., Shinagawa A., Yamamoto E., Seiki M. (1994) Nature 370, 61–65 [DOI] [PubMed] [Google Scholar]
- 11. Lichte A., Kolkenbrock H., Tschesche H. (1996) FEBS Lett. 397, 277–282 [DOI] [PubMed] [Google Scholar]
- 12. Cha H., Kopetzki E., Huber R., Lanzendörfer M., Brandstetter H. (2002) J. Mol. Biol. 320, 1065–1079 [DOI] [PubMed] [Google Scholar]
- 13. Tochowicz A., Goettig P., Evans R., Visse R., Shitomi Y., Palmisano R., Ito N., Richter K., Maskos K., Franke D., Svergun D., Nagase H., Bode W., Itoh Y. (2011) J. Biol. Chem. 286, 7587–7600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Itoh Y., Takamura A., Ito N., Maru Y., Sato H., Suenaga N., Aoki T., Seiki M. (2001) EMBO J. 20, 4782–4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cao J., Chiarelli C., Kozarekar P., Adler H. L. (2005) Thromb Haemost. 93, 770–778 [DOI] [PubMed] [Google Scholar]
- 16. van de Vijver M. J., He Y. D., van't Veer L. J., Dai H., Hart A. A., Voskuil D. W., Schreiber G. J., Peterse J. L., Roberts C., Marton M. J., Parrish M., Atsma D., Witteveen A., Glas A., Delahaye L., van der Velde T., Bartelink H., Rodenhuis S., Rutgers E. T., Friend S. H., Bernards R. (2002) N. Engl. J. Med. 347, 1999–2009 [DOI] [PubMed] [Google Scholar]
- 17. Wang Y., Klijn J. G., Zhang Y., Sieuwerts A. M., Look M. P., Yang F., Talantov D., Timmermans M., Meijer-van Gelder M. E., Yu J., Jatkoe T., Berns E. M., Atkins D., Foekens J. A. (2005) Lancet 365, 671–679 [DOI] [PubMed] [Google Scholar]
- 18. Pawitan Y., Bjöhle J., Amler L., Borg A. L., Egyhazi S., Hall P., Han X., Holmberg L., Huang F., Klaar S., Liu E. T., Miller L., Nordgren H., Ploner A., Sandelin K., Shaw P. M., Smeds J., Skoog L., Wedrén S., Bergh J. (2005) Breast Cancer Res. 7, R953–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Deryugina E. I., Quigley J. P. (2008) Histochem. Cell Biol. 130, 1119–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ribatti D., Nico B., Vacca A., Presta M. (2006) Nat. Protoc. 1, 85–91 [DOI] [PubMed] [Google Scholar]
- 21. Zucker S., Pei D., Cao J., Lopez-Otin C. (2003) Curr. Top. Dev. Biol. 54, 1–74 [DOI] [PubMed] [Google Scholar]
- 22. Bode W., Fernandez-Catalan C., Tschesche H., Grams F., Nagase H., Maskos K. (1999) Cell Mol. Life Sci. 55, 639–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pavlaki M., Cao J., Hymowitz M., Chen W. T., Bahou W., Zucker S. (2002) J. Biol. Chem. 277, 2740–2749 [DOI] [PubMed] [Google Scholar]
- 24. Cao J., Kozarekar P., Pavlaki M., Chiarelli C., Bahou W. F., Zucker S. (2004) J. Biol. Chem. 279, 14129–14139 [DOI] [PubMed] [Google Scholar]
- 25. Seiki M., Koshikawa N., Yana I. (2003) Cancer Metastasis Rev. 22, 129–143 [DOI] [PubMed] [Google Scholar]
- 26. Dufour A., Zucker S., Sampson N. S., Kuscu C., Cao J. (2010) J. Biol. Chem. 285, 35944–35956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lehti K., Allen E., Birkedal-Hansen H., Holmbeck K., Miyake Y., Chun T. H., Weiss S. J. (2005) Genes Dev. 19, 979–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Khaldoyanidi S. K., Glinsky V. V., Sikora L., Glinskii A. B., Mossine V. V., Quinn T. P., Glinsky G. V., Sriramarao P. (2003) J. Biol. Chem. 278, 4127–4134 [DOI] [PubMed] [Google Scholar]
- 29. Price J. E. (1996) Breast Cancer Res. Treat. 39, 93–102 [DOI] [PubMed] [Google Scholar]
- 30. Seraj M. J., Samant R. S., Verderame M. F., Welch D. R. (2000) Cancer Res. 60, 2764–2769 [PubMed] [Google Scholar]
- 31. Kirsch M., Schackert G., Black P. M. (2004) Cancer Treat Res. 117, 285–304 [DOI] [PubMed] [Google Scholar]
- 32. Itoh Y., Ito N., Nagase H., Evans R. D., Bird S. A., Seiki M. (2006) Mol. Biol. Cell 17, 5390–5399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kajita M., Itoh Y., Chiba T., Mori H., Okada A., Kinoh H., Seiki M. (2001) J. Cell Biol. 153, 893–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang P., Nie J., Pei D. (2004) J. Biol. Chem. 279, 51148–51155 [DOI] [PubMed] [Google Scholar]
- 35. Itoh Y., Ito N., Nagase H., Seiki M. (2008) J. Biol. Chem. 283, 13053–13062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li X. Y., Ota I., Yana I., Sabeh F., Weiss S. J. (2008) Mol. Biol. Cell 19, 3221–3233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sounni N. E., Rozanov D. V., Remacle A. G., Golubkov V. S., Noel A., Strongin A. Y. (2010) Int. J. Cancer 126, 1067–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Eisenach P. A., Roghi C., Fogarasi M., Murphy G., English W. R. (2010) J. Cell Sci. 123, 4182–4193 [DOI] [PubMed] [Google Scholar]
- 39. Cao J., Chiarelli C., Richman O., Zarrabi K., Kozarekar P., Zucker S. (2008) J. Biol. Chem. 283, 6232–6240 [DOI] [PubMed] [Google Scholar]
- 40. Remacle A. G., Rozanov D. V., Baciu P. C., Chekanov A. V., Golubkov V. S., Strongin A. Y. (2005) J. Cell Sci. 118, 4975–4984 [DOI] [PubMed] [Google Scholar]
- 41. Okamoto I., Kawano Y., Tsuiki H., Sasaki J., Nakao M., Matsumoto M., Suga M., Ando M., Nakajima M., Saya H. (1999) Oncogene 18, 1435–1446 [DOI] [PubMed] [Google Scholar]
- 42. Marrero-Diaz R., Bravo-Cordero J. J., Megías D., García M. A., Bartolomé R. A., Teixido J., Montoya M. C. (2009) Cell Motil. Cytoskeleton 66, 48–61 [DOI] [PubMed] [Google Scholar]
- 43. Suenaga N., Mori H., Itoh Y., Seiki M. (2005) Oncogene 24, 859–868 [DOI] [PubMed] [Google Scholar]
- 44. Anderegg U., Eichenberg T., Parthaune T., Haiduk C., Saalbach A., Milkova L., Ludwig A., Grosche J., Averbeck M., Gebhardt C., Voelcker V., Sleeman J. P., Simon J. C. (2009) J. Invest. Dermatol. 129, 1471–1482 [DOI] [PubMed] [Google Scholar]
- 45. Verma R. P., Hansch C. (2007) Bioorg. Med. Chem. 15, 2223–2268 [DOI] [PubMed] [Google Scholar]
- 46. Devy L., Huang L., Naa L., Yanamandra N., Pieters H., Frans N., Chang E., Tao Q., Vanhove M., Lejeune A., van Gool R., Sexton D. J., Kuang G., Rank D., Hogan S., Pazmany C., Ma Y. L., Schoonbroodt S., Nixon A. E., Ladner R. C., Hoet R., Henderikx P., Tenhoor C., Rabbani S. A., Valentino M. L., Wood C. R., Dransfield D. T. (2009) Cancer Res. 69, 1517–1526 [DOI] [PubMed] [Google Scholar]
- 47. Gálvez B. G., Matías-Román S., Albar J. P., Sánchez-Madrid F., Arroyo A. G. (2001) J. Biol. Chem. 276, 37491–37500 [DOI] [PubMed] [Google Scholar]
- 48. Sounni N. E., Devy L., Hajitou A., Frankenne F., Munaut C., Gilles C., Deroanne C., Thompson E. W., Foidart J. M., Noel A. (2002) FASEB J. 16, 555–564 [DOI] [PubMed] [Google Scholar]
- 49. D'Alessio S., Ferrari G., Cinnante K., Scheerer W., Galloway A. C., Roses D. F., Rozanov D. V., Remacle A. G., Oh E. S., Shiryaev S. A., Strongin A. Y., Pintucci G., Mignatti P. (2008) J. Biol. Chem. 283, 87–99 [DOI] [PubMed] [Google Scholar]
- 50. Rozanov D. V., Deryugina E. I., Monosov E. Z., Marchenko N. D., Strongin A. Y. (2004) Exp. Cell Res. 293, 81–95 [DOI] [PubMed] [Google Scholar]
- 51. Magenta G., Borenstein X., Rolando R., Jasnis M. A. (2008) BMC Cancer 8, 47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Giubellino A., Gao Y., Lee S., Lee M. J., Vasselli J. R., Medepalli S., Trepel J. B., Burke T. R., Jr., Bottaro D. P. (2007) Cancer Res. 67, 6012–6016 [DOI] [PubMed] [Google Scholar]
- 53. Dufour A., Sampson N. S., Zucker S., Cao J. (2008) J. Cell Physiol. 217, 643–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.