Abstract
Genic microsatellite markers, also known as functional markers, are preferred over anonymous markers as they reveal the variation in transcribed genes among individuals. In this study, we developed a total of 707 expressed sequence tag-derived simple sequence repeat markers (EST-SSRs) and used for development of a high-density integrated map using four individual mapping populations of B. rapa. This map contains a total of 1426 markers, consisting of 306 EST-SSRs, 153 intron polymorphic markers, 395 bacterial artificial chromosome-derived SSRs (BAC-SSRs), and 572 public SSRs and other markers covering a total distance of 1245.9 cM of the B. rapa genome. Analysis of allelic diversity in 24 B. rapa germplasm using 234 mapped EST-SSR markers showed amplification of 2 alleles by majority of EST-SSRs, although amplification of alleles ranging from 2 to 8 was found. Transferability analysis of 167 EST-SSRs in 35 species belonging to cultivated and wild brassica relatives showed 42.51% (Sysimprium leteum) to 100% (B. carinata, B. juncea, and B. napus) amplification. Our newly developed EST-SSRs and high-density linkage map based on highly transferable genic markers would facilitate the molecular mapping of quantitative trait loci and the positional cloning of specific genes, in addition to marker-assisted selection and comparative genomic studies of B. rapa with other related species.
Keywords: Brassica rapa, expressed sequence-derived SSRs, integrated map, polymorphism information content, transferability
1. Introduction
Brassica rapa (AA, 2n = 20) is an important diploid Brassica crop mainly grown for as a vegetable foodstuff, and to some extent for producing oilseed and fodder crops. Among the six cultivated Brassica species [the other five are the two diploids Brassica nigra (BB, n = 8) and B. oleracea (CC, n = 9) and the three amphidiploids B. juncea (AABB, n = 18), B. carinata (BBCC, n = 17), and B. napus (AACC, n = 19)], B. rapa has a comparatively small genome size (529 Mb), and has the second largest morphological and genetic diversity after B. oleracea. It is also one of the progenitor parents which contributed the A genome to the widely cultivated amphidiploid oilseed crops B. juncea and B. napus, as beautifully shown by U's triangle.1
During the last two decades, several genetic maps with conventional anonymous molecular markers, such as amplified fragment length polymorphisms (AFLPs), restriction fragment length polymorphisms (RFLPs), and genomic simple sequence repeats (SSRs), have been constructed in Brassica species, including B. rapa.2–7 These maps have been used to map, tag, and clone genetic loci [genes and/or quantitative trait loci (QTL)] that are associated with economically important traits, such as leaf traits, glucosinolates, seed coat colour, and other important agronomic traits.3,8–14 Further, the construction of a detailed genetic map helped to study comparative genome organization, evolution, and conservation among the Brassica species and with Arabidopsis thaliana, the closest related model plant to the Brassicaceae family.4,6 Comparative mapping between B. rapa and A. thaliana was used to identify and clone candidate genes at the QTL regions for flowering time,13,15 leaf hairiness,13 and other traits.14
However, a detailed high-density integrated genetic map combining many genetic maps developed from different populations and marker types has not been generated for B. rapa. The importance of high-density genetic maps in the understanding of genome organization, evolution, and the mapping and tagging of important QTL for molecular breeding and map-based cloning of economically important trait-related genes has created widespread interest for their development in many crop plants.16–22 Further, previously developed conventional markers are anonymous, laborious to genotype (e.g. AFLPs and RFLPs), less reproducible (e.g. random amplification of polymorphic DNA, RAPD), require more time for development (e.g. genomic SSRs), and, more importantly, are less transferable between species. Due to these disadvantages, these conventional markers are being replaced by SSRs or single nucleotide polymorphisms (SNPs) isolated from transcribed regions [such as complementary DNA, messenger RNA, and expressed sequence tags (ESTs)]. Recent advances in plant functional genomics projects are producing enormous amounts of ESTs which have been deposited in the National Center for Biotechnology Information (NCBI) database. These sequences from transcribed genes are assembled into unique gene sequences and used to design SSRs from genes with a unique identity and position in the genome.23–25 The co-dominant, multi-allelic, and high reproducibility nature besides amenable for high throughput marker analysis led to the rapid and economical expressed sequence tag-derived simple sequence repeat markers (EST-SSRs) development in several plant species since a large number of SSRs are found in coding regions.22–24,26–32
Although the draft genome sequence of the euchromatic regions of B. rapa is expected to become available soon from the Multinational Brassica rapa Genome Sequencing Project (MBrGSP), the development and mapping of more uniformly spaced high-density genic markers, such as unigene-derived microsatellites (UGMS) and intron polymorphic (IP) markers along with bacterial artificial chromosome (BAC)-derived SSRs, would facilitate the mapping of important traits and their utilization in molecular breeding. In addition, a high-density map of anonymous and genic markers would help in the correct alignment of gene-rich euchromatic and repetitive heterochromatic sequences in the B. rapa genome because the soon-to-be released draft B. rapa genome sequence covers only 384 Mb of the 529 Mb Brassica A genome (personal communication from MBrGSP).
In B. rapa, Parida et al.24 recently developed 347 unigene-derived SSR markers, suggesting that there are many more unidentified genic SSRs that would allow for the complete coverage of the B. rapa genome. This would help uniformly select genic SSR markers covering the total genome and facilitate the mapping, tagging, and identification of economically important genetic loci. Further, uniformly distributed markers would be useful for comparative mapping and evolutionary studies with other closely related Brassica species. Hence, the objectives of this study were to develop more EST-SSR markers, map the newly developed EST-SSRs along with the previously mapped BAC-derived SSRs, IP markers, and publicly available SSR markers into the Brassica rapa genome to construct a high-density gene-based updated integrated map, and transferability analysis of the mapped EST-SSRs markers in other Brassica relatives so that these markers could be used for comparative mapping between them.
2. Materials and methods
2.1. Plant materials
We used four B. rapa mapping populations to develop an integrated linkage map: CKDH, CRF2, PF2, and CSKF2. The CKDH population consisted of 78 double haploid lines derived from a cross between ‘Chiifu-401’ and ‘Kenshin’, which were earlier used to construct a reference genetic linkage map of B. rapa.5,7,33 The CRF2 population consisted of 190 F2 individuals derived from crossing the ‘Chiifu-401’ and ‘Rapid cycling B. rapa (RCBr)’ parental lines, and was previously used by Li et al.33 to construct a linkage map. The CSKF2 population consisted of 94 individual lines derived from crossing between the clubroot resistance cultivar ‘CR Shinki’ and a susceptible cultivar ‘94SK’. The fourth population, PF2, consisted of 144 F2 populations that were derived from crossing the diverse Chinese cabbage inbred lines ‘501’ with a large head and ‘601’ with a small head. In order to detect the polymorphic level and allelic frequencies of the newly mapped EST-SSRs, we selected 24 different B. rapa cultivars belonging to different sub-species and morphophytes which included Chinese cabbage, pak choi, and oil yielding types from the Korea Brassica Genome Resource Bank (Table 1, serial number 1–24). Different Brassica species and wild relatives collected from Centre for Genetic Manipulation of Crop Plants, Delhi University South Campus, India; Korea Brassica Genome Research Bank, Korea; and Leibniz institute of Plant Genetics and Crop Plant Research, Gatersleben, Germany were used for marker transferability analysis (Table 1).
Table 1.
List of different Brassica species and wild relatives used for allelic diversity and transferability analysis
| SL No | Name/accession number | Species | Source | SL no. | Name/accession number | Species | Source |
|---|---|---|---|---|---|---|---|
| 1 | Chiifu-401 | B. rapa ssp pekinensis | KBGRB | 36 | HC-17 | B. carinata | CGMCP |
| 2 | Kenshin | B. rapa ssp pekinensis | KBGRB | 37 | Sangam | B. nigra | CGMCP |
| 3 | 94sk | B. rapa ssp pekinensis | KBGRB | 38 | 94029 | B. nigra | CGMCP |
| 4 | 24020 | B. rapa ssp pekinensis | KBGRB | 39 | 28407 | B. tourniforti | IPK |
| 5 | 26021 | B. rapa ssp pekinensis | KBGRB | 40 | BRA 2850 | B. balearica | IPK |
| 6 | 26022 | B. rapa ssp pekinensis | KBGRB | 41 | BRA 1877 | B. barreilieri | IPK |
| 7 | 26028 | B. rapa ssp pekinensis | KBGRB | 42 | BRA 2922 | B. biovisina | IPK |
| 8 | 28053 | B. rapa ssp pekinensis | KBGRB | 43 | K 9825 | B. bourgeai | IPK |
| 9 | 28055 | B. rapa ssp pekinensis | KBGRB | 44 | K 6631 | B. cretica | IPK |
| 10 | cnu-28020 | B. rapa ssp pekinensis | KBGRB | 45 | BRA 2919 | B. desnottesii | IPK |
| 11 | cnu-28065 | B. rapa ssp pekinensis | KBGRB | 46 | K 9402 | B. depranensis | IPK |
| 12 | cnu-28072 | B. rapa ssp pekinensis | KBGRB | 47 | BRA 1039 | B. frutoiculosa | IPK |
| 13 | 25082 | B. rapa ssp chinensis | KBGRB | 48 | BRA 1810 | B. frutoiculosa | IPK |
| 14 | 25083 | B. rapa ssp chinensis | KBGRB | 49 | BRA 1169 | B. gravinae | IPK |
| 15 | 25084 | B. rapa ssp chinensis | KBGRB | 50 | BRA 2856 | B. incana | IPK |
| 16 | 25095 | B. rapa ssp chinensis | KBGRB | 51 | K 5997 | B. insularis | IPK |
| 17 | 25103 | B. rapa ssp chinensis | KBGRB | 52 | K 7635 | B. macrocarpa | IPK |
| 18 | 25106 | B. rapa ssp chinensis | KBGRB | 53 | K9242 | B. maurorum | IPK |
| 19 | 25110 | B. rapa ssp chinensis | KBGRB | 54 | BRA 1645 | B. repanda | IPK |
| 20 | 26109 | B. rapa ssp chinensis | KBGRB | 55 | K 6877 | B. rupestris | IPK |
| 21 | 27081 | B. rapa ssp chinensis | KBGRB | 56 | K 8823 | B. spinescens | IPK |
| 22 | Tetralocular | B. rapa ssp oleifera | CGMCP | 57 | BRA 1896 | B. villosa | IPK |
| 23 | YSPB | B. rapa ssp oleifera | CGMCP | 58 | LET | A. thaliana LE ecootype | KBGRB |
| 24 | Candle | B. rapa ssp oleifera | CGMCP | 59 | ETR | A. thaliana Col ecotype | KBGRB |
| 25 | Pusa kalyanai | B. rapa ssp oleifera | CGMCP | 60 | 28697 | Camelina sativa | KBGRB |
| 26 | RCBr | B. rapa (rapid cycling) | KBGRB | 61 | 26165 | Herba cichori | KBGRB |
| 27 | CNU 28003 | B. oleracea ssp capitata | KBGRB | 62 | 2659 | Diplotaxis muralis | KBGRB |
| 28 | CNU 28004 | B. oleracea ssp capitata | KBGRB | 63 | 28614 | Eruca staiva | KBGRB |
| 29 | Varuna | B. juncea | CGMCP | 64 | 28699 | Hesperis matronalis | KBGRB |
| 30 | Heera | B. juncea | CGMCP | 65 | 26056 | Moricandia arvensis | KBGRB |
| 31 | Donskaja | B. juncea | CGMCP | 66 | 28672 | Sinapis alba | KBGRB |
| 32 | TM 4 | B. juncea | CGMCP | 67 | 28597 | Raphanus staivus | KBGRB |
| 33 | Tapidor | B. napus | China | 68 | 26080 | Sisymprium leteum | KBGRB |
| 34 | Ningyou-7 | B. napus | China | 69 | 26093 | Lepidium apetalum | KBGRB |
| 35 | Car6 | B. carinata | CGMCP | 70 |
KBGRB—Korea Brassica Genome Resource Bank, Daejeon, Korea; CGMCP—Centre for Genetic Manipulation of Crop Plants, Delhi, India, IPK—Leibniz Institute of Plant Genetics and Crop Plant Research, Gatersleben, Germany; LET— A. thaliana landsberg erecta ecotype; ETR— A. thaliana columbia ecotype.
2.2. Searching for SSR-containing sequences and primer design
We downloaded a total of 182 703 B. rapa EST sequences from NCBI database (http://www.ncbi.nlm.nih.gov) and assembled using CAP334 to identify unigenes. The unigene sequences (singlets and contigs) were then searched for the presence of SSR motifs using the MIcro SAtellite identification tool (MISA) available at http://pgrc.ipk-gatersleben.de/misa/misa.html and sputnik software following the criteria described earlier by Hong et al.35 The contig or singleton sequences were used to design primers flanking the putative SSRs using Primer 3.36 The primer designing conditions were: 58–60°C melting temperature with a difference of only 1°C between each forward and reverse primer, 40–60% GC content, and 19–21 bp primer length and an estimated amplicon size of 150–400 bp. ORF Finder37 (http://bioinformatics.org/sms/orf_find.html) and UTRScan (http://utrdb.ba.itb.cnr.it/tool/utrscan) were used to find the location of repeat motifs in coding region and untranslated regions (5′UTR and 3′TR) or in open reading frames.
2.3. DNA extraction, marker analysis, and cloning of PCR amplicon
DNA was extracted from young expanded leaf samples collected from greenhouse-grown plants using an RBC Genomic DNA Extraction Kit (Real Biotech Corporation, Taipei, Taiwan). A total of 707 newly developed UGMS markers (prefixed by ACMP, hereafter referred to as EST-SSRs, Supplementary Table S1) were used for a polymorphism survey between the parental lines Chiifu-401, Kenshin, and RCBr. A total of 999 BAC-derived SSRs, which were previously developed in our laboratory (designated by ‘cnu’, ‘nia’, and ‘BRPGM’),7,33 272 IP markers developed and mapped by Panjabi et al.,6 and 707 new EST-SSRs were screened between CR Shinki and 94SK. In another experiment, a total of 450 newly developed unigene-derived SSR markers (prefixed by ‘sau_um’, unpublished), which were developed in Shenyang Agricultural University, and 651 public SSR markers5,38–47 were screened for polymorphisms between the parental lines ‘501’ and ‘601’ of the PF2 population. The PCR reaction conditions used by Li et al.33 were followed for the BAC-derived SSRs, IP markers, and the newly developed EST-SSRs. The PCR conditions for the EST-derived SSR markers were as follows: 5 min at 95°C; 36 cycles of 45 s at 95°C, 45 s at 55°C, and 45 s at 72°C; with a final step of 10 min at 72°C. PCR products were resolved in 8% polyacrylamide gel electrophoresis as described by Kim et al.7 The PCR amplicon from different species was cloned in pGEM-T Easy cloning vector (www.promega.com) according to the manufacturer's instruction and at least one clone were sequenced two times from each Brassica species.
2.4. Construction of linkage maps and diversity analysis
The four individual maps and the integrated genetic map were constructed with Joinmap version 4.048,49 using the same parameters as described by Li et al.33 The Kosambi mapping function was used to calculate map distances.50 Logarithm of the odds (LOD) scores of 4.0–8.0 was used to group markers. A recombination frequency <0.4 and a LOD score >1.0 were used to arrange the marker order. Common markers were used as ‘bridge’ markers to integrate the four maps using the function ‘Combine the Groups for Map Integration’. We used two approaches to integrate the four maps: first, we used the CKDH map as the reference map and sequentially integrated the other three maps in the order CRF2, PF2, and CSKF2; and secondly by simultaneously integrating the four maps to identify consistency. The final integrated linkage map was drawn using MapChart.51 Power Marker 3.1 was used to calculate the polymorphic information content (PIC) value and gene diversity. The PIC value was estimated according to the method of Botstein et al.52 The crucifer building blocks proposed by Schranz et al.53 were identified in B. rapa genetic map based on homology search of primers pairs against the Arabidopsis genome sequence as described previously by Kim et al.7
3. Results
3.1. Development of UGMS markers
We downloaded a total of 182 703 B. rapa EST sequences from NCBI database in April 2010 and alignment of these EST sequences gave 19 497 (18 931 contigs and 566 singlets) unigenes. Analyses in these unigenes identified 4174 microsatellite motifs in 3037 genes. Of these many unigenes containing one or more SSRs, we designed a total of 707 EST-SSR markers (Supplementary Table S1). Among the primer pairs designed, trinucleotide repeats were the highest (573, 81.05%) followed by di- (126, 17.82%) and tetranucleotide repeats (8, 1.13%), respectively (Table 2). Analysis of the location of the 707 SSR motifs in the sequence used to design primers showed that majority of them was located in the coding region (CDS, 491) compared to 5′UTR (107) and 3′UTR (109) (Supplementary Table S1). Of the 707 EST-SSR primer pairs, 691 (97.74%) produced repeatable and reliable amplifications of expected size in at least one line of the five B. rapa parental lines (Chiifu 401, Kenshin, Rapid cycling B. rapa, 94 SK and CR Shiki) screened, while 16 (2.26%) primer pairs either completely failed or led to weak amplifications and thus were excluded from further analysis.
Table 2.
Frequency and distribution of different repeat types used to design EST-SSR primer pairs in B. rapa
| Repeats | Number of repeat units |
Total | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | ≥14 | ||
| AC/GT | 1 | 1 | 2 | |||||||||
| AG/CT | 17 | 14 | 5 | 2 | 5 | 3 | 46 | |||||
| AT/TA | 9 | 3 | 2 | 1 | 2 | 1 | 2 | 20 | ||||
| CA/TG | 4 | 3 | 2 | 1 | 10 | |||||||
| GA/TC | 19 | 7 | 8 | 5 | 3 | 1 | 1 | 4 | 48 | |||
| AAC/GTT | 4 | 3 | 4 | 2 | 13 | |||||||
| AAG/CTT | 41 | 13 | 6 | 1 | 1 | 62 | ||||||
| ACA/TGT | 10 | 3 | 2 | 1 | 1 | 17 | ||||||
| ACC/GGT | 9 | 7 | 16 | |||||||||
| ACT/AGT | 2 | 2 | 1 | 5 | ||||||||
| AGA/TCT | 31 | 7 | 2 | 2 | 42 | |||||||
| AGC/GCT | 13 | 6 | 2 | 1 | 22 | |||||||
| AGG/CCT | 25 | 14 | 5 | 1 | 45 | |||||||
| ATA/ATT | 3 | 3 | ||||||||||
| ATC/GAT | 22 | 5 | 1 | 1 | 29 | |||||||
| ATG/CAT | 11 | 3 | 2 | 1 | 1 | 18 | ||||||
| CAA/TTG | 11 | 5 | 4 | 1 | 21 | |||||||
| CAC/GTG | 3 | 3 | 1 | 7 | ||||||||
| CAG/CTG | 8 | 3 | 2 | 1 | 14 | |||||||
| CCA/TGG | 17 | 7 | 1 | 1 | 26 | |||||||
| CCG/CGG | 11 | 3 | 1 | 15 | ||||||||
| CGA/TCG | 6 | 2 | 1 | 9 | ||||||||
| CGC/GCG | 5 | 3 | 8 | |||||||||
| CGT/GAC | 3 | 1 | 1 | 5 | ||||||||
| CTA/TAG | 3 | 1 | 1 | 5 | ||||||||
| CTC/GAG | 19 | 6 | 4 | 29 | ||||||||
| GAA/TTC | 32 | 10 | 5 | 1 | 1 | 2 | 1 | 1 | 53 | |||
| GCA/TGC | 14 | 3 | 2 | 19 | ||||||||
| GCC/GGC | 11 | 3 | 1 | 15 | ||||||||
| GGA/TCC | 19 | 10 | 4 | 1 | 1 | 35 | ||||||
| GTA/TAC | 2 | 2 | 4 | |||||||||
| TAA/TTA | 4 | 1 | 5 | |||||||||
| TCA/TGA | 17 | 11 | 2 | 1 | 31 | |||||||
| AAGG/CTTC | 2 | 2 | ||||||||||
| AGAA/TTAT | 2 | 2 | ||||||||||
| GTGC/TGGT | 2 | 2 | ||||||||||
| TACC/TGTT | 2 | 2 | ||||||||||
| Total | 364 | 134 | 107 | 39 | 23 | 14 | 12 | 5 | 1 | 2 | 6 | 707 |
3.2. Construction of individual maps
3.2.1. Updating the CKDH reference genetic map
The CKDH linkage map was adopted as the reference genetic linkage map by the MBrGSP. The version I CKDH reference genetic map of B. rapa was constructed by Choi et al.,5 while Kim et al.7 generated version II by incorporating more BAC-anchored SSRs, and this was further updated with the inclusion of 95 gene-based IP markers by Li et al.33 We screened 707 newly developed EST-SSR markers (prefix ACMP) for polymorphisms between the parental lines of the CKDH mapping population, Chiifu-401 and Kenshin, in the present study. However, only 99 (14%) of these markers were polymorphic between the 2 parental lines, and finally 95 EST-SSR markers were mapped to the 10 linkage groups of B. rapa. These 95 EST-SSR markers were distributed in all of the 10 B. rapa linkage groups, except in A2, and the number of markers ranged from 4 in A10 to 19 in A5. After adding the new EST-SSR markers, most of the previous markers were assigned in the same order without any major changes with respect to their position. The total length of the updated CKDH map was 1217.6 cM, which is 42.5 cM larger than the earlier map (1175.1 cM; Li et al.33). The linkage groups A3 (145.8 cM) and A6 (165.2 cM) increased by 10 cM compared to the map of Li et al.33 because of the addition of new EST markers. The average distance between adjacent markers decreased from 1.45 to 1.34 cM. The updated EST-SSRs CKDH map consisted of a total of 907 markers with 190 BAC-derived SSRs, 95 new EST-derived SSRs, 94 IP markers, and 528 other markers (Choi et al.,5 Table 2, Supplementary Table S2).
3.2.2. Updating the CRF2 linkage map
The CRF2 map was initially developed by Li et al.33 using BAC-derived SSR and IP markers. Of the 707 EST-SSRs screened for polymorphisms between the parental lines, Chiifu-401 and RCBr, we identified 144 pairs of polymorphic EST-SSRs, of which only 142 pairs could be used for genotyping and the construction of the linkage map. After excluding the distorted and ungrouped markers, a total of 129 new EST-SSRs were successfully integrated into the 10 linkage groups of B. rapa, giving a total length of 1119.5 cM. The distribution of the newly mapped EST-SSRs varied from 7 in linkage group A10 to 19 in A3. Compared to the previous CRF2 map,33 most of the BAC-SSRs and IP markers remained in the same order; however, two SSR loci and one IP locus in linkage group A4, one IP locus in A5, and one SSR locus in A9, all distorted markers, were deleted from the map as they reshuffled the marker order in these two linkage groups. The updated CRF2 linkage map now contains a total of 444 markers with 129 new EST-SSRs, 249 BAC-derived SSRs, and 66 IP loci (Supplementary Table S3). The length of the individual linkage groups ranged from 79.9 cM in A06 to 170.1 cM in A9. Although the total length of the genetic map did not significantly increase compared to the previous CRF2 map,33 the average distance between adjacent markers decreased from 3.47 to 2.53 cM.
3.2.3. Construction of the CSKF2 linkage map
The new CSKF2 population was developed by crossing the clubroot resistant cultivar CR Shinki and the susceptible 94SK line. For the construction of the genetic linkage map, we screened SSRs and IP markers for polymorphisms between the parental lines. Of the 707 EST-SSRs, 250 BRPGM-SSRs, 50 cnu_SSRs, 30 nia_SSRs, and 272 IP markers screened between the parental lines, and only 99 EST-SSRs, 28 BRPGM-SSRs, 19 cnu_SSRs, 6 nia_SSRs, and 19 IP makers were polymorphic. All markers, except the EST-SSRs, were previously described and some of them have already been mapped in the B. rapa genome.7,33 The previously mapped markers enabled us to align and integrate the different maps in the present study. We used the genotype data from a total of 173 markers to construct the linkage map, although only 161 markers [93 EST-SSR, 49 BAC-SSR, 17 IP, and 2 sequence characterized amplified region (SCAR) markers] could be grouped and assigned to the 10 linkage groups, covering 602.3 cM of the B. rapa genome (Supplementary Table S4). The number of markers in each linkage group ranged from 7 in A1 to 33 in A3, with an average distance of 3.74 cM between adjacent markers.
3.2.4. PF2 linkage map
The PF2 genetic map was constructed using 144 F2 lines derived from crossing between two diverse Chinese cabbage inbred lines, 501 with a large head and 601 with a small head, at Shenyang Agricultural University, China. This genetic map of B. rapa contains a total of 277 markers (72 EST-SSR loci, 154 genomic SSRs, and 1 leaf hairiness phenotypic marker) in the 10 linkage groups with total genome coverage of 908.4 cM. The average distance between adjacent markers was 4.02 cM (Supplementary Tables S1 and Table S5) and the shortest and longest linkage groups were A8 (74.9 cM) and A9 (122.0 cM).
3.3. Construction of the updated consensus genetic map of B. rapa
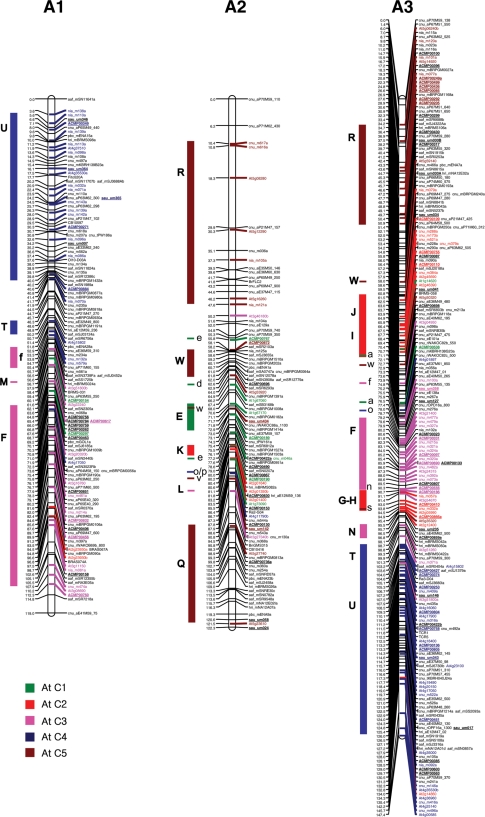
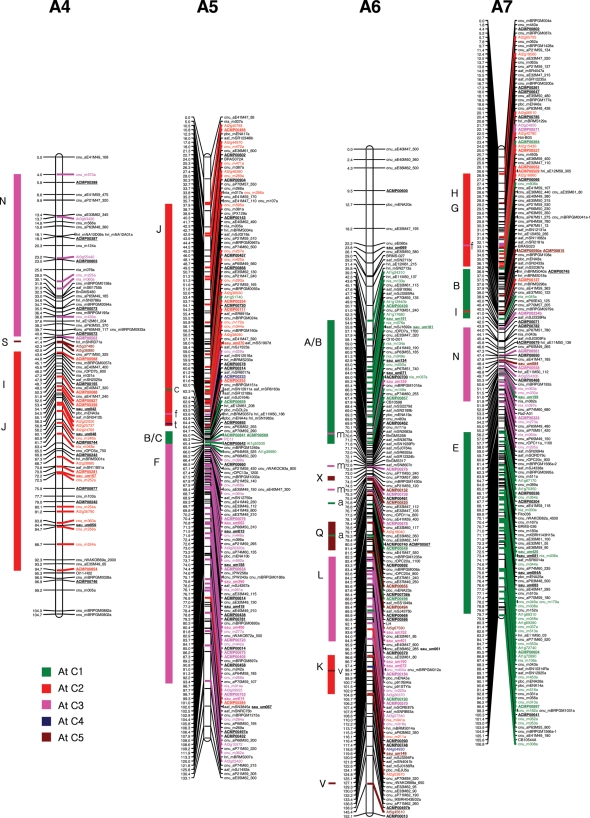
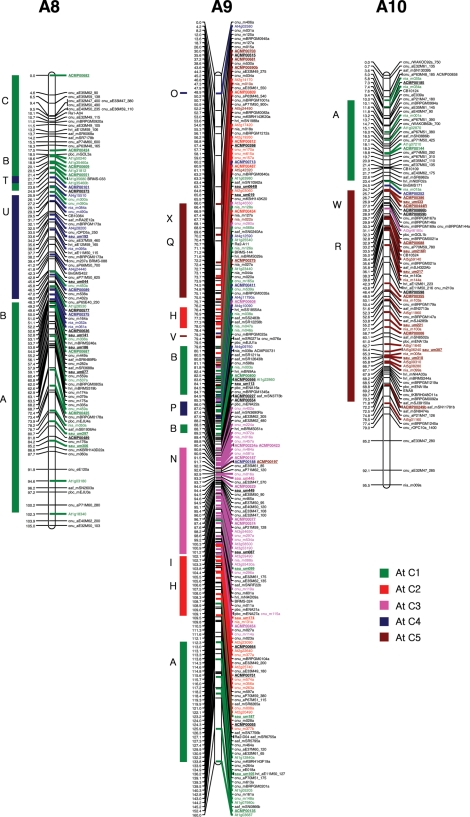
The four individual maps were integrated using the commonly mapped markers as bridge markers. In total, 241 bridge markers (including 66 EST-SSRs) were identified among at least two of the mapping populations. The distribution of bridge markers was 22 in A1, 12 in A2, 38 in A3, 13 in A4, 37 in A5, 19 in A6, 25 in A7, 17 in A8, 42 in A9, and 16 in A10, respectively. SSR and IP markers with more than one polymorphic locus in the same linkage groups were correctly identified for size and order before designating the common loci between the maps. A total of 1426 markers were mapped on the 10 linkage groups of B. rapa. The number of markers in the integrated map ranged from 97 markers in linkage group A2 to 209 markers in A3, and the length of the linkage groups varied from 95.5 cM in A10 to 160.0 cM in A9. The total length of the integrated consensus map was 1245.9 cM, with an average distance between adjacent markers of 0.87 cM (Table 3 and Fig. 1). The comparison of the four individual maps and the integrated map revealed a similar order of the markers, even though changes of order and the position of a few markers were observed within a 5 cM distance in some of the linkage groups, with the exception of five linkage groups (A3, A5, A7, A9, and A10) where an inversion of more than 10 cM was observed. The integrated map contained 306 new EST-derived SSR markers (prefixed by ACMP and sau-um), 395 BAC-derived SSRs, 153 IP markers, and 572 other markers (Table 3 and Fig. 1). The number of EST-derived SSRs mapped in the 10 linkage groups of B. rapa ranged from 16 in linkage group A2 to 50 in A3. The density of the updated EST-SSR-rich integrated map increased compared to the previously integrated CKDH and CRF2 maps by Li et al.33 since the average distance between marker loci decreased from 1.24 to 0.87 cM. The length of the integrated linkage groups was similar to the corresponding longest linkage groups of the component maps with a slight increase in map length, except for the linkage groups A1, A6, A8, and A10 where decreases of approximately 20, 13.1, 8.4, and 10 cM, respectively, were observed in comparison to the maximum lengths of the individual maps. The EST-SSR marker ACMP00682, which mapped to the top portion of the A08 linkage group, increased the map length by 4.8 cM. The large gaps observed in individual maps were reduced due to the increased marker density, and the final consensus integrated map had only one large gap (>10 cM) in the A2 linkage group.
Table 3.
Characteristics of the updated Brassica rapa integrated linkage map developed using four mapping populations
| Linkage group | Number of different marker types |
Total length (cM) | Average distance (cM) | ||||
|---|---|---|---|---|---|---|---|
| EST-derived SSRsa | BAC-derived SSRsb | IPc | Othersd | Total | |||
| A1 | 21 | 43 | 12 | 52 | 128 | 119.0 | 0.93 |
| A2 | 16 | 25 | 15 | 41 | 97 | 122.3 | 1.26 |
| A3 | 50 | 55 | 31 | 73 | 209 | 147.4 | 0.71 |
| A4 | 22 | 25 | 9 | 29 | 85 | 104.7 | 1.23 |
| A5 | 42 | 45 | 12 | 73 | 172 | 133.1 | 0.77 |
| A6 | 41 | 19 | 10 | 79 | 149 | 152.1 | 1.02 |
| A7 | 34 | 55 | 14 | 80 | 183 | 106.8 | 0.58 |
| A8 | 20 | 30 | 11 | 41 | 102 | 105.0 | 1.03 |
| A9 | 41 | 74 | 29 | 59 | 203 | 160.0 | 0.79 |
| A10 | 19 | 24 | 10 | 45 | 98 | 95.5 | 0.97 |
| Total | 306 | 395 | 153 | 572 | 1426 | 1245.9 | 0.87 |
Figure 1.
The distribution of EST–SSRs and other markers in the 10 integrated linkage groups (A1–A10) of Brassica rapa. BAC-derived SSR markers,7 EST–SSRs and IP markers6 containing representative blocks were highlighted by bold strokes. New EST-derived SSR markers are underlined. The colorful rectangular bars on left of the integrated linkage map indicates the crucifer building blocks homologous to the Arabidopsis thaliana (At) chromosomes (C1–C5).53
3.4. Identification of crucifer building blocks
With the addition of more markers, we could accurately resolve the B. rapa genome for collinearity blocks with A. thaliana chromosomes.53 Blast analysis of B. rapa BAC and EST sequences, wherefrom the SSRs were identified, helped to identify homologous Arabidopsis chromosomal blocks which, in turn, helped to identify conserved ancestral crucifer building blocks in the B. rapa genome (Fig. 1).53 We could confidently establish homologous blocks that were previously identified with slightly less stringent criteria due to low marker density,33 such as W and E blocks in linkage group A2, the B and C blocks in A5, the Q block in A6, the T block in A8, and the W block in A10. Furthermore, as the blocks were already identified in the corresponding homologous A genome chromosomes of B. juncea and B. napus, we could identify new blocks containing mostly one marker in the B. rapa A genome, and these could be regarded as probable blocks since the available evidence supports the highly conserved nature of the A chromosomes at gross level among the three species, despite their divergence a long time ago.4,6 The new probable conserved blocks identified in this study are the M block in linkage group A1, the V block in A2, the X block in A6 and A9, and the I block in A7 (Fig. 1, Supplementary Fig. S1). However, further addition of markers is necessary to accurately resolve these blocks, even though we used already established evidence from the corresponding A genome chromosome blocks in B. napus and B. juncea while considering these blocks.
3.5. Evaluation of the mapped EST-derived SSR markers for allelic diversity
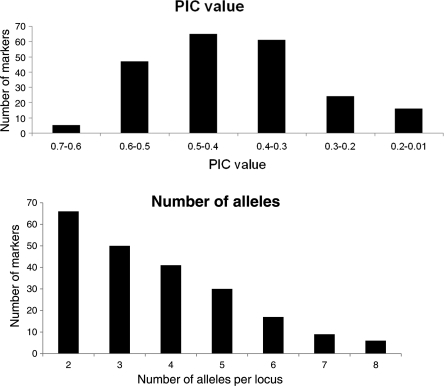
We selected 238 EST-SSRs (234 ACMP and 4 sau-um) that mapped to different B. rapa linkage groups to study their allelic diversity in 24 B. rapa genotypes belonging to the sub-species oleifera (oil type), pekinensis (Chinese cabbage), and chinensis (Pakchoi) type. Brassica rapa plants belonging to these sub-species are morphologically different with respect to leaf types, heading habits, and overall plant morphology. The PCR successfully amplified all of the 238 EST-SSRs markers in the 24 B. rapa genomes. The number of alleles amplified per EST-SSR ranged from 2 to 8, with an average of 2.9 (Fig. 2, Table 4). The majority of SSRs consisted of two alleles. The PIC value, which is a measure of allelic diversity, varied from 0.08 to 0.65, with an average of 0.40. The most commonly observed PIC values were between 0.4 and 0.5 (Table 4). The highest average PIC value was detected in the linkage group A1 (0.45) and the lowest one was in A9 (0.36).
Figure 2.
The distribution of PIC values and allele frequencies calculated from 238 EST-SSRs in 24 B. rapa germplasm.
Table 4.
Average number of alleles and the PIC values calculated for the mapped EST-derived SSRs in 10 linkage groups of B. rapa
| Linkage group | EST-markers number | Average PIC valuea | Average number of alleles |
|---|---|---|---|
| A1 | 17 | 0.45 | 3.7 |
| A2 | 12 | 0.40 | 3.0 |
| A3 | 40 | 0.37 | 2.8 |
| A4 | 22 | 0.40 | 3.0 |
| A5 | 33 | 0.42 | 2.7 |
| A6 | 29 | 0.41 | 2.7 |
| A7 | 28 | 0.40 | 2.6 |
| A8 | 13 | 0.37 | 2.8 |
| A9 | 31 | 0.36 | 2.5 |
| A10 | 13 | 0.44 | 2.8 |
| Total | 238 | 0.40 | 2.9 |
aPolymorphic information content (PIC).
3.6. Transferability of EST-SSRs in other cultivated and wild Brassica relatives
To assess the utility of EST-SSR loci across other cultivated and wild relatives of Brassica species, 167 EST-SSRs (146 EST-SSRs mapped and 21 EST-SSRs that were not mapped in the 10 linkage groups of B. rapa) (Supplementary Table S6) were used to amplify 48 germplasm belonging to 35 species of Brassicaceae family which includes five genotypes from B. rapa, two each from B. oleracea, B. nigra, B. napus, B. carinata and Arabidopsis thaliana, five from B. juncea and wild relatives (Table 1 and Fig. 3). Of the 167 primer pairs used to amplify in germplasm, cultivated Brassica species other than B. rapa showing 100% amplification were B. carinata, B. juncea and B. napus. Only one primer pair was not amplified in B. oleracea, while 17 primer pairs did not amplify in B. nigra. In Arabidopsis thaliana, 52 primer pairs did not show amplification. Among the wild species under Brassica genus, highest number of transferability was found in B. incana (amplified 161 out of 167 primer pairs) followed by B. bourgeaui and B. insularis (158 amplified in both the species). Brassica balearica DNA showed lowest number of primer pair amplification suggesting less cross-species transferable among them. Among the wild relative species, Herba chichori showed the highest number of amplification (163) and the lowest number for amplification was shown by Sysimprium leteum (71 primer pairs) (Supplementary Table S6).
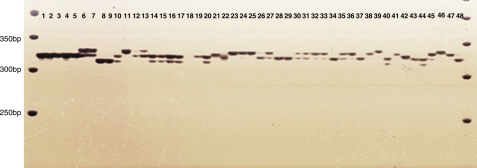
Figure 3.
Representative polyacrylamide gel picture of different alleles amplified with the primer ACMP00321 in 48 genotypes belonging to 35 different species. Serial numbers: 1–5, B. rapa; 6–7, B. oleracea; 8–9, B. nigra; 10–11, B. carinata; 12–13, B. napus, 14–17, B. juncea; 18–19, A. thaliana; 20, B. tourniforti; 21, B. balearica; 22, B. barreilieri; 23, B. bivoniana; 24, B. bourgeaui; 25, B. cretica; 26, B. desnottesii; 27, B. drepanensis; 28–29, B. fruticulosa; 30, B. gravinae; 31, B. incana; 32, B. insularis; 33, B. macrocarpa; 34, B. mourorum; 35, B. repanda; 36, B. rupestris; 37, B. spinescens; 38, B. villosa; 39, Camelina sativa; 40, Herba cichori; 41, Diplotaxis muralis; 42, Eruca sativa; 43, Hesperis matronalis; 44, Moricandia arvensis; 45, Sinapis alba; 46, Raphanus sativus; 47, Sisymprium leteum; 48, Lepidium apetalum. All species are listed in Table 1.
A total of 1326 SSR alleles were scored from 167 EST-SSRs primer pairs with an average of 7.94 alleles per primer pair across all 48 genotypes representing 35 species. The primer pair of ACMP00904 produced the highest number of alleles (17 bands), while two primer pairs namely ACMP00561 and ACMP00832 produced the lowest number of alleles (three bands) among the 48 germplasm. Most of the primer pairs showed the amplification of 5–6 alleles followed by primer pairs amplifying 7–8 and 9–10 alleles, respectively. There was a wide variation among species in the average number of alleles per primer pairs.
3.7. Sequence level comparison of SSR locus
To validate the sequence level conservation of SSRs across the species, at least one amplified product of EST-SSR ACMP00321 from 31 different species were cloned and sequenced. The EST-SSR primer pair for ACMP00321 was designed flanking four tri-nucleotide repeats (GAA). Although sequence alignment showed high conservation of nucleotide sequences from 31 different species, several single nucleotide substitutions and InDel polymorphism of 3-bp to 11-bp flanking the microsatellite repeats were observed besides variation in number of repeat motifs (Supplementary Fig. S2). Variations of repeat numbers were observed from four repeats to a maximum of six repeats. Brassica barreilieri and B. bourgeaui showed six numbers of repeats, while B. creteca, B. incana, and Camelina Sativa showed five numbers of repeats. Remaining species showed conserved four repeats.
4. Discussion
EST-SSRs are highly useful in molecular plant breeding and evolutionary studies since these markers are developed from transcribed region of the genes and are highly cross-species transferable. Although, recent study reported the development EST-SSR markers in B. rapa,24 the exact genomic location of the developed markers in the B. rapa genome were not determined by mapping, and there remained many more EST-SSRs to be identified owing to the large number of genes present in B. rapa genome (expected gene number is over 40 000, MBrGSP, personal communication). Therefore, in this study, we developed 707 new EST-SSRs, of which 691 (97.73%) primer pairs were successfully amplified the DNA fragments of expected sizes in the B. rapa genome. The majority of these EST-SSR markers were single locus markers, while only a few of them were duplicated and mapped to different linkage groups.
The EST-SSR markers were less polymorphic than the genomic SSRs as they were derived from genic regions. Of the 707 EST-SSRs, only 99 (14%) were polymorphic between the parental lines Chiifu-401 and Kenshin, whereas 311 SSRs (41.5%) from 749 BAC-SSRs were polymorphic.7 For the CRF2 population, 20% of the EST-SSRs were polymorphic between the parental lines Chiifu-401 and RCBr, which is also lower than for genomic SSRs (67%). Similarly, reduced levels of polymorphisms were detected between CR Shinki and 94SK (14%, the parental lines of the CSKF2 population) and between 501 and 601 (15.56%, the parental lines of the PF2 population). Similar results were also reported in pearl millet30 and soybean.20 These findings indicate that the SSRs located in coding regions were relatively more conserved than those in non-coding regions. Despite showing lower levels of polymorphisms, the EST-derived SSRs have much more potential than genomic SSRs to reveal the functional variation between individuals.
The updated high-density integrated map of B. rapa now contains a total of 1426 markers that include 414 new markers (306 EST-SSRs, 55 BAC-SSR, 10 IP, and 40 public SSR markers, 1 phenotypic marker, and 2 SCAR markers) and covers a total length of 1245.9 cM, which is almost similar to the earlier map (1262.0 cM; Li et al.33). We found a slight decrease in the map length due to the addition of more markers, thereby increasing the marker density from 1.27 cM in the previous map33 to 0.87 cM in the present one. The majority of the mapped EST-SSR markers were randomly distributed in the 10 linkage groups; however, a few were clustered in narrow regions of a few linkage groups, e.g. A3 (20–28 cM), A5 (59–62 cM), A8 (17–24 cM), and A10 (25–29 cM). These EST-SSR clusters may be indicative of gene-rich regions, although more gene-based markers are needed to fully characterize these regions.
We compared the updated integrated map and the component maps and found the overall order and positions of the markers to be same, despite observing minor local inversions within a 5 cM distance in many linkage groups, with the exception of a few markers with longer distance inversions (in A2, A3, A4, and A9). These kinds of local inversions were reported earlier in map integration in Arabidopsis, B. oleracea, lettuce, rapeseed, and many other plant species.54–60 The various reasons cited were: (i) mapping inaccuracies resulted from small mapping populations,55 (ii) closely spaced markers in one population,21 (iii) distorted segregation of markers, and (iv) real inversions. We believe that the addition of more common markers would help solve up to certain extent the local discrepancies observed in few linkage groups due to any one or combination of above-cited reasons. However, while comparing the mapped IP markers in our map with the B. juncea map,6 we observed the overall conservation of A genome chromosomes (data not shown) between the two species, which further supports the correct alignment of our map.
The addition of new BAC-SSRs, EST-SSRs, and IP markers in the updated consensus genetic map helped to confirm the accuracy of the previously identified cruciferous building blocks53 and to identify previously unidentified probable blocks33 in our linkage map by blast analysis against the A. thaliana genome sequence. We identified an additional five novel probable blocks (M in linkage group A1, V in A2, X in A6 and A9, and I in A7) compared to B. napus4 and B. juncea.6 However, additional markers are necessary to confirm these blocks since only one or a few markers were identified and the blocks were designated on the basis of the corresponding A chromosomes of B. juncea6 and B. napus.4
The analysis of allelic variation of the 238 mapped EST-SSRs in 24 B. rapa germplasm belonging to the sub-species oleifera, chinensis, and pekinensis revealed that the number of alleles ranged from 2 to 8. It is known that the observation of different allele frequencies of any given marker is closely related to its transferability among germplasm, as well as to the degree of variability within the marker locus.61 However, the most frequent number of alleles amplified per EST-SSRs was 2, suggesting that these markers were probably generated by insertion–deletion polymorphisms, as previously observed in soybean.20,62 We observed that the average PIC values of the mapped markers in the 10 linkage groups of B. rapa were 0.40 (range, 0.36–0.45), similar to the PIC of 0.40 observed in soybean by Hossain et al.61 and Hisano et al.;62 furthermore, they reported that the average PIC values of EST-derived markers were lower than for random genomic DNA-derived markers because of the highly conserved nature of the coding regions. We also expect lower PIC values for the EST-SSRs used in the present study than for the random and BAC-derived SSRs, even though we did not examine their PIC values, as the BAC-SSRs were more polymorphic than the EST-SSRs in the parental lines.
The advantage of high cross-species transferability of EST-SSRs can be widely exploited in comparative mapping studies to see the conservation and diversification of gene order in the related species and isolate the candidate genes from the target species using candidate genes. Cross-species transferability experiment analysis of newly developed EST-SSRs in 35 other cultivated and wild Brassica relatives showed varying number of amplification of EST-SSRs ranging from 100% in B. napus, B. juncea, and B. carinata to 45.50% in Sysimprium leteum. Varying rates of transferability of EST-SSR markers among related species or genera have been demonstrated in several studies.28,32 For example, all of the 61 EST-SSRs markers developed in Camellia sinensis were fully transferable to Camellia assamica and Camellia assamica ssp. Lasiocalyx, while it showed various rates of transferability to C. lutescens; C. irrawadiensis, and C. japonica.25 Raji et al.32 found cross-species amplification of 94% of cassava EST-SSRs in related wild species, while amplification of up to 100% of EST-SSRs designed in barley was observed in Hordeum chilense, and 76–100% amplification in different Triticum species.28 Further, comparison of DNA sequence from 31 species showed highly identical nature of the DNA sequences with only few SNPs and Indels among the species suggesting the highly conserved nature of the gene sequences even among the distantly related species. Although variation in repeat motifs was observed, Indels surrounding the SSR motifs were the main reason for getting sequence polymorphism between the species beside few SNPs observed throughout the sequences.
The development of an updated high-density integrated B. rapa genetic linkage map based on highly cross-species transferable gene-based markers such as EST-SSRs and IP markers would allow comparative genetic analysis between B. rapa and other Brassica crops. Further, the mapping of previously developed public SSRs in our genetic map would also facilitate the identification of candidate genes through the fine mapping of candidate QTL/gene regions for important traits as our map contains BAC-SSRs, EST-SSRs, and IP markers adjacent along with those markers. These could then be used for the marker-assisted selection of economically important trait loci/QTL in the respective populations.41,43,44,47 The soon-to-be available draft genome sequence of the euchromatic regions of the B. rapa genome does not diminish the importance of genetic linkage maps, as our map will facilitate the alignment of gene-rich euchromatic regions with less gene-dense repetitive regions, thereby helping to confirm and refine the integration of the genetic map with genomic sequences. The large number of gene-based markers mapped in the present study will promote the understanding of the comparative analysis of the Brassica genome at the structural and functional levels.
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Funding
This work was supported by grants from the Technology Development Program for Agriculture and Forestry, Ministry for Food, Agriculture, Forestry, and Fisheries (grant no. 607003-05), Republic of Korea; and National Natural Science Foundation of China (Grant No. 30771468). N.R. was partially supported by the National Research Foundation, Republic of Korea.
Supplementary Material
Acknowledgements
We are thankful to the Korea Brassica Genome Resource Bank for providing the CKDH, CRF2, and CSKF2 mapping populations, Centre for Genetic Manipulation of Crop Plants, Delhi University South Campus, India; and Leibniz Institute of Plant Genetics and Crop Plant Research, Gatersleben, Germany, for different Brassica and wild relatives germplasm; and Ms Kyeong Ahn Lee for technical support.
References
- 1.UN. Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn. J. Bot. 1935;7:389–452. [Google Scholar]
- 2.Teutonico R.A., Osborn T.C. Mapping of RFLP and quantitative trait loci in Brassica rapa and comparison to the linkage maps of B. napus, B. oleracea and Arabidopsis thaliana. Theor. Appl. Genet. 1994;89:885–94. doi: 10.1007/BF00224514. [DOI] [PubMed] [Google Scholar]
- 3.Snowdon R.J., Friedt W. Molecular markers in Brassica oilseed breeding: current status and future possibilities. Plant Breed. 2004;123:1–8. [Google Scholar]
- 4.Parkin I.A.P., Gulden S.M., Sharpe A.G., et al. Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana. Genetics. 2005;171:765–81. doi: 10.1534/genetics.105.042093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi S.R., Teakle G.R., Plaha P., et al. The reference genetic linkage map for the multinational Brassica rapa genome sequencing project. Theor. Appl. Genet. 2007;115:777–92. doi: 10.1007/s00122-007-0608-z. [DOI] [PubMed] [Google Scholar]
- 6.Panjabi P., Jagannath A., Bisht N., et al. Comparative mapping of Brassica juncea and Arabidopsis thaliana using Intron Polymorphism (IP) markers: homoeologous relationships, diversification and evolution of the A, B and C Brassica genomes. BMC Genomics. 2008;9:113. doi: 10.1186/1471-2164-9-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim H., Choi S.R., Bae J., et al. Sequenced BAC anchored reference genetic map that reconciles the ten individual chromosomes of Brassica rapa. BMC Genomics. 2009;10:432. doi: 10.1186/1471-2164-10-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramchiary N., Padmaja K.L., Sharma S., et al. Mapping of yield influencing QTL in Brassica juncea: implications for breeding of a major oilseed crop of dryland areas. Theor. Appl. Genet. 2007a;115:807–17. doi: 10.1007/s00122-007-0610-5. [DOI] [PubMed] [Google Scholar]
- 9.Ramchiary N., Bisht N.C., Gupta V., et al. QTL analysis reveals context-dependent loci for seed glucosinolate trait in the oilseed Brassica juncea: importance of recurrent selection backcross scheme for the identification of ‘true’ QTL. Theor. Appl. Genet. 2007b;116:77–85. doi: 10.1007/s00122-007-0648-4. [DOI] [PubMed] [Google Scholar]
- 10.Lou P., Zhao J.J., Kim J.S., et al. Quantitative trait loci for flowering time and morphological traits in multiple populations of Brassica rapa. J. Exp. Bot. 2007;58:4005–16. doi: 10.1093/jxb/erm255. [DOI] [PubMed] [Google Scholar]
- 11.Lou P., Zhao J., He H., et al. Quantitative trait loci for glucosinolate accumulation in Brassica rapa leaves. New Phytol. 2008;179:1017–32. doi: 10.1111/j.1469-8137.2008.02530.x. [DOI] [PubMed] [Google Scholar]
- 12.Lu G., Cao J., Yu X., Xiang X., Chen H. Mapping QTLs for root morphological traits in Brassica rapa L. based on AFLP and RAPD markers. J. Appl. Genet. 2008;49:23–31. doi: 10.1007/BF03195245. [DOI] [PubMed] [Google Scholar]
- 13.Li F., Kitashiba H., Inaba K., Nishio T. A Brassica rapa linkage map of EST-based SNP markers for identification of candidate genes controlling flowering time and leaf morphological traits. DNA Res. 2009;16:311–23. doi: 10.1093/dnares/dsp020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J., Lu Y., Yuan Y., et al. Map based cloning and characterization of a gene controlling hairiness and seed coat color traits in Brassica rapa. Plant Mol. Biol. 2009;69:553–63. doi: 10.1007/s11103-008-9437-y. [DOI] [PubMed] [Google Scholar]
- 15.Schranz M.E., Quijada P., Sung S.B., Lukens L., Amasino R.M., Osborn T.C. Characterization and effects of the replicated time gene FLC in Brassica rapa. Genetics. 2002;162:1457–68. doi: 10.1093/genetics/162.3.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marcel T.C., Varshney R.K., Barbieri M., et al. A high-density consensus map of barley to compare the distribution of QTLs for partial resistance to Puccinia hordei and of defence gene homologues. Theor. Appl. Genet. 2007;114:487–500. doi: 10.1007/s00122-006-0448-2. [DOI] [PubMed] [Google Scholar]
- 17.Singh K., Ghai M., Garg M., et al. An integrated molecular linkage map of diploid wheat based on a Triticum boeoticum × T. monococcum RIL population. Theor. Appl. Genet. 2007;115:301–12. doi: 10.1007/s00122-007-0543-z. [DOI] [PubMed] [Google Scholar]
- 18.Varshney R.K., Marcel T.C., Ramsay L., et al. A high density barley microsatellite consensus map with 775 SSR loci. Theor. Appl. Genet. 2007;114:1091–103. doi: 10.1007/s00122-007-0503-7. [DOI] [PubMed] [Google Scholar]
- 19.Vezzulli S., Troggio M., Coppola G., et al. A reference integrated map for cultivated grapevine (Vitis vinifera L.) from three crosses, based on 283 SSR and 501 SNP-based markers. Theor. Appl. Genet. 2008;117:499–511. doi: 10.1007/s00122-008-0794-3. [DOI] [PubMed] [Google Scholar]
- 20.Hwang T.Y., Sayama T., Takahashi M., et al. High-density integrated linkage map based on SSR markers in soybean. DNA Res. 2009;16:213–25. doi: 10.1093/dnares/dsp010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mace E.S., Rami J.-F., Bouchet S., et al. A consensus genetic map of sorghum that integrates multiple component maps and high-throughput Diversity Array Technology (DArT) markers. BMC Plant Biol. 2009;9:13. doi: 10.1186/1471-2229-9-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hong Y., Chen X., Liang X., et al. A SSR-based composite genetic linkage map for the cultivated peanut (Arachis hypogaea L.) genome. BMC Plant Biol. 2010;10:17. doi: 10.1186/1471-2229-10-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parida S.K., Anand Raj Kumar K., Dalal V., Singh N.K., Mohapatra T. Unigene derived microsatellite markers for the cereal genomes. Theor. Appl. Genet. 2006;112:808–17. doi: 10.1007/s00122-005-0182-1. [DOI] [PubMed] [Google Scholar]
- 24.Parida S.K., Yadava D.K., Mohapatra T. Microsatellites in Brassica unigenes: relative abundance,marker design, and use in comparative physical mapping and genome analysis. Genome. 2010;53:55–67. doi: 10.1139/g09-084. [DOI] [PubMed] [Google Scholar]
- 25.Sharma R.K., Bhardwaj P., Negi R., Mohapatra T., Ahuja P.S. Identification, characterization and utilization of unigene derived microsatellite markers in tea (Camellia sinensis L.) BMC Plant Biol. 2009;9:53. doi: 10.1186/1471-2229-9-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morgante M., Hanafey M., Powell W. Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nat Genet. 2002;30:194–200. doi: 10.1038/ng822. [DOI] [PubMed] [Google Scholar]
- 27.Varshney R.K., Graner A., Sorrells M.E. Genetic microsatellite markers in plants: features and applications. Trends Biotech. 2005;23:48–55. doi: 10.1016/j.tibtech.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 28.Castillo A., Budak H., Varshney R.K., Dorado G., Graner A., Hernandez P. Transferability and polymorphism of barley EST-SSR markers used for phylogenetic analysis in Hordeum chilense. BMC Plant Biol. 2008;8:97. doi: 10.1186/1471-2229-8-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishikawa G., Nakamura T., Ashida T., et al. Localization of anchor loci representing five hundred annotated rice genes to wheat chromosomes using PLUG markers. Theor. Appl. Genet. 2008;118:499–514. doi: 10.1007/s00122-008-0916-y. [DOI] [PubMed] [Google Scholar]
- 30.Senthilvel S., Jayashree B., Mahalakshmi V., et al. Development and mapping of simple sequence repeat markers for pearl millet from data mining of expressed sequence tags. BMC Plant Biol. 2008;8:119. doi: 10.1186/1471-2229-8-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaur S., Cogan N. O. I., Ye G., et al. Genetic map construction and QTL mapping of resistance to blackleg (Leptosphaeria maculans) disease in Australian canola (Brassica napus L.) cultivars. Theor. Appl. Genet. 2009;120:71–83. doi: 10.1007/s00122-009-1160-9. [DOI] [PubMed] [Google Scholar]
- 32.Raji A.A., Anderson J.V., Kolade1 O.A., Ugwu1 C.D., Dixon A.G., Ingelbrecht I.L. Gene-based microsatellites for cassava (Manihot esculenta Crantz): prevalence, polymorphisms, and cross-taxa utility. BMC Plant Biol. 2009;9:118. doi: 10.1186/1471-2229-9-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li X., Ramchiary N., Choi S.R., et al. Development of a high density integrated reference genetic linkage map for the multinational Brassica rapa Genome Sequencing Project. Genome. 2010;53:939–47. doi: 10.1139/G10-054. [DOI] [PubMed] [Google Scholar]
- 34.Huang X., Madan A. CAP3: a DNA sequence assembly program. Genome Res. 1999;9:868–77. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hong C.P., Piao Z.Y., Kang T.W., et al. Genomic distribution of simple sequence repeats in Brassica rapa. Mol. Cells. 2007;23:349–56. [PubMed] [Google Scholar]
- 36.Rozen S., Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Meth. Mol. Biol. 2000;132:365–86. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 37.Stothard P. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques. 2000;28:1102–4. doi: 10.2144/00286ir01. [DOI] [PubMed] [Google Scholar]
- 38.Lagercrantz U., Ellegren H., Andersson L. The abundance of various polymorphic microsatellite motifs differs between plants and vertebrates. Nucleic Acids Res. 1993;21:1111–5. doi: 10.1093/nar/21.5.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uzunova M.I., Ecke W. Abundance, polymorphism and genetic mapping of microsatellites in oilseed rape (Brassica napus L.) Plant Breed. 1999;118:323–6. [Google Scholar]
- 40.Lowe A.J., Jones A.E., Raybould A.F., Trick M., Moule C.L., Edwards K.J. Transferability and genome specificity of a new set of microsatellite primers among Brassica species of the U triangle. Mol. Ecol. Notes. 2002;2:7–11. [Google Scholar]
- 41.Lowe A.J., Moule C., Trick M., Edwards K. Efficient large-scale development of microsatellites for marker and mapping applications in Brassica crop species. Theor. Appl. Genet. 2004;108:1103–12. doi: 10.1007/s00122-003-1522-7. [DOI] [PubMed] [Google Scholar]
- 42.Suwabe K., Iketani H., Nunome T., Kage T., Hirai M. Isolation and characterization of microsatellites in Brassica rapa L. Theor. Appl. Genet. 2002;104:1092–8. doi: 10.1007/s00122-002-0875-7. [DOI] [PubMed] [Google Scholar]
- 43.Suwabe K., Tsukazaki H., Iketani H., et al. Simple sequence repeat-based comparative genomics between Brassica rapa and Arabidopsis thaliana: the genetic origin of clubroot resistance. Genetics. 2006;173:309–19. doi: 10.1534/genetics.104.038968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Piquemal J., Cinquin E., Couton F., et al. Construction of an oilseed rape (Brassica napus L.) genetic map with SSR markers. Theor. Appl. Genet. 2005;111:1514–23. doi: 10.1007/s00122-005-0080-6. [DOI] [PubMed] [Google Scholar]
- 45.Kim J.S., Chung T.Y., King G.J., et al. A sequence-tagged linkage map of Brassica rapa. Genetics. 2006;174:29–39. doi: 10.1534/genetics.106.060152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iniguez-Luy F.L., Vande Voort A., Osborn T.C. Development of a set of public SSR markers derived from genomic sequence of a rapid cycling Brassica oleracea L. genotype. Theor. Appl. Genet. 2008;117:977–85. doi: 10.1007/s00122-008-0837-9. [DOI] [PubMed] [Google Scholar]
- 47.Cheng X.M., Xu J., Xia S., et al. Development and genetic mapping of microsatellite markers from genome survey sequences in Brassica napus. Theor. Appl. Genet. 2009;118:1121–31. doi: 10.1007/s00122-009-0967-8. [DOI] [PubMed] [Google Scholar]
- 48.Stam P. Construction of integrated genetic linkage maps by means of a new computer package: JoinMap. Plant J. 1993;3:739–44. [Google Scholar]
- 49.Van Ooijen J.W., Voorrips R.E. JoinMap® Version 3.0: Software for the Calculation of Genetic Linkage Map. Wageningen, The Netherlands: Plant Research International; 2001. [Google Scholar]
- 50.Kosambi D.D. The estimation of map distance from recombination values. Ann. Eugen. 1944;12:172–5. [Google Scholar]
- 51.Voorrips R.E. MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002;93:77–8. doi: 10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
- 52.Botstein D., White R.L., Skolnick M., Davis R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980;32:314–31. [PMC free article] [PubMed] [Google Scholar]
- 53.Schranz M.E., Lysak M.A., Mitchell-Olds T. The ABC's of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends Plant Sci. 2006;11:535–42. doi: 10.1016/j.tplants.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 54.Lombard V., Delourme R. A consensus linkage map for rapeseed (Brassica napus L.): construction and integration of three individual maps from DH populations. Theor. Appl. Genet. 2001;103:491–507. [Google Scholar]
- 55.Hauge B.M., Hanley S.M., Cartinhour S., Cherry J.M., Goodman H.M. An integrated genetic/RFLP map of the Arabidopsis. Plant J. 1993;3:745–54. [Google Scholar]
- 56.Sewell M.M., Sherman B.K., Neale D.B. A consensus map for loblolly pine (Pinus taeda L.). I. construction and integration of individual linkage maps from two outbred three-generation pedigrees. Genetics. 1999;151:321–30. doi: 10.1093/genetics/151.1.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sebastian R.L., Howell E.C., King G.J., Marshall D.F., Kearsey M.J. An integrated AFLP and RFLP Brassica oleracea linkage map from two morphologically distinct doubled-haploid mapping populations. Theor. Appl. Genet. 2000;100:75–81. [Google Scholar]
- 58.Jeuken M., Wijk R., Peleman J., Lindhout P. An integrated interspecific AFLP map of lettuce (Lactuca) based on two L.sativa times L. saligna F2 populations. Theor. Appl. Genet. 2001;103:638–47. [Google Scholar]
- 59.Tani N., Takahashi T., Iwata H., et al. A consensus linkage map for Sugi (Cryptomeria japonica) from two pedigrees, based on microsatellites and expressed sequence tags. Genetics. 2003;165:1551–68. doi: 10.1093/genetics/165.3.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gustafson J.P., Ma X.F., Korzun V., Snape J.W. A consensus map of rye integrating mapping data from five mapping populations. Theor. Appl. Genet. 2009;118:793–800. doi: 10.1007/s00122-008-0939-4. [DOI] [PubMed] [Google Scholar]
- 61.Hossain K.G., Kawai H., Hayashi M., Hoshi M., Yamanaka N., Harada K. Characterization and identification of (CT)n microsatellites in soybean using sheared genomic libraries. DNA Res. 2000;7:103–10. doi: 10.1093/dnares/7.2.103. [DOI] [PubMed] [Google Scholar]
- 62.Hisano H., Sato S., Isobe S., et al. Characterization of the soybean genome using EST derived microsatellite markers. DNA Res. 2007;14:271–81. doi: 10.1093/dnares/dsm025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.