Abstract
It is now well known that vertebrates use multiple types of core promoter to accomplish differentiated tasks in Pol II-dependent transcription. Several transcriptional characteristics are known to be associated with core types, including distribution patterns of transcription start sites (TSSs) and selection between tissue-specific and constitutive expression profiles. However, their relationship to gene structure is poorly understood. In this report, we carried a comparative analysis of three Arabidopsis core types, TATA, GA, and Coreless, with regard to gene structure. Our genome-wide investigation was based on the peak TSS positions in promoters that had been identified in a large-scale experimental analysis. This analysis revealed that the types of core promoter are related with the room for promoters that is measured as the distance from the TSS to the end of the upstream gene, the distance from the TSS to the start position of the coding sequence (CDS), and the number and species of the cis-regulatory elements. Of these, it was found that the distance from the TSS to the CDS has a tight, inverse correlation to the expression level, and thus the observed relationship to the core type appears to be indirect. However, promoter length and preference of cis-elements are thought to be a direct reflection of core type-specific transcriptional initiation mechanisms.
Keywords: plant genome, core promoter, environmental response, 5′ UTR
1. Introduction
Pol II-dependent promoters of vertebrates are divided into two major groups: the TATA and CpG types. The former has sharp and peaky transcription start site (TSS) clusters with the peak TSS at a strict distance from the TATA box, and, in contrast, the latter has broad TSS clusters.1,2 The core type also affects the expression profile: the TATA and CpG types tend to show tissue-specific and constitutive expression profiles, respectively.3 In addition, the type of promoters has been reported to alter sequence diversity at the promoter region.4 Recent studies of the human genome have revealed that genes with TATA-type promoters have more compact gene structures than the ones with TATA-less promoters with respect to exon number, intron length, and mRNA length.5 These reports show that the core promoter type can influence not only transcriptional characteristics but also mutation rate and gene structure.
With regard to the organization of transcriptional regulatory elements, analysis of the human genome has revealed that distinct elements preferentially localize far upstream, at the promoter region, at the first intron, and at the 3′ region of genes.6 However, their relationship to core type is still poorly understood.
Looking at non-vertebrate promoters, a recent analysis of Drosophila melanogaster found both sharp and broad TSS clusters as in the case of mammalian ones, while Drosophila does have the TATA type but not the CpG type.7 These findings suggest that broad TSS clusters are not necessarily associated with the CpG type promoters in non-vertebrates. However, association of the broad type promoter with constitutive expression is conserved in Drosophila as well.7
The TATA box was found in plant promoters decades ago and has been thought to drive almost all plant promoters.8 Identification of a TATA-less promoter from tobacco9,10 suggested heterogeneity of plant core promoters, but it has only been recently that other core elements have been identified in plants.11,12 It is now known that TATA-type promoters account for only 20–30% of plant promoters12,13 as is the case in mammalian promoters.14
Recent studies on plant core promoters have revealed that higher plants do not have the CpG type,11 supporting the idea of vertebrate-specific possession of this type of promoter.15 Instead, a plant-specific core, the GA type, has been identified by a bioinformatics approach called LDSS analysis.11,12 Genome-wide quantitative TSS analysis of Arabidopsis has revealed that the sharp cluster shape of the TATA type is conserved between mammals and plants, and the broad clusters of CpG type in mammalian genomes are found in the GA type of Arabidopsis.12
In this report, we characterize three Arabidopsis core types, TATA, GA, and Coreless, with regard to their expression profiles and also their gene structure including promoter length and the distance from the TSS to the coding sequence (CDS). Results indicate that the core type does affect the gene structure as well as the expression profile. Furthermore, a new finding is that there is selective utilization of transcriptional regulatory elements in relation to the core type.
2. Materials and methods
All the bioinformatics analysis was done using homemade Perl scripts and Excel (Microsoft Japan, Tokyo). A total of 10 285 promoters that have quantitative TSS information in Arabidopsis thaliana (Col) were prepared in our previous study.12 The TSS information is supported by 158 237 TSS tags containing the Cap Signature from a single library, determined by the Cap Trapper-Massively Parallel Short Sequencing (CT-MPSS) methodology. For the rice analysis, 11 509 promoters for which there was full-length cDNA information were used.11 Promoter length was determined as the distance from the peak TSS of the promoter in question to the end of the gene model that locates upstream of the promoter. Versions used for the genome annotation are TAIR8 for Arabidopsis and RAP2 for rice. Nested genes were excluded from the measurements.
2.1. TSS information and core type
The position of the peak TSS for each TSS cluster identified by CT-MPSS analysis, the expression level and peak ratio of each TSS cluster, the core promoter type of each promoter, and the sequences of the Regulatory Element Groups (REGs) were determined in our previous reports.11,12,16 ‘Coreless’ promoters were defined in a previous report12 as the ones that do not have any TATA, Y Patch, GA, or CA elements at the expected positions. Otherwise mentioned, statistical examination of multiple populations was carried out by one-way analysis of variance (ANOVA) and Tukey–Kramer's test after log transformation of length. P-values of <0.05 under the assumption of non-biased distributions were considered as significant.
2.2. Utilization of microarray data
Accessions of microarray data used in Fig. 5 are as follows. Wound: response after 1 h TAIR_ME00330;17 HL: high-light treatment at 150 W/m2 for 3 h;18 drought: 1 h treatment, TAIR_ME00338;17 cold: 6 h treatment, E-GEOD-3326;19 Pseudomonas syringe pv tomato DC3000 for 6 h; ABA: 10 µM abscisic acid for 1 h, TAIR_ME00333; pathogen infection: P. syringe pv tomato DC3000 for 6 h, E-GEOD-3326; ABA: 10 µM abscisic acid for 1 h, TAIR_ME00333;20 auxin: 1 µM IAA for 3 h, TAIR_ME00336;20 CK: 1 µM zeatin for 3 h, TAIR_ME00356;20 JA: 10 µM methyl jasmonate for 3 h, TAIR_ME00337;20 SA: 10 µM salicylic acid (SA) for 3 h, TAIR_ME00364;20 H2O2: spraying 3% solution for 3 h.18 Genes that showed no expression (A/M Flags in the GeneChip data) were excluded from the analysis. Genes that did not have any TSS information were also excluded from the analysis.
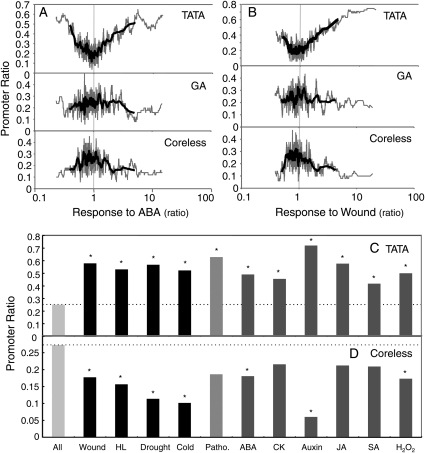
Figure 5.
Regulated expression and core promoter type. (A and B) The ratio of the core promoter types (TATA, GA, and Coreless) in relation to the response to ABA treatment or to wounding is shown. All the promoters were aligned according to their response, and the mean core ratio was calculated with a bin of 51 (grey) and 201 (black) promoters each. 1.0 on the horizontal axis indicates no response to the treatments. (C and D) The promoter ratio of the TATA and Coreless type is shown in relation to the type of gene response. Significantly different value from All, judged by Fisher's exact test, is shown with an asterisk.
Predicted transcriptional regulatory elements shown in Tables 1 and 3 (high RARf octamers) were determined based on microarray data.21 Analyses shown in Fig. 5A and B were achieved by scanning promoter ratios with bins of 51 (thin line) and 201 (thick line) promoters for average after sorting them according to their responses to ABA or wounding.
Table 1.
Core promoter type and octamer preference
| Octamer | Core: TATA |
Core: GA |
Coreless |
Total | |||
|---|---|---|---|---|---|---|---|
| Over-represented | Under-represented | Over-represented | Under-represented | Over-represented | Under-represented | ||
| CAa | 2 | 0 | 0 | 0 | 0 | 3 | 5 |
| GAa | 0 | 15 | 19 | 0 | 0 | 0 | 20 |
| TATAa | 86 | 0 | 0 | 3 | 0 | 90 | 137 |
| REGa | 9 | 118 | 20 | 5 | 14 | 9 | 614 |
| High RARfb | 196 | 184 | 73 | 105 | 177 | 102 | 7037 |
The number of octamer sequences with significantly over- or under-represented appearance ratios (whatever the values are) is shown.
aLDSS element. REG is a group of position-dependent sequences that are suggested to be transcriptional regulatory sequences.
bOther types of putative transcriptional regulatory sequences predicted from microarray data of ABA, auxin, BL, CK, ethylene, JA, SA, H2O2, drought, or DREB1Aox (RARf > 3.0). Core-related elements judged by LDSS analysis, including weak TATA elements and unidentified elements that show localized distribution (P < 0.05 under assumption of random distribution) with a peak position between −50 and +50 relative to the peak TSS, were removed from High RARf. The remaining sequences include some of the REGs, weak REG-like sequences (P < 0.05, peak position between −200 and −40) that had not been identified in our previous report,16 and position-independent putative regulatory elements.
Table 3.
Preferential distribution of high RARf octamers among three core-promoter types
| Core type | Over-represented | Under-represented |
|---|---|---|
| TATA-specific | 193 | 183 |
| GA-specific | 70 | 104 |
| Coreless-specific | 177 | 102 |
| TATA and GA | 3 | 1 |
| TATA and Coreless | 0 | 0 |
| GA and Coreless | 0 | 0 |
| TATA, GA, and Coreless | 0 | 0 |
The numbers of high RARf-octamers (putative transcriptional regulatory sequences) that are biased in each promoter type are shown. LDSS-positive core-related octamers are removed.
3. Results
3.1. Promoter length
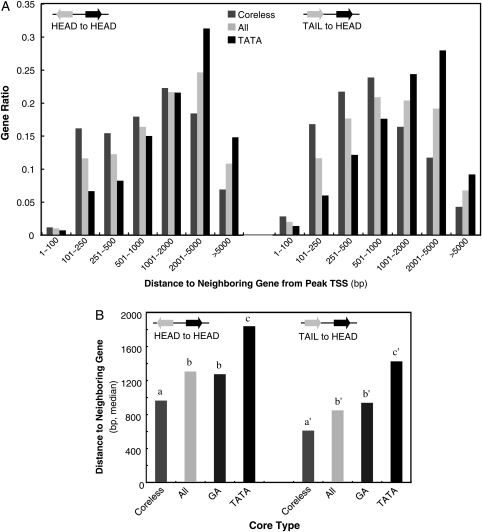
First, we analysed the relationship between core promoter type and the distance from the most major TSS of a gene to the end of the neighbouring upstream gene, that is an indication of room for promoters in the genome. This distance is not equal to the functional promoter length but longer and, technically to say, much easier to measure. We expect that the former distance in the compact Arabidopsis genome is reflected to the latter length. A total of 10 285 Arabidopsis genic promoters with experimentally identified peak TSSs12 were subjected to the analysis. The distance from the peak TSS to the end of the upstream gene model was measured for each gene, and the distribution of the length between the core types was determined. End of upstream gene model is the closer point of either start or endpoint of the transcribed region of the gene. As the distance varied considerably depending on the direction of the upstream gene, we analysed two situations as shown in Fig. 1A, head-to-head and tail-to-head cases. In general, the tail-to-head pattern had shorter length, and median values for head-to-head and tail-to-head were 1.30 and 0.85 kb, respectively (Fig. 1B). Statistical analysis revealed that these two populations are significantly different (P = 2e−11 in the Tukey–Kramer's test). The left graph of Fig. 1A (HEAD to HEAD) shows that the Coreless type, that is defined as a promoter group not containing either TATA, Y Patch, GA, or CA elements,12 is more abundant in the fractions of shorter length (1–1000 bp) than the average (‘All’ in the graph), while the TATA type is more abundant in the fractions of longer length (2001 to over 5000 bp). The same tendency is observed in the tail-to-head pattern as shown in the right graph. These inclinations are reflected in the median values for each promoter type (Fig. 1B), demonstrating a shorter length for the Coreless types and a longer one for the TATA type. The GA type showed a similar length to ‘All’.
Figure 1.
Promoter length and core promoter type. Promoters were divided into two groups according to the orientation of the upstream gene, and for each group, distribution of promoter length from the major TSS to the end of the upstream gene, defined by the gene model, is shown with respect to the core promoter types. The diagram in the graph indicates the direction of the genes, and the black arrow indicates the gene whose promoter length is analysed. Coreless promoters are identified as TATA-, GA-, Y-, and also CA-negative. (A) Distribution of promoter length is shown. The vertical axis indicates the ratio among promoters with a core type, and sum of gene ratios for Coreless, All, and TATA are 1.0, respectively. (B) The median promoter length is shown for each core promoter type. Statistically distinguished groups are labelled with different alphabetical letters over the bars.
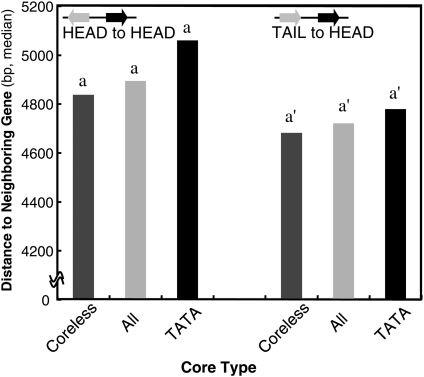
The Arabidopsis genome is tightly packed with an average gene length of 4.5 kb.22 Therefore, it was expected that there has been strong pressure towards shorter promoter lengths in the Arabidopsis genome, which provides an ideal situation to measure the required promoter length. The average gene length of the rice genome is 10 kb,23 and thus, in this case, less stringency for promoter length was expected. Although rice promoters showed the same tendencies as the Arabidopsis promoters, difference in the distance according to the core types was not statistically significant in the rice genome (Fig. 2).
Figure 2.
Promoter length and core promoter type in rice. The median promoter length in the rice genome is shown for each core promoter type. The diagram in the graph indicates the direction of genes, and the black arrow is the gene whose promoter length is analysed. Statistically distinguished groups are labelled with different alphabetical letters over the bars.
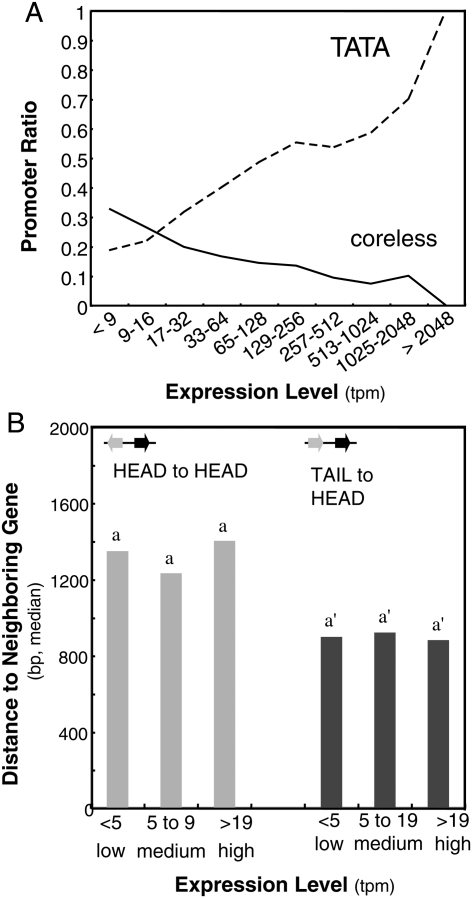
We previously confirmed in Arabidopsis a positive correlation between the TATA ratio and the expression level, as is the case in the mammalian TATA type.12 In this report, we analysed the relationship of the ratio of the Coreless promoters to the expression level, which is measured by counts of TSS tags in a library (tpm, tag per million). In this case, there was a negative correlation in contrast to the TATA ratio (Fig. 3A). These observations suggest the possibility that the different promoter lengths, according to the core type, are correlated with their expression level. We therefore directly compared promoter length with expression level (Fig. 3B) regardless of the core type, but as shown in the figure, no correlation was found. Thus, we concluded that the observed difference in promoter length between the core types is not an indirect result of expression level but a characteristic of the core type itself.
Figure 3.
Promoter length and expression level. (A) Promoter ratio according to expression ratio is shown. Tpm: tag per million in a TSS tag library.12 (B) For each combination of gene orientation, promoters were divided into three groups each according to their expression level. The promoter length of each category is shown. Statistically distinguished groups are labelled with different alphabetical letters over the bars.
3.2. Distance from TSS to CDS
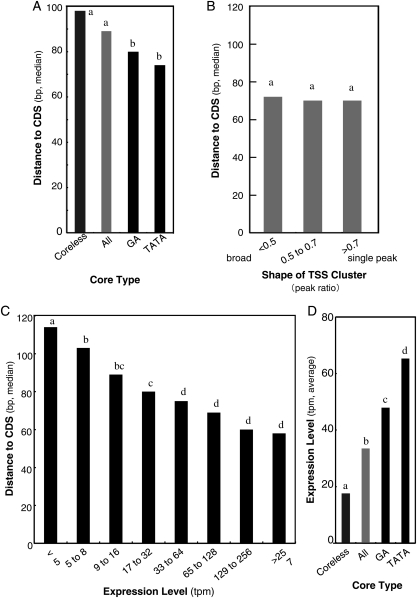
Another parameter of gene structure that is related to transcription is the distance from the peak TSS to the CDS. This distance represents the length of 5′ UTR when there is no intron in the region. We analysed the relationship of the core types to this length. As shown in Fig. 4A, the TATA type has a shorter distance while that of the Coreless type is longer. The former has a narrow and sharp TSS cluster12 that might shorten the distance from the TSS to the CDS in comparison to a broad TSS cluster. Another possibility is that this results from a direct relationship with expression level. These two possibilities were then investigated.
Figure 4.
Distance from TSS to CDS is tightly associated with expression level. (A) The distance from the major TSS to the downstream translation start site is shown for the core types. (B) The distance from the TSS to the CDS is shown in relation to the shape of the TSS cluster. (C) The distance from the TSS to the CDS is shown in relation to the expression level (tpm). (D) For each core type, the expression level (tpm) is shown. Statistically distinguished groups are labelled with different alphabetical letters over the bars.
Figure 4B and C shows the relationship between the distance to the CDS with the shape of the TSS cluster (grey graph on the left) and with the expression level (black graph on the right). The results show that this distance does not have any relationship with the shape of the TSS cluster but clear correlation with the expression level. Statistical analysis revealed that the latter correlation is significant.
We then analysed the relationship between the core types, distance to the CDS, and expression level (Fig. 4A and D). As expected, the figure revealed a clear mirror image between Fig. 4A and D, indicating that difference in the distance among the shown core types can be explained by difference in the expression level of each type. These results reveal that the relationship shown in Fig. 4A is not necessarily direct.
3.3. Regulated and constitutive expression
The results in our previous report, which were obtained with the aid of microarray data of light stress, drought stress, and H2O2 responses, suggested that the TATA is rich in stress-responsive promoters while the GA and Coreless types are rich in constitutive ones.12 Here, we extend the analysis with various other microarray data including those of several plant hormone and stress responses.
Figure 5A shows the promoter ratio of TATA, GA, and Coreless types with regard to response to ABA. A clear valley-shape distribution in the top graph indicates there is an enrichment of the TATA type among ABA-responsive promoters, including both positive and negative responses. While the TATA ratio of the no-response promoters (response to ABA is 1.0) is 0.195, the ratio increases to 0.484 and 0.508 where the response to ABA is 0.40 and 5.13 (ends of the thick lines in the graph), respectively, demonstrating a 2.5–2.6 fold increase in the TATA ratio. The middle graph of Fig. 5A shows the GA ratio, and this does not show any clear tendency. The bottom graph for the Coreless ratio shows a hill-shape distribution, meaning enrichment of the Coreless promoters at the no-response group to ABA. Figure 5B shows results of the same analysis but with the wound response, and the same tendency is observed.
The TATA and Coreless ratios were calculated for promoters that respond to various stresses and phytohormones (Fig. 5C and D, genes with >3.0- and <0.33-fold responses were selected as responsive genes), including wounding, high-light stress (HL), drought, cold, pathogen infection (P. syringe pv tomato DC3000), ABA, cytokinin (CK), auxin, jasmonic acid (JA), SA, and hydrogen peroxide (H2O2). ‘All’ in Fig. 5C and D means the average of all the Arabidopsis promoters. As shown in the graphs, the TATA ratio of the promoters that have positive and negative responses to the stresses and phytohormones are all higher than the average (‘All’), and the Coreless ratios are all lower than the average. From these results, we concluded that ‘regulated’ promoters are rich in the TATA type, and constitutive ones are rich in the Coreless type.
3.4. REG density
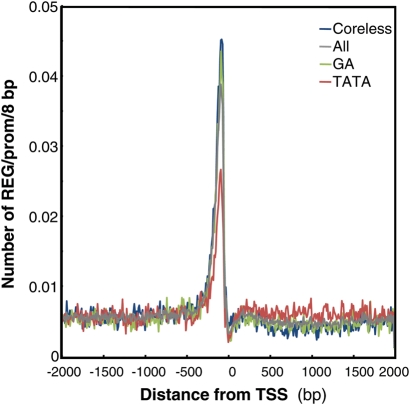
REG is a group of putative position-sensitive cis-regulatory elements identified form their distribution profiles in the promoter region. They have been identified as octamer elements that showed preferential appearance in −400 to −40 bp relative to the peak TSS. Because this group includes many reported cis-regulatory elements, we named it as REG.16 One unique characteristics of REG is no directional preference in the distribution profiles. From the Arabidopsis genome, 308 REGs have been identified.16 We next analysed the relationship between core type and REG density. Figure 6 shows the number of REGs per 8 bp in a promoter according to the promoter position for each core type. The analysis revealed that the GA and Coreless types have a 2-fold higher REG density than the TATA type.
Figure 6.
REG density and core promoter type. The distribution of REG according to the promoter position is shown for each core promoter type.
3.5. Preferred and avoided sequences for each core type
Figure 6 reveals that the GA and Coreless types have more REGs per promoter than the TATA type. One question this raises is whether the core type has any preference for REG species. We extended this question to various types of octamers representing core elements, REGs, and also other types of putative cis-regulatory elements extracted using microarray data, called high RARf octamers. This last category has been identified as overrepresented sequences in a promoter set showing transcriptional responses to several phytohormones and some environmental stresses, and includes many sequence motifs that are recognized by DNA-binding proteins.21 For each octamer category, appearance rates were compared between the total promoters in the genome and sets of promoters belonging to the individual core types. The probability of the observed difference under the assumption of random distribution was calculated regardless of the degree of difference in the appearance rates, and the number of octamers that showed P< 0.05 were counted. The identified octamers are thus preferentially found in promoters of a specific core type.
Table 1 shows the summary of the analysis. We have reported that TATA and GA elements have a mutually exclusive relationship in the Arabidopsis genome.12 This tendency is confirmed in the table: the TATA-type promoters have GA octamers that have significantly decreased appearance rates (Core: TATA∼Octamer: GA, 15 decreased octamers) where as no GA octamers were found for increased appearance rates (0 increased octamers), and vice versa (Core: GA∼Octamer: TATA, 3 decreased and 0 increased octamers).
Looking at the cis-related octamers (REG and high RARf), the table shows that there are preferred and avoided sequences for each core type. Consistent with the REG density analysis (Fig. 6), decreased REGs are found more in the TATA type than the increased ones (118 vs. 9) while the GA and Coreless types show the opposite tendency (20 vs. 5 for GA, and 14 vs. 9 for Coreless, respectively). In the case of the high RARf octamers, the number of preferred and avoided sequences is equal for each core type (196 vs. 184, 73 vs. 105, and 177 vs. 102 for TATA, GA, and Coreless, respectively).
The analysis shown in Table 1 identified high RARf sequences with biased appearance rates according to the core type. We then investigated whether these putative transcriptional regulatory elements have any specificity to core type. Results of REGs and high RARf octamers are shown in Tables 2 and 3, respectively. These results suggest the presence of core type-specific cis-regulatory elements whose function is dependent on the core type.
Table 2.
Preferential distribution of REG octamers among three core-promoter types
| Core type | Over-represented | Under-represented |
|---|---|---|
| TATA-specific | 8 | 115 |
| GA-specific | 19 | 3 |
| Coreless-specific | 14 | 8 |
| TATA and GA | 1 | 2 |
| TATA and Coreless | 0 | 1 |
| GA and Coreless | 0 | 0 |
| TATA, GA, and Coreless | 0 | 0 |
The number of REGs (putative transcriptional regulatory sequences) that are biased in each promoter type is shown.
4. Discussion
Table 4 summarizes the characteristics of the Arabidopsis core types. The TATA type is rich in promoters with regulated expression profiles, while the Coreless type is rich in constitutive promoters. The GA type was also suggested to be constitutive,12 but further analyses shown in Fig. 5 could not confirm this suggestion. Considering the other characteristics shown in the table, a clear contrast is found between the TATA and the Coreless types. The Coreless type has no recognizable core elements, but still functions as a genic promoter with constitutive expression. One of the essential questions about this promoter type is how the position and direction of transcriptional initiation is determined. These results do not offer an answer, but the presence of unique and abundant cis-elements, revealed by Fig. 6, Tables 2 and 3, might play a role, at least in determining the position. Another possibility for transcriptional initiation in the Coreless type is guidance by the open nucleosomal status. Because it can be made by nucleotide sequences much longer than ∼10 bp, this idea is not contradictory to the absence of any core elements in the Coreless type.
Table 4.
Characteristics of core promoter types
| TATA | GA | Coreless | |
|---|---|---|---|
| Promoter length | Long | — | Short |
| Distance from TSS to CDS | Short | Short | Long |
| REG density | Low | High | High |
| Expression level | Higha | Higha | Lowa |
| Expression profile | Regulateda | — | Constitutivea |
A summary of this work is shown.
aAlso supported by Yamamoto et al.12
The TATA type has characteristics of regulated gene expression. At the same time, Fig. 6 reveals that REG density of the promoter group is less than average. Currently, we do not have additional data explaining this apparent discrepancy. One possible explanation is that REG functions not only for transcriptional regulation but also for constitutive expression. Further studies would be necessary to understand this discrepancy.
Analysis of light activation of the TATA-less psaDb promoter has been suggested compatibility between the core type and some regulatory elements.10 While there is no clear evidence to support this hypothesis, the proposed model can be extended as a generalized question: do different core types require distinct sets of regulatory elements? Our analysis as shown in Tables 2 and 3 detected groups of putative regulatory elements that are preferentially used by specific core types. The presence of specific regulatory elements is not very surprising if we assume different transcriptional initiation mechanisms according to core types.
In spite of finding of specific regulatory elements, the majority of the examined elements do not show preference to specific core types (Table 1). This would imply that mechanisms for transcriptional initiation largely overlap among the three core types.
In this report, a close, negative relationship has been revealed between the distance from the TSS to the CDS and the expression level. There are at least three possible explanations of this relationship: transcriptional activation by closer CDSs, post-transcriptional mRNA stabilization by shorter 5′ UTRs, and an indirect relationship between two parameters with no functional relationship at all. The second possibility is unlikely because mRNA accumulation is thought to be determined primarily at the transcriptional level for nuclear-encoded genes. In order to assess the third possibility, we examined translational efficiency according to the distance from the TSS to the CDS on the assumption that genes requiring high expression would have high mRNA accumulation and high translation efficiency without any functional relationship between these two phenomena, and the latter correlates with a short distance from the TSS to the CDS. However, preliminary analysis of the translational efficiency with the aid of the ribosome-loading ratio24 did not show any significance in relation to the distance from the TSS to the CDS (data not shown). The lack of positive evidence for any of these three possibilities means that further study is required to reveal the explanation.
The suggested longer promoter length of the TATA type implies that the requirement for promoter length is more stringent for this promoter type. These different requirements of the core types would reflect different mechanisms of transcriptional initiation. Requirement of the differential promoter length between the TATA and the Coreless types is clear in the compact Arabidopsis genome (gene density: 4.9 kb/gene) and, although less obvious, it is still detectable in the rice genome (10.4 kb/gene). However, in large genomes, like the maize and human ones (both ∼100 kb/gene), detection of promoter length would be difficult. Hence, use of the Arabidopsis genome is advantageous when analysing promoter length requirements.
This report confirmed that individual core promoter types have distinct functional aspects in plants as well (Fig. 5), indicating differentiation of their biological roles. The TATA type is ubiquitously found in eukaryotes from yeast to human and rice, and their characteristics, that is regulated and high expression profiles and sharp shape of TSS clusters, are well conserved between plants and mammals, suggesting that it is essential for eukaryotes.
In addition to the TATA type, detection of the second type in both plants and vertebrates may suggest that there are some biological situations where the TATA type is inappropriate, that promote emergence of the second type of promoters. Current knowledge, including finding of this article, suggests that the TATA type is not good at performing low or constitutive expression patterns.3,12 This idea of the TATA type as a specialist, not an all-round player, is supported by the presence of some putative regulatory elements that tend to be avoided for the TATA type (Table 2 and 3), and a low ratio of the TATA type (10–30%) of promoters in an eukaryotic genome.12–14 Further comparison of various eukaryotic promoters would help understanding necessity of heterogeneity of core promoters in an eukaryotic genome.
5. Conclusion
Our genome-wide analysis has revealed that the core promoter type is related to gene structure, including room for promoters in the genome, the distance from the TSS to the CDS, and the number and species of the cis-regulatory elements. Although the relationship between the distance from the TSS to the CDS and the core type appears to be indirect, promoter length and the preference for cis-elements are suggested to reflect the respective transcriptional initiation mechanisms of the core type.
Funding
This work was supported in part by Grant-in-Aid for Scientific Research on Priority Areas ‘Comparative Genomics’ (Y.Y.Y. and J.O.), Scientific Research (B) (Y.Y.Y., Y.Y., M.H., and J.O.), Scientific Research (C) (Y.Y.Y.), Scientific Research on Innovative Areas (Y.Y.Y.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Acknowledgements
We would like to acknowledge Dr Masaki Ando for excellent advice on statistical tests.
References
- 1.Suzuki Y., Tsunoda T., Sese J., et al. Identification and characterization of the potential promoter regions of 1031 kinds of human genes. Genome Res. 2001;11:677–84. doi: 10.1101/gr.164001. doi:10.1101/gr.GR-1640R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carninci P., Sandelin A., Lenhard B., et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006;38:626–35. doi: 10.1038/ng1789. doi:10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- 3.Schug J., Schuller W.P., Kappen C., Salbaum J.M., Bucan M., Stoeckert C.J., Jr. Promoter features related to tissue specificity as measured by Shannon entropy. Genome Biol. 2005;6:R33. doi: 10.1186/gb-2005-6-4-r33. doi:10.1186/gb-2005-6-4-r33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taylor M.S., Kai C., Kawai J., Carninci P., Hayashizaki Y., Semple C.A. Heterotachy in mammalian promoter evolution. PLoS Genet. 2006;2:e30. doi: 10.1371/journal.pgen.0020030. doi:10.1371/journal.pgen.0020030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moshonov S., Elfakess R., Golan-Mashiach M., Sinvani H., Dikstein R. Links between core promoter and basic gene features influence gene expression. BMC Genomics. 2008;9:92. doi: 10.1186/1471-2164-9-92. doi:10.1186/1471-2164-9-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blanchette M., Bataille A.R., Chen X., et al. Genome-wide computational prediction of transcriptional regulatory modules reveals new insights into human gene expression. Genome Res. 2006;16:656–68. doi: 10.1101/gr.4866006. doi:10.1101/gr.4866006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoskins R.A., Landolin J.M., Brown J.B., et al. Genome-wide analysis of promoter architecture in Drosophila melanogaster. Genome Res. 2011;21:182–92. doi: 10.1101/gr.112466.110. doi:10.1101/gr.112466.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joshi C.P. An inspection of the domain between putative TATA box and translation start site in 79 plant genes. Nucleic Acids Res. 1987;15:6643–53. doi: 10.1093/nar/15.16.6643. doi:10.1093/nar/15.16.6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamamoto Y., Tsuji H., Obokata J. Structure and expression of a nuclear gene for the PSI-D subunit of photosystem I in Nicotiana sylvestris. Plant Mol. Biol. 1993;22:985–94. doi: 10.1007/BF00028971. doi:10.1007/BF00028971. [DOI] [PubMed] [Google Scholar]
- 10.Nakamura M., Tsunoda T., Obokata J. Photosynthesis nuclear genes generally lack TATA-boxes: a tobacco photosystem I gene responds to light through an initiator. Plant J. 2002;29:1–10. doi: 10.1046/j.0960-7412.2001.01188.x. doi:10.1046/j.0960-7412.2001.01188.x. [DOI] [PubMed] [Google Scholar]
- 11.Yamamoto Y.Y., Ichida H., Abe T., Suzuki Y., Sugano S., Obokata J. Differentiation of core promoter architecture between plants and mammals revealed by LDSS analysis. Nucleic Acids Res. 2007;35:6219–26. doi: 10.1093/nar/gkm685. doi:10.1093/nar/gkm685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamamoto Y.Y., Yoshitsugu T., Sakurai T., Seki M., Shinozaki K., Obokata J. Heterogeneity of Arabidopsis core promoters revealed by high density TSS analysis. Plant J. 2009;60:350–62. doi: 10.1111/j.1365-313X.2009.03958.x. doi:10.1111/j.1365-313X.2009.03958.x. [DOI] [PubMed] [Google Scholar]
- 13.Molina C., Grotewold E. Genome wide analysis of Arabidopsis core promoters. BMC Genomics. 2005;6:25. doi: 10.1186/1471-2164-6-25. doi:10.1186/1471-2164-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sandelin A., Carninci P., Lenhard B., Ponjavic J., Hayashizaki Y., Hume D.A. Mammalian RNA polymerase II core promoters: insights from genome-wide studies. Nat. Rev. Genet. 2007;8:424–36. doi: 10.1038/nrg2026. doi:10.1038/nrg2026. [DOI] [PubMed] [Google Scholar]
- 15.Butler J.E., Kadonaga J.T. The RNA polymerase II core promoter: a key component in the regulation of gene expression. Genes Dev. 2002;16:2583–92. doi: 10.1101/gad.1026202. doi:10.1101/gad.1026202. [DOI] [PubMed] [Google Scholar]
- 16.Yamamoto Y.Y., Ichida H., Matsui M., et al. Identification of plant promoter constituents by analysis of local distribution of short sequences. BMC Genomics. 2007;8:67. doi: 10.1186/1471-2164-8-67. doi:10.1186/1471-2164-8-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kilian J., Whitehead D., Horak J., et al. The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 2007;50:347–63. doi: 10.1111/j.1365-313X.2007.03052.x. doi:10.1111/j.1365-313X.2007.03052.x. [DOI] [PubMed] [Google Scholar]
- 18.Yamamoto Y.Y., Shimada Y., Kimura M., et al. Global classification of transcriptional responses to light stress in Arabidopsis thaliana. Endocytobio Cell Res. 2004;15:438–52. [Google Scholar]
- 19.Lee B.H., Henderson D.A., Zhu J.K. The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell. 2005;17:3155–75. doi: 10.1105/tpc.105.035568. doi:10.1105/tpc.105.035568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goda H., Sasaki E., Akiyama K., et al. The AtGenExpress hormone and chemical treatment data set: experimental design, data evaluation, model data analysis and data access. Plant J. 2008;55:526–42. doi: 10.1111/j.0960-7412.2008.03510.x. doi:10.1111/j.1365-313X.2008.03510.x. [DOI] [PubMed] [Google Scholar]
- 21.Yamamoto Y.Y., Yoshioka Y., Hyakumachi M., et al. Prediction of transcriptional regulatory elements for plant hormone responses based on microarray data. BMC Plant Biol. 2011;11:39. doi: 10.1186/1471-2229-11-39. doi:10.1186/1471-2229-11-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arabidopsis_Genome_Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. doi:10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 23.International_Rice_Genome_Sequencing_Project. The map-based sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. doi:10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- 24.Kawaguchi R., Bailey-Serres J. mRNA sequence features that contribute to translational regulation in Arabidopsis. Nucleic Acids Res. 2005;33:955–65. doi: 10.1093/nar/gki240. doi:10.1093/nar/gki240. [DOI] [PMC free article] [PubMed] [Google Scholar]