Abstract
Using ChIP Seq, we identified 556 and 467 putative STAT6 target sites in the Burkitt's lymphoma cell line Ramos and in the normal lung epithelial cell line BEAS2B, respectively. We also examined the positions and expression of transcriptional start sites (TSSs) in these cells using our TSS Seq method. We observed that 44 and 132 genes in Ramos and BEAS2B, respectively, had STAT6 binding sites in proximal regions of their previously reported TSSs that were up-regulated at the transcriptional level. In addition, 406 and 109 of the STAT6 target sites in Ramos and BEAS2B, respectively, were located in proximal regions of previously uncharacterized TSSs. The target genes identified in Ramos and BEAS2B cells in this study and in Th2 cells in previous studies rarely overlapped and differed in their identity. Interestingly, ChIP Seq analyses of histone modifications and RNA polymerase II revealed that chromatin formed an active structure in regions surrounding the STAT6 binding sites; this event also frequently occurred in different cell types, although neither STAT6 binding nor TSS induction was observed. The rough landscape of STAT6-responsive sites was found to be shaped by chromatin structure, but distinct cellular responses were mainly mediated by distinct sets of transcription factors.
Keywords: TSS Seq, ChIP Seq, IL-4, STAT6
1. Introduction
The signal transduction pathway initiated by IL-4 stimulation is involved in various biological phenomena, such as Th2 cell differentiation, immunoglobulin class switching in B cells and inflammatory responses in epithelial cells.1–3 Malfunctions of the IL-4 signalling pathway cause improper responses in these cell types that subsequently result in chronic allergic diseases such as asthma and atopic disorders.4,5 For example, in asthma, IL-4 is excessively produced by Th2 cells and induces differentiation and class switching of allergen-specific B cells towards IgE production.6,7 At the same time, IL-4 also initiates response cascades in various cell types, such as eosinophils, basophils, mast cells and epithelial cells.8 In particular, CCL11 (eotaxin-1) synthesis is enhanced in epithelial cells,9,10 and mucus-producing goblet cells become hyperactive and differentiate.11 Reflecting the pivotal roles of the IL-4 pathway in allergic responses, knockout mice deficient in signalling molecules, such as the IL-4 receptor and STAT6, show symptoms that resemble those of allergic disorders.12–14
In various cell types, the transcription factor (TF) STAT6 is a common downstream effector of the IL-4 signalling pathway. When bound by IL-4, IL-4 receptor α is phosphorylated by JAK kinases and recruits STAT6. The JAK kinases phosphorylate STAT6, induce its dimerization and facilitate its translocation to nucleus, where it acts as a transcriptional activator.15 Several dozen STAT6 target genes have been identified, such as IL-4,16 RETNLB,17 SOCS1,18,19 CD23A20 and CCL11. It has also been shown that the target sites of STAT6 have a consensus sequence: TTCNNNNGAA. However, a comprehensive view of the target genes of STAT6 remains elusive. In particular, previous studies mainly searched for STAT6 target genes in T cells21,22; target genes in other cell types (such as B cells or epithelial cells) have not been fully characterized.
Recently developed massively parallel sequencing technologies coupled with chromatin immunoprecipitation (ChIP Seq) have enabled comprehensive analyses of binding sites for TFs.23,24 Recent papers have reported the comprehensive identification of STAT6 target genes in Th2 cells in both mice and humans.22 ChIP Seq analysis has also been used to comprehensively assess chromatin status by monitoring changes in histone modification patterns.25–27 Indeed, for many genes in various cell types, epigenetic changes have been shown to play important roles in transcriptional regulation in response to cellular environmental changes. Moreover, we also developed a method to enable large-scale analysis of transcriptional start sites (which we termed TSS Seq) by combining our full-length cDNA technology (oligo-capping) with massively parallel sequencing.28,29 In this method, a sequence adaptor necessary for the sequencing reaction is introduced into the mRNA as a cap-replacing oligo so that the region immediately downstream of the TSS can be sequenced (TSS tag).30 We demonstrated that the position of a TSS and its transcriptional level can be analysed at the same time by digital counting of the TSS tags. Integrative analysis of epigenomic and transcriptomic data collectively revealed that the human gene transcriptome and its regulation are truly dynamic and depend on the cell type. Every cell seems to have a unique chromatin status, harbouring its own set of active promoters, and a large amount of transcription occurs even from intergenic regions.31,32 These results again suggest that it is essential to identify and characterize STAT6 target genes and their regulation in a wider variety of cell types to obtain a more complete view of the biological relevance of the IL-4 signalling pathway.
In this paper, we characterized STAT6 target genes and the chromatin status of the target sites by using ChIP Seq to evaluate STAT6, RNA polymerase II (pol II) and histone modifications. We further combined the ChIP Seq data with TSS Seq data to monitor the transcriptional consequences of STAT6 binding. We identified and characterized 44 putative STAT6 target genes in the Ramos cell line, which is a Burkitt's lymphoma cell line, and 132 in the BEAS2B cell line, which is a normal lung epithelial cell line. We also compared the STAT6 target genes identified in these cell types with recently reported STAT6 target genes in human Th2 cells. These results collectively illustrate that different cell types have their own mode of STAT6-mediated transcriptional regulation. Here, we report that STAT6 has diverse target gene repertoires depending on cell type.
2. Materials and methods
2.1. Sequence data
The short-read sequence archive data appearing in this paper are registered in GenBank/DDBJ under the following accession numbers: SRA008161, SRA008162, SRA008163, SRA008164, DRA000017, DRA000018, DRA000019, DRA000020, DRA000021, DRA000022, DRA000046, DRA000047, DRA000048, DRA000049, DRA000050, DRA000051, DRA000052, DRA000053, DRA000054, DRA000055, DRA000056, DRA000057, DRA000058, DRA000059, DRA000326, DRA000327, DRA000328, DRA000329, DRA000330, DRA000331, DRA000332, DRA000333, DRA000334, DRA000335, DRA000336, DRA000337, DRA000338, DRA000339, DRA000340 and DRA000341.
2.2. Cell culture
Cells of the human B lymphoma line Ramos were cultured in RPMI1640 medium (Nissui) supplemented with 10% fetal bovine serum, 0.3 g/l l-glutamine (GIBCO), 0.36% bicarbonate and 60 mg/l kanamycin (GIBCO). Human lung epithelial BEAS2B cells were maintained in Dulbecco's modified Eagle's medium: Nutrient Mixture F-12 (GIBCO) with 10% fetal bovine serum, 100 U/ml penicillin–100 μg/ml streptomycin (GIBCO) and 1 μg/ml puromycin (BD Biosciences). These cells were cultured at 37°C in a 5% CO2 incubator. Ramos and BEAS2B cells were stimulated by 50 ng/ml recombinant rhIL-4 (BD Biosciences). Ramos cells were stimulated 10 μg/ml human Anti-IgM (BETHYL). RNA interference was accomplished by transfecting cells with siGENOME SMARTpool STAT6 siRNA and siGENOME Non-Targeting siRNA #1 (Dharmacon). Short oligo-RNAs (50 nM) were transfected using 2 μl/ml Dharmafect 1 transfection reagent (Dharmacon) as recommended by the manufacturer.
2.3. Construction of the ChIP Seq library
For each IP, 5 × 107 cells were stimulated with rhIL-4 at 37°C for 30 min. Cells were fixed with a 0.5% formaldehyde solution (50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES)-KOH, pH 7.5, 100 mM NaCl, 1 mM ethylendiaminetetraacetic acid (EDTA), 0.5 mM ethylene glycol tetraacetic acid (EGTA) and 5.5% formaldehyde) at room temperature for 10 min. Glycine (150 mM) was added to quench the cross-linking. Cells were rinsed twice with cold-Phosphate Buffered Saline (PBS) and harvested. Each cell pellet was resuspended in 5 ml of Lysis Buffer 1 (50 mM HEPES-KOH pH 7.5, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40 and 0.25% Triton X-100). The cell lysate was rocked at 4°C for 10 min and spun at 1500 rpm for 5 min at 4°C. The supernatant was discarded. Each cell pellet was resuspended in 5 ml of Lysis Buffer 2 (10 mM Tris–HCl, pH 8.0, 200 mM NaCl, 1 mM EDTA and 0.5 mM EGTA). The cell lysate was rocked at room temperature for 10 min and spun at 1500 rpm for 5 min at 4°C. The supernatant was discarded. Each cell pellet was resuspended in 1 ml of Lysis Buffer 3 (10 mM Tris–HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% Na-deoxycholate and 0.5% N-lauroylsarcosine). The cell lysate was sonicated with 10 cycles of 30 s in ice water with a sonicator (TOMY SEIKO). Triton X-100 (100 μl of a 10% solution) was added to the sonicated cell lysate. The cell lysate was spun at 14 000 rpm for 10 min. The supernatant was transferred to a new tube, and 50 μl of the supernatant was saved as whole cell extract (WCE)-DNA. The WCE-DNA was stored at −20°C. The supernatant was transferred to 100 μl of magnetic beads (Dynal) that were bound to 10 μg of antibody. The samples were rotated at 4°C overnight and washed eight times with 1 ml of Wash Buffer and once with TE buffer containing 50 mM NaCl. Elution Buffer (200 μl) was added and eluted at 65°C for 15 min. The samples were spun down, and the supernatant was transferred to a new tube. The samples were incubated at 65°C overnight. WCE-DNA (50 μl) was thawed, and 150 μl of Elution Buffer was added to it. The samples were incubated at 65°C overnight. TE (200 μl) was added to the IP and WCE-DNA samples. RNase (8 μl of 10 mg/ml; Funakosi) was added. The samples were incubated at 37°C for 2 h, and 4 μl of 20 mg/ml proteinase K (Takara) was added. The samples were incubated at 55°C for 2 h. Phenol chloroform extraction and ethanol precipitation were performed. Samples destined for ChIP Seq by Illumina GAIIx were prepared according to the manufacturer's instructions. Anti-STAT-6 antibody (Santa Cruz, M-20), anti-RNA pol II antibody (Abcam, ab817), anti-H3K4me3 antibody (ab1012) and anti-H3Ac antibody (Millipore, 06–599) were used for the indicated experiments.
2.4. Construction of the TSS Seq library
Cells were seeded 48 h before stimulation. At 24 h after seeding, the cells were transfected with STAT6 targeting and control siRNA and cultured for 24 h for the indicated experiments. At 24 h after transfection, the transfected cells were stimulated with IL-4 and control 0.1% Bovine serum albumin (BSA) PBS solution. At 24 h after stimulation, the cells were harvested, and RNA was extracted using an RNeasy Kit (Qiagen). Fifty micrograms of the obtained total RNA was subjected to oligo-capping with some modifications of the original protocol; BAP-TAP-treated total RNAs were ligated with 1.2 μg of RNA oligo (5′- AAUGAUACGGCGACCACCGAGAUCUACACUCUUUCCCUACACGACGCUCUUCCGAUCUGG-3′). After the DNase I treatment (TaKaRa), polyA + RNA was selected using oligo-dT powder (Collaborative). First-strand cDNA was synthesized from 10 pmol of random hexamer primer (5′-CAAGCAGAAGACGGCATACGANNNNNNC-3′) using Super Script II (Invitrogen) at 12°C for 1 h and 42°C overnight. Template RNA was degraded by alkaline treatment. One-fifth of the first-strand cDNA was used as the PCR template. For the PCR, a Gene Amp PCR kit (PerkinElmer) was used with the PCR primers 5′-AATGATACGGCGACCACCGAG-3′ and 5′-CAAGCAGAAGACGGCATACG-3′ under the following reaction conditions: 15 cycles of 94°C for 1 min, 56°C for 1 min and 72°C for 2 min. The PCR fragments were size-fractionated by 12% polyacrylamide gel electrophoresis, and the fraction at 150–250 bp was recovered. The quality and the quantity of the obtained single-stranded first-strand cDNA were assessed using a BioAnalyzer (Agilent).
2.5. Deep sequencing and data processing of the ChIP Seq and TSS Seq libraries
Approximately 1 ng of the size-fractionated cDNA was used for sequencing using an Illumina GAIIx. Statistics of the tags generated for each experiment are summarized in Tables 1–3 and Supplementary Fig. S1. The generated sequence tags were mapped onto the human genomic sequence (hg18 as of UCSC Genome Browser) using the sequence alignment Programme Eland (Illumina). Unmapped or redundantly mapped sequences were removed from the data set, and only uniquely mapped sequences without any mismatches were used for further analyses. Positions relative to RefSeq genes were calculated based on the respective genomic coordinates. Genomic coordinates of exons and the protein-coding regions of the RefSeq transcripts are as described in hg18.
Table 1.
Identification of STAT6 targets: summary of the ChIP Seq analyses of STAT6 and pol II
| ChIP (STAT6) | ChIP (Pol II) | |
|---|---|---|
| Ramos | ||
| # total reads | 19 099 520 | 21 903 761 |
| # IL-4(+) peaks | 600 | 17 431 |
| # IL-4(−) peaks | 73 | 19 239 |
| BEAS2B | ||
| # IP total reads | 18 981 052 | 21 709 675 |
| # IL-4(+) peaks | 773 | 13 457 |
| # IL-4(−) peaks | 314 | 8174 |
The numbers of peaks detected using the described parameters are shown. The numbers of peaks detected using different parameters are shown in Supplementary Fig. S1.
Table 2.
Identification of STAT6 targets: summary of TSS Seq analysis
| TSS Seq (IL-4 (+)) | TSS Seq (IL-4 (−)) | |
|---|---|---|
| # Ramos total reads | 15 268 493 | 15 759 415 |
| #BEAS2B total reads | 14 818 816 | 11 628 747 |
| #BEAS2B (siSTAT6) | 14 122 877 | 13 789 254 |
| Total reads | 44 210 186 | 41 177 416 |
Table 3.
Identification of STAT6 targets: number of identified STAT6 binding sites in the indicated populations
| Ramos (pol II (+); TSS (+)) | BEAS2B (pol II (+); TSS (+)) | |
|---|---|---|
| #total | 556 (50) | 467 (124) |
| Promoter | 150 (30) | 358 (121) |
| Alternative promoter | 205 (12) | 33 (1) |
| Intergenic | 201 (8) | 76 (2) |
The number of STAT6 binding sites at which pol II binding and TSS induction were simultaneously observed (‘STAT6 targets’) is shown in parenthesis.
For the ChIP Seq analysis, the binding sites of STAT6, RNA pol II and histone modifications were identified based on the short-read tag information as follows: the region encompassed by each mapped tag sequence was extended to 120 bp, which reflects the insertion sites of sample DNA fragments. For each genomic position, the number of overlapping extended tags was counted. Based on the calculated tag information, the sum of the included tags was evaluated to determine whether more than a 10-fold difference between the IP and the WCE was present. Genomic regions in which positive enrichment of the tags continued for more than 121 bp were then selected. The statistical significance for this selection procedure compared with the background rate was evaluated using the Poisson probabilities as previously described in the reference.24
For the TSS Seq analysis, the TSS tags were clustered into 500-bp bins. Representative TSSs were selected as the position from which the largest number of TSS tags was mapped. The selected representative TSSs were used for further analyses. When the genomic coordinates of a representative TSS was located within −10 to +1 kb of the 5′-end of a RefSeq transcript model, it was associated with the RefSeq gene as the TSS of its authentic promoter. For digital TSS-tag count analysis, TSS-tag counts were normalized as parts per million tags and used for further comparisons.
2.6. Other computational procedures
For Gene Ontology (GO) term analysis, GO terms were associated with the identified STAT6 target genes using loc2go at NCBI. GO term enrichment was evaluated by calculating hypergeometric distributions. The statistical significance between the expression levels and their fold inductions in Ramos and BEAS2B cells was evaluated by the Wilcoxon signed-rank test. To analyse the gene expression changes of the TFs in Ramos and BEAS2B cells, 140 TFs were selected in TRANSFAC (Rel. 2010.1). Fold changes of the expression levels in response to IL-4 stimulation were evaluated by digital TSS-tag counts. The consensus sequences of the TFs were evaluated using MATCH33 with cut-off values determined using minFP, which minimizes false-positive results. The enrichment of the detected putative binding sites in the active target and silent targets in Ramos and BEAS2B cells was evaluated by calculating hypergeometric distributions. Putative TF binding sites that were significantly enriched (P < 0.05) in the respective groups were selected.
2.7. Gene-specific PCR
For real-time PCR analysis of independent genes, template DNA was prepared using essentially the same protocol used for the short-read tag library construction. PCR primers were designed using Primer3 (http://frodo.wi.mit.edu/primer3/). The PCR primers used in this study are shown in Supplementary Table S1. Semi-quantitative Reverse Transcription (RT)–PCR was performed using an ABI 7900HT (ABI).
2.8. Electrophoresis Mobility Shift Assay (EMSA)
BEAS2B cells were stimulated with rhIL-4 at 37°C for 30 min. Cells were suspended in 400 μl of Buffer A (10 mM HEPES-KOH, pH 7.8, 10 mM KCl, 0.1 mM EDTA, 0.1% NP-40). Cells were spun at 5000 rpm for 1 min at 4°C. The supernatant was discarded. Cell pellet was resuspended in 100 μl of Buffer C (50 mM HEPES-KOH, pH 7.8, 420 mM KCl, 0.1 mM EDTA, 5 mM MgCl2, 20% Glycerol and 1 mM sodium orthovanadate). The cell lysate was rotated at 4°C for 30 min and spun at 15000 rpm for 15 min at 4°C. The supernatant was used as nuclear extract. Remaining procedures were performed using the LightShift Chemiluminescent EMSA kit (Thermo SCIENTIFIC). The DNA probe sequences used in this study are shown in Supplementary Table S1.
3. Results and discussion
3.1. Identification of putative STAT6 binding sites in Ramos and BEAS2B cells
To identity STAT6 binding sites, we generated a total of 19 099 520 and 18 981 052 36-bp sequence ChIP Seq tags in Ramos and BEAS2B cells, respectively (Tables 1–3). We selected genomic regions at which ChIP Seq tags were enriched at the estimated false detection rate of P < 5e−6 (assuming a Poisson distribution of background tags). We further selected the sites at which no such enrichment was observed in the absence of IL-4. In total, we identified 556 and 467 STAT6 binding sites that were specifically detected in IL-4-stimulated Ramos and BEAS2B cells, respectively (Fig. 1A and B). The validation analysis using real-time RT–PCR for individual genes is presented in Supplementary Fig. S1A and B. Details of the experimental and computational procedures are also described in the ‘Material and methods’ and in the Supplementary figures. See Supplementary Fig. S1C and D for results using different parameters for the computational target identification procedure and for the GenBank accession numbers of the short-read tag data sets.
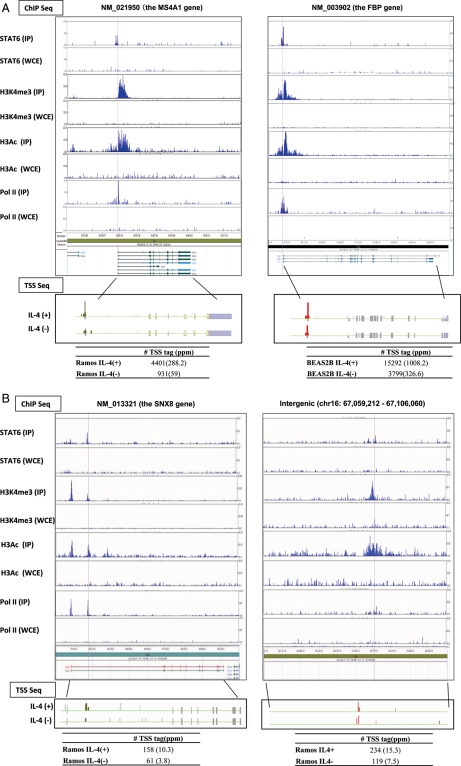
Figure 1.
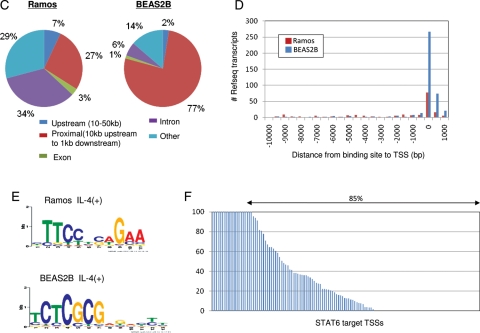
Identification of STAT6 target RefSeq genes. (A) Examples of identified STAT6 targets in authentic RefSeq promoter regions in Ramos cells (left panel) and BEAS2B cells (right panel). STAT6 binding sites are indicated by red lines. Expression changes evaluated by digital TSS-tag counts are also shown in the bottom margin. (B) STAT6 targets in putative alternative promoter regions (left panel) and in intergenic regions (right panel) in Ramos cells. Upper and lower panels present the results from ChIP Seq and TSS Seq analyses, respectively. The IP used for each ChIP Seq analysis is shown in the margin. WCE, whole cell extract used as a background control. (C) Frequency of STAT6 binding sites relative to RefSeq regions in Ramos cells (left panel) and BEAS2B cells (right panel). (D) Distance between STAT6 binding sites and 5′-ends of RefSeq transcript models in Ramos cells (red bar) and BEAS2B cells (blue bar). (E) Consensus sequences deduced from STAT6 binding sites in Ramos cells (upper panel) and BEAS2B cells (lower panel). (F) Fold induction of the STAT6 target TSSs evaluated by digital TSS-tag counts in STAT6-knockdown BEAS2B cells. The y-axis indicates the relative fold induction compared with that in wild-type BEAS2B cells. TSSs indicated by arrows showed reduced induction in knockdown cells.
We analysed the genomic positions of the identified STAT6 binding sites (Fig. 1C and D). In BEAS2B cells, 358 (77%) sites were located within −10 to +1 kb of a RefSeq gene region (the 5′-end of the most upstream RefSeq transcript model was designated as zero). In contrast, in Ramos cells, 150 (27%) of the binding sites were located in these regions, whereas 37% were located in internal gene regions (Fig. 1C). MEME analysis34 of the binding sites in Ramos cells detected the consensus sequence TTCNNNNGAA, which matches previous studies.35,36 However, a different consensus sequence, TCTCGCG, was detected in BEAS2B cells (Fig. 1E). For the results of the validation analysis in the representative cases, see Supplementary Fig. S2. These results may indicate that STAT6 employs different modes of transcriptional activation in these cells.
3.2. TSS Seq analysis and ChIP Seq analysis of RNA pol II
To directly analyse the transcriptional consequences of STAT6 binding, particularly in Ramos and BEAS2B cells, we constructed and analysed TSS Seq libraries using the same materials. We expected that TSS Seq analysis would allow us to associate the binding sites in intergenic regions and genic regions distant from the 5′-end of the authentic RefSeq transcript models with TSSs of previously uncharacterized transcripts. We generated a total of 31 027 908 and 26 447 563 36 bp single-end read TSS tags in Ramos and in BEAS2B cells, respectively. Of these, 22 227 238 (72%) and 20 964 221 (78%) of the tags mapped within the 5′-end exon of RefSeq genes and upstream proximal regions in Ramos and BEAS2B cells, respectively, indicating that the estimated accuracy of the TSS tags for determining TSS positions was ∼86% (also see Supplementary Fig. S3C). The mapped tags were further clustered into bins of 500 to represent putative promoter groups (TSS clusters, TSCs). Consistent with our previous results,28 although the numbers of the TSCs were generally large, those having TSS tags of >1 ppm (corresponding to one copy per cell, assuming every cell has 1 million mRNA copies) represented a very minor population (Supplementary Fig. S3D for statistics). In total, there were 33 196 and 41 359 TSCs with expression levels of >1 ppm in IL-4-stimulated Ramos and BEAS2B cells, respectively. To further ensure the validity of the TSS Seq analysis, we also analysed pol II binding sites by ChIP Seq. We generated and analysed 21 903 762 and 21 709 675 36-bp single-end read tags and identified a total of 17 431 and 13 457 pol II binding sites with an estimated false detection rate of P < 0.001 (Tables 1–3) in Ramos and BEAS2B cells, respectively (Supplementary Fig. S1E–G). Of these, ∼63, 20 and 17% overlapped TSC regions located in the 5′-ends, internal parts and intergenic regions, respectively, of the RefSeq genes.
We associated the STAT6 ChIP Seq data with the TSS Seq data and the pol II ChIP Seq data. As summarized in Tables 4 and 5, in BEAS2B cells, among 358 putative STAT6 binding sites that were identified within RefSeq gene regions, 132 (37%) had TSCs that were induced by at least 2-fold by IL4 stimulation and pol II binding sites in their proximal region (−10 to 1 kb compared with the binding site). In nine cases, there were neither TSCs nor pol II binding signals in the surrounding regions, but we found them in other locations in the same RefSeq gene region. To determine whether these TSCs are associated with the STAT6 binding site, we conducted a TSS Seq analysis using 27 912 131 TSS tags in STAT6-knockdown BEAS2B cells (Table 2; also see Supplementary Fig. S3E–G). In 85% of the STAT6 target TSSs, the transcriptional induction indicated by digital TSS Seq-tag counts was repressed in STAT6 knockdown cells, indicating that the observed transcriptional induction at the associated TSCs was directly mediated by STAT6 (Fig. 1F). Altogether, we identified 44 and 132 STAT6 target TSCs located at the 5′-ends of RefSeq genes, 45 and 9 TSCs previously uncharacterized TSCs (putatively alternative promoters of the same gene) and 34 and 6 TSCs located in intergenic regions in Ramos and BEAS2B cells, respectively. Full lists of the identified STAT6 targets in the respective categories are shown in Supplementary Table S2. For the remainder of the 435 and 320 STAT6 binding sites, there was no induced TSC associated with the binding site (see the following sections for detailed characterization). For the results of the similar analyses using antigen-stimulated Ramos cells, see Supplementary Fig. S4A–E.
Table 4.
Epigenetic regulation of STAT6-mediated transcriptional activation: summary of ChIP Seq analyses of histone modifications
| ChIP (H3K4me3) | ChIP (H3Ac) | |
|---|---|---|
| Ramos | ||
| # total reads | 130 135 923 | 123 883 144 |
| # IL-4(+) peaks | 27 364 | 46 294 |
| # IL-4(−) peaks | 26 128 | 49 953 |
| BEAS2B | ||
| # IP total reads | 117 611 512 | 124 648 546 |
| # IL-4(+) peaks | 24 281 | 37 480 |
| # IL-4(−) peaks | 23 485 | 32 777 |
Table 5.
Epigenetic regulation of STAT6-mediated transcriptional activation: the frequency of the STAT6 targets having the indicated signals are shown for the indicated populations
| Promoter |
Alternative promoter |
Intergenic |
||||
|---|---|---|---|---|---|---|
| Ramos | BEAS2B | Ramos | BEAS2B | Ramos | BEAS2B | |
| #total | 150 | 358 | 205 | 33 | 201 | 76 |
| H3K4me3 (+) | 124 (83%) | 327 (91%) | 19 (9%) | 3 (9%) | 16 (8%) | 7 (9%) |
| H3Ac (+) | 131 (87%) | 321 (90%) | 91 (44%) | 2 (6%) | 105 (52%) | 10 (13%) |
| Pol II (+) | 116 (77%) | 313 (87%) | 47 (23%) | 6 (18%) | 51 (25%) | 25 (33%) |
| TSC induction (+) | 44 (29%) | 132 (37%) | 45 (22%) | 9 (27%) | 32 (16%) | 6 (8%) |
3.3. Characterization of STAT6 targets
We identified STAT6 targets in multiple categories (see also Supplementary Fig. S5 for other examples).
3.3.1. protein-coding gene targets
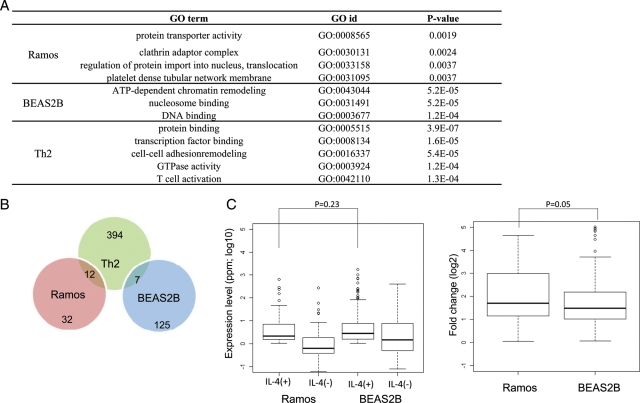
In Ramos and BEAS2B cells, 44 and 132 RefSeq genes were identified as STAT6 target genes, respectively, for which binding of STAT6 to the 5′-end region was observed together with the presence and induction of TSC and pol II binding (Fig. 1A). Previously reported target genes such as the SOCS1 in BEAS2B cells and low-affinity Fc receptor genes (CD23A) in Ramos cells are included, as are some uncharacterized genes. For example, the MS4A1 gene (NM_021950) was identified as a STAT6 target gene in Ramos cells. MS4A1 is a human B-lymphocyte surface molecule, and in vitro studies with an MS4A1 monoclonal antibody showed that this gene plays important roles in the regulation of B cell activation and proliferation.37,38 Moreover, this antibody is clinically used as an anti-rheumatic drug.39,40 Identification of a STAT6-mediated pathway of MS4A1 induction may present a target for the design of novel drugs. In general, GO term41 enrichment analysis showed that GO terms associated with protein secretion and trafficking molecules were enriched among the STAT6 targets in Ramos cells. However, in BEAS2B cells, GO terms associated with transcription and chromatin remodelling were enriched (Fig. 2A). STAT6 may have different repertoires of targets in Ramos and BEAS2B cells.
Figure 2.
Comparison of STAT6 target genes between Ramos and BEAS2B cells. (A) GO terms associated with the STAT6 target genes identified in Ramos cells (top panel), BEAS2B cells (middle panel) and Th2 cells (bottom panel). (B) Overlap of the STAT6 target genes in Ramos cells (red), BEAS2B cells (blue) and Th2 cells (green). (C) Expression levels (left panel) and fold induction (right panels) in response to IL-4 stimulation evaluated by digital TSS-tag counts. Statistical significance between the indicated populations is shown in the margin. STAT6 target genes in Ramos and BEAS2B cells.
3.3.2. Putative alternative promoter targets
In 45 cases in Ramos cells and 9 cases in BEAS2B cells, STAT6 binding sites were found to be associated with previously uncharacterized putative alternative promoters of protein-coding genes (left panel; Fig. 1B). For example, a putative STAT6-responsive alternative promoter was identified in the second intron of the GAB1 gene (Supplementary Fig. S5). GAB1 is tyrosine-phosphorylated upon stimulation by various cytokines, growth factors and antigen receptors. Phosphorylated GAB1 interacts with phosphatidylinositol 3-kinase (PI3K), and the N-terminal PH domain of GAB1 binds to 3-phosphoinositide (PI3).42 Putative transcripts from the alternative promoter of GAB1 lack the PH domain, which may reduce PI3K lymphocyte activation via immunoreceptors and cytokine receptors by competing as a PI3 docking-deficient isoform.
3.3.3. Putative non-protein coding gene targets
Among the STAT6 binding sites associated with intergenic TSCs, one TSC overlapped a previously identified putative ncRNA gene (‘NR’ gene) according to the UCSC Genome Browser. In one additional case, an intergenic TSC was associated with a fully sequenced full-length cDNA, suggesting that STAT6 intergenic targets are mostly uncharacterized transcripts.43–45 The right panel in Fig. 1B presents one such case. Although the molecular functions of these intergenic transcripts remain elusive, this is the first report to indicate the possibility that STAT6 also regulates the expression of so-called putative non-coding RNA transcripts.
3.4. Diverse regulation of STAT6 targets in different cell types
We compared the STAT6 target genes identified in Ramos cells with those in BEAS2B cells and found that they did not overlap at all (Fig. 2B). Also, as shown in Figs 1 and 2, STAT6 target genes showed distinct characteristics between Ramos and BEAS2B cells. Specifically, the target genes had different consensus binding sequences, different relative binding site positions compared with the RefSeq region and different associated GO terms. We also included in the comparison STAT6 target sites in human Th2 cells that were identified in a recent study.22 The reported consensus binding sequence of TTCNNNNGAA in Th2 cells was similar to the consensus sequence we identified in Ramos cells. However, the reported target genes in Th2 cells very rarely overlapped with those in Ramos or BEAS2B cells. In contrast, the expression and induction levels of the STAT6 target genes in Ramos and BEAS2B cells were nearly equivalent (Fig. 2C).
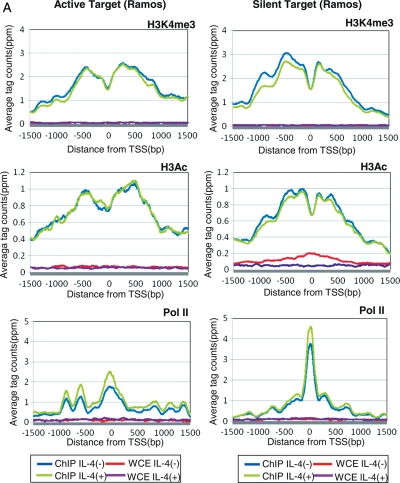
Having observed that different cells have different repertoires of STAT6 targets even when they are similarly activated, we speculated that this differential regulation of STAT6 activation may be mediated by distinct chromatin structures in distinct cell types. We examined changes in histone modification patterns in response to IL-4 stimulation, namely trimethylation of lysine 4 (H3K4me3) and acetylation (H3Ac) of histone 3, using ChIP Seq analysis of a total of 496 279 125 36-bp single-end read tags (Table 4). These histone modifications are thought to be representative markers of active chromatin.46 As shown in Fig. 3A, we observed that H3K4me3 and H3Ac were similarly enriched around the TSSs of STAT6 target genes (STAT6-positive, pol II binding-positive and TSS induction-positive genes) both in Ramos and BEAS2B cells, suggesting that active transcription occurs at the identified target sites. Interestingly, in both cell types, active histone markers were already present around the target sites prior to IL-4 stimulation. It is possible that, upon IL-4 stimulation, STAT6 is recruited to the chromatin that is already programmed to be bound by STAT6 or other TFs (left panels; Fig. 3A and B; also see Fig. 4B for a schematic representation).
Figure 3.
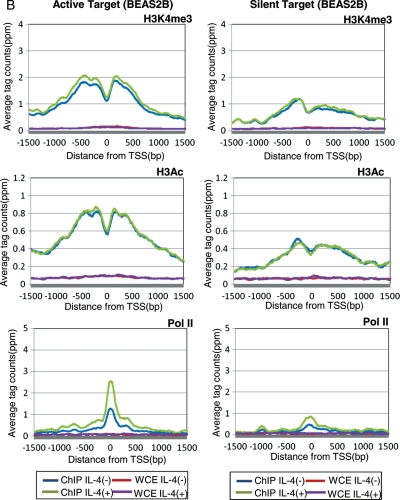
Status of histone modifications and pol II binding in the proximal regions of STAT6 target TSSs. (A) Distribution of the averaged ChIP Seq tags for H3K4me3 (top panels), H3Ac (middle panels) and pol II (bottom panels) in Ramos cells. Data from active target genes (STAT6 binding plus TSS induction in Ramos cells) are shown in the left panels, and data from silent target genes (STAT6 binding plus TSS induction in BEAS2B cells but both negative in Ramos cells) are shown in the right panels. Blue, green, red and purple lines indicate the results for the IP (IL-4 (+)), IP (IL-4 (−)), WCE (IL-4 (+)) and WCE (IL-4 (−)) experiments, respectively. On the x-axis, the position of the associated TSS is designated as zero. (B) Results of an analysis similar to that shown in (A) in BEAS2B cells.
Figure 4.
Distinct regulation of STAT6 targets in different cell types. (A) Fold change of TF genes in response to IL-4 stimulation in Ramos cells (red bar) and BEAS2B cells (blue bar) evaluated by digital TSS-tag counts. TFs were ordered by their fold induction in Ramos cells on the x-axis. (B) Schematic representation of a model for distinct STAT6 activation in Ramos cells (upper panels) and BEAS2B cells (lower panels). Inferred chromatin status and pol II binding are shown for the active targets in the left panel and for the silent targets in the right panel. Green circles, STAT6; purple circles, pol II; yellow and orange circles, putative additional factors in Ramos and BEAS2B cells, respectively; left and right columns, active and silent chromatin, respectively.
We further examined the histone modification patterns in Ramos cells in regions that are STAT6 target regions in BEAS2B cells but not in Ramos cells (we denote these as ‘silent targets’ hereafter). Interestingly, active histone markers were also observed in Ramos cells even though these regions were silent in this cell type; neither STAT6 binding nor the presence of TSS tags was observed (right panels; Fig. 3A). Pol II recruitment was even observed in these regions (right bottom panel; Fig. 3A). Although they are in a cellular context in which no transcriptional induction occurs, these sites seem to be resting in a prepared state until STAT6 is recruited.
3.5. Distinct modes of STAT6-mediated transcriptional induction in different cell types
We also noticed that numerous STAT6 binding sites in Ramos cells did not consequently induce transcription even though STAT6 binding was clearly detected by ChIP Seq and confirmed by real-time PCR (Tables 4 and 5; also see Supplementary Fig. S6 for examples). We found that a majority of these STAT6 binding sites also resided within H3K4me3- and H3Ac-positive regions and that pol II was frequently recruited. Thus, only the induction of the TSS was absent in these regions (Table 5). Similar observations were also made regarding putative alternative promoter binding sites of STAT6 and intergenic targets of STAT6. In these cases, some additional factors seemed to be missing, which hampered the full activation of transcription.
In BEAS2B cells, we also found a number of STAT6 binding sites lacking transcriptional induction, which is similar to the case in Ramos cells (Table 5). However, in BEAS2B cells, active histone markers were found to be less prevalent around the silent target regions (STAT6 target sites in Ramos cells), and pol II was not bound in these regions (right panels; Fig. 3B). In the active target sites, active histone markers were already present prior to STAT6 binding, but pol II recruitment occurred only after IL-4 stimulation (left panels; Fig. 3B). Results of a similar analysis for Th2 target regions in Ramos and BEAS2B cells are shown in Supplementary Fig. S7. It is possible that STAT6-mediated transcriptional activation consists of multiple regulatory steps ranging from the formation of active chromatin to the recruitment of STAT6 and transcriptional activation, each of which requires different factors that may be cell-specific (Fig. 4B).
3.6. Transcriptional regulation of Ramos and BEAS2B cells
To search for factors missing in Ramos and BEAS2B cells that drive transcriptional activation in other cells, we analysed the transcriptional induction of genes encoding TFs in response to IL-4 stimulation by counting TSS tags. For this purpose, we selected 183 previously functionally characterized TF genes from TRANSFAC.47 As shown in Fig. 4A, ∼45 (25%) of the TF genes were either up- or down-regulated in Ramos or BEAS2B cells. Although similar numbers of TF genes were up- or down-regulated, they rarely overlapped between cell types (P = 0.8). We further examined which particular TFs had consensus binding sites enriched in the population of STAT6 target genes, although they were diluted in the surrounding regions of the STAT6 binding sites at which no transcriptional induction occurred. In BEAS2B cells, we found that the predicted binding sites of FAC1, which catalyses nucleosome sliding as a component of the NURF complex,48,49 were enriched (P < 3e-5) in the former population. The expression of FAC1 was induced in response to IL-4 by 23-fold in BEAS2B cells, whereas it was repressed by 50-fold in Ramos cells. On the other hand, in Ramos cells, the predicted binding sites of CEBP-γ were found to be enriched around the transcription-inducing STAT6 binding sites (P < 0.02), and transcription level of CEBP-γ was induced 27-fold upon IL-4 stimulation. These TFs may be responsible for the cell-specific regulation of STAT6 target genes. A complete list of the identified TFs in the respective cell types and their statistical significance is shown in Supplementary Fig. S8. These TFs should be the first targets in any attempt to elucidate the TFs that cooperatively regulate the transcription of STAT6 targets in a cell-specific manner.
3.7. Conclusions
In this paper, we describe the characterization of STAT6 target genes in Ramos and BEAS2B cells using various types of massively parallel sequencing analyses. Among the 44 and 132 STAT6 target genes identified in these two respective cell types, several previously uncharacterized target genes were included (Supplementary Table S2). The target genes identified in this study reside in various functional categories in different cell types and expand the opportunities to treat IL-4-related allergic diseases by offering potential pharmacological targets.
It should also be noted that putative alternative promoters were among the STAT6 targets identified in this study. Because the potential transcripts from these alternative promoters lack or have different N-terminal protein-coding regions in many cases (exemplified in Fig. 1B), they are likely to have diversified protein functions. Although elucidating the detailed biological roles of these targets will require further in-depth functional assays, it is likely that STAT6 also utilizes these previously poorly characterized transcripts in addition to canonical protein-coding transcripts for the fine tuning of target gene function. We also identified STAT6 target genes in intergenic regions of RefSeq transcripts that are likely to drive non-protein coding transcripts. Multi-layered regulation mediated by canonical promoters of protein-coding genes, alternative promoters and non-coding RNAs may further increase the complexity of the responses to STAT6-mediated IL-4 stimulation in various cell types, thereby collectively orchestrating immune responses in humans. One of the greatest advantages of integrating ChIP Seq data with TSS Seq data may be that previously uncharacterized molecules could be analysed in a uniform platform.
We also found that different cell types have different repertoires of STAT6 targets. In Ramos cells, genes mainly involved in protein secretion and protein trafficking (such as MS4A1 and CCR650) were identified as STAT6 targets, suggesting that one of the main biological roles of STAT6 activation is to change patterns of secretory proteins and/or cell surface proteins during the maturation of B cells. On the other hand, in BEAS2B cells, genes involved in the alteration of transcriptional programs (such as PTRH251 and FUBP152) were found to be enriched. We further compared the newly identified STAT6 targets in Ramos and BEAS2B cells with recently identified STAT6 targets in Th2 cells. We found that neither overlapped regarding functional categories, suggesting that STAT6 targets bring distinct cellular consequences depending on cell types (Fig. 2).
We attempted to identify how different STAT6 transcriptional activation programmes are realized in different cell types. We found that the chromatin status and pol II binding patterns were different between Ramos and BEAS2B cells (Fig. 3). In BEAS2B cells, the formation of active chromatin and recruitment of pol II were specific to active STAT6 target sites. Pol II recruitment occurred only upon IL-4 stimulation. In contrast, in Ramos cells, this event sometimes occurred even around silent STAT6 targets, which are active in different cell types but silent in this cell type. Different cell types seem to have different modes of STAT6-mediated target activation. We therefore searched for factors that may be necessary for full transcriptional activation of STAT6 targets in Ramos and BEAS2B cells. As putative cooperative factors for STAT6, we identified FAC1 (which is a component of the nucleosome sliding complex NURF) in BEAS2B cells and CEBP-γ (which is a transcriptional activator53) in Ramos cells. The identification of different categories of TFs in different cell types may indicate that transcriptional induction mediated by STAT6 requires modification of the chromatin structure in BEAS2B cells, whereas the recruitment of synergetic TFs is more important in Ramos cells. Distinct cellular responses may be mediated by the intrinsic chromatin status and unique sets of TFs, and the factors that play the most important roles may be different depending on cell type.
We also found that STAT6 binding did not always result in transcriptional activation. On the contrary, the targets that were transcriptionally activated upon STAT6 binding were relatively few in both Ramos and BEAS2B cells. This feature of STAT6 binding was also observed in Th2 cells by other groups.22 There seems to be complex regulation of transcriptional induction even after the formation of active chromatin and the recruitment of TFs. The advantage of using massively parallel sequencing as a common platform is that genome-wide data can be generated simultaneously for different steps of transcriptional activation, ranging from the binding of STAT6 and histone modification to pol II binding and transcriptional induction. It will be important to determine which regulatory step is the most important for each gene to fully understand STAT6-mediated transcriptional activation mechanisms. Further in-depth analyses of the identified target genes will also be necessary to understand the mechanisms whereby these factors in their respective cell types collectively induce necessary and/or harmful immune responses in healthy and diseased individuals. Biochemical and phenotypic analyses at the cellular and organismal levels are still beyond the reach of genome-wide approaches. However, the target gene catalogue identified and characterized in this study in different cell types should serve as a valuable resource.
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Funding
This work was supported by grants from the New Energy and Industrial Technology Development Organization (NEDO) project of the Ministry of Economy, Trade and Industry (METI) of Japan, the Japan Key Technology Center project of METI of Japan, and a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science and Technology of Japan. This research is supported by the Japan Society for the Promotion of Science (JSPS) through its ‘Funding Program for World-Leading Innovative R&D on Science and Technology (FIRST Program)’.
Supplementary Material
Acknowledgements
We are grateful to K. Abe, E. Sekimori and K. Imamura for technical support.
References
- 1.Kaplan M.H., Schindler U., Smiley S.T., Grusby M.J. Stat6 is required for mediating responses to IL-4 and for development of Th2 cells. Immunity. 1996;4:313–19. doi: 10.1016/s1074-7613(00)80439-2. [DOI] [PubMed] [Google Scholar]
- 2.Ikizawa K., Kajiwara K., Koshio T., Matsuura N., Yanagihara Y. Inhibition of IL-4 receptor up-regulation on B cells by antisense oligodeoxynucleotide suppresses IL-4-induced human IgE production. Clin. Exp. Immunol. 1995;100:383–89. doi: 10.1111/j.1365-2249.1995.tb03710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Renauld J.C. New insights into the role of cytokines in asthma. J. Clin. Pathol. 2001;54:577–89. doi: 10.1136/jcp.54.8.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shirakawa I., Deichmann K.A., Izuhara I., Mao I., Adra C.N., Hopkin J.M. Atopy and asthma: genetic variants of IL-4 and IL-13 signalling. Immunol. Today. 2000;21:60–4. doi: 10.1016/s0167-5699(99)01492-9. [DOI] [PubMed] [Google Scholar]
- 5.Kuperman D.A., Schleimer R.P. Interleukin-4, interleukin-13, signal transducer and activator of transcription factor 6, and allergic asthma. Curr. Mol. Med. 2008;8:384–92. doi: 10.2174/156652408785161032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lloyd C.M., Hessel E.M. Functions of T cells in asthma: more than just T(H)2 cells. Nat. Rev. 2010;10:838–48. doi: 10.1038/nri2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holgate S.T., Polosa R. Treatment strategies for allergy and asthma. Nat. Rev. 2008;8:218–30. doi: 10.1038/nri2262. [DOI] [PubMed] [Google Scholar]
- 8.Prussin C., Metcalfe D.D. 5. IgE, mast cells, basophils, and eosinophils. J. Allergy Clin. Immunol. 2006;117:S450–56. doi: 10.1016/j.jaci.2005.11.016. [DOI] [PubMed] [Google Scholar]
- 9.Hoeck J., Woisetschlager M. STAT6 mediates eotaxin-1 expression in IL-4 or TNF-alpha-induced fibroblasts. J. Immunol. 2001;166:4507–15. doi: 10.4049/jimmunol.166.7.4507. [DOI] [PubMed] [Google Scholar]
- 10.Matsukura S., Stellato C., Plitt J.R., et al. Activation of eotaxin gene transcription by NF-kappa B and STAT6 in human airway epithelial cells. J. Immunol. 1999;163:6876–83. [PubMed] [Google Scholar]
- 11.Dabbagh K., Takeyama K., Lee H.M., Ueki I.F., Lausier J.A., Nadel J.A. IL-4 induces mucin gene expression and goblet cell metaplasia in vitro and in vivo. J. Immunol. 1999;162:6233–7. [PubMed] [Google Scholar]
- 12.Mohrs M., Ledermann B., Kohler G., Dorfmuller A., Gessner A., Brombacher F. Differences between IL-4- and IL-4 receptor alpha-deficient mice in chronic leishmaniasis reveal a protective role for IL-13 receptor signaling. J. Immunol. 1999;162:7302–8. [PubMed] [Google Scholar]
- 13.Takeda K., Tanaka T., Shi W., et al. Essential role of Stat6 in IL-4 signalling. Nature. 1996;380:627–30. doi: 10.1038/380627a0. [DOI] [PubMed] [Google Scholar]
- 14.Igaz P., Toth S., Falus A. Biological and clinical significance of the JAK-STAT pathway; lessons from knockout mice. Inflamm. Res. 2001;50:435–41. doi: 10.1007/PL00000267. [DOI] [PubMed] [Google Scholar]
- 15.Hebenstreit D., Wirnsberger G., Horejs-Hoeck J., Duschl A. Signaling mechanisms, interaction partners, and target genes of STAT6. Cytok. Growth Factor Rev. 2006;17:173–88. doi: 10.1016/j.cytogfr.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 16.Georas S.N., Cumberland J.E., Burke T.F., Chen R., Schindler U., Casolaro V. Stat6 inhibits human interleukin-4 promoter activity in T cells. Blood. 1998;92:4529–38. [PubMed] [Google Scholar]
- 17.Stutz A.M., Pickart L.A., Trifilieff A., Baumruker T., Prieschl-Strassmayr E., Woisetschlager M. The Th2 cell cytokines IL-4 and IL-13 regulate found in inflammatory zone 1/resistin-like molecule alpha gene expression by a STAT6 and CCAAT/enhancer-binding protein-dependent mechanism. J. Immunol. 2003;170:1789–96. doi: 10.4049/jimmunol.170.4.1789. [DOI] [PubMed] [Google Scholar]
- 18.Hebenstreit D., Luft P., Schmiedlechner A., et al. IL-4 and IL-13 induce SOCS-1 gene expression in A549 cells by three functional STAT6-binding motifs located upstream of the transcription initiation site. J. Immunol. 2003;171:5901–7. doi: 10.4049/jimmunol.171.11.5901. [DOI] [PubMed] [Google Scholar]
- 19.Dickensheets H., Vazquez N., Sheikh F., et al. Suppressor of cytokine signaling-1 is an IL-4-inducible gene in macrophages and feedback inhibits IL-4 signaling. Genes Immun. 2007;8:21–7. doi: 10.1038/sj.gene.6364352. [DOI] [PubMed] [Google Scholar]
- 20.Tinnell S.B., Jacobs-Helber S.M., Sterneck E., Sawyer S.T., Conrad D.H. STAT6, NF-kappaB and C/EBP in CD23 expression and IgE production. Int. Immunol. 1998;10:1529–38. doi: 10.1093/intimm/10.10.1529. [DOI] [PubMed] [Google Scholar]
- 21.Chen Z., Lund R., Aittokallio T., Kosonen M., Nevalainen O., Lahesmaa R. Identification of novel IL-4/Stat6-regulated genes in T lymphocytes. J. Immunol. 2003;171:3627–35. doi: 10.4049/jimmunol.171.7.3627. [DOI] [PubMed] [Google Scholar]
- 22.Elo L.L., Jarvenpaa H., Tuomela S., et al. Genome-wide profiling of interleukin-4 and STAT6 transcription factor regulation of human Th2 cell programming. Immunity. 2010;32:852–62. doi: 10.1016/j.immuni.2010.06.011. [DOI] [PubMed] [Google Scholar]
- 23.Johnson D.S., Mortazavi A., Myers R.M., Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 24.Robertson G., Hirst M., Bainbridge M., et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat. Methods. 2007;4:651–7. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 25.Barski A., Cuddapah S., Cui K., et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 26.Robertson A.G., Bilenky M., Tam A., et al. Genome-wide relationship between histone H3 lysine 4 mono- and tri-methylation and transcription factor binding. Genome Res. 2008;18:1906–17. doi: 10.1101/gr.078519.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Z., Zang C., Rosenfeld J.A., et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsuchihara K., Suzuki Y., Wakaguri H., et al. Massive transcriptional start site analysis of human genes in hypoxia cells. Nucleic Acids Res. 2009;37:2249–63. doi: 10.1093/nar/gkp066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sathira N., Yamashita R., Tanimoto K., et al. Characterization of transcription start sites of putative non-coding RNAs by multifaceted use of massively paralleled sequencer. DNA Res. 2010;17:169–83. doi: 10.1093/dnares/dsq007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suzuki Y., Sugano S. Construction of a full-length enriched and a 5’-end enriched cDNA library using the oligo-capping method. Methods Mol. Biol. 2003;221:73–91. doi: 10.1385/1-59259-359-3:73. [DOI] [PubMed] [Google Scholar]
- 31.Carninci P., Kasukawa T., Katayama S., et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–63. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 32.Carninci P., Sandelin A., Lenhard B., et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006;38:626–35. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- 33.Kel A.E., Gossling E., Reuter I., Cheremushkin E., Kel-Margoulis O.V., Wingender E. MATCH: a tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2003;31:3576–9. doi: 10.1093/nar/gkg585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bailey T.L., Boden M., Buske F.A., et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–8. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schindler U., Wu P., Rothe M., Brasseur M., McKnight S.L. Components of a Stat recognition code: evidence for two layers of molecular selectivity. Immunity. 1995;2:689–97. doi: 10.1016/1074-7613(95)90013-6. [DOI] [PubMed] [Google Scholar]
- 36.Ehret G.B., Reichenbach P., Schindler U., et al. DNA binding specificity of different STAT proteins. Comparison of in vitro specificity with natural target sites. J. Biol. Chem. 2001;276:6675–88. doi: 10.1074/jbc.M001748200. [DOI] [PubMed] [Google Scholar]
- 37.Tedder T.F., Boyd A.W., Freedman A.S., Nadler L.M., Schlossman S.F. The B cell surface molecule B1 is functionally linked with B cell activation and differentiation. J. Immunol. 1985;135:973–9. [PubMed] [Google Scholar]
- 38.Tedder T.F., Forsgren A., Boyd A.W., Nadler L.M., Schlossman S.F. Antibodies reactive with the B1 molecule inhibit cell cycle progression but not activation of human B lymphocytes. Eur. J. Immunol. 1986;16:881–7. doi: 10.1002/eji.1830160802. [DOI] [PubMed] [Google Scholar]
- 39.Smith M.R. Rituximab (monoclonal anti-CD20 antibody): mechanisms of action and resistance. Oncogene. 2003;22:7359–68. doi: 10.1038/sj.onc.1206939. [DOI] [PubMed] [Google Scholar]
- 40.Glennie M.J., French R.R., Cragg M.S., Taylor R.P. Mechanisms of killing by anti-CD20 monoclonal antibodies. Mol. Immunol. 2007;44:3823–37. doi: 10.1016/j.molimm.2007.06.151. [DOI] [PubMed] [Google Scholar]
- 41.Ashburner M., Ball C.A., Blake J.A., et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sarmay G., Angyal A., Kertesz A., Maus M., Medgyesi D. The multiple function of Grb2 associated binder (Gab) adaptor/scaffolding protein in immune cell signaling. Immunol. Lett. 2006;104:76–82. doi: 10.1016/j.imlet.2005.11.017. [DOI] [PubMed] [Google Scholar]
- 43.Ota T., Suzuki Y., Nishikawa T., et al. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat. Genet. 2004;36:40–5. doi: 10.1038/ng1285. [DOI] [PubMed] [Google Scholar]
- 44.Kimura K., Wakamatsu A., Suzuki Y., et al. Diversification of transcriptional modulation: large-scale identification and characterization of putative alternative promoters of human genes. Genome Res. 2006;16:55–65. doi: 10.1101/gr.4039406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wakaguri H., Yamashita R., Suzuki Y., Sugano S., Nakai K. DBTSS: database of transcription start sites, progress report 2008. Nucleic Acids Res. 2008;36:D97–101. doi: 10.1093/nar/gkm901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 47.Wingender E., Dietze P., Karas H., Knuppel R. TRANSFAC: a database on transcription factors and their DNA binding sites. Nucleic Acids Res. 1996;24:238–41. doi: 10.1093/nar/24.1.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wysocka J., Swigut T., Xiao H., et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodeling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 49.Li H., Ilin S., Wang W., et al. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–5. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Williams I.R. CCR6 and CCL20: partners in intestinal immunity and lymphorganogenesis. Ann. N. Y. Acad. Sci. 2006;1072:52–61. doi: 10.1196/annals.1326.036. [DOI] [PubMed] [Google Scholar]
- 51.Jan Y., Matter M., Pai J.T., et al. A mitochondrial protein, Bit1, mediates apoptosis regulated by integrins and Groucho/TLE corepressors. Cell. 2004;116:751–62. doi: 10.1016/s0092-8674(04)00204-1. [DOI] [PubMed] [Google Scholar]
- 52.Liu J., Akoulitchev S., Weber A., et al. Defective interplay of activators and repressors with TFIH in xeroderma pigmentosum. Cell. 2001;104:353–63. doi: 10.1016/s0092-8674(01)00223-9. [DOI] [PubMed] [Google Scholar]
- 53.Nishizawa M., Wakabayashi-Ito N., Nagata S. Molecular cloning of cDNA and a chromosomal gene encoding GPE1-BP, a nuclear protein which binds to granulocyte colony-stimulating factor promoter element 1. FEBS Lett. 1991;282:95–7. doi: 10.1016/0014-5793(91)80452-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.