Abstract
A linkage map of expressed sequence tag (EST)-based markers in radish (Raphanus sativus L.) was constructed using a low-cost and high-efficiency single-nucleotide polymorphism (SNP) genotyping method named multiplex polymerase chain reaction–mixed probe dot-blot analysis developed in this study. Seven hundred and forty-six SNP markers derived from EST sequences of R. sativus were assigned to nine linkage groups with a total length of 806.7 cM. By BLASTN, 726 markers were found to have homologous genes in Arabidopsis thaliana, and 72 syntenic regions, which have great potential for utilizing genomic information of the model species A. thaliana in basic and applied genetics of R. sativus, were identified. By construction and analysis of the genome structures of R. sativus based on the 24 genomic blocks within the Brassicaceae ancestral karyotype, 23 of the 24 genomic blocks were detected in the genome of R. sativus, and half of them were found to be triplicated. Comparison of the genome structure of R. sativus with those of the A, B, and C genomes of Brassica species and that of Sinapis alba L. revealed extensive chromosome homoeology among Brassiceae species, which would facilitate transfer of the genomic information from one Brassiceae species to another.
Keywords: comparative genomics, Raphanus sativus, SNP genotyping, synteny, chromosome homoeology
1. Introduction
Angiosperms evolved from a common ancestral genome that underwent duplications, rearrangements, and mutations in succeeding generations. Phylogenetic relatedness of different species correlates with a degree of synteny between their genomes.1,2 Comparative genomics contributes greatly to understanding the basic processes of genome evolution from or to related organisms within a phylogenetic framework3–5 and applying model species genome information to the study of related organisms.6,7 Comparative mapping studies in the grass family (Poaceae) have pioneered the field of plant comparative genomics.3,8 The collinear genomic regions between domesticated cereals and forage crops have been detected by such studies, which have opened the way to efficient map-based cloning and enabled inference of the basic organization of the ancestral grass genome.
The dicot family Brassicaceae consists of 338 genera with ca. 3700 species in 25 tribes,9 and their chromosome number varies greatly from n = 4 to 128.10 Comparative genomics in Brassicaceae has largely focused on direct comparisons between the model species Arabidopsis thaliana and the species of interest, especially, the agronomically important Brassica crops. Comparative linkage mapping among three diploid Brassica species, i.e. Brassica nigra, Brassica oleracea, and Brassica rapa,6 and that between A. thaliana and three Brassica species11 have suggested that genomes of the diploid Brassica species are composed of a triplicated and rearranged ancestral genome. Additional evidence for the genome triplication in Brassica has been shown by nucleotide sequencing12,13 and cytogenetic methods.14,15 However, some genomic regions are present in less or more than three copies in Brassica genomes.7,16 Furthermore, ∼30–40% of genes in the syntenic regions of A. thaliana have been lost in their counterparts of the Brassica genomes,13,16 and genomic rearrangements and gene duplications have occurred in A. thaliana after the divergence of Brassica and Arabidopsis,17,18 making the comparative maps between A. thaliana and Brassica species complicated.7,11,19
A Brassicaceae ancestral karyotype (n = 8) has been previously deduced from comparative genetic maps of two n = 8 species in the tribe Camelineae, i.e. Capsella rubella and Arabidopsis lyrata, with A. thaliana.5,20–22 Arabidopsis thaliana and its closely related species with six or seven chromosome pairs have been suggested to be derived from the ancestral karyotype through a similar mechanism of chromosome reduction.23 Based on a set of 21 conserved genomic regions within the Arabidopsis genome identified in Brassica napus by Parkin et al.,19 Schranz et al.5 have proposed a set of 24 genomic blocks within the Brassicaceae ancestral karyotype. These 24 genomic blocks represent the conserved segments among the Brassicaceae ancestral karyotype, Arabidopsis, and Brassica. Using these genomic blocks, the homoeologous relationship and evolution of the A, B, and C Brassica genomes have been studied,24 and a conserved chromosome (AK1) of the Brassicaceae ancestral karyotype has been revealed in Sinapis alba.25
More recently, Mandáková and Lysak4 have proposed a Proto-Calepineae karyotype (n = 7), which has been suggested to be descended from the Brassicaceae ancestral karyotype and to be the karyotype of the common progenitor of the tribes Calepineae, Conringieae, and Noccaeeae, and the Brassicaceae lineage II, including the tribes Brassiceae, Isatideae, Sisymbrieae, and Eutremeae. Two to three copies of genomic blocks associated closely in the Proto-Calepineae karyotype have also been found in the Brassica genome,5,19 suggesting that the Proto-Calepineae karyotype has undergone the whole-genome triplication in the clade leading to the tribe Brassiceae. However, the genomic evolutionary process from the primary paleo-hexaploid ancestor to the present Brassiceae species is not clear. Reconstruction of karyotypes of different species using the common genomic blocks of the Brassicaceae ancestral karyotype is expected to solve this problem.
Radish (Raphanus sativus L., 2n = 2x = 18) belonging to the tribe Brassiceae and closely related to B. rapa is an important commercial crop that is grown and consumed all over the world, especially in eastern Asia. Raphanus sativus has great variations in the root shape from round and thick with a diameter of more than 30 cm to thin and long with a length of more than 2 m. The thick roots are commonly harvested as vegetables, while there are some cultivars used as leafy vegetables, silique vegetables, or oil crops. As a genetic map with DNA markers can be utilized in applied genetics and breeding, several genetic linkage maps have been constructed in R. sativus26–29 based on Amplified Fragment Length Polymorphism (AFLP), Random Amplified Polymorphism (RAPD), and Simple Sequence Repeats (SSR) markers, and quantitative trait loci (QTLs) for shape and colour of roots, club-root resistance, and cyst nematode resistance have been detected.27,29,30 However, since the AFLP and RAPD markers are anonymous and SSR markers are usually located in non-coding sequences, the comparative genomics of R. sativus with other species has not been performed, and utilization of the genome information of Arabidopsis and B. rapa in the study of R. sativus is limited.
Recently, abundant information on expressed sequence tag (EST) sequences of radish has been published on the radish sequence database (Radish DB; http://radish.plantbiology.msu.edu). As single-nucleotide polymorphisms (SNPs) are the most abundant type of DNA polymorphism in genomes, in order to construct a linkage map of R. sativus, we explored these EST sequences to design primer pairs for specific amplification of genes, and identified SNPs for production of EST-based SNP markers by determining nucleotide sequences of two lines used for production of an F2 population. Genotyping of EST-based SNP markers of F2 plants was performed using dot-blot-SNP analysis combining multiplex polymerase chain reaction (PCR)31 and mixed probe hybridization,32 and a high-density linkage map was constructed to reveal synteny with the genome of A. thaliana. Furthermore, the genome structure of R. sativus was reconstructed using the 24 genomic blocks of the Brassicaceae ancestral karyotype and was compared with those of other Brassiceae species.
2. Materials and methods
2.1. Plant materials
Two radish lines self-pollinated for three generations from ‘Sayatori 26704’ (National Institute of Vegetable and Tea Science, Japan) and ‘Aokubi S-h’ (Takii Seed Co., Japan), respectively, were used. ‘Sayatori 26704’ (hereafter ‘Sayatori’) is a seedpod vegetable with a very thin and small root like a rat tail cultivated in South Asia. ‘Aokubi S-h’ (hereafter ‘Aokubi’) is a Japanese radish with a long and thick root. F1 hybrids were produced by crossing ‘Aokubi’ with ‘Sayatori’, and F2 seeds were obtained by selfing with bud pollination of a single F1 hybrid plant.
2.2. Analysis of DNA polymorphism between ‘Aokubi’ and ‘Sayatori’
Genomic DNAs of the ‘Aokubi’ and ‘Sayatori’ inbred lines were extracted from leaves by the CTAB method.33 For designing primer pairs, ∼9000 unigenes were selected from the RS2 library of the Radish Database (http://radish.plantbiology.msu.edu). The exon/intron junction and the UTR region of the unigenes were predicted by the MAEZATO system, which aligns a target EST sequence with a possible homologous Arabidopsis cDNA.34 Forward primers were designed within an exon region and reverse primers were designed within a region containing a predicted 3′-UTR using Primer3 plus (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi). As flowering time is important for the production of vegetables in R. sativus, the coding sequence of an FLC1 homologue in B. rapa35 was also used for designing primers. PCR was conducted with these primer pairs using genomic DNA of the ‘Aokubi’ and ‘Sayatori’ inbred lines as templates. A 20 µl reaction mixture contained 20 ng of plant genomic DNA, 10 pmol of each primer, 1× ExTaq buffer, 2 nmol of each dNTP, and 0.5 U of Taq DNA polymerase (ExTaq, Takara Biomedicals, Japan). The thermal cycling condition was 35 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 2 min. A 4 µl PCR product was electrophoresed on 1.2% agarose gel, and a single fragment amplified by PCR was sequenced by the Sanger method.36 The sequences were aligned using SEQUENCHER version 4.7 (Gene Codes Corporation, MI, USA) to identify SNPs. Sequences having SNPs between ‘Aokubi’ and ‘Sayatori’ were used for designing bridge probes37 for dot-blot-SNP analysis.
2.3. Dot-blot-SNP analysis
Genomic DNAs from 189 plants of the F2 population were prepared using the modified CTAB method.38 In order to achieve cost-effective and high-efficiency SNP genotyping, a dot-blot-SNP method combining multiplex PCR31 and a mixed probe37 was developed and named multiplex PCR–mixed probe (MPMP) dot-blot analysis. First, 36 primer pairs of SNP markers were grouped into six by MultiPLX 2.031 and were assigned to six lines in a table. Secondly, hybridization conditions of probes of the markers were predicted as described by Shiokai et al.32 using the DINAMelt web server (http://mfold.rna.albany.edu/?q=DINAMelt/Hybrid2),39 and the probes having similar hybridization conditions in different primer groups were placed into the same column to design an MPMP table (an example being shown in Supplementary Table S1). Multiplex PCR was conducted in a 10 µl reaction mixture containing 4 ng plant genomic DNA, 10 pmol of each primer mixed as designed in the MPMP table, 1× KAPATaq EXtra buffer (without Mg2+), 1.75 mM MgCl2, 0.3 mM of each dNTP, and 0.25 U of DNA polymerase (KAPATaq Extra, KAPABIOSYSTEMS, Boston, MA, USA). Thermal cycling conditions were as follows: 1 min denaturation at 94°C, 35 cycles of 94°C for 30 s, 56°C for 30 s, and 72°C for 1 min, and a final extension at 72°C for 2 min. Amplified DNA was denatured in a solution of 0.4 N NaOH and 10 mM EDTA, and dot-blotted onto a nylon membrane (Nytran, Pall, NY, USA) using Multi-pin Blotter (Atto, Tokyo, Japan). Probes in the same columns of the MPMP table were mixed and hybridized together with digoxigenin-labelled probes having sequences complementary to the bridge probe.37 Allele-specific signals were detected according to Shiokai et al.37 This MPMP dot-blot analysis using 36 EST-SNP markers was compared with dot-blot-SNP analysis using a single EST-SNP marker.
2.4. Linkage analysis
Linkage analysis was carried out using the JoinMap 4.0 software (Kyazma B.V., Wageningen, The Netherlands).40 The SNP markers were grouped into nine linkage groups (LGs) at high LOD threshold (≥6). Marker order was subsequently determined by a regression mapping algorithm on the basis of a minimum LOD score of 1.0 and a recombination threshold of 0.4 in each LG. Recombination values were converted to genetic distance (cM) using the Kosambi mapping function.41 Each map was graphically visualized with MapChart.42 The EST-based SNP markers were named <Rs> <EST name or gene name> <s>.
2.5. Comparison with the A. thaliana genome sequences
Sequences in the A. thaliana genome homologous with the loci of EST-based SNP markers in the linkage map were surveyed using the BLASTN program of the National Centre for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) with a threshold value of E < 10−20. Syntenic regions were identified according to the conserved collinearity of the markers of R. sativus and corresponding homologous genes of A. thaliana. A single non-collinear homologue in a syntenic region was ignored.
3. Results and discussion
3.1. SNP identification and production of DNA markers
To identify SNPs between the parental lines, i.e. ‘Sayatori’ and ‘Aokubi’, 4709 primer pairs were designed and used for amplification of coding regions of genes containing 3′-untranslated regions, and single DNA fragments in both parental lines were amplified by PCR with 3576 primer pairs and sequenced. After trimming ambiguous sequences at both ends and excluding the sequences with a quality score <90% (Sequencher, http://www.genecodes.com/), 2290 sequences, covering 791.9 kb, were obtained (Supplementary Table S2). Among them, 1465 (64.0%) DNA fragments showed nucleotide polymorphism. The frequency of variable bases, i.e. SNPs and indels, was 1/72 bp, and the frequency of SNPs was 1/107 bp. As continuous SNPs or indels can be designed as one marker, such sites were regarded as one SNP site or one indel site. The frequencies of SNP sites and indel sites were 1/115 and 1/520 bp, respectively. These results suggest that the use of SNP markers enables construction of a high-density EST-based genetic map. Of the 1465 sequences having SNPs or indels between the parental lines, we randomly selected 1410 fragments for designing dot-blot-SNP markers.
3.2. Dot-blot-SNP analysis and mapping
Since dot-blot-SNP analysis allows hybridization of only a fully complementary sequence without hybridization of a mismatched sequence, this method ensures accuracy of genotyping,37 and enables genotyping of multiple-copy genes in polyploid species, e.g. B. napus, and diploid species having ancestral genome replication, e.g. B. rapa and R. sativus. In MPMP dot-blot-SNP analysis, more than half of the makers showed allele-specific signals in the first round of probe hybridization under the conditions predicted by the method of Shiokai et al.32 In the second round of probe hybridization, conditions were adjusted according to the signals shown in the first round. At most, three rounds of probe hybridization were conducted. The results of the genotyping data were consistent with the dot-blot-SNP method using a single SNP marker. In this way, the MPMP dot-blot-SNP method was found to speed up the genotyping and to reduce the cost of analysis. Of the 1410 dot-blot-SNP markers, 881 yielded clear dot-blot signals with distinct differences between SNP alleles.
Using 772 dot-blot-SNP markers among the 881 SNP markers, genotypes of 189 F2 plants were analysed. By linkage analysis with the JoinMap 4.0 software, 746 were assigned to nine LGs. Primer sequences, probe sequences, and hybridization conditions of the 746 markers are listed in Supplementary Table S3. Nine LGs were designated as LG1–LG9 in decreasing order of map distances (cM) (Fig. 1). The longest group LG1 consisting of 88 markers and the shortest one LG9 consisting of 75 markers have lengths of 126.5 and 62.7 cM, respectively. The total length covered by the linkage map was 806.7 cM with an average distance of intervals between markers of 1.1 cM. Based on the physical length of 530 Mb in R. sativus,43 1 cM was estimated to be 657 kb.
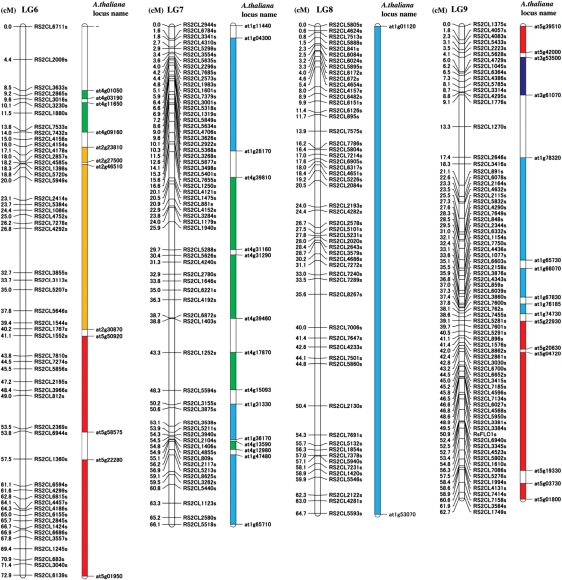
Figure 1.
A linkage map of R. sativus with a comparative map of A. thaliana. Nine LGs are labelled as LG1–LG9 in the order of length. Marker positions (in cM) are shown on the left side and the corresponding marker names are shown on the right side of each LG. Each locus was tested for homology with A. thaliana using BLAST and segments of two or more markers showing homology with A. thaliana in collinearity are regarded as syntenic regions, which are shown to the right of the R. sativus LGs as coloured vertical bars. The A. thaliana chromosomes are coloured according to Parkin et al.,19 and are shown at the bottom right of the figure. Except for both ends of each syntenic region, the names of identified homologous loci in A. thaliana are not shown, but are shown in Supplementary Table S4.
Many ESTs, which were used for developing the markers in the present study, have high similarity to genes with known or hypothetical functions. For example, on the shortest LG9, RsFLC1 amplified by the primers designed from BrFLC1 (accession no. AY115678.1) was mapped. LG9 also contained RS2CL4436, a homologue of AtPPa in the S locus of B. rapa (AB257127.1), and Rs2CL2115, a homologue of the B. rapa disease resistance gene BrTN3 (FJ842847.1). Furthermore, ESTs homologous to A. thaliana genes involved in cadmium ion transmembrane transportation (NM_104680.2), salt stress response (NM_106207.3), cold and drought response (NM_122163.2), sucrose synthesis (NM_122090.3), and amino acid biosynthesis (NM_001036839.2) were mapped on LG9. Other LGs also contain many interesting genes.
3.3. Synteny between R. sativus and A. thaliana
As protein-coding regions in the genomes are conserved in related species more than intergenic regions, EST-based markers are useful for comparative mapping between them. We searched for genes homologous with these markers in the A. thaliana genome by BLASTN. The results are shown in Supplementary Table S4. Under a significance threshold of E < 10−20, A. thaliana loci homologous with 726 R. sativus markers were identified, these homologous loci covering nearly the whole genome of A. thaliana. The five chromosomes of A. thaliana were divided into many segments and distributed to various regions of the R. sativus map (Fig. 1). There were 72 syntenic regions, mostly containing at least three markers and having conserved collinearity with the homologues in A. thaliana. The whole LG8 showed collinearity to more than half of chromosome 1 of A. thaliana. This synteny information is considered to be useful for utilizing the genome information of A. thaliana in the study of R. sativus and may speed up map-based cloning, as shown in B. rapa.7,44
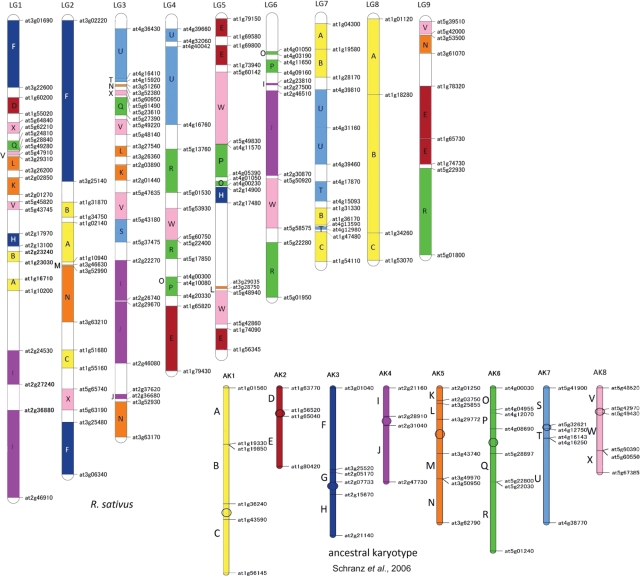
The 24 genomic blocks within the ancestral karyotype proposed by Schranz et al.5 have been identified in many species of the family Brassicaceae4,16,23–25,45 and can contribute to build a unified comparative genomics system in the Brassicaceae. The genome structure of R. sativus was reconstructed by these genomic blocks (Fig. 2). Except for the G block, all the blocks were found in the genome of R. sativus. The absence of G block in the present map is possibly due to its shortness and closeness to the AK3 centromere of the Brassicaceae ancestral karyotype.5 Half of the genomic blocks within the Brassicaceae ancestral karyotype, including A, B, C, E, F, I, J, O, P, R, U, and W, were triplicated. Some parts of A, U, and E blocks were revealed to have been further replicated, resulting in four, four, and six copies, respectively. It has also been reported that half of the genomic blocks are triplicated, while others occur only once or twice in B. rapa genome.16 These results indicate recent replication and loss of blocks during genomic evolutionary processes from the paleohexaploid to the present Brassiceae species. Some neighbouring blocks, e.g. I/J, A/B, and O/P, in the Brassicaceae ancestral karyotype were not separated in the genome of R. sativus. One chromosome (AK1) of the Brassicaceae ancestral karyotype comprising A, B, and C blocks was found to be completely conserved, forming LG8 of R. sativus, as the case of S05 in S. alba.25 The other copies of AK1 may have incurred genomic rearrangement and dispersed in LG1, LG2, and LG7. The blocks of V, K, L, Q, and X from different chromosomes of the Brassicaceae ancestral karyotype associated to form parts of LG1 and LG3. Similarly, the blocks of R, W, O, and P associated to form parts of LG4 and LG6, and possibly LG5. These V/K/L/Q/X, R/W, and O/P block associations have been detected in the Proto-Calepineae karyotype (Supplementary Fig. S1),4 and found to be replicated in the A, B, and C genomes of Brassica species.4,5,19,24 In S. alba, although V/K/L/Q/X block association has not been observed, probably due to the small number of markers used, R/W block association has been found to be triplicated.25 These findings reinforce the hypothesis on the origin of the Brassicaceae lineage II from the common Proto-Calepineae karyotype.4
Figure 2.
Genome structure of R. sativus based on 24 genomic blocks within the Brassicaceae ancestral karyotype. The genomic blocks of the Brassicaceae ancestral karyotype, shown on the bottom right, are labelled by the letters A to X. One of the eight colours corresponds to each chromosome. Boundaries of the blocks are defined by their flanking locus name of A. thaliana, according to Schranz et al.5 The position of each genomic block in R. sativus was defined according to the comparative map of R. sativus and A. thaliana in Fig. 1.
3.4. Identification of homoeology between the genomes in Brassiceae species
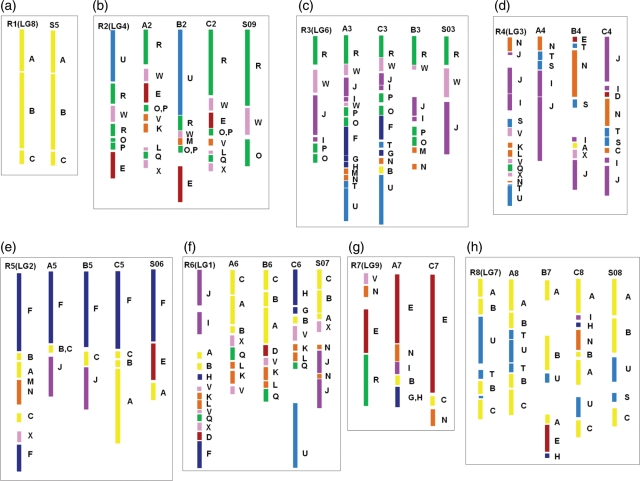
In terms of the 24 genomic blocks within the Brassicaceae ancestral karyotype, the structures of the A, B, and C genomes of Brassica species and that of S. alba have been reconstructed.5,16,24,25 The chromosome organization of R. satiuvs was compared with those of the four genomes using these genomic blocks. Eight groups were formed with the extent of homoeology between them, as shown in Fig. 3. Except for LG5 of R. sativus, in which only short regions such as W/E and O/P were shared by other chromosomes, all the chromosomes in R. sativus were inferred to have corresponding homoeologous chromosomes in other species. Large homoeologous regions could be observed in each set of homoeologous chromosomes. According to the homoeology shared between them, we assign a new nomenclature for the R. satiuvs LGs, i.e. R1, R2, R3, R4, R5, R6, R7, R8, and R9 to LG8, LG4, LG6, LG3, LG2, LG1, LG9, LG7, and LG5, respectively. Only R1 (LG8) and S05 are homoeologous to AK1, which was rearranged in the Brassica A, B, and C genomes (Fig. 3a). The structure of R2 (LG4) is nearly the same as that of B2, while the U block is not shared by A2, C2, and S09 (Fig. 3b). Since the Rapa/Oleracea lineage including R. sativs and the Nigra lineage are considered to have diverged ∼7.9 Mya,14 U/R/W/O/P/E shared by both B. nigra and R. sativus might be of a more ancestral Brassiceae chromosome. The genomic blocks in R3 (LG6) could be found in all the other homoeologous chromosomes except that I, P, and O blocks were not detected in S03 of S. alba (Fig. 3c). Block association N/S/T/I/J was conserved in all genomes except for S. alba (Fig. 3d), although rearrangements may have occurred in R. sativus, B. nigra, and B. oleracea. Block association F/C/J was shared by A5 and B5 (Fig. 3e), while F/C/B/A was conserved in R5 (LG2) and C5. Since the A, B, and C genomic blocks originally neighboured, formation of F/C/J needs more arrangement than that of A/B/C/F, indicating A/B/C/F to be more ancestral than F/C/J. Block association A/B/C was conserved in A6, B6, and S07 (Fig. 3f), and block association V/K/L/Q/X was shared by all homoeologous chromosomes except for S07. N/E block association in R7 (LG9) was found to be shared by A7 and C7 (Fig. 3g), while it was not detected in B. nigra and S. alba, revealing that it was formed after the split time between the Rapa/Oleracea lineage and the Nigra lineage. The A, B, C, T, and U genomic blocks were present in both R8 (LG7) and A8 (Fig. 3h), and A, B, U, and C blocks were also detected in C8 and S08, indicating a high syntenic relationship between them.
Figure 3.
Comparative block arrangements in the genomes of R. sativus, S. alba, and the A, B, and C genomes of Brassica species. Eight homoeologous chromosome sets [from (a) to (h)] were grouped with the extent of homoeology between them. LG5 of R. sativus is not shown, because a corresponding homoeologous chromosome in other species was not identified. The block arrangements in the A, B, and C genomes of Brassica species are based on Panjabi et al.,24 except that A3 and A8 are based on Mun et al.24 and Trick et al.,48 respectively. The block arrangements in S. alba are based on Nelson et al.25
Since the A, B, and C genomes of Brassica species are of major economic and calorific importance, contributing to a lot of world-wide edible vegetables and oil crops, great progress has been achieved in their genetics and genomics studies (www.brassica.info), especially in the A genome (http://www.brassica-rapa.org/BRGP/index.jsp). The chromosome A3 in B. rapa has been sequenced using the traditional Sanger technology.46 Recently, next-generation sequencing technology has also been used to determine the genome sequence of B. rapa, and 183 scaffolds have been anchored onto the chromosomes, the total coverage of the genome being 88.9%.47 The homoeology shared between R. sativus and the A, B, and C genomes of Brassica species indicates that genomic study of R. sativus would benefit from the rich genomic information available in Brassica species and inversely may also promote the study of Brassica species.
4. Conclusion
High frequency of SNPs and a high-efficiency SNP genotyping method developed in the present study facilitated the construction of a high-density R. sativus linkage map with EST-based SNP markers. Establishment of syntenic relationship between R. sativus and A. thaliana based on these EST markers would be greatly beneficial for identification and positional cloning of genes involved in important agronomic traits in R. sativus. Identification of the chromosome homoeology in Brassiceae species based on 24 genomic blocks of the ancestral karyotype would facilitate transfer of available genomic information between the homoeologous chromosomes to speeding up the exploration of genetic resources in Brassiceae species. Note: After this manuscript was submitted, a paper by Shirasawa et al. (An EST-SSR linkage map of Raphanus sativus and comparative genomics of the Brassicaceae) was published. LG1 (R6), LG2 (R5), LG3 (R4), LG4 (R2), LG5 (R9), LG6 (R3), LG7 (R8), LG8 (R1), and LG9 (R7) in the present study are identified to correspond to LG2, LG3, LG4, LG5, LG6, LG8, LG7, LG9, and LG1 of Shirasawa et al., respectively.
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Funding
This work was supported by the Program for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry (BRAIN), Japan.
Supplementary Material
Acknowledgements
We are grateful to Mr Uno for management of plants and Dr Ashutosh for revising a manuscript.
References
- 1.Gale M.D., Devos K.M. Plant comparative genetics after 10 years. Science. 1998;282:656–9. doi: 10.1126/science.282.5389.656. [DOI] [PubMed] [Google Scholar]
- 2.Tang H., Bowers J.E., Wang X., Ming R., Alam M., Paterson A.H. Synteny and collinearity in plant genomes. Science. 2008;320:486–8. doi: 10.1126/science.1153917. [DOI] [PubMed] [Google Scholar]
- 3.Gale M.D., Devos K.M. Comparative genetics in the grasses. Proc. Natl Acad. Sci. USA. 1998;95:1971–4. doi: 10.1073/pnas.95.5.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mandáková T., Lysak M.A. Chromosomal phylogeny and karyotype evolution in x = 7 crucifer species (Brassicaceae) Plant Cell. 2008;20:2559–70. doi: 10.1105/tpc.108.062166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schranz M., Lysak M., Mitchell-Olds T. The ABC's of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends Plant Sci. 2006;11:535–42. doi: 10.1016/j.tplants.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 6.Lagercrantz U., Lydiate D.J. Comparative genome mapping in Brassica. Genetics. 1996;144:1903–10. doi: 10.1093/genetics/144.4.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li F., Kitashiba H., Inaba K., Nishio T. A Brassica rapa linkage map of EST-based SNP markers for identification of candidate genes controlling flowering time and leaf morphological traits. DNA Res. 2009;16:311–23. doi: 10.1093/dnares/dsp020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moore G., Devos K.M., Wang Z., Gale M.D. Cereal genome evolution. Grasses, line up and form a circle. Curr. Biol. 1995;5:737–9. doi: 10.1016/s0960-9822(95)00148-5. [DOI] [PubMed] [Google Scholar]
- 9.Al-Shehbaz I.A., Beilstein M.A., Kellogg E.A. Systematics and phylogeny of the Brassicaceae (Cruciferae): an overview. Plant Syst. Evol. 2006;259:89–120. [Google Scholar]
- 10.Warwick S.I., Al-Shehbaz I.A. Brassicaceae: chromosome number index and database on CD-Rom. Plant Syst. Evol. 2006;259:237–48. [Google Scholar]
- 11.Lagercrantz U. Comparative mapping between Arabidopsis thaliana and Brassica nigra indicates that Brassica genomes have evolved through extensive genome replication accompanied by chromosome fusions and frequent rearrangements. Genetics. 1998;150:1217–28. doi: 10.1093/genetics/150.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang T.J., Kim J.S., Kwon S.J., et al. Sequence-level analysis of the diploidization process in the triplicated FLOWERING LOCUS C region of Brassica rapa. Plant Cell. 2006;18:1339–47. doi: 10.1105/tpc.105.040535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Town C.D., Cheung F., Maiti R., et al. Comparative genomics of Brassica oleracea and Arabidopsis thaliana reveal gene loss, fragmentation, and dispersal after polyploidy. Plant Cell. 2006;18:1348–59. doi: 10.1105/tpc.106.041665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lysak M.A., Koch M.A., Pecinka A., Schubert I. Chromosome triplication found across the tribe Brassiceae. Genome Res. 2005;15:516–25. doi: 10.1101/gr.3531105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lysak M.A., Cheung K., Kitschke M., Bures P. Ancestral chromosomal blocks are triplicated in Brassiceae species with varying chromosome number and genome size. Plant Physiol. 2007;145:402–10. doi: 10.1104/pp.107.104380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mun J.H., Kwon S.J., Yang T.J., et al. Genome-wide comparative analysis of the Brassica rapa gene space reveals genome shrinkage and differential loss of duplicated genes after whole genome triplication. Genome Biol. 2009;10:R111. doi: 10.1186/gb-2009-10-10-r111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho K., O'Neill C.M., Kwon S.J., et al. Sequence-level comparative analysis of the Brassica napus genome around two stearoyl-ACP desaturase loci. Plant J. 2010;61:591–9. doi: 10.1111/j.1365-313X.2009.04084.x. [DOI] [PubMed] [Google Scholar]
- 18.Lysak M.A., Lexer C. Towards the era of comparative evolutionary genomics in Brassicaceae. Plant Syst. Evol. 2006;259:175–98. [Google Scholar]
- 19.Parkin I.A., Gulden S.M., Sharpe A.G., et al. Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana. Genetics. 2005;171:765–81. doi: 10.1534/genetics.105.042093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yogeeswaran K., Frary A., York T.L., et al. Comparative genome analyses of Arabidopsis spp.: inferring chromosomal rearrangement events in the evolutionary history of A. thaliana. Genome Res. 2005;15:505–15. doi: 10.1101/gr.3436305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuittinen H., de Haan A.A., Vogl C., et al. Comparing the linkage maps of the close relatives Arabidopsis lyrata and A. thaliana. Genetics. 2004;168:1575–84. doi: 10.1534/genetics.103.022343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koch M.A., Kiefer M. Genome evolution among cruciferous plants: a lecture from the comparison of the genetic maps of three diploid species—Capsella rubella, Arabidopsis lyrata subsp. petraea, and A. thaliana. Am. J. Bot. 2005;92:761–7. doi: 10.3732/ajb.92.4.761. [DOI] [PubMed] [Google Scholar]
- 23.Lysak M.A., Berr A., Pecinka A., Schmidt R., McBreen K., Schubert I. Mechanisms of chromosome number reduction in Arabidopsis thaliana and related Brassicaceae species. Proc. Natl Acad. Sci. USA. 2006;103:5224–9. doi: 10.1073/pnas.0510791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Panjabi P., Jagannath A., Bisht N.C., et al. Comparative mapping of Brassica juncea and Arabidopsis thaliana using Intron Polymorphism (IP) markers: homoeologous relationships, diversification and evolution of the A, B and C Brassica genomes. BMC Genomics. 2008;9:113. doi: 10.1186/1471-2164-9-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nelson M.N., Parkin I.A., Lydiate D.J. The mosaic of ancestral karyotype blocks in the Sinapis alba L. genome. Genome. 2011;54:33–41. doi: 10.1139/G10-097. [DOI] [PubMed] [Google Scholar]
- 26.Bett K.E., Lydiate D.J. Genetic analysis and genome mapping in Raphanus. Genome. 2003;46:423–30. doi: 10.1139/g03-026. [DOI] [PubMed] [Google Scholar]
- 27.Kamei A., Tsuro M., Kubo N., et al. QTL mapping of clubroot resistance in radish (Raphanus sativus L.) Theor. Appl. Genet. 2010;120:1021–7. doi: 10.1007/s00122-009-1230-z. [DOI] [PubMed] [Google Scholar]
- 28.Tsuro M., Suwabe K., Kubo N., Matsumoto S., Hirai M. Construction of a molecular linkage map of radish (Raphanus sativus L.), based on AFLP and Brassica-SSR markers. Breed. Sci. 2005;55:107–11. [Google Scholar]
- 29.Budahn H., Peterka H., Mousa M.A., Ding Y., Zhang S., Li J. Molecular mapping in oil radish (Raphanus sativus L.) and QTL analysis of resistance against beet cyst nematode (Heterodera schachtii) Theor. Appl. Genet. 2009;118:775–82. doi: 10.1007/s00122-008-0937-6. [DOI] [PubMed] [Google Scholar]
- 30.Tsuro M., Suwabe K., Kubo N., Matsumoto S., Hirai M. Mapping of QTLs controlling root shape and red pigmentation in radish, Raphanus sativus L. Breed. Sci. 2008;58:55–61. [Google Scholar]
- 31.Kaplinski L., Andreson R., Puurand T., Remm M. MultiPLX: automatic grouping and evaluation of PCR primers. Bioinformatics. 2005;21:1701–2. doi: 10.1093/bioinformatics/bti219. [DOI] [PubMed] [Google Scholar]
- 32.Shiokai S., Kitashiba H., Nishio T. Prediction of the optimum hybridization conditions of dot-blot-SNP analysis using estimated melting temperature of oligonucleotide probes. Plant Cell Rep. 2010;29:829–34. doi: 10.1007/s00299-010-0867-z. [DOI] [PubMed] [Google Scholar]
- 33.Murray M.G., Thompson W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–5. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fujii H., Ogata T., Sugiyama A., et al. Development of software to improve the design efficiency of theintron-spanning PCR primer from EST utilizing genomic information of Arabidopsis thaliana. Abstracts of the International Conference ‘Plant & Animal Genomes XVIII Conference’; San Diego, CA, USA: Sherago International, Inc.; 2010. [Google Scholar]
- 35.Schranz M.E., Quijada P., Sung S.B., Lukens L., Amasino R., Osborn T. C. Characterization and effects of the replicated flowering time gene FLC in Brassica rapa. Genetics. 2002;162:1457–68. doi: 10.1093/genetics/162.3.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanger F., Donelson J.E., Coulson A.R., Kossel H., Fischer D. Use of DNA polymerase I primed by a synthetic oligonucleotide to determine a nucleotide sequence in phage fl DNA. Proc. Natl Acad. Sci. USA. 1973;70:1209–13. doi: 10.1073/pnas.70.4.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shiokai S., Shirasawa K., Sato Y., Nishio T. Improvement of the dot-blot-SNP technique for efficient and cost-effective genotyping. Mol. Breed. 2010;25:179–85. [Google Scholar]
- 38.Doyle J.J., Doyle J.L. Isolation of plant DNA from fresh tissue. Focus. 1990;12:13–15. [Google Scholar]
- 39.Markham N.R., Zuker M. DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res. 2005;33:W577–81. doi: 10.1093/nar/gki591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Ooijen J.W. JoinMap 4.0, Software for the Calculation of Genetic Linkage Maps in Experimental Populations. Wageningen, The Netherlands: Kyazma, B.V.; 2006. [Google Scholar]
- 41.Kosambi D.D. The estimation of map distance from recombination values. Ann. Eugen. 1994;12:172–75. [Google Scholar]
- 42.Voorrips R.E. MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002;93:77–8. doi: 10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
- 43.Marie D., Brown S.C. A cytometric exercise in plant DNA histograms, with 2C values for 70 species. Biol. Cell. 1993;78:41–51. doi: 10.1016/0248-4900(93)90113-s. [DOI] [PubMed] [Google Scholar]
- 44.Li F., Kitashiba H., Nishio T. Association of sequence variation in Brassica GLABRA1 orthologs with leaf hairiness. Mol. Breed., 2010 doi:10.1007/s11032-010-9508-z (Online) [Google Scholar]
- 45.Schranz M.E., Windsor A.J., Song B.H., Lawton-Rauh A., Mitchell-Olds T. Comparative genetic mapping in Boechera stricta, a close relative of Arabidopsis. Plant Physiol. 2007;144:286–98. doi: 10.1104/pp.107.096685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mun J.H., Kwon S.J., Seol Y.J., et al. Sequence and structure of Brassica rapa chromosome A3. Genome Biol. 2010;11:R94. doi: 10.1186/gb-2010-11-9-r94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y., Sun S., Liu B., et al. A sequence-based genetic linkage map as a reference for Brassica rapa pseudochromosome assembly. BMC Genomics. 2011 doi: 10.1186/1471-2164-12-239. doi:10.1186/1471-2164-12-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trick M., Kwon S.J., Choi S.R., et al. Complexity of genome evolution by segmental rearrangement in Brassica rapa revealed by sequence-level analysis. BMC Genomics. 2009;10:539. doi: 10.1186/1471-2164-10-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.