Abstract
Uridine phosphorylase (UPP) catalyzes the reversible conversion of uridine to uracil and ribose-1-phosphate and plays an important pharmacological role in activating fluoropyrimidine nucleoside chemotherapeutic agents such as 5-fluorouracil and capecitabine. Most vertebrate animals, including humans, possess two homologues of this enzyme (UPP1 & UPP2), of which UPP1 has been more thoroughly studied and is better characterized. Here, we report two crystallographic structures of human UPP2 (hUPP2) in distinctly active and inactive conformations. These structures reveal that a conditional intramolecular disulfide bridge can form within the protein that dislocates a critical phosphate-coordinating arginine residue (R100) away from the active site, disabling the enzyme. In vitro activity measurements on both recombinant hUPP2 and native mouse UPP2 confirm the redox sensitivity of this enzyme, in contrast to UPP1. Sequence analysis shows that this feature is conserved among UPP2 homologues and lacking in all UPP1 proteins due to the absence of a necessary cysteine residue. The state of the disulfide bridge has further structural consequences for one face of the enzyme that suggest UPP2 may have additional functions in sensing and initiating cellular responses to oxidative stress. The molecular details surrounding these dynamic aspects of hUPP2 structure and regulation provide new insights as to how novel inhibitors of this protein may be developed with improved specificity and affinity. As uridine is emerging as a promising protective compound in neuro-degenerative diseases, including Alzheimer’s and Parkinson’s, understanding the regulatory mechanisms underlying UPP control of uridine concentration is key to improving clinical outcomes in these illnesses.
Keywords: Uridine phosphorylase, Redox regulation, Protein structure, Conformational change, 5-Benzylacyclouridine, X-ray crystallography
1. Introduction
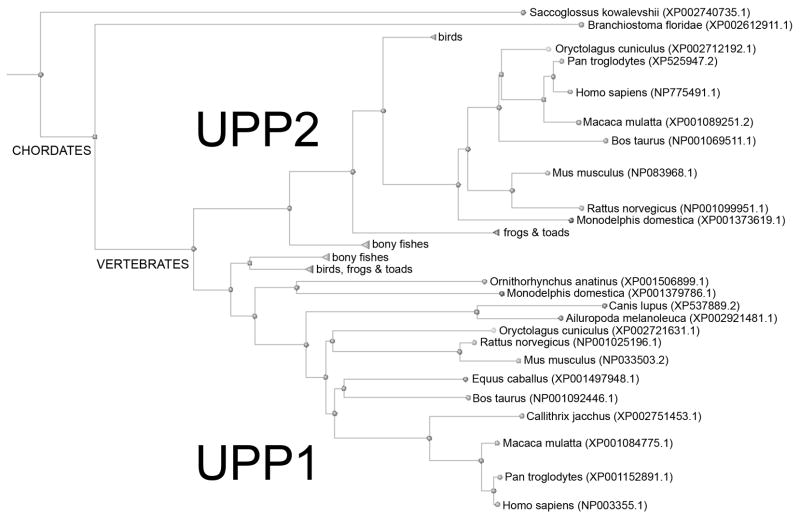
Uridine phosphorylase (UPP; EC 2.4.2.3) catalyzes the reversible phosphorolysis of uridine and chemically-related compounds to uracil and ribose-1-phosphate, implicating it in both pyrimidine salvage and regulation of uridine homeostasis (Cappiello et al., 1998; Cao and Pizzorno, 2004; Tozzi et al., 2006). It is a member of the nucleoside phosphorylase-1 super-family of proteins, but shares a degree of substrate overlap with thymidine phosphorylase (TYMP). The enzyme is ubiquitously distributed amongst animals and bacteria, with most vertebrates possessing two versions of the phosphorylase, UPP1 (Watanabe and Uchida, 1995) and UPP2 (Johansson, 2003). Phylogentic analysis suggests that this duplication was an early event in vertebrate evolution and has been followed by sporadic loss of UPP2 in some organisms (Fig. 1). While the two human homologues retain ~62% sequence identity, human UPP1 (hUPP1) was the first identified, cloned, and characterized, and consequently has been the focus of the majority of research (Liu et al., 1998; Russell et al., 2001; Cao et al., 2002, 2005). These investigations of hUPP1 function were initially spurred by the discovery of its central role in the activation of fluorinated pyrimidine nucleoside analogues, such as 5-fluorouracil (5-FU), used in cancer chemotherapy (Peters et al., 1986). Subsequently it was shown that certain tumors have increased levels of hUPP1 activity, possibly contributing to the anti-neoplastic tissue selectivity of fluoropyrimidines (Liu et al., 1998; Kawamura et al., 2006). Other research has explored the ability of UPP inhibitors to raise cellular uridine concentrations to mitigate the toxic effects of fluoropyrimidines on healthy tissues during treatment (Chu et al., 1984; Al Safarjalani et al., 2006). Molecules such as 5-benzylacyclouridine (BAU) (Niedzwicki et al., 1982) that selectively block hUPP activity have been analyzed as a means to increase the maximum tolerated dosage and therapeutic index of 5-FU by boosting uridine levels for cyto-protection when properly sequenced (Pizzorno et al., 1998).
Fig. 1.
Phylogenetic tree of all known vertebrate UPP sequences. The presence of two homologues of UPP appears to have arisen early in vertebrates and is retained in most mammals, though some individual animals seem to have subsequently lost UPP2.
Not as much is known about the specific role of human UPP2 (hUPP2). Early studies showed that its tissue distribution was more limited than that of hUPP1, restricted predominantly to the kidneys, with some expression in the liver and spleen (Johansson, 2003). In contrast, mouse UPP2 (mUPP2) is found highly expressed in the liver and to a lesser amount in the kidneys. More recently, mUPP2 was shown to be a target of regulation by several hepatic nuclear receptor agonists including liver X receptors, farnesoid X receptor, and peroxisome proliferator-activated receptor-α (Zhang et al., 2004; Kong et al., 2009). These findings imply an unexpected role for UPP2 in hepatic lipogenesis or cholesterol transport. Further it was discovered that both the genes for hUPP2 and mUPP2 contain hepatic nuclear factor-4α (HNF-4α) responsive expression elements. Given HNF-4α’s essential role in the expression of genes involved in various hepatic metabolic functions, these findings further suggest a possible connection between uridine and lipid metabolism via UPP2.
Our understanding of the fundamental catalytic mechanisms underlying UPP activity was founded on the structural analysis of bacterial UPPs, first with E. coli UPP (Morgunova et al., 1995; Burling et al., 2003; Caradoc-Davies et al., 2004; Bu et al., 2005) followed by research on the closely-related S. typhimurium homologue (Dontsova et al., 2005; Lashkov et al., 2009, 2010). Only recently have multiple structures of the human enzyme, hUPP1 (Roosild et al., 2009; Roosild and Castronovo, 2010), its bovine homologue, bUPP1 (Paul et al., 2010), and a UPP from the parasitic protozoa, Trypanosoma brucei (Larson et al., 2010), revealed key differences between prokaryotic and eukaryotic variations of this enzyme. Most strikingly, eukaryotic enzymes have dimeric biological assemblies while prokaryotic proteins are hexameric. This difference in quaternary structure appears to enhance the conformational flexibility of the former, increasing the dynamic mobility of the enzyme’s domains relative to each other (Roosild et al., 2009). It may also have important implications regarding the formation of protein-protein interactions with other cellular elements, as the eukaryotic enzymes have substantially more exposed surface area per amino acid chain as a result of the dissolution of the bacterial trimer-of-dimers assembly interface.
Here, we present two high resolution crystallographic structures of hUPP2 bound to the classical inhibitor BAU. These structures reveal a novel redox-dependent inactivation mechanism for the enzyme through the formation of an intramolecular disulfide bridge that distorts the positioning of an arginine residue critical for catalysis of uridine phosphorylysis. Corroborating activity assays confirm the redox sensitivity of UPP2 family proteins, which conserve the cysteine residues underlying this mechanism. The elucidation of this regulatory mechanism has important implications toward understanding the function of this second UPP homologue in humans. Additionally, it suggests new approaches for designing inhibitors of this protein with improved selectivity and efficacy. These insights regarding the structural dynamics of hUPP2 also implicate the protein as a potential cellular sensor of oxidative stress and suggest the possibility of new relationships between uridine degradation and lipid metabolism.
2. Materials and methods
2.1. Protein production and purification
A construct for the recombinant expression of hUPP2 in E. coli was designed based on the domain boundaries observed in the structures of hUPP1 (Roosild et al., 2009). This included replacement of the first twenty residues of hUPP2 with the cloning artifact ‘MKHHHHHHHHGGLVPRGSS’ providing an N-terminal, thrombin-cleavable, metal affinity chromatography purification tag. Three C-terminal residues, ‘LCD’, were also omitted. This construct was made in pExpress411 plasmid (DNA 2.0) codon-optimized for bacterial expression. Production and isolation of hUPP2 were conducted following standard laboratory protocols for recombinant bacterial protein expression and purification. In brief, freshly transformed BL21(DE3) E. coli colonies were cultured in Terrific Broth and induced with 0.1 mM isopropyl-β-D-thiogalactopyranoside at an O.D. of 1.0. Growth was continued overnight at 18 °C. Cells were harvested and resuspended in 50 mM Tris buffer pH 8.0, 300 mM KCl, 10% glycerol with 20 mM imidazole. The bacteria were then disrupted by sonication on ice and membranes with other insoluble material were pelleted by high speed centrifugation (100,000x g). Recombinant hUPP2 was subsequently purified from the resulting supernatant using Ni-NTA affinity chromatography and batch eluted with 500 mM imidazole added to the sonication buffer above. Following overnight digestion with thrombin at 4 °C, further purification was conducted using gel filtration chromatography over Superdex 200 resin equilibrated in 300 mM KCl, 50 mM Tris buffer pH 8.0 with 1 mM Tris (2-carboxy-ethyl) phosphine or 1 mM dithiothreitol (DTT). Typical yields of protein exceed 20 mgs per liter of culture. The final sample was verified to be homogenous by SDS-PAGE experiments and used directly for crystallization. For enzymatic activity assays, hUPP1 was produced as previously reported (Roosild et al., 2009).
2.2. Crystallization
Purified hUPP2 at 6 mgs/ml was supplemented with 1 mM BAU and subjected to crystallization screening using the JCSG+ suite (Qiagen) in both room temperature and cold room conditions. Large clusters of thin rod-shaped crystals, from which individual crystals could be carefully extracted, formed in conditions optimized to 2 M ammonium sulfate, 100 mM Bis-Tris buffer pH 5.5, at 4 °C. Thick, diamond-shaped crystals were grown in condition #76 of the JCSG+ suite (20% polyvinylpyrrolidone K15, 100 mM Co(II)Cl2, 100 mM Tris buffer pH 8.5) at 4 °C, but proved recalcitrant to improvement through optimization using stock solutions. Therefore, crystals grown directly from screening were used for diffraction analysis. Crystals were frozen by submersion in liquid nitrogen after a few seconds incubation in cryoprotectant containing the crystallization constituents supplemented with 25% glycerol.
2.3. Data collection/processing and structure determination
Data was collected at SSRL beamline 7-1 as summarized in Table 1. A complete, high quality dataset to 2.0 Å resolution was collected for the rod-shaped crystals of C2 space group. The diamond-shaped crystals diffracted to better than 1.5 Å resolution, resulting in a high quality 1.54 Å dataset of P43212 symmetry. Diffraction data was processed and reduced by the HKL2000 package with Denzo and Scalepack (Otwinowski and Minor, 1997). Molecular replacement phasing of the data was successful through Molrep (CCP4, 1994) using homology models of hUPP2 monomers based on the structure of hUPP1 with BAU as a search model (PDB ID 3EUF) (Roosild et al., 2009). Solution phases were sufficient to resolve unambiguous density for unmodeled BAU ligands (one per protein chain) and a sulfate/phosphate ion in the active site of each protein chain in the rod shaped crystals. Rounds of model building and refinement were performed using Coot (Emsley and Cowtan, 2004) and Refmac (CCP4, 1994). For both structures there was a lack of electron-density for the residual N-terminal cloning artifact residues ‘GSS’, and additional N-terminal residues ‘G’ (diamond) or ‘GKR’ (rod). For four of six protein chains in the rod crystals, the electron-density map covering residues 85–90 lacked adequate density for structural modeling of this region. The final structures were refined with Refmac to an Rfactor/Rfree of 20.7%/24.2% (rod) or 19.2%/21.2% (diamond), with approximately 91% of residues in most favorable regions of the Ramachandran plot as analyzed by Procheck (Laskowski et al., 1993). The models were further validated using Molprobity (Davis et al., 2007), both scoring in the 97th percentile. Structure figures were rendered using ICM Browser-Pro (Molsoft) or LigPlot (Wallace et al., 1995). The atomic coordinates and structure factors (PDB IDs 3P0E & 3P0F) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics (http://www.rcsb.org/).
Table 1.
Crystallographic data collection and model refinement statistics
| Active hUPP2 | Inactive hUPP2 | |
|---|---|---|
| Diffraction Data: | ||
| Source | SSRL 7-1 | SSRL 7-1 |
| λ | 0.98 Å | 0.98 Å |
| Space Group | C2 | P43212 |
| Cell constants | a=275.34 Å | a=59.70 Å |
| b=56.14 Å | b=59.70 Å | |
| c=141.39 Å | c=189.31 Å | |
| β=96.4° | ||
| Mosacity | 0.5 | 0.3 |
| Resolution | 50-2.00 Å (2.07-2.00 Å) | 50-1.54 Å (1.60-1.54 Å) |
| Rsym | 15.3% (40.0%) | 5.5% (38.0%) |
| I/σ | 9.1 (1.5) | 13.2 (1.8) |
| Completeness | 92.4% (54.3%) | 99.1% (93.1%) |
| Model Refinement: | ||
| Number of reflections | 134248 | 51304 |
| Number of monomers/A.U. | 6 | 1 |
| Atoms/A.U. | 14052 | 2632 |
| Protein | 13543 | 2305 |
| Ligand | 150 | 23 |
| Water | 359 | 304 |
| Rcryst | 20.7% | 19.2% |
| Rfree | 24.2% | 21.2% |
| Rmsd bond lengths | 0.015 Å | 0.013 Å |
| Rmsd bond angles | 1.48° | 1.38° |
| Ramachandran statistics | ||
| Most favored regions | 91.1% | 91.1% |
| Additionally allowed regions | 8.9% | 8.9% |
| Molprobity Score (percentile) | 1.49 (97%) | 1.21 (97%) |
| PDB ID | 3P0E | 3P0F |
2.4. In vitro hUPP2 activity assay
The catalytic activity of hUPP2 was determined by measuring the absorbance change at 280 nm caused by hydrolysis of uridine to uracil (ΔA = 2100 M−1cm−1) as described for hUPP1 (Renck et al., 2010) using a Perkin Elmer Lambda 650 UV-Vis spectrophotometer. All assays were performed at 25 °C, in a buffer of 50 mM Tris pH 8.0, 300 mM KCl, and at a protein concentration of 40 μg/ml. Uridine and inorganic phosphate were provided just prior to UV recording at concentrations of 0.3 mM and 1 mM, respectively. The averages of three independent measurements are reported.
2.5. Native mouse liver UPP2 activity assay
Livers from TYMP/UPP1 double knockout mice (López et al., 2009) were isolated and flash frozen in liquid nitrogen. Tissues were homogenized in 50 mM Tris-HCl (pH 7.5) without DTT, using a tissuemizer Ultra-Turrax (IKA). UPP2 activity was measured by monitoring conversion of [3H]uridine to [3H]uracil using TLC chromatographic separation as previously described (Liu et al., 1998) with the following modifications. Where indicated, Tris-HCl tissue lysates were prepared and handled in a glove box saturated with high purity nitrogen gas to maintain an anaerobic environment. All aqueous buffer solutions were also cleared of dissolved oxygen by bubbling nitrogen gas through them for 30 minutes prior to use. Tissue homogenates were cleared by centrifugation at 14,000 RPM for 15 minutes at 4 °C. The UPP2 reactions were separated on silica TLC plates using an 85:15:5 mixture of chloroform, methanol, and acetic acid, respectively. The mean values of 8–16 measurements are shown.
3. Results
3.1. hUPP2 structure
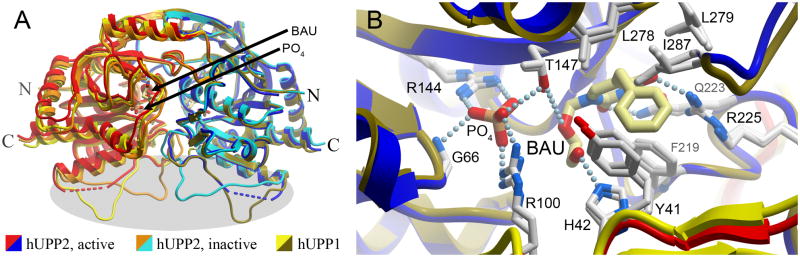
Recombinant hUPP2 purified from E. coli was crystallized under two different conditions producing rod-shaped (C2) and diamond-shaped (P43212) crystals (Table 1). Both structures were solved through Molecular Replacement phasing using monomeric homology models of hUPP2 derived from the structure of hUPP1 with bound BAU (PDB ID 3EUF). Globally, as expected, both structures of hUPP2 are highly similar to each other and hUPP1 (Fig. 2A), including retention of a dimeric biological assembly (even though one crystal form possesses three dimers in the asymmetric unit while the other has only half of a dimer, with the reciprocal partnering chain on a crystallographic two-fold symmetry axis). Overall backbone Cα root mean square deviations between the two structures or in comparison with hUPP1 are less than 0.7 Å, excluding a loop region comprising amino acids 83–102 (77–96 in hUPP1). Analysis of the electron density distribution at the enzyme’s active site reveals density in composite omit maps consistent with bound BAU for both structures and for phosphate (or sulfate) ion only in the C2 crystals. Close comparison of the active sites of the two enzymes reveals further the remarkably strict retention of both residue identities and positions in this part of the protein (Fig. 2B, 3). This is consistent with the highly similar kinetic characterization of both enzymes for uridine reported earlier: Km 189 μM vs. 76 μM and Vmax/Km 0.95 vs. 1.13 for hUPP1 compared to hUPP2 (Liu et al., 1998; Johansson, 2003). In fact, every residue in contact with either the BAU inhibitor or the phosphate ligand is identical in the two homologues. That even the hydrophobic residues that interact with the benzyl moiety of BAU, but don’t form contacts with natural substrates (L278, L279, and I287), are conserved both in identity and position strongly suggest that this inhibitor binds both proteins with equal affinity. Notably, the diamond-shaped crystals diffracted X-rays particularly well (beyond 1.5 Å), in part due to the presence of two surface-exposed cobalt binding sites, one of which is formed at an artificial crystal contact point between two hUPP2 dimers (Fig. S1). There is currently no evidence that either of these positions have metal affinity in their native environments, but rather were likely induced by the high concentration of Co(II)Cl2 (100 mM) used in the crystallization buffer.
Fig. 2.
Comparison of the structure of hUPP2 with hUPP1. (A) Overlay of two alternate structures of hUPP2 bound to BAU with the structure of hUPP1 bound to BAU (PDB ID 3EUF) reveals the strict retention of both overall fold and all secondary structural elements. The position of the inhibitor and binding site for inorganic phosphate proximate to the dimer interface are indicated. For this figure, the right-side monomers of the dimeric enzymes (blue, turquoise, gold) were least-squares aligned producing slightly greater deviations in the backbone traces of the partnering chains, consistent with inherent interdomain flexibility within these proteins (Roosild et al., 2009; Roosild and Castronovo, 2010). However, substantial variation in the main chain conformation is limited to a single, surface-exposed loop on one face of the enzyme (grey highlight). (B) Comparison of the active site structures of hUPP2 and hUPP1 illustrates strict retention of both the identity and positioning of every residue that contacts the enzymes’ substrates. Amino acid labels correspond to the hUPP2 primary sequence with the equivalent hUPP1 residue number being six less.
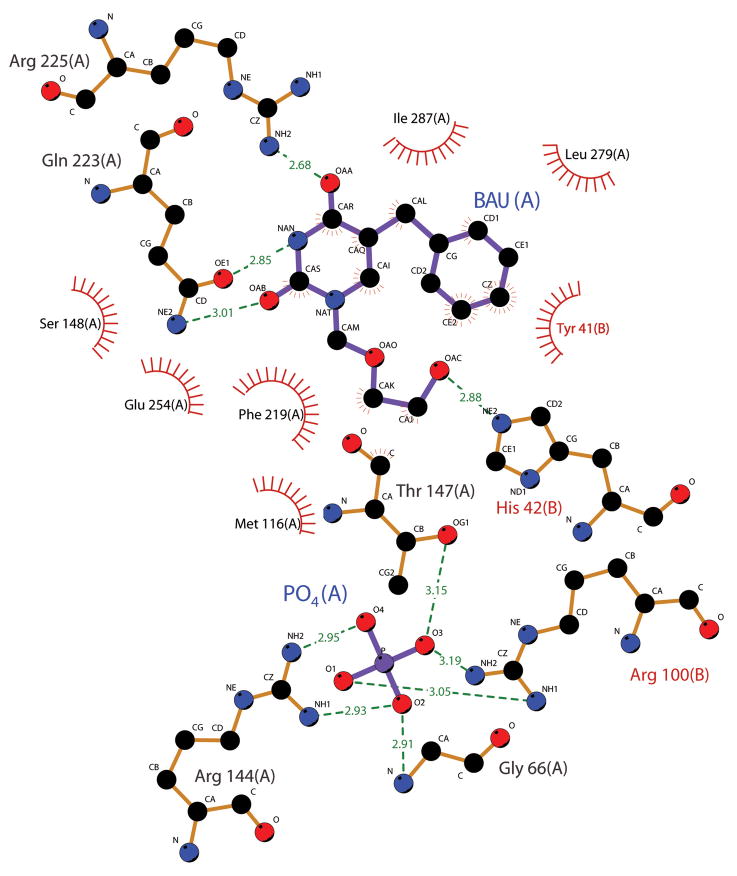
Fig. 3.
Depiction of hUPP2 ligand-interacting residues. Residues forming energetically favorable interactions with either the inhibitor BAU or the phosphate substrate are illustrated using Ligplot (Wallace et al., 1995). All residues shown are strictly conserved in identity and position between hUPP2 and hUPP1.
3.2. Intramolecular disulfide bridge formation in hUPP2
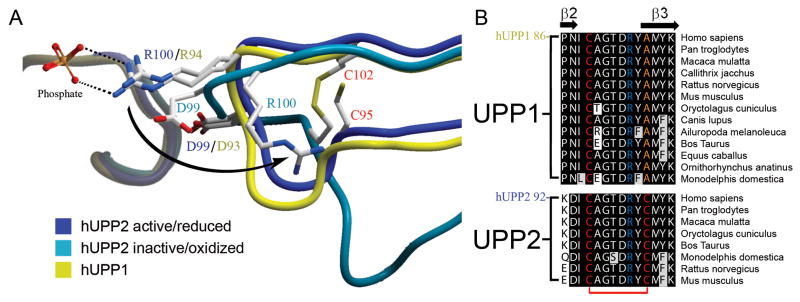
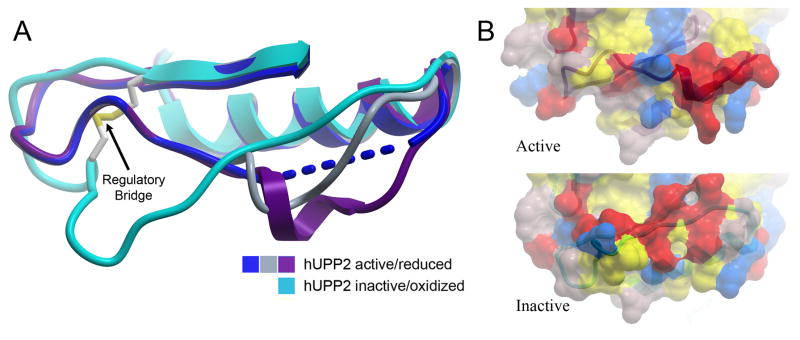
Surprisingly, the two structures of hUPP2 have dramatic differences in the positioning of an arginine residue (R100) that is critical for the coordination of phosphate in the enzyme’s active site (Fig. 4A). In the C2 crystals, this residue is in the same conformation observed for hUPP1 and other UPPs of known structure and associates with a phosphate ion in the substrate binding pocket. However, in the tetragonal crystals, R100 is displaced to a position far removed from the catalytic groove of the enzyme and the phosphate binding site is unoccupied. The repositioning of the neighboring residue, D99, closer to the active site likely further disables the protein’s capacity to bind the negatively-charged phosphate substrate. Further analysis reveals that the impetus for this conformational change stems from the formation of a disulfide bridge between residues C95 and C102 that kinks the intervening loop that harbors the catalytically important arginine. Thus, hUPP2 appears redox sensitive, assuming a catalytically active conformation when these two cysteine residues are in a reduced state, but converting to an inactive, disabled state upon oxidative formation of a covalent linkage between these two thiol-bearing residues. Remarkably, sequence analysis suggests that this mechanism of regulation is a conserved feature of UPP2 enzymes and absent from UPP1 homologues, as the second cysteine necessary for disulfide bridge formation is present only in the former family of proteins (Fig. 4B).
Fig. 4.
Redox inactivation of hUPP2. (A) Alternate structures of hUPP2 reveal two different conformations for the loop bearing a critical phosphate coordinating residue (R100). In one case, the structure is virtually identical to that seen in hUPP1 and other uridine phosphorylases (blue vs. yellow). However, in the second case, the formation of a disulfide bridge between C95 and C102 (red) contorts the shape of this loop and twists the key arginine residue away from the active site (turquoise). Additionally, an acidic residue (D99) is shifted closer to the phosphate binding site, which is unoccupied in this structure. These structural changes suggest that this is a catalytically incompetent conformation of this enzyme. (B) Alignment of all known mammalian UPP sequences shows that the second of the cysteine residues needed for disulfide bridge formation is invariably conserved among UPP2 proteins, but absent from all UPP1 enzymes.
3.3. Oxidative inactivation of hUPP2
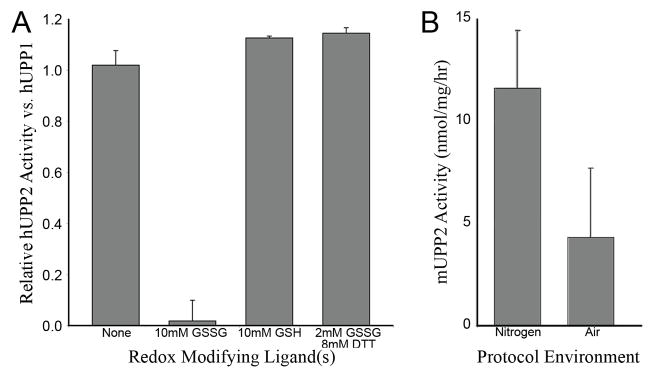
To confirm the redox sensitivity of hUPP2, the in vitro activity of recombinant hUPP2 and hUPP1 was compared in the presence of redox modifying chemicals. Enzyme activity was assayed by monitoring the rate of change of sample absorbance at 280 nm resulting from the conversion of uridine to uracil in the presence of enzyme and inorganic phosphate. hUPP2 activity was rapidly reduced upon exposure to oxidized glutathione (GSSG) to undetectable levels (Fig. 5A) while hUPP1 activity remained essentially unchanged by this treatment. In contrast, relative to hUPP1, hUPP2 activity was slightly increased upon exposure to reduced gluthione (GSH), probably due to the reactivation of a small portion of the enzyme that had become oxidized during the process of purification. Co-administration of DTT with GSSG completely protected enzyme activity, suggesting that the inhibition produced by GSSG results from its oxidation of hUPP2 and not an alternative substrate-competitive mechanism. Further corroboration of the redox regulation of UPP2 was sought through analysis of native mouse UPP2. The livers of TYMP/UPP1 double knock-out mice, in which the only known remaining uridine phosphorolytic enzyme is UPP2, were assessed for uridine catabolism under atmospheric and anaerobic conditions (Fig. 5B). Uridine phosphorylase activity was nearly double in those liver samples protected from oxygen exposure.
Fig. 5.
Redox sensitivity of UPP2. (A) The relative activity of recombinant hUPP2, normalized to hUPP1, upon exposure to various redox modifying compounds is graphed. hUPP2 activity is inactivated upon exposure to oxidized glutatione (GSSG) and slightly enhanced by reduced glutathione (GSH). Other reducing agents, such as dithiothreitol (DTT), can protect hUPP2 from GSSG inactivation, suggesting the observed effect is due to its redox properties and not through some other mechanism of molecular inhibition. (B) The measured activity of native mouse UPP2 is graphed, varying the composition of atmosphere used during enzyme isolation from liver homogenates and subsequent activity analysis. These results obtained with native enzyme confirm the oxidation sensitivity of this family of proteins.
4. Discussion
Human uridine phosphorylase activity has been extensively studied by clinical researchers due to its role in activating chemotherapeutic fluoropyrimidine nucleosides (Peters et al., 1986; Cao and Pizzorno, 2004) and maintaining equilibrated nucleotide and nucleoside pools for DNA replication and repair (López et al., 2009). More recently it has been shown that most higher organisms, including humans, possess two versions of this enzyme, although the evolutionary advantage imparted by this duplicity is unknown. Our structural analysis of hUPP2 shows that there is no measurable difference in the active sites of the two enzymes that would generate a meaningful alteration in substrate specificity or catalytic mechanism between the two homologues. Thus more likely, the benefit to the organism stems from differences in the expression of the two genes and regulation of the proteins they yield. To this end, we have identified a novel mechanism of regulation of UPP2 that involves conditional, redox-dependent formation of a disulfide bridge between two cysteine residues that bracket a loop bearing a catalytically-critical substrate-coordinating residue. Formation of this covalent linkage imparts a conformational distortion upon the intervening loop that disables phosphate binding by the enzyme and, in turn, its ability to generate uracil from uridine. The redox sensitivity of UPP2 was confirmed both in recombinant hUPP2 and in native mUPP2 through activity assays and appears to be a conserved feature of UPP2 family proteins. This result has important implications for other research on UPP2, as it suggests the possibility that measured activities of this enzyme may be underestimated if adequate care to minimize oxygen exposure is not taken. Less clear is the evolutionary advantage imparted by the inactivation of UPP activity under conditions of oxidative stress. Recently, a link has been discovered between UPP2 and hepatic lipogenesis, based on the enzyme’s expression being under the control of HNF-4α, a central regulator of hepatic metabolism (Zhang et al., 2004; Kong et al., 2009). Given also that a crucial metabolic end product of uridine catabolism is β-alanine, a precursor to fatty acid synthesis, it may be that disruption of the supply of uridine metabolites may mitigate cellular damage upon exposure to oxidative stress by moderating concurrent lipid biosynthesis. Alternately, tissue uridine concentrations can be elevated by blocking UPP activity (Chu et al., 1984) and uridine has been shown to have cyto-protective properties (Choi et al., 2006, 2008). Thus raising endogenous uridine levels in reaction to a redox imbalance may be a general cellular defensive response and possibly not limited to liver tissue. Notably, emerging discoveries that uridine has the ability to increase the size of neurites (Chorna et al., 2009; Pooler et al., 2009), accelerate phosphatide synthesis (Richardson et al., 2003; Wurtman et al., 2006) and increase the levels of the neurofilament proteins NF-M and NF-70 (Pooler et al., 2009), appear to coincide with the limited tissue distribution of UPP2. To investigate these possibilities, further studies will need to be undertaken which can assess the redox state of intracellular UPP2 in vivo and quantitatively measure the changes that manifest in response to various growth and stress signals.
While clearly UPP2 possesses the same catalytic capacity as UPP1, it is intriguing to consider whether UPP2 may serve additional functions beyond its enzymatic activity. The conditional formation of the regulatory disulfide bridge has long-range consequences for the structure of the lengthy loop preceding it (Fig. 6A). This region of the protein is inherently the most mobile, and in the active structure of hUPP2, four of six protein chains have portions too disordered for modeling. However, in the two remaining domains, crystallographic contacts stabilize and induce helical secondary structure in this region. This architecture is vastly different from that observed in the inactivated structure, where this loop is stretched and elongated. Thus, the redox state of the two cysteine residues imparts dramatic structural changes to an extended region of the surface of hUPP2 (Fig. 6B). Given also the dimeric nature of the enzyme and the fact that the reciprocal loops in question lie on the same face of the biological assembly (Fig. 2A), it is easy to envision the redox dependent association of hUPP2 with other protein elements. Conceivably, hUPP2 may act as a sensor for oxidative stress, dissociating from and releasing a responsive regulatory element as a consequence of oxidation.
Fig. 6.
Long-range structural consequences of the state of the regulatory disulfide bridge. (A) Of six independent protein chains in the asymmetric unit of crystals of active hUPP2, four are too disordered for modeling in part of the loop region preceding the disulfide bridge (blue). The remaining two chains reveal partially helical structure (grey & purple), due to stabilization through protein-protein contacts with crystallographic-symmetry-related domains. These conformations differ substantially from the redox inactivated structure of hUPP2, in which this region of the protein is flat and elongated (turquoise). (B) Surface representation of the same region, colored by polarity and comparing active and inactive structures, illustrates the degree to which this face of the enzyme is altered, depending upon its redox state.
The nature of the disulfide bridge inactivation mechanism discovered in UPP2 may hold additional lessons for identifying oxidation sensitive regulatory elements in other enzymes, or engineering similar systems in artificial proteins. While thiol-mediated oxidation-regulated enzymes are well documented, most of these proteins harbor catalytic cysteines that must be reduced for activity (Brandes et al., 2009). Examples of where disulfide bridges form and break for enzyme regulation are scarcer and typically still involve one cysteine residue of the pair being catalytically necessary, such as in the case of Cdc25B phosphatase (Buhrman et al., 2005), but interesting parallels can be seen in recently reported studies of cyclophilin A (Motohashi et al., 2003; Gourlay et al., 2007) and the thiol-mediated dimerization of Src protein tyrosine kinases (Kemble and Sun, 2009). Mechanisms more analogous of the conditional oxidative regulation of an enzyme through transient bridging of cysteine residues proximal to those critical for catalysis, as seen here, can be found from the field of protein engineering. Redox sensitive lysozyme has been created by fashioning a disulfide bridge across the peptidoglycan binding pocket of the enzyme, restricting access of substrate to the enzyme (Matsumura and Matthews, 1989). Similar results have also been obtained for a designed, mutant form of troponin C (Grabarek et al., 1990) and bacterial malate dehydrogenase (Muslin et al., 1995). Even the application of artificial adjacent cysteine residues to create redox sensitive structural distortions that reduce enzyme activity has been successfully engineered (Park and Raines, 2001). Nevertheless, it now appears clear that nature devised this ingenious mechanism for sensing and responding to redox changes long ago.
Following the solution of the two reported structures of hUPP2, a third structure of hUPP2 was deposited in the Protein Data Bank by the Structural Genomics Consortium (Structural Genomics Consortium, 2010; PDB ID 2XRF). While crystallized in a unique trigonal space group, this structure is nearly identical to our active conformation of hUPP2, with a backbone Cα root mean square deviation of 0.6 Å. While it was co-crystallized with uridine, the active site was found to hold only uracil and a molecule of glycerol from the crystallization solution in approximately the ribose sugar binding position. The most interesting feature of this third structure is that they found a magnesium ion bound within the regulatory loop on which the critical R100 residue resides (Fig. S2). It is likely that the high concentration of magnesium chloride used for crystallization (200 mM) may have artificially driven occupation of the site by this metal, as there is no evidence of any electron density in this position in either of our hUPP2 structures. However, the fact that the protein was able to adopt a supportive geometry for metal coordination in this key region provides further corroboration of its inherent conformational flexibility. Also notable is that the loop at the back of hUPP2’s active site (residues 282–290) subtly closes around the smaller uracil molecule, relative to the bulkier BAU, exactly as extensively described for hUPP1 (Roosild and Castronovo, 2010).
This research holds important implications for the design of the next generation of UPP inhibitors. Previous investigations have molecularly targeted UPP activity to boost plasma and tissue uridine levels in order to rescue normal tissues from cytotoxic fluoronucleotides used in the treatment of neoplastic growths (Pizzorno et al., 1998; Al Safarjalani et al., 2006). The structures of hUPP2 suggest the possibility that this enzyme might be selectively inhibited over hUPP1 by chemically stabilizing the inactive conformation reported herein. It is noteworthy, that hUPP1 is often found elevated in tumor samples (Liu et al., 1998; Kawamura et al., 2006), but hUPP2 has generally been found to be absent from such cells (Johansson, 2003). Thus, if a homologue-selective inhibitor could be developed, 5-FU activation might be curtailed and uridine levels elevated to protective concentrations in healthy hUPP2-expressing cells, without reducing the activation or efficacy of the chemotherapeutic agent within cancerous, hUPP1-overexpressing tissue. Given additionally, recent reports indicating that traditional inhibitors such as BAU may also affect other enzymes, such as human aldehyde oxidase (Klecker et al., 2006), compounds with enhanced selectivity may be the key to creating improved regimens with tolerable side-effects. Beyond cancer, elevating uridine concentrations in combination with docosahexaenoic acid administration is being explored as a potential treatment for both Alzheimer’s disease (Holguin et al., 2008) and Parkinson’s disease (Cansev et al., 2008). Uridine has also been found to be cyto-protective in astrocytes exposed to metabolic stresses, such as ischemia (Choi et al., 2006, 2008). Thus, development of selective inhibitors, capable of discriminating between the two human homologues of UPP so as to raise endogenous uridine levels in a specific subset of tissues, may yield novel methods for the treatment of an array of diseases.
Supplementary Material
Cobalt coordination in the inactive hUPP2 structure. Residues coordinating cobalt (II) ions in the crystals of inactive hUPP2 are illustrated. Electron density from 2Fo-Fc maps is contoured at the sigma level indicated. (A) One cobalt ion is bound at a crystal contact formed between symmetry-related monomers from two different biological assemblies, with each domain contributing a single histidine residue, producing a 19σ peak in the 2Fo-Fc map. (B) A second cobalt ion binds to the surface of hUPP2, as revealed by a 10σ peak in the 2Fo-Fc map. Red spheres represent ordered water molecules. Bond lengths are shown in angstroms.
Magnesium binding by hUPP2. Comparison of two structures of hUPP2, both in the enzymatically-active state, reveal the potential of the regulatory loop bearing the two disulfide bridge-forming cysteine residues and the phosphate binding arginine (R100) to also bind magnesium. The metal ion is coordinated primarily through interaction with backbone carbonyl groups, but also associates with the side chain of residue D93. Bond lengths are shown in angstroms.
Acknowledgments
This work was supported by funds to TPR from the Nevada INBRE Program of the National Center for Research Resources (P20 RR-016464) and to GP from NIH-NCI (R01 CA-102121). This work was conducted in part at the Stanford Synchrotron Radiation Laboratory (SSRL) which is funded by the Department of Energy, Office of Biological and Environmental Research. We thank members of the staff of SSRL for assistance in crystallographic data collection.
Abbreviations
- UPP
uridine phosphorylase
- TYMP
thymidine phosphorylase
- hUPP1
human uridine phosphorylase 1
- 5-FU
5-fluorouracil
- BAU
5-benzylacyclouridine
- hUPP2
human uridine phosphorylase 2
- mUPP2
mouse uridine phosphorylase 2
- HNF-4α
hepatic nuclear factor-4α
- DTT
dithiothreitol
- GSSG
oxidized glutathione
- GSH
reduced glutathione
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Al Safarjalani ON, Rais R, Shi J, Schinazi RF, Naguib FN, et al. Modulation of 5-fluorouracil host-toxicity and chemotherapeutic efficacy against human colon tumors by 5-(phenylthio)acyclouridine, a uridine phosphorylase inhibitor. Cancer Chemother Pharmacol. 2006;58:692–698. doi: 10.1007/s00280-006-0213-x. [DOI] [PubMed] [Google Scholar]
- Brandes N, Schmitt S, Jakob U. Thiol-based redox switches in eukaryotic proteins. Antioxid Redox Signal. 2009;11:997–1014. doi: 10.1089/ars.2008.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu W, Settembre EC, el Kouni MH, Ealick SE. Structural basis for inhibition of Escherichia coli uridine phosphorylase by 5-substituted acyclouridines. Acta Crystallogr D Biol Crystallogr. 2005;61:863–872. doi: 10.1107/S0907444905007882. [DOI] [PubMed] [Google Scholar]
- Buhrman G, Parker B, Sohn J, Rudolph J, Mattos C. Structural mechanism of oxidative regulation of the phosphatase Cdc25B via an intramolecular disulfide bond. Biochemistry. 2005;44:5307–5316. doi: 10.1021/bi047449f. [DOI] [PubMed] [Google Scholar]
- Burling FT, Kniewel R, Buglino JA, Chadha T, Beckwith A, et al. Structure of Escherichia coli uridine phosphorylase at 2.0 Å. Acta Crystallogr D Biol Crystallogr. 2003;59:73–76. doi: 10.1107/s0907444902018929. [DOI] [PubMed] [Google Scholar]
- Cansev M, Ulus IH, Wang L, Maher TJ, Wurtman RJ. Restorative effects of uridine plus docosahexaenoic acid in a rat model of Parkinson’s disease. Neurosci Res. 2008;62:206–209. doi: 10.1016/j.neures.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D, Russell RL, Zhang D, Leffert JJ, Pizzorno G. Uridine phosphorylase (−/−) murine embryonic stem cells clarify the key role of this enzyme in the regulation of the pyrimidine salvage pathway and in the activation of fluoropyrimidines. Cancer Res. 2002;62:2313–2317. [PubMed] [Google Scholar]
- Cao D, Pizzorno G. Uridine phosophorylase: an important enzyme in pyrimidine metabolism and fluoropyrimidine activation. Drugs Today (Barc) 2004;40:431–443. doi: 10.1358/dot.2004.40.5.850491. [DOI] [PubMed] [Google Scholar]
- Cao D, Leffert JJ, McCabe J, Kim B, Pizzorno G. Abnormalities in uridine homeostatic regulation and pyrimidine nucleotide metabolism as a consequence of the deletion of the uridine phosphorylase gene. J Biol Chem. 2005;280:21169–21175. doi: 10.1074/jbc.M412343200. [DOI] [PubMed] [Google Scholar]
- Cappiello M, Mascia L, Scolozzi C, Giorgelli F, Ipata PL. In vitro assessment of salvage pathways for pyrimidine bases in rat liver and brain. Biochim Biophys Acta. 1998;1425:273–281. doi: 10.1016/s0304-4165(98)00071-3. [DOI] [PubMed] [Google Scholar]
- Caradoc-Davies TT, Cutfield SM, Lamont IL, Cutfield JF. Crystal structures of Escherichia coli uridine phosphorylase in two native and three complexed forms reveal basis of substrate specificity, induced conformational changes and influence of potassium. J Mol Biol. 2004;337:337–354. doi: 10.1016/j.jmb.2004.01.039. [DOI] [PubMed] [Google Scholar]
- Choi JW, Yoo BK, Shin CY, Ryu MK, Ryu JH, et al. Uridine prevents the glucose deprivation-induced death of immunostimulated astrocytes via the action of uridine phosphorylase. Neurosci Res. 2006;56:111–118. doi: 10.1016/j.neures.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Choi JW, Shin CY, Choi MS, Yoon SY, Ryu JH, et al. Uridine protects cortical neurons from glucose deprivation-induced death: possible role of uridine phosphorylase. J Neurotrauma. 2008;25:695–707. doi: 10.1089/neu.2007.0409. [DOI] [PubMed] [Google Scholar]
- Chorna NE, Santiago-Perez LI, Erb L, Seye CI, Neary JT, et al. P2Y receptors activate neuroprotective mechanisms in astrocytic cells. J Neurochem. 2004;91:119–132. doi: 10.1111/j.1471-4159.2004.02699.x. [DOI] [PubMed] [Google Scholar]
- Chu MY, Naguib FN, Iltzsch MH, el Kouni MH, Chu SH, et al. Potentiation of 5-fluoro-2′-deoxyuridine antineoplastic activity by the uridine phosphorylase inhibitors benzylacyclouridine and benzyloxybenzylacyclouridine. Cancer Res. 1984;44:1852–1856. [PubMed] [Google Scholar]
- Collaborative Computational Project Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dontsova MV, Gabdoulkhakov AG, Molchan OK, Lashkov AA, Garber MB, et al. Preliminary investigation of the three-dimensional structure of Salmonella typhimurium uridine phosphorylase in the crystalline state. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2005;61:337–340. doi: 10.1107/S1744309105007463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Gourlay LJ, Angelucci F, Baiocco P, Boumis G, Brunori M, et al. The three-dimensional structure of two redox states of cyclophilin A from Schistosoma mansoni. Evidence for redox regulation of peptidyl-prolyl cis-trans isomerase activity. J Biol Chem. 2007;282:24851–24857. doi: 10.1074/jbc.M702714200. [DOI] [PubMed] [Google Scholar]
- Grabarek Z, Tan RY, Wang J, Tao T, Gergely J. Inhibition of mutant troponin C activity by an intra-domain disulphide bond. Nature. 1990;345:132–5. doi: 10.1038/345132a0. [DOI] [PubMed] [Google Scholar]
- Holguin S, Martinez J, Chow C, Wurtman R. Dietary uridine enhances the improvement in learning and memory produced by administering DHA to gerbils. FASEB J. 2008;22:3938–3946. doi: 10.1096/fj.08-112425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M. Identification of a novel human uridine phosphorylase. Biochem Biophys Res Commun. 2003;307:41–46. doi: 10.1016/s0006-291x(03)01062-3. [DOI] [PubMed] [Google Scholar]
- Kawamura K, Takiguchi N, Wada A, Takenobu H, Kimura H, et al. Up-regulated expression of the uridine phosphorylase gene in human gastric tumors is correlated with a favorable prognosis. Anticancer Res. 2006;26:4647–4651. [PubMed] [Google Scholar]
- Kemble DJ, Sun G. Direct and specific inactivation of protein tyrosine kinases in the Src and FGFR families by reversible cysteine oxidation. Proc Natl Acad Sci USA. 2009;106:5070–5075. doi: 10.1073/pnas.0806117106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klecker RW, Cysyk RL, Collins JM. Zebularine metabolism by aldehyde oxidase in hepatic cytosol from humans, monkeys, dogs, rats, and mice: influence of sex and inhibitors. Bioorg Med Chem. 2006;14:62–66. doi: 10.1016/j.bmc.2005.07.053. [DOI] [PubMed] [Google Scholar]
- Kong X, Fan H, Liu X, Wang R, Liang J, et al. Peroxisome proliferator-activated receptor gamma coactivator-1alpha enhances antiproliferative activity of 5′-deoxy-5-fluorouridine in cancer cells through induction of uridine phosphorylase. Mol Pharmacol. 2009;76:854–860. doi: 10.1124/mol.109.056424. [DOI] [PubMed] [Google Scholar]
- Larson ET, Mudeppa DG, Gillespie JR, Mueller N, Napuli AJ, et al. The crystal structure and activity of a putative trypanosomal nucleoside phosphorylase reveal it to be a homodimeric uridine phosphorylase. J Mol Biol. 2010;396:1244–1259. doi: 10.1016/j.jmb.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lashkov AA, Gabdoulkhakov AG, Shtil AA, Mikhailov AM. Crystallization and preliminary X-ray diffraction analysis of Salmonella typhimurium uridine phosphorylase complexed with 5-fluorouracil. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2009;65:601–603. doi: 10.1107/S1744309109016133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lashkov AA, Zhukhlistova NE, Gabdoulkhakov AH, Shtil AA, Efremov RG, et al. The X-ray structure of Salmonella typhimurium uridine nucleoside phosphorylase complexed with 2,2′-anhydrouridine, phosphate and potassium ions at 1.86 A resolution. Acta Crystallogr D Biol Crystallogr. 2010;66:51–60. doi: 10.1107/S0907444909044175. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Moss DS, Thornton JM. Main-chain bond lengths and bond angles in protein structures. J Mol Biol. 1993;231:1049–1067. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
- Liu M, Cao D, Russell R, Handschumacher RE, Pizzorno G. Expression, characterization, and detection of human uridine phosphorylase and identification of variant uridine phosphorolytic activity in selected human tumors. Cancer Res. 1998;58:5418–5424. [PubMed] [Google Scholar]
- López LC, Akman HO, García-Cazorla A, Dorado B, Martí R, et al. Unbalanced deoxynucleotide pools cause mitochondrial DNA instability in thymidine phosphorylase-deficient mice. Hum Mol Genet. 2009;18:714–722. doi: 10.1093/hmg/ddn401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura M, Matthews BW. Control of enzyme activity by an engineered disulfide bond. Science. 1989;243:792–794. doi: 10.1126/science.2916125. [DOI] [PubMed] [Google Scholar]
- Morgunova EY, Mikhailov AM, Popov AN, Blagova EV, Smirnova EA, et al. Atomic structure at 2.5 Å resolution of uridine phosphorylase from E. coli as refined in the monoclinic crystal lattice. FEBS Lett. 1995;367:183–187. doi: 10.1016/0014-5793(95)00489-v. [DOI] [PubMed] [Google Scholar]
- Motohashi K, Koyama F, Nakanishi Y, Ueoka-Nakanishi H, Hisabori T. Chloroplast cyclophilin is a target protein of thioredoxin. Thiol modulation of the peptidyl-prolyl cis-trans isomerase activity. J Biol Chem. 2003;278:31848–31852. doi: 10.1074/jbc.M304258200. [DOI] [PubMed] [Google Scholar]
- Muslin EH, Li D, Stevens FJ, Donnelly M, Schiffer M, Anderson LE. Engineering a domain-locking disulfide into a bacterial malate dehydrogenase produces a redox-sensitive enzyme. Biophys J. 1995;68:2218–2223. doi: 10.1016/S0006-3495(95)80430-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedzwicki JG, Chu SH, el Kouni MH, Rowe EC, Cha S. 5-benzylacyclouridine and 5-benzyloxybenzylacyclouridine, potent inhibitors of uridine phosphorylase. Biochem Pharmacol. 1982;31:1857–1861. doi: 10.1016/0006-2952(82)90488-9. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Park C, Raines RT. Adjacent cysteine residues as a redox switch. Protein Eng. 2001;14:939–942. doi: 10.1093/protein/14.11.939. [DOI] [PubMed] [Google Scholar]
- Paul D, O’Leary SE, Rajashankar K, Bu W, Toms A, et al. Glycal formation in crystals of uridine phosphorylase. Biochemistry. 2010;49:3499–3509. doi: 10.1021/bi902073b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters GJ, Laurensse E, Leyva A, Lankelma J, Pinedo HM. Sensitivity of human, murine, and rat cells to 5-fluorouracil and 5′-deoxy-5-fluorouridine in relation to drug-metabolizing enzymes. Cancer Res. 1986;46:20–28. [PubMed] [Google Scholar]
- Pizzorno G, Yee L, Burtness BA, Marsh JC, Darnowski JW, et al. Phase I clinical and pharmacological studies of benzylacyclouridine, a uridine phosphorylase inhibitor. Clin Cancer Res. 1998;4:1165–1175. [PubMed] [Google Scholar]
- Pooler AM, Guez DH, Benedictus R, Wurtman RJ. Uridine enhances neurite outgrowth in nerve growth factor-differentiated pheochromocytoma cells. Neuroscience. 2005;134:207–214. doi: 10.1016/j.neuroscience.2005.03.050. [DOI] [PubMed] [Google Scholar]
- Renck D, Ducati RG, Palma MS, Santos DS, Basso LA. The kinetic mechanism of human uridine phosphorylase 1: Towards the development of enzyme inhibitors for cancer chemotherapy. Arch Biochem Biophys. 2010;497:35–42. doi: 10.1016/j.abb.2010.03.004. [DOI] [PubMed] [Google Scholar]
- Richardson UI, Watkins CJ, Pierre C, Ulus IH, Wurtman RJ. Stimulation of CDP-choline synthesis by uridine or cytidine in PC12 rat pheochromocytoma cells. Brain Res. 2003;971:161–167. doi: 10.1016/s0006-8993(03)02333-3. [DOI] [PubMed] [Google Scholar]
- Roosild TP, Castronovo S, Fabbiani M, Pizzorno G. Implications of the structure of human uridine phosphorylase 1 on the development of novel inhibitors for improving the therapeutic window of fluoropyrimidine chemotherapy. BMC Struct Biol. 2009;9:14. doi: 10.1186/1472-6807-9-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roosild TP, Castronovo S. Active site conformational dynamics in human uridine phosphorylase 1. PLoS One. 2010;5:e12741. doi: 10.1371/journal.pone.0012741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell RL, Cao D, Zhang D, Handschumacher RE, Pizzorno G. Uridine phosphorylase association with vimentin. Intracellular distribution and localization. J Biol Chem. 2001;276:13302–13307. doi: 10.1074/jbc.M008512200. [DOI] [PubMed] [Google Scholar]
- Structural Genomics Consortium. Human uridine phosphorylase 2 in complex with uracil. RCSB Protein Data Bank ID 2XRF. 2010 Epub Sept. 29, 2010. [Google Scholar]
- Tozzi MG, Camici M, Mascia L, Sgarrella F, Ipata PL. Pentose phosphates in nucleoside interconversion and catabolism. FEBS J. 2006;273:1089–1101. doi: 10.1111/j.1742-4658.2006.05155.x. [DOI] [PubMed] [Google Scholar]
- Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- Watanabe S, Uchida T. Cloning and expression of human uridine phosphorylase. Biochem Biophys Res Commun. 1995;216:265–272. doi: 10.1006/bbrc.1995.2619. [DOI] [PubMed] [Google Scholar]
- Wurtman RJ, Ulus IH, Cansev M, Watkins CJ, Wang L, Marzloff G. Synaptic proteins and phospholipids are increased in gerbil brain by administering uridine plus docosahexaenoic acid orally. Brain Res. 2006;1088:83–92. doi: 10.1016/j.brainres.2006.03.019. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Repa JJ, Inoue Y, Hayhurst GP, Gonzalez FJ, et al. Identification of a liver-specific uridine phosphorylase that is regulated by multiple lipid-sensing nuclear receptors. Mol Endocrinol. 2004;18:851–862. doi: 10.1210/me.2003-0285. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cobalt coordination in the inactive hUPP2 structure. Residues coordinating cobalt (II) ions in the crystals of inactive hUPP2 are illustrated. Electron density from 2Fo-Fc maps is contoured at the sigma level indicated. (A) One cobalt ion is bound at a crystal contact formed between symmetry-related monomers from two different biological assemblies, with each domain contributing a single histidine residue, producing a 19σ peak in the 2Fo-Fc map. (B) A second cobalt ion binds to the surface of hUPP2, as revealed by a 10σ peak in the 2Fo-Fc map. Red spheres represent ordered water molecules. Bond lengths are shown in angstroms.
Magnesium binding by hUPP2. Comparison of two structures of hUPP2, both in the enzymatically-active state, reveal the potential of the regulatory loop bearing the two disulfide bridge-forming cysteine residues and the phosphate binding arginine (R100) to also bind magnesium. The metal ion is coordinated primarily through interaction with backbone carbonyl groups, but also associates with the side chain of residue D93. Bond lengths are shown in angstroms.