Abstract
In the retina of adult teleosts, stem cells are sustained in two specialized niches: the ciliary marginal zone (CMZ) and the microenvironment surrounding adult Müller glia. Recently, Müller glia were identified as the regenerative stem cells in the teleost retina. Secreted signaling molecules that regulate neuronal regeneration in the retina are largely unknown. In a microarray screen to discover such factors, we identified midkine-b (mdkb). Midkine is a highly conserved heparin-binding growth factor with numerous biological functions. The zebrafish genome encodes two distinct midkine genes: mdka and mdkb. Here, we describe the cellular expression of mdka and mdkb during retinal development and the initial, proliferative phase of photoreceptor regeneration. The results show that in the embryonic and larval retina mdka and mdkb are expressed in stem cells, retinal progenitors and neurons in distinct patterns that suggest different functions for the two molecules. Following the selective death of photoreceptors in the adult, mdka and mdkb are co-expressed in horizontal cells and proliferating Müller glia and their neurogenic progeny. These data reveal that Mdka and Mdkb are signaling factors present in the retinal stem cell niches in both embryonic and mature retinas, and that their cellular expression is actively modulated during retinal development and regeneration.
Keywords: mdka, mdkb, growth factors, development, regeneration, neurogenesis, zebrafish
Introduction
Midkine is a secreted heparin binding growth factor that is highly conserved throughout the animal kingdom (Kadomatsu et al. 1988, Kadomatsu et al. 1990, Winkler et al., 2003, Kadomatsu and Muramatsu, 2004). Numerous functions have been ascribed to this molecule: neurogenic, mitogenic, neurotrophic, chemotactic, fibrinolytic and anti-apoptotic (for review see Muramatsu 2002, Kadomatsu and Muramatsu, 2004). In mammals, Midkine is expressed in many tissues during embryonic development, most prominently in the developing neural tube and at epithelial–mesenchymal boundaries (Mitsiadis et al., 1995).
In mammalian tissues injury elevates the expression of Midkine (Obama et al. 1998, Sakakima et al. 2006; Jochheim-Richter et al., 2006, Kikuchi-Horie et al., 2004; Miyashiro et al., 1998), and in zebrafish the expression of the ‘a’ paralog of Midkine is increased in regenerating heart and fin (Lien et al., 2006; Schebesta et al., 2006). These results suggest a role for Midkine in regeneration and repair of vertebrate tissues (Ohta et al. 1999). Supporting this idea, mice deficient for Midkine show decreased regenerative capacity following partial hepatectomy (Ochiai et al. 2004), and in albino rats, intravitreal injection of recombinant Midkine promotes survival of photoreceptors following light-induced injury (Unoki et al., 1994, Masuda et al. 1995).
The zebrafish genome contains two midkine genes, mdka and mdkb, encoding proteins that share 68% amino acid identity (Winkler et al., 2003). In early embryos, the two midkines have distinct patterns of expression and biological actions (Winkler and Moon 2001, Winkler et al., 2003, Schäfer et. et al. 2005, Liedtke and Winkler, 2008).
The zebrafish retina develops from cells of the anterior neural plate that form the optic cups, which by 24 hours post-fertilization (hpf) are well developed and consist of proliferating neuroepithelial cells (Varga et al., 1999, Schmitt and Dowling, 1994, Hitchcock and Raymond, 2004; Stenkamp, 2007). Neuronal differentiation begins at 28-32 hpf within a precocious patch, ventro-nasal to the optic stalk (Burrill and Easter, 1994, Hu and Easter, 1999, Schmitt and Dowling, 1994, Schmitt and Dowling, 1999). Retinal cell differentiation and lamination proceed at a fast pace in sequential waves that originate in the ventro-nasal patch and sweep dorsally and then temporally through the different layers. By 72 hpf, the retina is fully laminated and functional (Schmitt and Dowling, 1999, Easter et al. 1996, Hu and Easter, 1999).
After the initial differentiation of the retinal neuroepithelium, new neurons are added to the retina throughout the life of the animal. This neurogenesis persists in two regions, specialized niches that harbor stem cells and their immediate progeny: in the ciliary marginal zone (CMZ), at the border between differentiated retina and the iris, and in the differentiated retina, where resident stem cells give rise to a lineage of cells that exclusively generates rod photoreceptors (Raymond et al., 2006, Hitchcock et al., 2004). Within the CMZ, developmental time is spatially recapitulated, stem cells are located peripherally in the CMZ, adjacent to the iris, and progenitors with increasingly restricted competence are located more centrally. This spatial pattern is evidenced by a regionalized expression of genes that sequentially specify cellular identities (Raymond et al., 2006, see also Harris and Perron, 1998).
The teleost retina can regenerate photoreceptors and neurons in a process that generally recapitulates cellular and molecular events during late retinogenesis (Raymond et al. 2006, Otteson and Hitchcock, 2003, Hitchcock et al., 2004). Recent studies identified Müller glia as stem cells responsible for both persistent rod genesis (see above) and neuronal regeneration (Bernardos et al,, 2007, Yurco and Cameron, 2005, Kassen et al., 2007, Raymond et al., 2006, Fausett and Goldman, 2006; Fimbel et al., 2007; Thummel et al., 2008). Following the death of extant neurons, Müller glia de-differentiate, reenter the cell cycle and give rise to multipotent progenitors, which continue to proliferate, migrate and differentiate to replace the missing neurons. Genes that guide developmental neurogenesis are re-expressed in regenerative Müller glia and their progeny (Raymond et al., 2006 and the references therein). Little is known, however, about secreted signaling molecules regulating these regenerative events.
We identified mdkb in a screen for genes whose expression is regulated by the selective death and regeneration of photoreceptors. As a first step to understand the role of Midkines in the retina, we analyzed the cellular expression of mdkb and its paralog, mdka, during both retinal development and adult photoreceptor regeneration. During development, mdka is expressed in stem cells and progenitors, transiently expressed in developing Müller glia and constitutively expressed in horizontal cells. In contrast, mdkb is expressed by newly postmitotic cells and constitutively expressed by retinal ganglion and amacrine cells. During regeneration, in addition to their constitutive patterns of expression, both midkines are expressed in horizontal cells and proliferating Müller glia and their neurogenic progeny. This study describes for the first time the expression of these two secreted factors in the developing, adult and regenerating retina and establishes the foundation for future studies to investigate the function of these molecules.
Materials and Methods
Care of zebrafish and embryos
Zebrafish (Danio rerio; 6-8 months old, 2-3 cm long) were purchased from suppliers and maintained in aquaria at 28.5°C with a 10/14-hour dark/light cycle. Three strains were used: albino, wild type, and Tg(gfap:GFP)mi2000 (to identify Müller glia; a gift from Dr. Pamela Raymond). Embryos were generated by natural mating at light onset and reared at 28.5°C in embryonic rearing solution (Westerfield, 2000). Procedures for handling animals were approved by the University of Michigan’s Committee for the Use and Care of Animals.
Light treatments
Two light-lesion paradigms were used in this study. Each selectively kills photoreceptors, which induces proliferation and photoreceptor regeneration that follows a common time-course. First, for the gene chip analysis, albino zebrafish (University of Oregon, Eugene, OR) were treated as described previously (Vihtelic and Hyde, 2000; Vihtelic et al., 2006). Second, to induce photoreceptor death in pigmented fish, wild-type animals were dark adapted for 8 days and exposed to constant fluorescent light (26000-31000 lux). Control animals were sacrificed immediately following dark adaptation, and experimental animals were sacrificed at 24hrs, 48hrs, 72hrs, 5days, 7 days or 10 days after light onset. Additional animals were returned to normal lighting and sacrificed at 14, 21 or 28 days after light onset. This experiment was performed in triplicate.
RNA isolation and Microarray analysis
Oligonucleotide microarrays were used to identify genes that were differentially expressed after light-induced photoreceptor death. For these experiments, retinas were sampled at a single time point, 72hrs after light onset, which is after the death of photoreceptors and at a time when the retina is replete with injury-induced photoreceptor progenitors (Vihtelic and Hyde, 2000). Control animals were maintained in normal light conditions. After animals were sacrificed, retinas were dissected free from the surrounding ocular structures, and retinal RNA was isolated and processed in 8 separate pools (4 control, 4 experimental; 12 retinas each). From each pool of RNA, probes were synthesized and hybridized to a single chip. Briefly, total RNA was amplified to yield double-stranded antisense RNA (aRNA), which was biotinylated using the Affymetrix GeneChip Expression IVT Labeling Kit (Affymetrix, Santa Clara, CA). Ten micrograms of labeled aRNA were fragmented and hybridized to Zebrafish Genome Arrays (Affymetrix). Chips were scanned using the GeneChip Scanner 3000 (Affymetrix). The fluorescence intensity readouts were sorted into CHP files with the Affymetrix Microarray Suite v5.0. The array data described in this publication will be deposited in NCBI’s Gene Expression Omnibus (Edgar et al., 2002) (http://www.ncbi.nlm.nih.gov.proxy.lib.umich.edu/geo/).
A false discovery rate confidence interval (FDR-CI) was used to identify statistically significant changes in fluorescence intensity as described (Benjamini and Yekutieli, 2005, Hero et al., al. 2004). This approach employs robust multiarray averaging (RMA) to normalize the data, assigns fold-intensity differences and utilizes a statistical method that provides a false discovery rate-adjusted confidence interval (FDR-CI) for each differentially-labeled probe set (Hero et al., al. 2004; see also Yoshida et al., 2002). This analysis generated a rank-ordered list of probe sets that showed 2-fold or greater changes in fluorescence intensity on the chip images. Analysis of gene ontologies was performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (Dennis et al., 2003; http://david.abcc.ncifcrf.gov).
RNA isolation and quantitative RT-PCR (QRT-PCR)
For QRT-PCR, both albino and wild type zebrafish were sacrificed, and eyecups were removed and retinas dissected and carefully separated from the retinal pigment epithelium. RNA was isolated from control and light-lesioned albino zebrafish in experiments independent of those used for the microarray screen. Four retinas per group from 3-4 different zebrafish were pooled. Retinas were homogenized and RNA was isolated according to the manufacturers protocol (Ambion, Austin, TX). RNA was quantified with a spectrophotometer, and RNA quality was assessed on ethydium bromide stained agarose gels. 500 ng of total RNA was used to synthesize cDNA following the manufacturers protocol (Superscript II, Invitrogen, Carlsbad, CA). The resulting first-strand reaction was diluted 1:4 and used as a template for the subsequent QRT-PCR reaction (iQ™ SYBR® Green Supermix Bio-Rad, Hercules, CA) in the iCycler Real-Time PCR detection System (Bio-Rad). The following amplification and melt curve analysis protocol was used: 95°C 3min, 40×(95°C: 20s, 57°C: 20s, 72°C: 30s), 95°C: 1min, 90×55°C: 10s. The following primers were used: rhodopsin, forward- 5′ agcccatacgaatacccaca and reverse- 5′ agcttcttgtgctcgatgg; opsin-1, forward- 5′ aaaccacaagggaagcaatg and reverse- 5′ ttgtgctggcaaacagagtc; proliferating cellular nuclear antigen (pcna), forward- 5′ catccagacacttagagctgaaga and reverse- 5′ ctggtctgtgagagcttgatgtt; connexin 52.6, forward- 5′ tggacagatggtacctttgcc and reverse- 5′ gttgtctggaatggaccttcg (Zoidl et. al. 2004); mdka, forward- 5′ tgaagttttgttactgagctttgtg and reverse- 5′ agccagtgtacataagtgtgtgtgt; mdkb, forward- 5′ gctgttgtaatttgtagcaggtttt and reverse- 5′ cattcaatctcgttgtcatttacag; 18s rRNA forward- 5′ tcggctaccacatccaaggaaggcagc and reverse- 5′ ttgctggaattaccgcggctgctggca. The threshold cycle (Ct) was determined by the iCycler using the maximum curvature approach and then maintained constant for subsequent runs. Relative levels of gene expression were determined by the ΔΔCt method using either connexin 52.6 or 18s rRNA as an endogenous reference. Specificity of the amplification products was verified by agarose gel electrophoresis.
Tissue preparation, in situ hybridization and immunohistochemistry
At selected developmental times, embryos and larvae were fixed by immersion in 4% paraformaldehyde in 0.1M phosphate buffer, cryoprotected in 20% sucrose, frozen and cryosectioned. At set times following light treatment, adult fish were anesthetized in 0.05% 3-aminobenzoic acid-ethyl ester (Sigma), eyes were enucleated, lenses removed and eyecups were processed identically to the embryos and larvae.
Sense and antisense riboprobes for mdka or mdkb were synthesized from plasmids containing the full-length mdka and mdkb cDNAs (gift from Dr. Christoph Winkler [Winkler et al., 2003]; accession numbers NM_131070 [mdka] and NM_131716 [mdkb]). Plasmids were linearized and digoxigenin- (DIG) labeled riboprobes were generated by in vitro transcription using the DIG RNA Labeling kit (Roche Diagnostics, Indianapolis, IN). In situ hybridization was performed as described previously (Hitchcock and Kakuk-Atkins, 2004) using DIG-labeled probes and Fast Red (Roche Diagnostics, Indianapolis, IN) as the enzymatic substrate. The enzymatic reaction was monitored using a fluorescence microscope and stopped after 2-5 hours, when the signal was distinct and the background low. Fresh Fast Red staining solution was prepared and added to the slides each hour. The anti-DIG antibodies are Fab fragments isolated from sheep immunized against the whole digoxigenin molecule and conjugated with alkaline phosphatase (Hoffman-La Roche, Inc., Nutley, NJ, catalogue number 11093274910). To combine in situ hybridization with immunohistochemistry, the Fast Red color reaction was first allowed to develop, then slides were processed immediately for immunohistochemistry using standard procedures.
Other commercially available antibodies were also used in this study. The antibody against Proliferating Cell Nuclear Antigen (PCNA; Sigma-Aldrich, St. Louis, MO) was used at a 1:1000 dilution. This antibody was isolated from mice immunized with recombinant rat PCNA protein and validated by its capacity to immunoprecipitate PCNA protein from whole cell extracts (Waseem and Lane, 1990). In our study, tissues stained with this antibody produce a pattern that is identical to that previously reported (e.g., Raymond et al., 2006). Antibodies against Green Fluorescent Protein (GFP; ab6556; Abcam Inc., Cambridge, MA) were used at a 1:1000 dilution. These antibodies were raised in rabbit against purified recombinant GFP made in Escherichia coli. In Western blots, this antibody recognizes a single 67 kDa band in lysate from whole cells expressing GFP-fusion protein. Again, tissues stained with this antibody produce a pattern that is identical to that previously reported (Bernardos et al., 2007). The Prox-1 antibodies were raised in rabbit against a synthetic peptide from the C-terminus of mouse Prox-1 (EIFKSPNCLQELLHE), which in Western Blots recognizes a single 83kDa band, and used at a 1:2000 dilution (AB5475, Chemicon International, al. Temecula, CA). Zebrafish Prox-1 possesses 84% identity at the amino acid level with the mouse homologue (Glasgow and Tomarev, 1998), and this antibody has been shown to label Prox-1 expressing cells in zebrafish (Ober et. al., 2006). In addition, all sections were also stained with the fluorescent nuclear stain, 4,6-diamidino-2-phenylindole dihydrochloride (DAPI, Invitrogen-Molecular Probes, Eugene, OR). Prior to PCNA immunostaining, sections were processed for antigen retrieval as previously described (Raymond et al., 2006). Secondary antibodies, were goat, anti-rabbit or goat, anti-mouse and were used at a dilution of 1:500 (Invitrogen-Molecular Probes, Eugene, OR).
Photographic images
Images were taken with a Nikon DMX 1200 digital camera mounted on a Nikon Eclipse E800 epifluorescent compound microscope. Adobe Photoshop CS2 (Adobe Systems, San Jose, CA) was used to construct the figures. The layer tool was used to generate image overlays, and the channel mixer tool was used to change the red signal to magenta. In some images, the clone stamp tool was used to remove unwanted scale bars. Images in Figure 2 and Supplemental Figure 1 were taken with an AxioCam mRM digital camera and a Zeiss Axio Imager epifluorescent compound microscope (Carl Zeiss Microimaging, Thornwood, NY). Images were false colored using the Zeiss AxioVision 4.0 software and exported into Adobe Photoshop CS2 (Adobe Systems, San Jose, CA) and treated as described above.
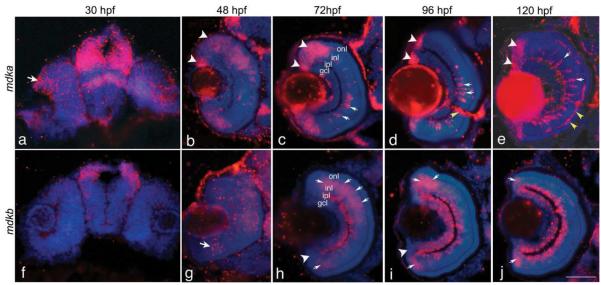
Figure 2. The cellular expression of mdka and mdkb during retinal development.
Panels a-e are in situ hybridizations that illustrate the retinal expression of mdka at 30-120 hpf, respectively. The white arrowheads identify mdka-expressing cells in the circumferential marginal zone and retina. The white arrows in panels c-e identify the columnar cells within the inner nuclear layer that express mdka. The yellow arrowhead in panel d identifies the optic nerve. The yellow arrowheads in panel e identify presumptive horizontal cells. Panels f-j are in situ hybridizations that illustrate the expression of mdkb at 30-120 hpf, respectively. The arrow in panel g identifies the ventronasal patch. The arrows in panels g-j demarcate regions of transient mdkb expression. The arrowheads in panels h and i identify the circumferential marginal zone, which does not express mdkb. Note the prominent expression of mdkb within the gcl and inl in panels h-j. onl, outer nuclear layer; inl, inner nuclear layer; ipl, inner plexiform layer; gcl, ganglion cell layer. Scale bar equals 50μm.
Results
Expression Analyses
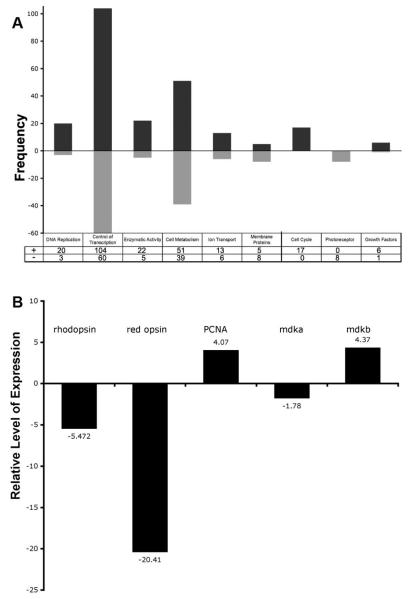
The RMA/FDR-CI identified 671 experimental probe sets that showed a 2-fold or greater difference in fluorescence intensity (hereafter referred to as ‘gene expression’) when compared to control probe sets. To characterize the global changes in gene expression, the differentially expressed genes were submitted to DAVID (Dennis et al., 2003) for functional annotation clustering. Table 1 lists gene ontology biological process terms that were statistically significantly overrepresented within this dataset. This analysis showed that many of the differentially expressed genes are associated with DNA replication and, very broadly, enzyme activity and cellular metabolism. Another ontology group in this collection, ‘visual perception,’ corresponds to genes encoding opsins. Functional categories were also identified from the gene ontology analysis, and after removing the entries lacking annotation, the number of genes that showed an increase or decrease in expression within each category was determined (Fig. 1A). Genes known to control the mitotic cell cycle, to regulate photoreceptor physiology, or to encode growth factors were separately evaluated. As anticipated, the expression of each of the 17 cell cycle control genes is increased, which reflects the accretion of mitotic photoreceptor progenitors, whereas the expression of each of the 8 photoreceptor-specific genes is decreased, reflecting the selective death of rods and cones. Genes encoding seven differentially-expressed growth factors (midkine b [see below], granulin a, granulin 1, granulin 2, dickkopf 1, galectin 1-like 2, matrixmetalloproteinase 9, follistatin) were also identified. The direction of change in the expression for four of these genes was independently validated by in situ hybridization (data not shown; see Results). Finally, quantitative RT-PCR (QRT-PCR) was performed for genes encoding Rhodopsin, Red Opsin, PCNA, Mdka and Mdkb (Figure 1B; Supplemental Fig. 1). In the light lesioned retinas, the levels of mRNA encoding rhodopsin and red opsin were reduced, whereas the levels of mRNA encoding pcna and mdkb were increased. The microarray analysis did not detect a change in the expression of mdka, whereas the QRT-PCR revealed a modest decrease in the level of mRNA encoding this protein.
Table 1.
Gene ontology (GO) process terms that were statistically significantly overrepresented among the differentially expressed genes
| GO Biological Process Term | p-value |
|---|---|
| intracellular part | 1.80E-06 |
| intracellular | 5.20E-05 |
| enzyme regulator activity | 1.30E-04 |
| DNA-dependent DNA replication | 3.60E-04 |
| DNA replication initiation | 6.10E-04 |
| protein polymerization | 1.30E-03 |
| cysteine-type endopeptidase activity | 2.00E-03 |
| DNA replication | 2.80E-03 |
| intracellular non-membrane-bound organelle | 6.40E-03 |
| non-membrane-bound organelle | 6.40E-03 |
| chromosomal part | 6.70E-03 |
| calcium-dependent phospholipid binding | 7.20E-03 |
| DNA metabolic process | 8.60E-03 |
| enzyme inhibitor activity | 1.00E-02 |
| chromatin | 1.40E-02 |
| intracellular organelle | 1.40E-02 |
| DNA-dependent ATPase activity | 1.60E-02 |
| nucleosome | 1.70E-02 |
| chromosome | 2.30E-02 |
| cysteine-type peptidase activity | 2.50E-02 |
| nucleoside-triphosphatase activity | 2.90E-02 |
| microtubule-based movement | 2.90E-02 |
| cytoskeleton-dependent intracellular transport | 2.90E-02 |
| lipid binding | 3.30E-02 |
| translation elongation factor activity | 3.30E-02 |
| deoxyribonucleotide metabolic process | 3.50E-02 |
| GTPase activity | 4.10E-02 |
| response to stress | 4.10E-02 |
| protease inhibitor activity | 4.10E-02 |
| pyrophosphatase activity | 3.90E-02 |
| GTPase activity | 4.10E-02 |
| hydrolase activity | 4.20E-02 |
| hydrolase activity, acting on acid anhydrides | 4.30E-02 |
| sensory perception of light stimulus | 4.50E-02 |
| visual perception | 4.50E-02 |
| ATPase activity | 4.50E-02 |
| biogenic amine metabolic process | 4.80E-02 |
| nucleosome assembly | 4.90E-02 |
Figure 1. Graphical representation of differentially-expressed genes.
Panel A illustrates functional gene categories and the number of genes in each category showing increased (+) or decreased (−) expression at 72 hrs after light onset. Genes within the left-most five categories resulted from the analysis of functional gene ontologies, whereas right-most three categories, cell cycle, photoreceptor, growth factors, were selected by hand from the list of differentially-expressed genes. Panel B illustrates the QRT-PCR results for five genes, rhodopsin, red opsin, PCNA, mdka and mdkb. Values above or below each bar represent the fold change in expression for each gene at 72hrs after light onset.
mdka and mdkb are expressed in distinct cells during retinal development
To determine the cellular pattern of expression of mdka and mdkb in the developing retina, in situ hybridization was performed on tissue sections from embryos and larvae between 24 and 120 hpf (Fig. 2). Sense probes did not show any specific hybridization (Supplemental Fig. 2). Neither mdka nor mdkb transcripts are detected in the eye at 24 hpf (data not shown), consistent with previous observations (Winkler and Moon, 2001; Winkler et al., 2003). However, retinal expression of mdka is detected at 30 hpf. At this time, a low level of expression is present throughout the retinal neuroepithelium and more intense expression is observed at the retinal margin, presaging the site of the CMZ (Fig. 2a). At 48 hpf, mdka is expressed at the retinal margin and more broadly in the INL (Fig. 2b), but mdka is not expressed in the ventral and central retina where differentiated cells and laminae are present. Between 48 hpf and 72 hpf, mdka expression becomes progressively restricted to the retinal margin. At 72 hpf, mdka expression appears centrally in columnar cells spanning the INL with morphology suggestive of Müller glia (Fig. 2c). mdka expression in these radially oriented cells persists through 120 hpf (Fig. 2c-e) when mdka transcription begins in presumptive horizontal cells (Fig. 2e, see below). Interestingly, in addition to this dynamic pattern of expression in the neural retina, mdka is strongly expressed in cells within the nascent optic nerve (Fig.2d) and the lens epithelium.
The spatial pattern of expression for mdkb is distinctly different from mdka and suggests this gene is transcribed in newly postmitotic cells that are integrated into the inner retinal layers. mdkb is first expressed at 48 hpf in a ventro-nasal patch and more broadly in the laminated central retina (Fig.2g). At 72 hpf, mdkb is expressed in differentiated ganglion and amacrine cells, which straddle the inner plexiform layer (IPL) (Fig.2 h) and in a broad annulus of cells central to the CMZ (Fig. 2h). Between 96 hpf and 120 hpf, expression persists within the inner nuclear and ganglion cell layers, whereas the annulus of mdkb expression becomes progressively restricted to cells just central to the CMZ (Fig. 2 h-j). In contrast to mdka, mdkb is not expressed by retinal progenitors in the CMZ (Fig. 2h, i). mdkb appears not to be expressed in cells destined for the outer nuclear layer. These data suggest that mdkb is expressed by inner retinal cells as they differentiate and constitutively expressed by mature ganglion and amacrine cells. This pattern of expression is maintained in the adult (see below).
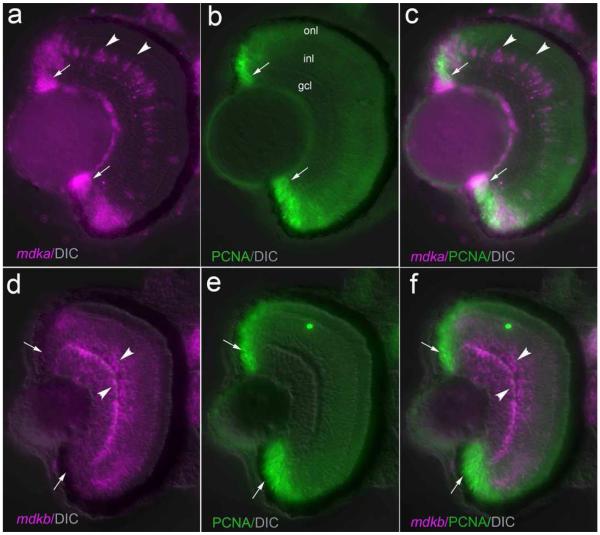
To analyze the expression of mdka and mdkb relative to proliferating cells, we combined in situ hybridization with immunohistochemistry for PCNA, a cofactor of DNA polymerases that is expressed during the late G1, S and early G2 phases of the cell cycle (Kurki et al., 1986; Moldovan et al., 2007). At 72hpf, when both mature and proliferating cells are present in the retina, mdka is expressed in PCNA-positive cells within the CMZ (Fig.3 a, b, c). In contrast, mdkb is co-expressed with PCNA in only a few cells at the interface between the CMZ and the differentiated retina (Fig.3 d, e, f).
Figure 3. mdka is expressed by retinal progenitors and mdkb is expressed by differentiated cells.
Panel a is an in situ hybridization that illustrates the expression of mdka at 72 hpf. Panel b is the same section as in panel a, but immunostained with antibodies against proliferating cell nuclear layer. Panel c is the digital overlay of panels a and b. Panel d is an in situ hybridization showing the expression of mdkb at 72 hpf. Panel e is the same section as in panel d, but immunostained with antibodies against proliferating cell nuclear antigen. Panel f is the digital overlay of panels d and e. In each panel, the white arrows identify the circumferential marginal zone in, whereas the arrowheads identify cells expressing mdka (panels a and c) and mdkb (panels d and f) within central retina. onl, outer nuclear layer; inl, inner nuclear layer; ipl, inner plexiform layer; gcl, ganglion cell layer. Scale bar equals 50μm.
Together, these data show that, during early retinal development, mdka is expressed in mitotic retinal progenitors, whereas mdkb is expressed in newly postmitotic cells. In addition, mdka is transiently expressed in presumptive Müller glia (see next section). Finally, both genes are constitutively expressed in subsets of neurons within the inner retinal layers.
mdka is transiently expressed in Müller glia and constitutively expressed in horizontal cells
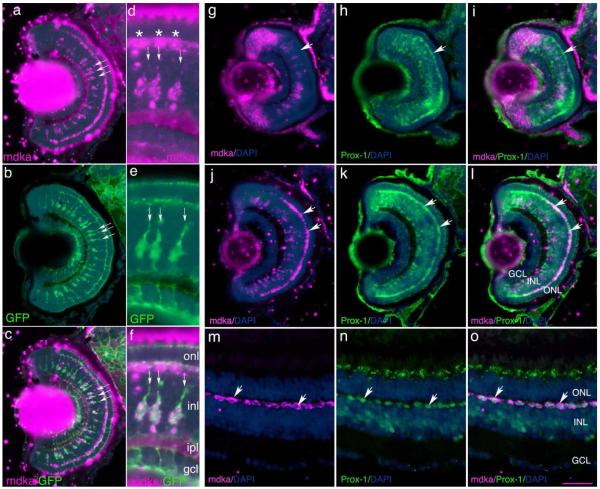
During larval development, the mdka-expressing cells in the INL have centrally located large nuclei with mdka mRNA in radially-extending processes (Fig. 2 c, d, e; Fig. 3 a; Fig. 4 a). This morphology suggests mdka is expressed by Müller glia. To test this speculation, we combined in situ hybridization with immunostaining for green fluorescent protein (GFP) on retinal sections from Tg(gfap:GFP)MI2001 transgenic zebrafish (Bernardos and Raymond, 2006). These fish express GFP under the control of the glial fibrillary acidic protein (gfap) regulatory elements, which in the retina selectively marks Müller glia. The mdka-positive cells in the INL (Fig. 4 a, c, d, f) overlay precisely with GFP-positive cells (Fig. 4 b, c, e, f), confirming that mdka is transcribed in Müller glia.
Figure 4. mdka is expressed in Müller glia.
Panels a and d illustrate in situ hybridizations of mdka expression in Tg(gfap:GFP)Mi2001 fish at 120 hpf. Panels b and e illustrate Müller glia immunostained with antibodies against green fluorescent protein. Panel c is the digital overlay of panels a and b; panel f is the digital overlay of panels d and e. In each panel, the three arrows in identify the same three Müller glia. In panel d, the asterisks identify mdka expression in presumptive horizontal cells. Panel g is an in situ hybridizations showing the expression of mdka at 72 hpf. Panel h is the same section as in panel g, but immunostained with antibodies against Prox1. Panel i is the digital overlay of panels g and h. Note that at 72hpf, horizontal cells synthesize Prox1, but do not yet express mdka. Panel j is an in situ hybridization showing the expression of mdka at 120hpf. Panel k is the same section as in panel j, but immunostained with antibodies against Prox1. Panel l is the digital overlay of panels j and k. Note the co-localization of mdka mRNA and Prox1 protein. Panel m is an in situ hybridization showing the expression of mdka in the adult retina. Panel n is the same section as in panel j, but immunostained with antibodies against Prox1. Panel o is the digital overlay of panels d and e. Arrows in j-o identify horizontal cells that express mdka and are immunostained for Prox1. onl, outer nuclear layer; inl, inner nuclear layer; gcl, ganglion cell layer; DAPI, nuclear stain 4,6-diamidino-2-phenylindole dihydrochloride. Scale bar equals 50μm; onl, outer nuclear layer; inl, inner nuclear layer; ipl, inner plexiform layer; gcl, ganglion cell layer; ipl: inner plexiform layer. Scale bar equals 50μm.
To test our inference that mdka is expressed by horizontal cells, we combined in situ hybridization and antibodies against Prox-1. Prox-1, a homeodomain protein required for horizontal cell development (Dyer et al., 2003) and is expressed in chicks and mammals by horizontal, amacrine and bipolar cells (Dyer et al., 2003; Belecky-Adams et al., 1997; Edqvist and Hallbook, 2004). At 72 hpf, Prox-1 labels the narrow band of horizontal cell bodies at the outer boundary of the INL, though mdka expression in these cells is not detected at this time (Fig. 2c and 4g-i). At 120 hpf, however, mdka expression co-localizes with Prox-1 immunostaining in the horizontal cells (Fig. 4j-l). Note that mdka expression is reduced or absent in the dorsal retina, which is less mature than ventral retina, showing that mdka is not expressed by newly differentiated horizontal cells. In the adult retina, mdka is constitutively expressed in Prox-1-positive horizontal cells (Fig. 4m-o).
During photoreceptor regeneration, both midkine genes are expressed by proliferating Müller glia and photoreceptor progenitors
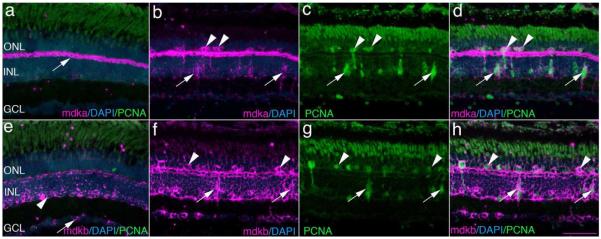
To investigate the expression of the midkine genes during photoreceptor regeneration, in situ hybridization was used to compare the cellular patterns of mdka and mdkb expression in control and experimental retinas. The patterns of midkine expression in adult, control retinas are similar to that observed at 120 hpf: mdka is expressed in horizontal cells (Fig. 4m, 5a and Supplemental Fig. 2), whereas mdkb is expressed by cells in the vitreal aspect of the INL and the GCL (Fig. 5e and Supplemental Fig.2). In the adult, unlesioned retina, neither probe labels dividing cells (Supplemental Fig. 3). Housing wild-type, pigmented fish in constant bright light results in the thinning of the outer nuclear layer, due to the death of photoreceptors, and the corresponding decrease in mRNA encoding opsin genes. The anatomical loss of photoreceptors is fully restored after fish are returned to a normal lighting environment, though mRNA levels do not return to normal levels (Supplemental Figure 1). 72hrs after light onset corresponds to the peak of injury-induced proliferation and the peak of PCNA expression (Vihtelic and Hyde, 2000; Supplemental Fig. 1), therefore, we sampled this time point to examine the relationship between the expression of mdka and mdkb and the injury-induced cell proliferation (Fig. 5). In the light-lesioned retinas, the expression of mdka persists in horizontal cells and is newly expressed by proliferating, PCNA-positive photoreceptor progenitors in both the inner and outer nuclear layers (Fig. 5b-d). In contrast to mdka, the domain of mdkb expression expands from the inner tier of (amacrine) cells within the INL to include all cells of the INL, including horizontal cells (Fig. 5f-h; Supplemental Fig. 4). In addition, mdkb is expressed in PCNA-positive photoreceptor progenitors. Light lesions using the adult Tg(gfap:GFP)MI2001 zebrafish confirm that proliferating cells expressing mdka and mdkb are Müller glia and their mitotic progeny (data not shown). Together, data from the in situ hybridizations show that photoreceptor death induces changes in the cellular pattern of mdka expression and a marked increase in the number of cells that express mdkb. Further, following photoreceptor death, and in contrast to that observed in developing and adult retinas, the expression of both midkines becomes coincident in horizontal cells and photoreceptor progenitors.
Figure 5. In the light-lesioned retina, mdka and mdkb are expressed by horizontal cells and injury-induced photoreceptor progenitors.
Panel a is an in situ hybridization showing the expression of mdka in a control retina. The white arrow identifies mdka-expressing horizontal cells. Panel b is an in situ hybridization showing mdka expression in a retina following 72 hrs of light exposure. Panel c is the same section as in panel b, immunostained with antibodies against PCNA. Panel d is the digital overlay of panels b and c. Arrowheads and arrows in panels b-d identify double-labeled cells in the ONL and INL, respectively. Panel e illustrates an in situ hybridization showing the expression of mdkb in a control retina. Panel f is an in situ hybridization showing mdkb expression in a retina following 72 hrs of light exposure. The arrowhead and arrow identify mdkb-expressing cells in the inner and outer nuclear layers, respectively. Panel g is the same section as in panel f, immunostained with antibodies against PCNA. Panel h illustrates the digital overlay of panels f and g. In panels b-d and f-h, arrowheads and arrows identify double-labeled cells in the ONL and INL, respectively. ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer; PCNA, Proliferating Cellular Nuclear Antigen; DAPI: nuclear stain 4,6-diamidino-2-phenylindole, dihydrochloride. Scale bar equals 50μm.
Discussion
Light-induced death and regeneration of photoreceptors in zebrafish is a well-described phenomenon (Vihtelic and Hyde, 2000). Photoreceptors die between 24 and 48hrs after light onset. Overlapping with the death of photoreceptors, injury-induced proliferation commences at about 24hrs and continues for the next several days, enlarging a pool of photoreceptor progenitors. From this pool of mitotic cells, the first regenerated photoreceptors begin to differentiate at about 4 days post injury (Bernardos et al., 2007). We used oligonucleotide microarrays to identify genes that are differentially expressed at 72hrs after light onset, near the peak of the proliferative phase. The light-lesion model was selected, because, 1) it selectively kills photoreceptors without introducing ancillary injury to the eye; 2) we infer that degeneration of only two cell types (rods and cones) will result in less complex changes in the retinal transcriptome; and 3) the selective death of photoreceptors, while sparing the other cell types, models aspects of photoreceptor degeneration in mouse genetic models and human disease. The specific goal of the array experiment was to identify growth factors expressed by the regenerative Müller glia and their progeny that we could hypothesize function to regulate aspects of photoreceptor regeneration. Several growth factors were identified in the array screen, and we chose to characterize the developmental and injury-induced expression of mdkb, which was shown to be significantly upreguated both in the microarrays and QRT-PCR analyses, and its paralog, mdka, which we show is also expressed in the retina and upregulated in regenerating heart and fin (Lien et al., 2006; Schebesta et al., 2006).
During retinal development, the expression of the two midkines is differentially regulated, with each gene exhibiting distinct temporal and spatial patterns of cellular expression. During embryonic retinal development, midkines are expressed in adjacent, non-overlapping domains, which, as retinal neurons differentiate, become restricted to annuli at the retinal margin. Double labeling with antibodies against PCNA showed that mdka is expressed by retinal progenitors, whereas mdkb is expressed in nearby, newly postmitotic cells. The adjacent cellular expression of these paralogous genes in the early retinal neuroepithelium is similar to their spatial patterns of expression in the neural tube. In early embryos, mdka and mdkb are expressed at the forebrain/midbrain and midbrain/hindbrain boundaries, where they form contiguous, non-overlapping domains and function to pattern the neural tube (Winkler et al., 2003).
During larval retinal development, each midkine is expressed in a spatial pattern that persists into adulthood. Mdka is expressed by mitotic cells in the CMZ and horizontal cells in the INL. In the optic tectum of adult zebrafish, mdka transcripts are present in the subventricular zone (Winkler et al., 2003), a site of persistent neurogenesis (Marcus et al., 1999). In mammals, Midkine is expressed in neural progenitors, in both the normal and injured brain, and functions to promote proliferation (Kikuchi-Horie et al., al. 2004, Zou et al. 2006). Our finding that mdka is expressed in the neurogenic CMZ of the zebrafish retina suggets, mdka is a component of neural stem cell compartments in vertebrates. The persistent expression of mdka in horizontal cells is intriguing. Horizontal cells are among the first retinal neurons to differentiate, and it has been proposed that they play a pioneering role in the development of photoreceptors (Messersmith and Redburn, 1990, Hagedorn et al., 1998). In contrast to mdka, mdkb is expressed in newly postmitotic cells destined for the inner retina. The initial expression of mdkb follows the wave of differentiation, and this pattern of expression is then recapitulated among each generation of newly-postmitotic cells as they exit the CMZ. mdkb is then also constitutively expressed by amacrine cells and cells in the ganglion cell layer.
Several extrinsic signaling molecules have been identified that regulate development of the teleost retina. IGFs and wnts regulate proliferation of retinal progenitors, whereas sonic hedgehog, FGFs, retinoic acid and Notch-mediated signaling regulate aspects of neuronal differentiation (Vinokuthmar et al., 2008; Stenkamp, 2007 and citations therein). The function of the Midkines in the teleost retina is not yet established, however, based on the tight spatial association of the two Midkines with proliferating and differentiating retinal neurons, respectively, it can be speculated that Mdka is a molecular component of the retinal stem cell niche (Raymond et al., 2006) and may play a role in regulating proliferation, whereas Mdkb may play a role in aspects of neuronal differentiation.
During photoreceptor regeneration, both genes show altered patterns of cellular expression. The expression of mdkb expands from the inner tier of the INL to include all neurons of the INL. In addition, mdkb is expressed in the injury-induced progenitors. The marked expansion in the number of cells within the INL that express mdkb can account for the upregulation of mdkb expression detected by the microarrays and the QRT-PCR. In contrast, the expression of mdka showed no significant change in the microarrays, and the QRT-PCR showed that mdka expression is slightly down regulated, even though the cellular expression of mdka in the injured retina is consistently altered. There is no obvious explanation for these seemingly contradictory results, though it should be noted that the circadian clock regulates mdka expression (Calinescu et al., in preparation), and this could influence either the microarray or QRT-PCR analyses. Nonetheless, the coincident expression of both midkines in horizontal cells and injury-induced photoreceptor progenitors, patterns of expression distinct from that observed in the developing retina, suggests that each gene product may have a common function during retinal injury and repair. In zebrafish, mdka expression is upregulated during regeneration of the fin and heart (Schebasta et al., 2006; Lien et al., 2006). The cellular expression of mdka in the regenerating fin has not yet been described. In the regenerating heart, mdka is expressed in cells that surround the wound site, but it is not expressed in the proliferative cardiomyocytes that replace the ventricular tissue (Lien et al., 2006). These results and the data reported here show that Midkine paralogs can be differentially regulated during the regeneration of specific tissues, but that both Midkines function as injury-induced growth factors. Further, both midkines may play an essential role in injured tissues with the capacity to regenerate to full anatomical integrity and functional recovery.
In summary, the expression of mdka and mdkb is actively and differentially regulated during retinal development and injury-induced photoreceptor regeneration. During development, the two midkine genes are expressed in distinct populations of cells – stem cells, retinal progenitors and mature neurons, suggesting these secreted molecules subserve different functions. Following retinal injury, the spatial pattern of midkine expression is altered and both genes are expressed in horizontal cells and Müller glia and their neurogenic offspring. This is the first description of the cellular expression of midkines in the vertebrate retina, and this study adds Mdka and Mdkb to the molecular signature of Müller glia that exhibit features of neural stem cells. Across the vertebrate phylum, Midkine is upregulated following injury, and in humans Midkine is integral to the growth of numerous carcinomas. An emerging concept in biology is that the same regulatory proteins control common cellular events during development, tissue repair and carcinogenesis (Beachy et al., 2004; Gardiner, 2005). Our studies support this concept and add Mdka and Mdkb to the family of signaling molecules present in the developing and regenerating retina.
Supplementary Material
Table 1. Differentially-expressed genes at 72hrs after light onset, rank ordered by log2 difference in average fluorescence intensity between control and experimental chips (see column M).
Supplemental Figure 1. Housing pigmented zebrafish to intense bright fluorescent light induces photoreceptor loss followed by regeneration. Panels a-e illustrate Nissl-stained sections from control retina (a) and retinas exposed to bright, constant light for 24hrs (b), 72hrs (c), 10 days or (d) 10 days following by 18 days of recovery. Panels f-g illustrate quantitative measurements of the photoreceptor, outer nuclear and rod nuclear layers, respectively (see panel a), normalized to the thickness of the inner nuclear layer. Note that there is a significant decrease in the thickness of the various layers starting at 72h of light exposure. Panels i-k illustrate analysis of rhodopsin, red opsin and PCNA expression by quantitative real-time PCR. ROS, rod outer segments; OLM, outer limiting membrane; OPL, outer plexiform layer; IPL, inner plexiform layer; ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer; PL, photoreceptor layer; rod ONL, rod outer nuclear layer. Scale bar equals 50μm, *p<0.05, **p<0.01.
Supplemental Figure 2. Sense probes do not produce specific cellular labeling. Panels a-d illustrate in situ hybridizations comparing mdka antisense (a, c) and sense (b, d) probes. Panels a and b are from a 96hpf larva and panels c and d are from an adult retina. Panels e-f illustrate in situ hybridizations comparing mdkb antisense (e, g) and sense (f, h) probes. Panels e and f are from a 96hpf larva, whereas panels g and h are from an adult. Arrows and arrowheads illustrate specific cellular expression when using antisense probes. GCL – ganglion cell layer; INL – inner nuclear layer; ONL – outer nuclear layer.
Supplemental Figure 3. In the adult retina mdka or mdkb is not expressed in dividing cells. Panels a and d illustrate in situ hybridizations that show the cellular expression of mdka and mdkb, respectively, in the adult retina. Panels b and e are the same sections as in a and d, following immunostaining with antibodies against PCNA, and illustrate PCNA-positive cells (rod progenitors) (arrowheads) within the outer nuclear layer. Panels c and f are digital overlays. Note that the PCNA immunostaining does not co-localize with the expression of midkines. ONL – outer nuclear layer; INL – inner nuclear layer; GCL – ganglion cell layer.
Supplemental Figure 4. Time course of mdka and mdkb expression during and after the light lesion. Panels a-h illustrate the time course of the injury-induced expression of mdka, whereas panels i-p illustrate the time course of the injury-induced expression of mdkb. Note that the expansion of mdkb expression into the vitreal inner nuclear layer begins by 24hrs after light onset (cf. panel I with Fig. 5a) and returns to normal only at 14 days after the onset of the light exposure. In contrast, the expression of mdka varies relatively little over the same time course. onl, outer nuclear layer; inl inner nuclear layer; gcl, ganglion cell layer. Scale bar equals 50μm.
Acknowledgments
We thank Laura Kakuk-Atkins and Mitch Gillett for excellent technical assistance. We thank Dr. Christoph Winkler for the gift of plasmids encoding mdka and mdkb, Dr. Pamela Raymond for the gift of the Tg(gfap:GFP)MI2001 fish, Dr. Philip Gage for reading earlier versions of this manuscript. We also thank Rachel Harding for technical assistance. This work was supported by NIH grants T32-DE07057 (AAC) and P30-EY07003 and R01-EY07060 (PFH) and a Senior Scientific Investigator Award from Research to Prevent Blindness, Inc. (PFH).
Literature cited
- Beachy PA, Karhadkar SS, Berman DM. Tissue repair and stem cell renewal in carcinogenesis. Nature. 2004;432:324–31. doi: 10.1038/nature03100. [DOI] [PubMed] [Google Scholar]
- Belecky-Adams T, Tomarev S, Li HS, Ploder L, McInnes RR, Sundin O, Adler R. Pax-6, Prox 1, and Chx10 homeobox gene expression correlates with phenotypic fate of retinal precursor cells. Invest Ophthalmol Vis Sci. 1997;38:1293–303. [PubMed] [Google Scholar]
- Benjamini Y, Yekutieli D. Quantitative trait Loci analysis using the false discovery rate. Genetics. 2005;1:783–90. doi: 10.1534/genetics.104.036699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardos RL, Raymond PA. GFAP transgenic zebrafish. Gene Expr Patterns. 2006;6:1007–13. doi: 10.1016/j.modgep.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Bernardos RL, Barthel LK, Meyers JR, Raymond PA. Late-stage neuronal progenitors in the retina are radial Muller glia that function as retinal stem cells. J Neurosci. 2007;27:7028–40. doi: 10.1523/JNEUROSCI.1624-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrill JD, Easter SS., Jr The first retinal axons and their microenvironment in zebrafish: cryptic pioneers and the pretract. J Neurosci. 1995;15:2935–47. doi: 10.1523/JNEUROSCI.15-04-02935.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig SEL, Calinescu A-A, Hitchcock PF. Identification of the molecular signatures integral to regenerating photoreceptors in the retina of the zebrafish. J. Ocular Biol. Diseases Informat. 2008 doi: 10.1007/s12177-008-9011-5. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biology. 2003;4:P3. [PubMed] [Google Scholar]
- Dyer MA, Livesey FJ, Cepko CL, Oliver G. Prox1 function controls progenitor cell proliferation and horizontal cell genesis in the mammalian retina. Nat Genet. 2003;34:53–8. doi: 10.1038/ng1144. [DOI] [PubMed] [Google Scholar]
- Easter SS, Jr, Nicola GN. The development of vision in the zebrafish (Danio rerio) Dev Biol. 1996;180:646–63. doi: 10.1006/dbio.1996.0335. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–10. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edqvist PH, Hallbook F. Newborn horizontal cells migrate bi-directionally across the neuroepithelium during retinal development. Development. 2004;131:1343–51. doi: 10.1242/dev.01018. [DOI] [PubMed] [Google Scholar]
- Fausett BV, Goldman D. A role for alpha1 tubulin-expressing Müller glia in regeneration of the injured zebrafish retina. J Neurosci. 2006;26:6303–13. doi: 10.1523/JNEUROSCI.0332-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fimbel SM, Montgomery JE, Burket CT, Hyde DR. Regeneration of inner retinal neurons after intravitreal injection of ouabain in zebrafish. J Neurosci. 2007;27:1712–24. doi: 10.1523/JNEUROSCI.5317-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner DM. Ontogenetic decline of regenerative ability and the stimulation of human regeneration. Rejuvenation Res. 2005;8:141–53. doi: 10.1089/rej.2005.8.141. [DOI] [PubMed] [Google Scholar]
- Glasgow E, Tomarev SI. Restricted expression of the homeobox gene prox-1 in developing zebrafish. Mech Dev. 1998;76:175–8. doi: 10.1016/s0925-4773(98)00121-x. [DOI] [PubMed] [Google Scholar]
- Hagedorn M, Mack AF, Evans B, Fernald RD. The embryogenesis of rod photoreceptors in the teleost fish retina, Haplochromis burtoni. Brain Res Dev Brain Res. 1998;108:217–27. doi: 10.1016/s0165-3806(98)00051-0. [DOI] [PubMed] [Google Scholar]
- Harris WA, Perron M. Molecular recapitulation: the growth of the vertebrate retina. Int J Dev Biol. 1998;42:299–304. [PubMed] [Google Scholar]
- Hero AO, Fleury G, Mears AJ, Swaroop A. Multicriteria gene screening for analysis of differential expression with DNA microarrays. EURASIP JASP. 2004:43–52. [Google Scholar]
- Hitchcock P, Kakuk-Atkins L. The basic helix-loop-helix transcription factor neuroD is expressed in the rod lineage of the teleost retina. J Comp Neurol. 2004;477:108–17. doi: 10.1002/cne.20244. [DOI] [PubMed] [Google Scholar]
- Hitchcock P, Ochocinska M, Sieh A, Otteson D. Persistent and injury-induced neurogenesis in the vertebrate retina. Prog Retin Eye Res. 2004;23:183–94. doi: 10.1016/j.preteyeres.2004.01.001. [DOI] [PubMed] [Google Scholar]
- Hitchcock PF, Raymond PA. The teleost retina as a model for development and regeneration biology. Zebrafish. 2004;1:257–27. doi: 10.1089/zeb.2004.1.257. [DOI] [PubMed] [Google Scholar]
- Hu M, Easter SS. Retinal neurogenesis: the formation of the initial central patch of postmitotic cells. Dev Biol. 1999;207:309–21. doi: 10.1006/dbio.1998.9031. [DOI] [PubMed] [Google Scholar]
- Iwasaki W, Nagata K, Hatanaka H, Inui T, Kimura T, Muramatsu T, Yoshida K, Tasumi M, Inagaki F. Solution structure of midkine, a new heparin-binding growth factor. EMBO J. 1997;16:6936–46. doi: 10.1093/emboj/16.23.6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jochheim-Richter A, Rüdrich U, Koczan D, Hillemann T, Tewes S, Petry M, Kispert A, Sharma AD, Attaran F, Manns MP, Ott M. Gene expression analysis identifies novel genes participating in early murine liver development and adult liver regeneration. Differentiation. 2006;74:167–73. doi: 10.1111/j.1432-0436.2006.00066.x. [DOI] [PubMed] [Google Scholar]
- Kadomatsu K, Tomomura M, Muramatsu T. cDNA cloning and sequencing of a new gene intensely expressed in early differentiation stages of embryonic carcinoma cells and in mid-gestation period of mouse embryogenesis. Biochem. Biophys. Res. Commun. 1988;151:1312–1318. doi: 10.1016/s0006-291x(88)80505-9. [DOI] [PubMed] [Google Scholar]
- Kadomatsu K, Huang RP, Suganuma T, Murata F, Muramatsu T. A retinoic acid responsive gene MK found in the teratocarcinoma system is expressed in spatially and temporally controlled manner during mouse embryogenesis. J Cell Biol. 1990;110:607–16. doi: 10.1083/jcb.110.3.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadomatsu K, Muramatsu T. Midkine and pleiotrophin in neural development and cancer. Cancer Lett. 2004;204(2):127–43. doi: 10.1016/S0304-3835(03)00450-6. [DOI] [PubMed] [Google Scholar]
- Kassen SC, Ramanan V, Montgomery JE, T Burket C, Liu CG, Vihtelic TS, Hyde DR. Time course analysis of gene expression during light-induced photoreceptor cell death and regeneration in albino zebrafish. Dev Neurobiol. 2007;67:1009–31. doi: 10.1002/dneu.20362. [DOI] [PubMed] [Google Scholar]
- Kikuchi-Horie K, Kawakami E, Kamata M, Wada M, Hu JG, Nakagawa H, Ohara K, Watabe K, Oyanagi K. Distinctive expression of midkine in the repair period of rat brain during neurogenesis: immunohistochemical and immunoelectron microscopic observations. J Neurosci Res. 2004;75:678–87. doi: 10.1002/jnr.20015. [DOI] [PubMed] [Google Scholar]
- Kojima S, Inui T, Muramatsu H, Suzuki Y, Kadomatsu K, Yoshizawa M, Hirose S, Kimura T, Sakakibara S, Muramatsu T. Dimerization of midkine by tissue transglutaminase and its functional implication. J Biol Chem. 1997;272:9410–6. doi: 10.1074/jbc.272.14.9410. [DOI] [PubMed] [Google Scholar]
- Kurki P, Vanderlaan M, Dolbeare F, Gray J, Tan EM. Expression of proliferating cell nuclear antigen (PCNA)/cyclin during the cell cycle. Exp Cell Res. 1986;166:209–19. doi: 10.1016/0014-4827(86)90520-3. [DOI] [PubMed] [Google Scholar]
- Liedtke D, Winkler C. Midkine-b regulates cell specification at the neural plate border in zebrafish. Dev Dyn. 2008;237:62–74. doi: 10.1002/dvdy.21384. [DOI] [PubMed] [Google Scholar]
- Lien C-L, Schebesta M, Makino S, Weber GJ, Keating MT. Gene expression analysis of zebrafish heart regeneration. PLOS Biol. 2006;4:1386–1396. doi: 10.1371/journal.pbio.0040260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus RC, Delaney CL, Easter SS., Jr Neurogenesis in the visual system of embryonic and adult zebrafish (Danio rerio) Vis Neurosci. 1999;16:417–24. doi: 10.1017/s095252389916303x. [DOI] [PubMed] [Google Scholar]
- Masuda K, Watanabe I, Unoki K, Ohba N, Muramatsu T. Functional Rescue of photoreceptors from the damaging effects of constant light by survival-promoting factors in the rat. Invest Ophthalmol Vis Sci. 1995;36:2142–6. [PubMed] [Google Scholar]
- Messersmith EK, Redburn DA. Kainic acid lesioning alters development of the outer plexiform layer in neonatal rabbit retina. Int J Dev Neurosci. 1990;8:447–61. doi: 10.1016/0736-5748(90)90077-f. [DOI] [PubMed] [Google Scholar]
- Muramatsu T. Midkine and pleiotrophin: two related proteins involved in development, survival, inflammation and tumorigenesis. J Biochem. 2002;132:359–71. doi: 10.1093/oxfordjournals.jbchem.a003231. [DOI] [PubMed] [Google Scholar]
- Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–79. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Miyashiro M, Kadomatsu K, Ogata N, Yamamoto C, Takahashi K, Uyama M, Muramatsu H, Muramatsu T. Midkine expression in transient retinal ischemia in the rat. Curr Eye Res. 1998;17:9–13. doi: 10.1076/ceyr.17.1.9.5257. [DOI] [PubMed] [Google Scholar]
- Mitsiadis TA, Salmivirta M, Muramatsu T, Muramatsu H, Rauvala H, Lehtonen E, Jalkanen M, Thesleff I. Expression of the heparin-binding cytokines, midkine (MK) and HB-GAM (pleiotrophin) is associated with epithelial-mesenchymal interactions during fetal development and organogenesis. Development. 1995;121:37–51. doi: 10.1242/dev.121.1.37. [DOI] [PubMed] [Google Scholar]
- Obama H, Biro S, Tashiro T, Tsutsui J, Ozawa M, Yoshida H, Tanaka H, Muramatsu T. Myocardial infarction induces expression of midkine, a heparin-binding growth factor with reparative activity. Anticancer Res. 1998;18:145–52. [PubMed] [Google Scholar]
- Ober EA, Verkade H, Field HA, Stainier DY. Mesodermal Wnt2b signalling positively regulates liver specification. Nature. 2006;442(7103):688–91. doi: 10.1038/nature04888. [DOI] [PubMed] [Google Scholar]
- Ochiai K, Muramatsu H, Yamamoto S, Ando H, Muramatsu T. The role of midkine and pleiotrophin in liver regeneration. Liver Int. 2004;24:484–91. doi: 10.1111/j.1478-3231.2004.0990.x. [DOI] [PubMed] [Google Scholar]
- Ohta S, Muramatsu H, Senda T, Zou K, Iwata H, Muramatsu T. Midkine is expressed during repair of bone fracture and promotes chondrogenesis. J Bone Miner Res. 1999;14:1132–44. doi: 10.1359/jbmr.1999.14.7.1132. [DOI] [PubMed] [Google Scholar]
- Otteson DC, Hitchcock PF. Stem cells in the teleost retina: persistent neurogenesis and injury-induced regeneration. Vision Res. 2003 Apr;43:927–36. doi: 10.1016/s0042-6989(02)00400-5. [DOI] [PubMed] [Google Scholar]
- Qiu L, Escalante CR, Aggarwal AK, Wilson PD, Burrow CR. Monomeric midkine induces tumor cell proliferation in the absence of cell-surface proteoglycan binding. Biochemistry. 2000;39:5977–87. doi: 10.1021/bi991519e. [DOI] [PubMed] [Google Scholar]
- Raymond PA, Barthel LK, Bernardos RL, Perkowski JJ. Molecular characterization of retinal stem cells and their niches in adult zebrafish. BMC Dev Biol. 2006;6:36. doi: 10.1186/1471-213X-6-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakima H, Kamizono T, Matsuda F, Izumo K, Ijiri K, Yoshida Y. Midkine and its receptor in regenerating rat skeletal muscle after bupivacaine injection. Acta Histochem. 2006;108:357–64. doi: 10.1016/j.acthis.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Schäfer M, Rembold M, Wittbrodt J, Schartl M, Winkler C. Medial floor plate formation in zebrafish consists of two phases and requires trunk-derived Midkine-a. Genes Dev. 2005;19:897–902. doi: 10.1101/gad.336305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schebesta M, Lien CL, Engel FB, Keating MT. Transcriptional profiling of caudal fin regeneration in zebrafish. ScientificWorldJournal. 2006;1:38–54. doi: 10.1100/tsw.2006.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt EA, Dowling JE. Early eye morphogenesis in the zebrafish, Brachydanio rerio. J Comp Neurol. 1994 Jun 22;344:532–42. doi: 10.1002/cne.903440404. [DOI] [PubMed] [Google Scholar]
- Schmitt EA, Dowling JE. Early retinal development in the zebrafish, Danio rerio: light and electron microscopic analyses. J Comp Neurol. 1999;404:515–36. [PubMed] [Google Scholar]
- Stenkamp DL. Neurogenesis in the fish retina. Int Rev Cytol. 2007;259:173–224. doi: 10.1016/S0074-7696(06)59005-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thummel R, Kassen SC, Montgomery JE, Enright JM, Hyde DR. Inhibition of Muller glial cell division blocks regeneration of the light-damaged zebrafish retina. Dev Neurobiol. 2008;68:392–408. doi: 10.1002/dneu.20596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unoki K, Uehara F, Muramatsu T, Ohba N. Rescue of photoreceptors from the damaging effects of constant light by midkine, a retinoic acid-responsive gene product. Invest Ophthalmol Vis Sci. 1994;35:4063–8. [PubMed] [Google Scholar]
- Varga ZM, Wegner J, Westerfield M. Anterior movement of ventral diencephalic precursors separates the primordial eye field in the neural plate and requires cyclops. Development. 1999;126:5533–46. doi: 10.1242/dev.126.24.5533. [DOI] [PubMed] [Google Scholar]
- Vihtelic TS, Hyde DR. Light-induced rod and cone cell death and regeneration in the adult albino zebrafish (Danio rerio) retina. J Neurobiol. 2000;44:289–307. doi: 10.1002/1097-4695(20000905)44:3<289::aid-neu1>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Vihtelic TS, Soverly JE, Kassen SC, Hyde DR. Retinal regional differences in photoreceptor cell death and regeneration in light-lesioned albino zebrafish. Exp Eye Res. 2006;82:558–75. doi: 10.1016/j.exer.2005.08.015. [DOI] [PubMed] [Google Scholar]
- Vinothkumar S, Rastegar S, Takamiya M, Ertzer R, Strähle U. Sequential and cooperative action of Fgfs and Shh in the zebrafish retina. Dev Biol. 2008;314:200–14. doi: 10.1016/j.ydbio.2007.11.034. [DOI] [PubMed] [Google Scholar]
- Waseem NH, Lane DP. Monoclonal antibody analysis of the proliferating cell nuclear antigen (PCNA). Structural conservation and the detection of a nucleolar form. J. Cell Sci. 1990;96:121–129. doi: 10.1242/jcs.96.1.121. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The zebrafish book. A guide for the laboratory use of zebrafish (Danio rerio) fourth ed Univ. of Oregon Press; Eugene: 2000. [Google Scholar]
- Winkler C, Moon RT. Zebrafish mdk2, a novel secreted midkine, participates in posterior neurogenesis. Dev Biol. 2001;229:102–18. doi: 10.1006/dbio.2000.9967. [DOI] [PubMed] [Google Scholar]
- Winkler C, Schafer M, Duschl J, Schartl M, Volff JN. Functional divergence of two zebrafish midkine growth factors following fish-specific gene duplication. Genome Res. 2003;13:1067–81. doi: 10.1101/gr.1097503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Mears AJ, Friedman JS, Carter T, He S, Oh E, Jing Y, Farjo R, Fleury G, Barlow C, Hero AO, Swaroop A. Expression profiling of the developing and mature Nrl-/- mouse retina: identification of retinal disease candidates and transcriptional regulatory targets of Nrl. Hum Mol Genet. 2004;13:1487–503. doi: 10.1093/hmg/ddh160. [DOI] [PubMed] [Google Scholar]
- Yurco P, Cameron DA. Responses of Müller glia to retinal injury in adult zebrafish. Vision Res. 2005;45:991–1002. doi: 10.1016/j.visres.2004.10.022. [DOI] [PubMed] [Google Scholar]
- Zou P, Muramatsu H, Miyata T, Muramatsu T. Midkine, a heparin-binding growth factor is expressed in neural precursor cells and promotes their growth. J Neurochem. 2006;99:1470–9. doi: 10.1111/j.1471-4159.2006.04138.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table 1. Differentially-expressed genes at 72hrs after light onset, rank ordered by log2 difference in average fluorescence intensity between control and experimental chips (see column M).
Supplemental Figure 1. Housing pigmented zebrafish to intense bright fluorescent light induces photoreceptor loss followed by regeneration. Panels a-e illustrate Nissl-stained sections from control retina (a) and retinas exposed to bright, constant light for 24hrs (b), 72hrs (c), 10 days or (d) 10 days following by 18 days of recovery. Panels f-g illustrate quantitative measurements of the photoreceptor, outer nuclear and rod nuclear layers, respectively (see panel a), normalized to the thickness of the inner nuclear layer. Note that there is a significant decrease in the thickness of the various layers starting at 72h of light exposure. Panels i-k illustrate analysis of rhodopsin, red opsin and PCNA expression by quantitative real-time PCR. ROS, rod outer segments; OLM, outer limiting membrane; OPL, outer plexiform layer; IPL, inner plexiform layer; ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer; PL, photoreceptor layer; rod ONL, rod outer nuclear layer. Scale bar equals 50μm, *p<0.05, **p<0.01.
Supplemental Figure 2. Sense probes do not produce specific cellular labeling. Panels a-d illustrate in situ hybridizations comparing mdka antisense (a, c) and sense (b, d) probes. Panels a and b are from a 96hpf larva and panels c and d are from an adult retina. Panels e-f illustrate in situ hybridizations comparing mdkb antisense (e, g) and sense (f, h) probes. Panels e and f are from a 96hpf larva, whereas panels g and h are from an adult. Arrows and arrowheads illustrate specific cellular expression when using antisense probes. GCL – ganglion cell layer; INL – inner nuclear layer; ONL – outer nuclear layer.
Supplemental Figure 3. In the adult retina mdka or mdkb is not expressed in dividing cells. Panels a and d illustrate in situ hybridizations that show the cellular expression of mdka and mdkb, respectively, in the adult retina. Panels b and e are the same sections as in a and d, following immunostaining with antibodies against PCNA, and illustrate PCNA-positive cells (rod progenitors) (arrowheads) within the outer nuclear layer. Panels c and f are digital overlays. Note that the PCNA immunostaining does not co-localize with the expression of midkines. ONL – outer nuclear layer; INL – inner nuclear layer; GCL – ganglion cell layer.
Supplemental Figure 4. Time course of mdka and mdkb expression during and after the light lesion. Panels a-h illustrate the time course of the injury-induced expression of mdka, whereas panels i-p illustrate the time course of the injury-induced expression of mdkb. Note that the expansion of mdkb expression into the vitreal inner nuclear layer begins by 24hrs after light onset (cf. panel I with Fig. 5a) and returns to normal only at 14 days after the onset of the light exposure. In contrast, the expression of mdka varies relatively little over the same time course. onl, outer nuclear layer; inl inner nuclear layer; gcl, ganglion cell layer. Scale bar equals 50μm.