Abstract
The discovery of RNA interference has revealed complex roles for small RNAs in regulating gene expression and cellular physiology. Small RNAs have been demonstrated to be involved in post-transcriptional suppression of translation, targeted degradation of messenger RNAs, and transcriptional suppression via epigenetic modifications of histones and DNA. In fission yeast, RNAi mediates suppression of centromeric transcripts, whereas in plants, transcriptional gene silencing appears to be primarily an antiviral mechanism. In mammals, the well annotated functional role of RNAi is primarily post-transcriptional, but there is increasing evidence that this mechanism can also work to suppress or modulate gene transcription, although it is not clear what primary function this serves. We overview, compare, and contrast the transcriptional silencing pathways in yeast, plants, and mammals in this article. This minireview is intended to provide the reader with a framework of how the RNAi machinery appears to be universally involved in various aspects of transcriptional regulation with discussions of similarities and differences in the components and mechanisms of achieving transcriptional silencing.
Keywords: DNA Methylation, Gene Expression, Histone Methylation, RNA Interference (RNAi), Transcription Repressor, Argonaute, RNA Interference, Epigenetic, siRNA
Introduction
The phenomenon of siRNA-triggered transcriptional gene silencing (TGS)2 is conserved across different phyla. In the fission yeast Schizosaccharomyces pombe, siRNA TGS is associated closely with heterochromatin-related gene silencing (1). In the plant Arabidopsis thaliana, siRNA-mediated TGS is associated with the establishment and maintenance of DNA methylation (RNA-directed DNA methylation) (2). In mammalian cells, studies of siRNA-triggered TGS have thus far focused only on the transcriptional silencing of protein-encoding genes via the application of promoter-targeted siRNAs (3). Although different organisms have developed diversified protein players in this process, some commonly shared features exist, especially the argonaute proteins. In general, the mechanism of siRNA TGS is composed of three mutually related parts: 1) the biogenesis and amplification of siRNA triggers, 2) molecular interactions involving an Argonaute Protein and a noncoding or coding RNA, and 3) the accompanied epigenetic changes such as histone modifications and DNA methylation.
Biogenesis and Amplification of siRNA Triggers
Genetic studies have contributed to establishing a correlation between RNAi and TGS (1, 2). Post-TGS (PTGS) and TGS share some of the same machinery to produce the siRNA triggers (4). In S. pombe, the major RNAi proteins for siRNA production during PTGS and TGS are identical, including the argonaute protein Ago1, the RNase III enzyme Dicer (Dcr1), and an RNA-dependent RNA polymerase (Rdp1), all of which are derived from single-copy genes (5). In contrast, the plant A. thaliana has developed a more complex system in which the argonaute proteins Ago4 and Ago6, the nuclear Dicer enzyme DCL2-4, and the RNA-dependent RNA polymerase Rdr2 are all required for TGS (6–9). The siRNA triggers for TGS in plants are mainly 24 nucleotides in length, which distinguishes them from the 22-nucleotide-long microRNAs (miRNAs) and other small RNAs (10). In mammalian cells, the first demonstrations of TGS were artificially induced using chemically synthesized siRNAs or siRNAs generated from polymerase (Pol) III-expressed short hairpin RNA transcripts (11–15). To date, the only mammalian endogenous small RNAs that have been demonstrated to trigger TGS are miRNAs (16, 17). For convenience, we will refer to all of the small RNA-triggered chromatin modifications, including heterochromatin-associated gene silencing in S. pombe, DNA methylation-mediated inhibition of gene repression, and miRNA/siRNA-triggered heterochromatin formation, as TGS.
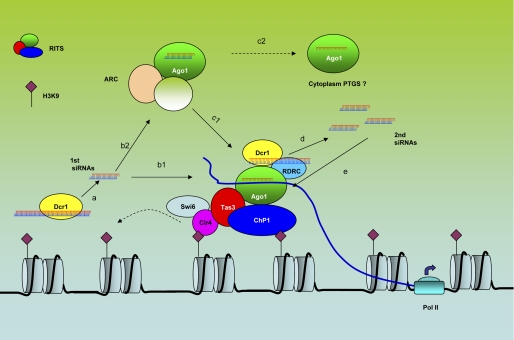
Several studies have revealed that the biogenesis of TGS siRNAs in S. pombe is coupled with a sophisticated siRNA amplification process. The centromeric repeat-originated siRNAs are used here as an example to illustrate this process (Fig. 1). In the centromeric region, bidirectional transcripts are continually generated, which anneal to form dsRNAs (18, 19). Although in low abundance, these dsRNAs are processed by dcr1 to generate the initial siRNAs. Once these are loaded into Ago1, these siRNAs direct the RITS (RNA-induced initiation of TGS) complex, containing Ago1, Chp1, and Tas3, to the centromeric region by base pairing with the local nascent transcripts. Simultaneously, RITS recruits the RNA-directed RNA polymerase complex (RDRC), which generates more dsRNAs using the nascent transcripts as the templates. These newly generated dsRNAs are then processed into the secondary siRNAs by Dcr1, increasing the local concentration of siRNAs. This model is supported by both genetic and biochemical evidence that shows that RDRC and RITS are mutually required for localizing to the centromeric region and for the biogenesis of centromeric siRNAs (19–21). Importantly but somewhat surprisingly, the production of centromeric siRNAs is also dependent on Clr4, which catalyzes the methylation of histone H3K9. RITS recognizes the H3K9me mark through one of its components, Chp1, which has a chromodomain (19). In turn, localization of RITS to the heterochromatin promotes the production and amplification of additional siRNAs.
FIGURE 1.
siRNA-directed TGS in S. pombe. Step a, low-abundance bidirectional transcripts are generated from the centromeric repeats, and these pair with the complementary strand derived from the processed siRNAs (primary siRNAs) that are produced by Dcr1. Step b1, the siRNAs are incorporated into Ago1 in the RITS complex, which is recruited to the centromeric region, where it targets local noncoding transcripts (blue lines). Step b2, alternatively, siRNAs can be loaded into Ago1 in ARC. In this complex, the siRNAs are maintained as double-stranded structures. Step c1, Ago1 in ARC is transferred to the RITS complex and recruited to the centromeric region. Step c2, the Ago1-siRNA complex can potentially shuttle to the cytoplasm for PTGS. Step d, RITS and RDRC are mutually dependent on each other for enrichment in the centromeric region, where RITS and RDRC physically interact. RDRC generates double-stranded RNAs, which are processed to secondary siRNAs by Dcr1. Step e, secondary siRNAs are more abundant than primary siRNAs and can efficiently direct more RITS complex to the centromeric region.
The centromeric siRNAs are 22–25 nucleotides in length and are the products of Dicer (19, 22, 23). A study in which the Ago1-associated siRNAs were sequenced proved that most of these siRNAs map to the centromeric region in S. pombe (24, 25). Additionally, these centromeric siRNAs originate from both strands, without bias. Alternatively, some of these centromeric siRNAs might be the products of the structured unidirectional RNAs generated from the centromeric repeats (26). A parallel scheme for siRNA generation and amplification in plants has not been reported. However, several lines of evidence indicate that plants might exploit a similar strategy. First, the plant Rdr2 enzyme is homologous to Rdp1 in S. pombe and has been shown to be indispensible for TGS in plants (6). An A. thaliana mutant strain, drm1/2-cmt3 (DRM (domains rearranged methylase), CMT3 (chromomethylase 3)), in which DNA methylation activity is absent, has a greatly reduced abundance of chromatin-targeting siRNAs (27–29). Thus, there is a requirement for the generation of siRNAs for the establishment of DNA methylation. The secondary siRNAs, which are the amplification products of Rdr2, are also detected in a silencing trigger system (30). A striking and unique feature of siRNA production in plants is the involvement of two atypical polymerases, Pol IV and Pol V (31, 32). These two atypical polymerases, together with many other RNAi-associated proteins, are found to co-localize in the nucleolar siRNA-processing center termed the cajal body (33, 34). Of note, these siRNA-processing centers do not overlap with the DNA methylation sites.
To date, in mammals, promoter-targeting siRNA triggers have either been delivered with cationic lipids and the siRNAs have been chemically synthesized siRNAs, or they have been produced from plasmids using promoter-expressed shRNAs. Two features of siRNA-mediated TGS in mammalian cells should be noted. First, not all the synthesized promoter-targeted siRNAs or shRNAs induce TGS. In fact, two closely related siRNAs can have dramatically different efficiencies. Second, except for a single report (35), it is uncertain how long the TGS effect can last because siRNAs in particular will be diluted as cell division continues. To this point, in plant cells, it was found that DNA methylation is reduced, and gene silencing is released as the siRNA triggers are removed (36). Thus, in mammals, TGS triggered by exogenous siRNAs differs in duration and potency compared with TGS mediated by endogenous siRNAs in plants and yeast.
Molecular Interaction Networks Centered on Argonaute Proteins and Noncoding RNAs
In S. pombe, the central role of Ago1 in TGS is well understood, as it is central in connecting siRNA biogenesis with histone modifications. For instance, in Ago1 mutants, the siRNA abundance is greatly reduced with a concomitant loss of H3K9 methylation (22). In addition, Ago1 slicing activity is required for TGS (37). A recent study showed the presence of another Ago1 complex, named the argonaute siRNA chaperone (ARC), in S. pombe (38). Interestingly, the slicing activity of Ago1 in this complex is inhibited by a protein partner, Arb1. When in this complex, the Ago1 siRNAs are trapped in a precursor state and are unable to trigger TGS.
The importance of RNA Pol II in S. pombe TGS was first demonstrated in a study showing that the heterochromatin remodeling via TGS is compromised in a Pol II mutant (39). Follow-up research highlighted the interaction between RITS and the nascent centromeric repeat transcripts, which act as a platform for the assembly of the TGS machinery (40). In this study, RITS was tethered to a reporter gene locus via an λN-peptide, which tethered it to Tas3, which binds to its cognate RNA-binding site BoxB. Strikingly, tethering RITS to the nascent transcripts resulted in the generation of siRNAs from the reporter gene and resulted in the generation of siRNAs, histone H3 Lys-9 methylation, and Swi6 (a kind of chromodomain protein in yeast) binding.
In the plant A. thaliana, a different argonaute family member, Ago4, was found to interact or co-localize with various important components in TGS, including RNA Pol IV, RNA Pol V, and RNA Pol II. As mentioned above, Pol IV and Pol V are two atypical RNA polymerases uniquely conserved in the plant kingdom (41). It has been demonstrated that Pol IV acts in concert with Ago4 upstream of the generation of siRNAs, whereas Pol V and Ago4 function together downstream of siRNA production (42–44). Ago4 binds to Pol V to mediate the production of noncoding transcripts in the intergenic region required for silencing of overlapping or adjacent genes (33, 45). However, further studies showed that Pol II and not Pol V is responsible for transcribing the noncoding RNA scaffold required for assembling the TGS complex (46). This alternative model is supported by a recent report showing that Pol II is associated with Ago4 at the TGS target sites in the nucleus (47). The same study also identified a new protein, RDM1, which binds single strands of methylated DNA that are unwound during Pol V- or Pol II-mediated transcription. The role of RDM1 may be to direct the TGS effector complex to the siRNA complementary target sites.
To date, in mammals, both Ago1 and Ago2, along with RNA Pol II, have been shown to be involved in siRNA-directed TGS (13, 14). A direct interaction between the Pol II C-terminal domain and Ago1 was also detected (14, 15). This interaction may be conserved in yeast, plants, and animals. However, it is not clearly understood how Ago1 or Ago2 associates with the Pol II-generated noncoding transcripts to form the TGS effector complex.
Histone Modification and DNA Methylation
In S. pombe, Clr4 (a H3K9 methyltransferase) is directly involved in the initial H3K9 methylation as well as its spreading. The major components of this process include Clr4, Swi6, RITS, RDRC, and a CenH DNA element. The CenH region acts as the nucleation site, which contains repeat sequences that are homologous to the centromeric region. RITS and RDRC work in concert with Clr4 to establish the initial H3K9me mark in the CenH region (48). However, the spreading of this mark to outside the nucleation site requires Swi6 and Clr4. Clr4 can catalyze the methylation of H3K9 and concomitantly bind to this marker via its chromodomain (49). Swi6 is involved via recruitment of the histone deacetylase complex to promote the spreading of H3K9 methylation. The RITS component Tas3 promotes H3K9me spreading via interaction with Chp1 (50).
In plants, DNA methylation occurs in three different sequence contexts: CpG, CpNpG, and CpNpN (where N represents A, T, or C) (2). CpNpN is an asymmetrical methylation site. During RNA-directed DNA methylation (TGS in plants), de novo methylation is mediated by three methyltransferases: methyltransferases 1 and 2 (DRM1/2) and chromomethyltransferase 3 (27, 28, 51). The maintenance of DNA CpG methylation requires another methyltransferase, Met1 (52). In addition to methyltransferases, chromatin-remodeling proteins such as DDM1 (decreased in DNA methylation 1), DRD1 (defective in RNA-directed DNA methylation 1) (53), and CLSY1 (CLASSY 1; also known as CHR38, a putative chromatin-remodeling factor) have also been shown to be involved in plant TGS.
Histone modification has been shown to function cooperatively with DNA methylation for gene silencing, as indicated by the silencing of the SUP locus in A. thaliana (54). In this model, the maintenance of CpNpG methylation is dependent on the H3K9 methyltransferase Kryptonite, which catalyzes H3K9me, which then allows LHP1 (like heterochromatin protein 1; homologous to metazoan HP1) to bind. In turn, LHP1 recruits CMT3 to maintain the methylation state of CpNpG sites (55). In mammalian cells, there is not yet a good paradigm for siRNA-induced chromatin modifications. For instance, a siRNA targeting the EF1A promoter was shown to induce DNA methylation (11), whereas in a different study, siRNAs did not trigger DNA methylation of the CDH promoter (12). These contradictory results are most likely due to the different cell models and experimental condition used in these studies. Additionally, the local chromatin structure in which the promoter resides might play a major role in this process, as do the number and density of CpG motifs.
siRNA-triggered Transcriptional Gene Activation and Promoter-associated Noncoding Transcripts
Up to now, siRNA-triggered transcriptional gene activation (TGA) has been described only in human cell lines (56–58). Li et al. (56) first reported that some promoter-targeting siRNAs can trigger the activation of the downstream gene. This phenomenon was shown to be sequence-specific, and it involved Ago2. The transcriptional activation was shown to be accompanied by an increase in the levels of the positive histone marker H3K4me and was not related to an IFN response. In another study, siRNA-triggered TGA was proposed to be mediated through targeting of a promoter-associated antisense transcript that is the target for the Ago2-siRNA complex (58). The mechanism by which Ago2 is involved in TGA is currently unknown. One study suggested that TGA is induced by Ago2-mediated cleavage of the promoter-associated antisense transcripts, which releases repression of the promoter (59). According to this model, siRNA-triggered TGA is more likely to be a post-transcriptional process that takes place in either the nucleus or cytoplasm. However, both TGA and TGS were triggered by siRNAs targeting the progesterone receptor promoter, suggesting that both TGA and TGS are closely related processes (57).
Another striking finding related to siRNA-mediated TGS or TGA is the complexity of the transcriptome in eukaryotic cells (29, 60, 61). In human cells, several RNA species were found to be associated with known gene loci in a genome-wide analysis, which include promoter-associated RNAs, terminus-associated small RNAs, and promoter upstream transcripts (62–64). It will be important to determine whether or not these newly described RNA species are associated with TGA or TGS in human cells, perhaps acting as scaffolds for argonaute binding.
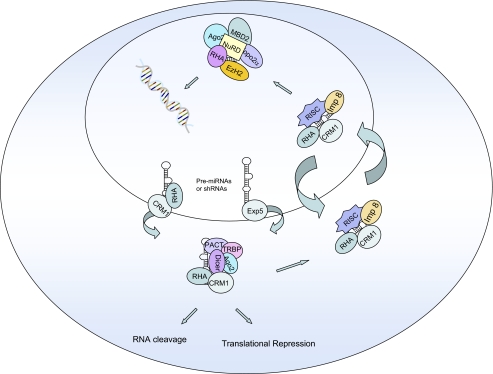
Two studies have demonstrated that miRNAs trigger TGS of nuclear genes via targeting promoter regions (16, 17). These findings suggest that miRNAs and perhaps siRNAs and Ago proteins traffic between the cytoplasm and nucleus. Recent findings from our laboratory demonstrate that such trafficking indeed occurs via the CRM1 transport carrier, and Ago1, Ago2, EzH2, Topo2α, and Mta are in a complex that traffics between the cytoplasm and nucleus (Fig. 2) (65). Thus, cytoplasmically processed miRNAs or siRNAs (from shRNA precursors) can enter the nucleus via this mechanism to trigger TGS (Fig. 2).
FIGURE 2.
CRM1-mediated trafficking on Ago proteins and chromatin-remodeling proteins in mammalian cells. The small RNAs associated with the Ago proteins could be miRNAs or siRNAs. RHA, RNA helicase A; RISC, RNA-induced silencing complex; PACT, protein kinase R-activating protein; TRBP, HIV-1 TAR RNA-binding protein; NuRD, nucleosome-remodeling deacetylase.
Future Directions
In comparison with plants and yeast, studies of the mechanism of TGS in mammalian cells are still at an early stage. Three directions may assist in expanding our knowledge of the mechanistic aspects of TGS in mammalian cells. The first is to profile the genome-wide association of Ago1/2 by using ChIP-on-chip and DNA sequencing. However, such experiments have to be designed with caution to differentiate authentic signals from background noise. The use of an Ago1 or Ago2 knock-out cell line would serve as an excellent negative control. In addition, such experiments require specific anti-Ago1 versus anti-Ago2 antibodies. Establishment of a genome-wide map of Ago1 and Ago2 chromatin association patterns would be invaluable for better understanding of TGS and TGA in mammalian cells. Furthermore, it should be possible to identify the relationship between Ago-binding sites and other epigenetic markers such as methylated histones and/or DNA methylation patterns and methyl DNA-binding proteins.
The second suggested direction is to focus on the gene loci whose promoter regions overlap with or are included in noncoding RNA transcripts. As discussed above, noncoding RNAs seem to act as sources of siRNAs as well as scaffolds for TGS in both plants and yeast.
The third direction is to learn from studies of TGS in the other systems. For example, the HxK model, widely used in plant TGS studies, can be helpful in addressing TGS mechanisms in mammalian cells (66). H constructs express the target promoter sequences as the TGS triggers, whereas in the K construct, the target promoter controls the expression of a reporter gene. TGS can be studied in plant progeny after two parent transgenic plants, carrying the H vector and the K vector, respectively, are crossed. Such a model has the advantage of freely manipulating both the target and TGS trigger. Finally, as new TGS-associated proteins are identified in other organisms, it will be important to determine whether homologs exist in mammalian cells and, if so, whether or not these are associated with TGS or TGA.
It is safe to state that small RNA-triggered TGS and now TGA are exciting mechanisms for modulating gene expression. Understanding the details of these processes will add to our knowledge of the consequences of these phenomena in development and ultimately in diseases such as cancer.
This work was supported, in whole or in part, by National Institutes of Health Grants AI29329 and HL07470 (to J. J. R.). This is the fifth article in the Thematic Minireview Series on Epigenetics. This minireview will be reprinted in the 2011 Minireview Compendium, which will be available in January, 2012.
- TGS
- transcriptional gene silencing
- PTGS
- post-TGS
- miRNA
- microRNA
- Pol
- polymerase
- RDRC
- RNA-directed RNA polymerase complex
- ARC
- argonaute siRNA chaperone
- TGA
- transcriptional gene activation.
REFERENCES
- 1. Grewal S. I. S. (2010) Curr. Opin. Genet. Dev. 20, 134–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Matzke M., Kanno T., Daxinger L., Huettel B., Matzke A. J. (2009) Curr. Opin. Cell Biol. 21, 367–376 [DOI] [PubMed] [Google Scholar]
- 3. Morris K. V. (2008) Curr. Top. Microbiol. Immunol. 320, 211–224 [DOI] [PubMed] [Google Scholar]
- 4. Matzke M. A., Birchler J. A. (2005) Nat. Rev. 6, 24–35 [DOI] [PubMed] [Google Scholar]
- 5. Sigova A., Rhind N., Zamore P. D. (2004) Genes Dev. 18, 2359–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xie Z., Johansen L. K., Gustafson A. M., Kasschau K. D., Lellis A. D., Zilberman D., Jacobsen S. E., Carrington J. C. (2004) PLoS Biol. 2, E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zilberman D., Cao X., Jacobsen S. E. (2003) Science 299, 716–719 [DOI] [PubMed] [Google Scholar]
- 8. Henderson I. R., Zhang X., Lu C., Johnson L., Meyers B. C., Green P. J., Jacobsen S. E. (2006) Nat. Genet. 38, 721–725 [DOI] [PubMed] [Google Scholar]
- 9. Zheng X., Zhu J., Kapoor A., Zhu J. K. (2007) EMBO J. 26, 1691–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hamilton A., Voinnet O., Chappell L., Baulcombe D. (2002) EMBO J. 21, 4671–4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Morris K. V., Chan S. W., Jacobsen S. E., Looney D. J. (2004) Science 305, 1289–1292 [DOI] [PubMed] [Google Scholar]
- 12. Ting A. H., Schuebel K. E., Herman J. G., Baylin S. B. (2005) Nat. Genet. 37, 906–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Janowski B. A., Huffman K. E., Schwartz J. C., Ram R., Nordsell R., Shames D. S., Minna J. D., Corey D. R. (2006) Nat. Struct. Mol. Biol. 13, 787–792 [DOI] [PubMed] [Google Scholar]
- 14. Kim D. H., Villeneuve L. M., Morris K. V., Rossi J. J. (2006) Nat. Struct. Mol. Biol. 13, 793–797 [DOI] [PubMed] [Google Scholar]
- 15. Weinberg M. S., Villeneuve L. M., Ehsani A., Amarzguioui M., Aagaard L., Chen Z. X., Riggs A. D., Rossi J. J., Morris K. V. (2006) RNA 12, 256–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Younger S. T., Corey D. R. (March 22, 2011) Nucleic Acids Res. 10.1093/nar/gkr155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim D. H., Saetrom P., Snøve O., Jr., Rossi J. J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 16230–16235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Martienssen R. A. (2003) Nat. Genet. 35, 213–214 [DOI] [PubMed] [Google Scholar]
- 19. Verdel A., Jia S., Gerber S., Sugiyama T., Gygi S., Grewal S. I., Moazed D. (2004) Science 303, 672–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Motamedi M. R., Verdel A., Colmenares S. U., Gerber S. A., Gygi S. P., Moazed D. (2004) Cell 119, 789–802 [DOI] [PubMed] [Google Scholar]
- 21. Sugiyama T., Cam H., Verdel A., Moazed D., Grewal S. I. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 152–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Volpe T. A., Kidner C., Hall I. M., Teng G., Grewal S. I., Martienssen R. A. (2002) Science 297, 1833–1837 [DOI] [PubMed] [Google Scholar]
- 23. Reinhart B. J., Bartel D. P. (2002) Science 297, 1831. [DOI] [PubMed] [Google Scholar]
- 24. Cam H. P., Sugiyama T., Chen E. S., Chen X., FitzGerald P. C., Grewal S. I. (2005) Nat. Genet. 37, 809–819 [DOI] [PubMed] [Google Scholar]
- 25. Hansen K. R., Burns G., Mata J., Volpe T. A., Martienssen R. A., Bähler J., Thon G. (2005) Mol. Cell. Biol. 25, 590–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Simmer F., Buscaino A., Kos-Braun I. C., Kagansky A., Boukaba A., Urano T., Kerr A. R., Allshire R. C. (2010) EMBO Rep. 11, 112–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cao X., Aufsatz W., Zilberman D., Mette M. F., Huang M. S., Matzke M., Jacobsen S. E. (2003) Curr. Biol. 13, 2212–2217 [DOI] [PubMed] [Google Scholar]
- 28. Cao X., Jacobsen S. E. (2002) Curr. Biol. 12, 1138–1144 [DOI] [PubMed] [Google Scholar]
- 29. Lister R., O'Malley R. C., Tonti-Filippini J., Gregory B. D., Berry C. C., Millar A. H., Ecker J. R. (2008) Cell 133, 523–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Daxinger L., Kanno T., Bucher E., van der Winden J., Naumann U., Matzke A. J., Matzke M. (2009) EMBO J. 28, 48–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Herr A. J., Jensen M. B., Dalmay T., Baulcombe D. C. (2005) Science 308, 118–120 [DOI] [PubMed] [Google Scholar]
- 32. Kanno T., Huettel B., Mette M. F., Aufsatz W., Jaligot E., Daxinger L., Kreil D. P., Matzke M., Matzke A. J. M. (2005) Nat. Genet. 37, 761–765 [DOI] [PubMed] [Google Scholar]
- 33. Li C. F., Pontes O., El-Shami M., Henderson I. R., Bernatavichute Y. V., Chan S. W., Lagrange T., Pikaard C. S., Jacobsen S. E. (2006) Cell 126, 93–106 [DOI] [PubMed] [Google Scholar]
- 34. Pontes O., Li C. F., Nunes P. C., Haag J., Ream T., Vitins A., Jacobsen S. E., Pikaard C. S. (2006) Cell 126, 79–92 [DOI] [PubMed] [Google Scholar]
- 35. Hawkins P. G., Santoso S., Adams C., Anest V., Morris K. V. (2009) Nucleic Acids Res. 37, 2984–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Aufsatz W., Mette M. F., van der Winden J., Matzke A. J., Matzke M. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, Suppl. 4, 16499–16506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Irvine D. V., Zaratiegui M., Tolia N. H., Goto D. B., Chitwood D. H., Vaughn M. W., Joshua-Tor L., Martienssen R. A. (2006) Science 313, 1134–1137 [DOI] [PubMed] [Google Scholar]
- 38. Buker S. M., Iida T., Bühler M., Villén J., Gygi S. P., Nakayama J., Moazed D. (2007) Nat. Struct. Mol. Biol. 14, 200–207 [DOI] [PubMed] [Google Scholar]
- 39. Kato H., Goto D. B., Martienssen R. A., Urano T., Furukawa K., Murakami Y. (2005) Science 309, 467–469 [DOI] [PubMed] [Google Scholar]
- 40. Bühler M., Verdel A., Moazed D. (2006) Cell 125, 873–886 [DOI] [PubMed] [Google Scholar]
- 41. Pikaard C. S., Haag J. R., Ream T., Wierzbicki A. T. (2008) Trends Plant Sci. 13, 390–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang X., Henderson I. R., Lu C., Green P. J., Jacobsen S. E. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 4536–4541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pontier D., Yahubyan G., Vega D., Bulski A., Saez-Vasquez J., Hakimi M. A., Lerbs-Mache S., Colot V., Lagrange T. (2005) Genes Dev. 19, 2030–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Haag J. R., Pontes O., Pikaard C. S. (2009) PLoS ONE 4, e4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. El-Shami M., Pontier D., Lahmy S., Braun L., Picart C., Vega D., Hakimi M. A., Jacobsen S. E., Cooke R., Lagrange T. (2007) Genes Dev. 21, 2539–2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zheng B., Wang Z., Li S., Yu B., Liu J. Y., Chen X. (2009) Genes Dev. 23, 2850–2860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gao Z., Liu H. L., Daxinger L., Pontes O., He X., Qian W., Lin H., Xie M., Lorkovic Z. J., Zhang S., Miki D., Zhan X., Pontier D., Lagrange T., Jin H., Matzke A. J., Matzke M., Pikaard C. S., Zhu J. K. (2010) Nature 465, 106–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hall I. M., Shankaranarayana G. D., Noma K., Ayoub N., Cohen A., Grewal S. I. (2002) Science 297, 2232–2237 [DOI] [PubMed] [Google Scholar]
- 49. Zhang K., Mosch K., Fischle W., Grewal S. I. (2008) Nat. Struct. Mol. Biol. 15, 381–388 [DOI] [PubMed] [Google Scholar]
- 50. Li H., Motamedi M. R., Yip C. K., Wang Z., Walz T., Patel D. J., Moazed D. (2009) Mol. Cell 34, 155–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lindroth A. M., Cao X., Jackson J. P., Zilberman D., McCallum C. M., Henikoff S., Jacobsen S. E. (2001) Science 292, 2077–2080 [DOI] [PubMed] [Google Scholar]
- 52. Kankel M. W., Ramsey D. E., Stokes T. L., Flowers S. K., Haag J. R., Jeddeloh J. A., Riddle N. C., Verbsky M. L., Richards E. J. (2003) Genetics 163, 1109–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kanno T., Mette M. F., Kreil D. P., Aufsatz W., Matzke M., Matzke A. J. (2004) Curr. Biol. 14, 801–805 [DOI] [PubMed] [Google Scholar]
- 54. Johnson L., Cao X., Jacobsen S. (2002) Curr. Biol. 12, 1360–1367 [DOI] [PubMed] [Google Scholar]
- 55. Jackson J. P., Lindroth A. M., Cao X., Jacobsen S. E. (2002) Nature 416, 556–560 [DOI] [PubMed] [Google Scholar]
- 56. Li L. C., Okino S. T., Zhao H., Pookot D., Place R. F., Urakami S., Enokida H., Dahiya R. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 17337–17342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Janowski B. A., Younger S. T., Hardy D. B., Ram R., Huffman K. E., Corey D. R. (2007) Nat. Chem. Biol. 3, 166–173 [DOI] [PubMed] [Google Scholar]
- 58. Schwartz J. C., Younger S. T., Nguyen N. B., Hardy D. B., Monia B. P., Corey D. R., Janowski B. A. (2008) Nat. Struct. Mol. Biol. 15, 842–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Morris K. V., Santoso S., Turner A. M., Pastori C., Hawkins P. G. (2008) PLoS Genet. 4, e1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wilhelm B. T., Marguerat S., Watt S., Schubert F., Wood V., Goodhead I., Penkett C. J., Rogers J., Bähler J. (2008) Nature 453, 1239–1243 [DOI] [PubMed] [Google Scholar]
- 61. Katayama S., Tomaru Y., Kasukawa T., Waki K., Nakanishi M., Nakamura M., Nishida H., Yap C. C., Suzuki M., Kawai J., Suzuki H., Carninci P., Hayashizaki Y., Wells C., Frith M., Ravasi T., Pang K. C., Hallinan J., Mattick J., Hume D. A., Lipovich L., Batalov S., Engström P. G., Mizuno Y., Faghihi M. A., Sandelin A., Chalk A. M., Mottagui-Tabar S., Liang Z., Lenhard B., Wahlestedt C. (2005) Science 309, 1564–1566 [DOI] [PubMed] [Google Scholar]
- 62. Seila A. C., Calabrese J. M., Levine S. S., Yeo G. W., Rahl P. B., Flynn R. A., Young R. A., Sharp P. A. (2008) Science 322, 1849–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kapranov P., Cheng J., Dike S., Nix D. A., Duttagupta R., Willingham A. T., Stadler P. F., Hertel J., Hackermüller J., Hofacker I. L., Bell I., Cheung E., Drenkow J., Dumais E., Patel S., Helt G., Ganesh M., Ghosh S., Piccolboni A., Sementchenko V., Tammana H., Gingeras T. R. (2007) Science 316, 1484–1488 [DOI] [PubMed] [Google Scholar]
- 64. Preker P., Nielsen J., Kammler S., Lykke-Andersen S., Christensen M. S., Mapendano C. K., Schierup M. H., Jensen T. H. (2008) Science 322, 1851–1854 [DOI] [PubMed] [Google Scholar]
- 65. Castanotto D., Lingeman R., Riggs A. D., Rossi J. J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 21655–21659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mette M. F., Aufsatz W., van der Winden J., Matzke M. A., Matzke A. J. M. (2000) EMBO J. 19, 5194–5201 [DOI] [PMC free article] [PubMed] [Google Scholar]