Abstract
Induction of endotoxin tolerance leads to a reduced inflammatory response after repeated challenge by LPS and is important for resolution of inflammation and prevention of tissue damage. Enterobacterial LPS is recognized by the TLR4 signaling complex, whereas LPS of some non-enterobacterial organisms is capable of signaling independently of TLR4 utilizing TLR2-mediated signal transduction instead. In this study we report that Porphyromonas gingivalis LPS, a TLR2 agonist, fails to induce a fully endotoxin tolerant state in a human monocytic cell line (THP-1) and mouse bone marrow-derived macrophages. In contrast to significantly decreased production of human IL-8 and TNF-α and, in mice, keratinocyte-derived cytokine (KC), macrophage inflammatory protein-2 (MIP-2), and TNF-α after repeated challenge with Escherichia coli LPS, cells repeatedly exposed to P. gingivalis LPS responded by producing less TNF-α but sustained elevated secretion of IL-8, KC, and MIP-2. Furthermore, in endotoxin-tolerant cells, production of IL-8 is controlled at the signaling level and correlates well with NF-κB activation, whereas TNF-α expression is blocked at the gene transcription level. Interferon β plays an important role in attenuation of chemokine expression in endotoxin-tolerized cells as shown in interferon regulatory factor-3 knock-out mice. In addition, human gingival fibroblasts, commonly known not to display LPS tolerance, were found to be tolerant to repeated challenge by LPS if pretreated with interferon β. The data suggest that the inability of the LPS-TLR2 complex to induce full endotoxin tolerance in monocytes/macrophages is related to diminished production of interferon β and may partly explain the involvement of these LPS isoforms in the pathogenesis of chronic inflammatory diseases.
Keywords: Chemokines, Cytokine, Endotoxin, Interferon, Toll-like receptors (TLR), Chronic Inflammation, Endotoxin Tolerance
Introduction
Detection of pathogen-associated molecular patterns by Toll-like receptors (TLRs)2 expressed on innate immune cells triggers a robust and essential inflammatory reaction. Inflammation as a well coordinated process that comprises increased vascular permeability, migration of polymorphonuclear leukocytes, monocytes, and lymphocytes into affected tissues, and activation of cells to secrete inflammatory mediators is essential for host defense (1). If it is well controlled and resolved in a timely manner, it benefits the host by elimination of the invading pathogen. Otherwise, prolonged or excessive inflammation leads to chronicity and tissue damage (2).
Toll-like receptors represent a family of evolutionarily highly conserved transmembrane molecules that act as pathogen recognition receptors. To date, 13 mammalian TLRs have been identified, and each appears to be required for responses to a different class of infectious pathogen (3). Almost immediately after microbes invade, microbial products signal through TLRs, broadly distributed on immune cells, activating these cells to produce proinflammatory cytokines, interferons, histamine, and antimicrobial peptides (4). Toll-like receptor signaling represents a principal molecular pathway for host innate immunity (5). All members of the TLR superfamily signal in a similar manner and activate common signaling pathways, most notably those leading to the activation of the transcription factors NF-κB and IRF (6).
TLR signaling may be divided into two distinct pathways; one leading to the MyD88-dependent arm-triggering expression of proinflammatory cytokines and the other leading to the MyD88-independent arm (TRIF/TRAM-mediated) responsible for interferon type I production (7). Myeloid differentiation factor 88 (MyD88)-dependent signaling is common to all the TLRs except TLR3, which exclusively utilizes the myeloid differentiation factor 88-independent pathway. TLR4 is unique in that it can trigger both the MyD88-dependent and MyD88-independent pathways (8). Signaling through the TLR4 pathway is one of the principal molecular mechanisms for the detection of Gram-negative pathogens and their LPS by host immune cells (9). The rapid response against LPS can be of benefit to the host in moderate levels by promoting inflammation and priming the immune system to eradicate the invading pathogens. However, an excessive response to LPS, which is not properly resolved, can lead to chronic inflammatory conditions (10).
One of the mechanisms involved in the regulation of the inflammatory response is the phenomenon of endotoxin tolerance. Endotoxin tolerance is a protective mechanism in which repeated exposure of host immune cells to endotoxin results in repressed expression of proinflammatory cytokines (11). This phenomenon was attributed to the monocyte/macrophage lineage of the human immune system and was reproduced in animal models and reported in humans (12). On the other hand, human skin and gingival fibroblasts, which do not display LPS tolerance or negative regulators of inflammatory response, were shown to sustain an inflammatory response in the presence of virulence factors (13). Most known regulatory mechanisms target the TLR signaling pathway and thus broadly inhibit multiple aspects of the inflammatory response (14). In addition to this robust signaling-based control mechanism, an elegant gene-specific regulatory mechanism exists to allow individual aspects of the TLR-induced response and genes to be differentially regulated (15). Over the past several years, many negative regulators of TLRs have been identified, and this negative regulation is achieved at multiple levels ranging from extracellular decoy receptors (soluble TLR4 and TLR2) and membrane-bound suppressors (ST2, SIGIRR) to intracellular inhibitors (IRAKM, SOCS1, NOD2, TOLLIP) and epigenetic control of gene expression (16).
To date TLR4 seems to be the most heavily regulated of all the TLRs (17). The reason for this might be related to the potential, extreme toxicity of TLR4 signaling. Cells of myeloid lineage are capable of recognizing picomolar quantities of LPS and respond via several signal transduction cascades with the release of a wide range of proinflammatory cytokines (18). Enterobacterial LPS is recognized by a signaling complex comprising at least CD14, TLR4, and MD-2 (19). However, LPS of non-enterobacterial organisms, such as Porphyromonas gingivalis, Bacteroides fragilis, Chlamydia trachomatis, and Helicobacter pylori are capable of signaling independent of TLR4 and utilizing TLR2-mediated signal transduction instead (20). Interestingly, all of these bacteria, in which LPS activate the TLR2 signaling mechanism, are involved in the pathogenesis of chronic inflammatory diseases: periodontitis, inflammatory bowel disease, urogenital infection, and gastric ulcers, respectively (21–24). The ability of these bacteria to cause chronic inflammation could be a consequence of less defined LPS-TLR2 signaling control mechanisms and differential induction of endotoxin tolerance by TLR4 and TLR2 agonists. Our aim was to examine differences in the mechanism of induction of endotoxin tolerance by canonical Escherichia coli LPS, a TLR4 agonist, and LPS isoforms, which activate TLR2.
We have already reported that impaired immune tolerance to P. gingivalis LPS is responsible for neutrophil-dominated chronic inflammation seen in periodontitis (25). In the current study we show that in contrast to E. coli LPS-induced tolerance, which is characterized by the down-regulation of human IL-8 and TNF-α and mouse KC, MIP-2, and TNF-α production, P. gingivalis LPS-pretreated human monocytes and mouse bone marrow-derived macrophages remained able to secret IL-8 and KC and MIP-2, respectively, but production of TNF-α was significantly decreased. Because E. coli LPS, as a TLR4 agonist, activates both MyD88 and TRIF pathways and is connected to successful down-regulation of cytokine/chemokine production in endotoxin-tolerant cells and TLR2 signals only through MyD88 pathway, we hypothesized that different control mechanisms for chemokine and TNF-α production exist in endotoxin-tolerant cells and that IFN-β plays a pivotal role in the control of the NF-κB signaling cascade and chemokine secretion.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
LPS from E. coli O55:B5 was obtained from Sigma. LPS from P. gingivalis ATCC 33277 was obtained from InvivoGen, San Diego, CA. MALDI-TOF mass spectrometry of this P. gingivalis LPS revealed predominant peaks corresponding to penta-acylated diphosphorylated lipid A isoform, already proven to be a TLR2 agonist (26). Recombinant human IFN-β was purchased from Peprotech, and low endotoxin azide-free-purified anti-human IFN-β antibody was from BioLegend. Mouse IgG1 isotype control to anti-IFN-β antibody was obtained from R&D Systems. Rabbit anti-IκB-α IgG was from Cell Signaling, and goat anti-rabbit IgG HRP-conjugated antibody was from Santa Cruz Biotechnology.
Cell Culture
Human THP-1 monocytes were obtained from the European Collection of Cell Cultures (ECACC) and maintained in RPMI 1640 medium supplemented with 10% fetal calf serum, 2 mm l-glutamine, penicillin (100 units/ml), and streptomycin (100 μg/ml) (Invitrogen). THP-1 cells were cultured at 37 °C, 100% humidity, and 5% CO2 at 5 × 105 cells/ml density. Bone marrow-derived macrophages were prepared from wild-type and IRF3 knock-out C57BL/6 mice (a kind gift from Prof. T. Taniguchi, University of Tokyo) and cultured for 1 week in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FCS, 1% penicillin/streptomycin, and 1% l-glutamine. Proliferation was driven by granulocyte macrophage-colony-stimulating factor derived from L929 supernatant. Human gingival fibroblasts (HGFs) were established from explants of healthy gingival tissues obtained during routine clinical procedures as described previously (27). The study was approved by the Research Ethics Committee of Northern Ireland, participant information sheets were provided, and written informed consent was obtained from patients wishing to participate in the study. HGFs were cultured to confluence in DMEM (Invitrogen) supplemented with 10% heat-inactivated fetal calf serum, 2 mm l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin in a humidified atmosphere of 5% CO2 at 37 °C at 8 × 104 cells/ml density. The cells between the 5th and 13th passages were used for assays. Twenty-four hours before treatment the medium was changed to 1% FCS DMEM supplemented with the same concentrations of antibiotics and glutamine.
Induction of Endotoxin Tolerance and Cytokine Measurement
A monocyte sepsis model was used for induction of endotoxin tolerance (28). Briefly, THP-1 cells were treated with 1 μg/ml E. coli or P. gingivalis LPS for 24 h, washed 3 times with serum-free medium, and re-treated once with the same concentration of LPS for 4 h. Mouse bone marrow-derived macrophages were treated with 100 ng/ml LPS. Human gingival fibroblasts were treated with 10 ng/ml E. coli LPS and 10 μg/ml P. gingivalis LPS. Control cells were incubated in medium alone and re-treated in the same way as pretreated cells. Cell-free supernatants from tolerized and naïve (preincubated in medium alone) cells were collected after 4 h, and IL-8, KC, MIP-2, and mouse TNF-α were measured by ELISA kits according to the manufacturer's instructions (R&D Systems, Abingdon, UK) as was human TNF-α (PeproTech EC Ltd., London, UK). IFN-β was measured in the cell supernatants after 24 h of treatment by ELISA (PBL Interferon Source). Cell viability after 24 h or pretreatment was confirmed by MTT assay (29).
Western Blot Analysis
THP-1 cells (5 × 105/ml) pretreated with medium or LPS for 24 h were restimulated for the indicated time periods noted in the figures and then lysed on ice for 30 min in 35 μl of lysis buffer (20 mm Tris, pH 7.4, 150 mm NaCl, 1 mm EDTA, 1% Nonidet P-40, 0.1% SDS, 0.5% sodium deoxycholate, 1 mm sodium orthovanadate, 1 mm sodium flouride, 1 mm PMSF, and proteinase inhibitor coctail (Roche Applied Science)). Cell debris was pelleted by centrifugation, and supernatants were collected and stored at −80 °C until assayed. Thirty micrograms of total cellular protein were denatured at 100 °C for 5 min in loading buffer (60 mm Tris, 2.5% SDS, 10% glycerol, 5% mercaptoethanol, 0.01% bromphenol blue) and subjected to 10% SDS-PAGE. Gels were transferred to nitrocellulose membranes and blocked with PBS containing 0.05% Tween 20 (PBS-T) and 5% nonfat milk powder for 1 h. After washing in PBS-T, membranes were probed with rabbit anti-IκB-α IgG (1:1000) (Cell Signaling) for 2 h at room temperature. Membranes were then washed with PBS-T and incubated with a polyclonal secondary goat anti-rabbit IgG HRP Ab (1:2000; Santa Cruz Biotechnology) for 1 h at room temperature. After three washes in PBS-T, membranes were developed using ECL substrate (GE Healthcare) according to the manufacturer's protocol (Thermo Scientific).
Quantitative Real-time PCR
Total RNA from control and tolerized THP-1 cells treated with 1 μg/ml E. coli and P. gingivalis LPS for 4 h was prepared using RNeasy kit (Qiagen) according to the manufacturer's instructions. One microgram of total RNA was reverse-transcribed for 1 h at 42 °C using an oligo(dT)12–18 primer (Invitrogen) and the SuperScript II RT kit (Invitrogen). Quantitative real-time PCR was performed using the SYBR Green PCR core reagents mix (Applied Biosystems) containing 1× SYBR Green PCR buffer, 3 mm MgCl2, 100 μm dATP, dCTP, and dGTP, 200 μm deoxyuridine triphosphate, 0.025 units/μl AmpliTaq Gold DNA polymerase, 0.01 units/μl AmpErase uracil N-glycosylase, and 2 pmol/μl gene-specific forward and reverse primers designed by the Eurofins MWG Operon: IL-8 forward (CTTGTCATTGCCAGCTGTGT) and reverse (TGACTGTGGAGTTTTGGCTG); TNF-α forward (TGGCCAATGGCGTGGAGCTG) and reverse (AGACGGCGATGCGGCTGATG). The reaction conditions were as follows: 2 min at 50 °C (1 cycle), 10 min at 95 °C (1 cycle), 15 s at 95 °C, and 1 min at 60 °C (40 cycles). Gene-specific PCR products were amplified using an Applied Biosystems PRISM 7500 detection system (PerkinElmer Life Sciences). Samples were normalized using the 18 S ribosomal unit as a housekeeping gene. Three replicates for each experimental point were performed, and differences were assessed with the two-tailed Student's t test. Results are expressed as the relative -fold changes of the stimulated over the control group, which was used as a calibrator.
Statistical Analysis
Differences between the means of treatments were analyzed by the Student's t test using GraphPad Prism Version 4 (GraphPad Software, San Diego, CA). Differences between multiple treatments were compared by one-way analysis of variance followed by Tukey's post test. Values are expressed as the mean ± S.E. A value of p < 0.05 was considered to represent a statistically significant difference (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
RESULTS
Effect of E. coli and P. gingivalis LPS Retreatment on IκB-α Degradation in THP-1 Cells
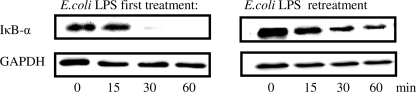
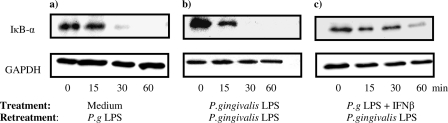
Because activation of NF-κB is indispensable for proinflammatory gene activation, we detected IκB-α degradation in THP-1 cells. Control and either E. coli LPS-tolerized or P. gingivalis LPS-tolerized THP-1 cells were treated with the same LPS for 15, 30, and 60 min, and IκB-α was detected by Western blot. 1 μg/ml E. coli LPS induced rapid degradation of IκB-α in control human monocytes, but in E. coli LPS-tolerized cells re-treated with the same LPS (1 μg/ml), IκB-α degradation was significantly reduced (Fig. 1).
FIGURE 1.
IκB-α degradation in THP-1 cells after the first and repeated challenge with 1 μg/ml E. coli LPS. Control and E. coli LPS tolerized THP-1 cells were treated with 1 μg/ml E. coli LPS for the indicated time points. Whole cell lysates were immunoblotted for IκB-α and GAPDH as a loading control. Blots are representative of three separate experiments.
Degradation of IκB-α in THP-1 cells treated with 1 μg/ml P. gingivalis LPS occurred with similar kinetics to monocytes treated with E. coli LPS. However, repeated treatment with P. gingivalis LPS failed to induce significantly reduced degradation of IκB-α, indicating that in contrast to E. coli LPS-tolerized cells, these monocytes were still responsive (Fig. 2).
FIGURE 2.
IκB-α degradation in THP-1 cells after the first and repeated challenge with 1 μg/ml P. gingivalis LPS. Control and P. gingivalis LPS-tolerized THP-1 cells were treated with 1 μg/ml P. gingivalis LPS for the indicated time points. Whole cell lysates were immunoblotted for IkB-α and GAPDH as a loading control. Blots are representative of three separate experiments.
Effect of E. coli and P. gingivalis LPS Retreatment on TNF-α and IL-8 Expression in THP-1 Cells
Due to the involvement of the NF-κB pathway in the expression of both TNF-α and IL-8 genes, we examined transcription of both genes in control and tolerized monocytes. Both E. coli and P. gingivalis LPS induced a significant increase in TNF-α and IL-8 mRNA in THP-1 cells after the first treatment. In E. coli LPS-re-treated cells transcription of both genes was significantly reduced; however, in P. gingivalis LPS-re-treated cells IL-8 expression remained high, whereas TNF-α expression significantly dropped (Table 1).
TABLE 1.
Differential regulation of IL-8 and TNF-α mRNA expression in E. coli and P. gingivalis LPS-tolerized cells
RT-PCR analysis of TNF-α and IL-8 gene expression in control and tolerized THP-1 cells shows selective inhibition of TNF-α only in P. gingivalis LPS-tolerized cells. THP-1 cells were pretreated with 1 μg/ml concentrations of either E. coli LPS or P. gingivalis LPS and re-treated with the same LPS. Numbers show -fold increase in TNF-α and IL-8 expression by control and pretreated THP-1 cells after 4 h of LPS retreatment. 18 S mRNA was used for the internal control. Values represent the -fold increase ± S.E. of three different experiment.
| Cytokine | Medium/Medium | Medium/E. coli LPS | E. coli LPS/E. coli LPS | Medium/P. gingivalis LPS | P. gingivalis LPS/P. gingivalis LPS |
|---|---|---|---|---|---|
| IL-8 | 1 | 943.4 ± 83.2 | 214.8 ± 38.6*** | 815.2 ± 61.7 | 796.0 ± 53.4 |
| TNF-α | 1 | 114.7 ± 28.3 | 23.3 ± 7.9*** | 107.4 ± 18.4 | 27.8 ± 4.9*** |
***, p < 0.001.
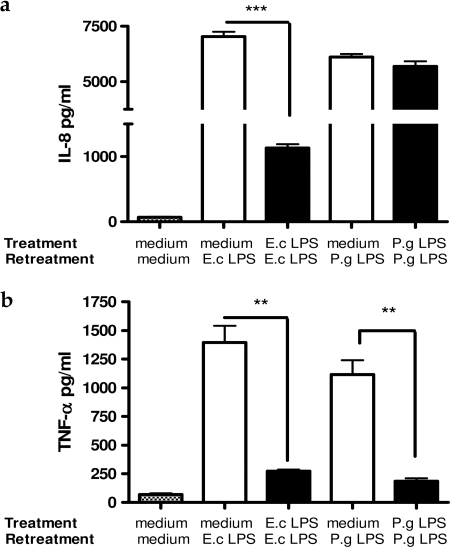
In agreement with detected levels of TNF-α and IL-8 mRNA after initial challenge with 1 μg/ml E. coli LPS, THP-1 cells responded by producing high levels of IL-8 and TNF-α. Repeated challenge of THP-1 cells with the same concentration of E. coli LPS significantly decreased production of both cytokines (p < 0.01) (Fig. 3, a and b). THP-1 cells treated with 1 μg/ml P. gingivalis LPS responded in a similar manner as cells treated with E. coli LPS. However, repeated challenge with P. gingivalis LPS almost completely abolished production of TNF-α (p < 0.01), whereas IL-8 concentration remained as high as it was after the first challenge (Fig. 3, a and b).
FIGURE 3.
Production of IL-8 (a) and TNF-α (b) by THP-1 cells after the first and repeated challenge by 1 μg/ml E. coli (E.c) LPS or P. gingivalis (P.g) LPS. Values represent the mean ± S.E. of n = 3.
Taken together, these results suggest the existence of different regulatory mechanisms for TNF-α and IL-8 secretion in endotoxin-re-treated monocytes. IL-8 secretion in tolerized cells is signaling-dependent, whereas production of TNF-α is controlled at the transcriptional level. Furthermore, TLR2-induced endotoxin tolerance is only partial, with persistently high secretion of IL-8 but reduced production of TNF-α.
Evaluation of E. coli LPS and P. gingivalis LPS Cross-tolerance
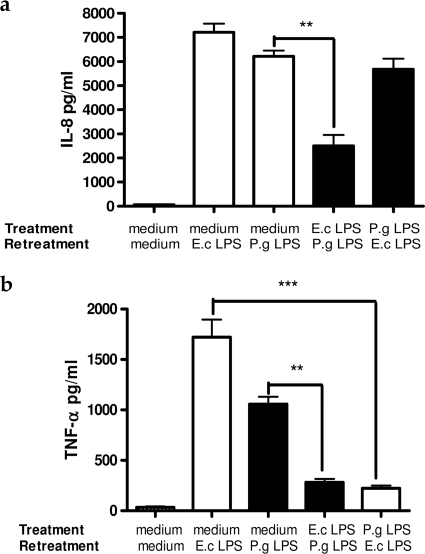
To examine if biologically active substances secreted during 24 h of pretreatment influence the induction of endotoxin tolerance, cross-tolerance between E. coli and P. gingivalis LPS was examined. THP-1 cells were tolerized with E. coli LPS and then re-treated with P. gingivalis LPS and vice versa. After 4 h of re-treatment, the production of IL-8 and TNF-α was measured in the cell supernatants. E. coli LPS induced cross-tolerance after subsequent exposure to P. gingivalis LPS, and the concentrations for both IL-8 and TNF-α were significantly lower in comparison to control cells. Interestingly, in the cells tolerized with P. gingivalis LPS and re-treated with E. coli LPS, TNF-α production dropped significantly, but IL-8 levels remained at similar levels to those observed in control cells (Figs. 4, a and b).
FIGURE 4.
IL-8 (a) and TNF-α (b) production by THP-1 cells cross-tolerized with E. coli (E.c) LPS and P. gingivalis (P.g) LPS. THP-1 cells were pretreated with 1 μg/ml E. coli LPS for 24 h and re-treated with 1 μg/ml P. gingivalis LPS for 4 h or vice versa. Control THP-1 cells were incubated in medium alone and re-treated with 1 μg/ml E. coli or P. gingivalis LPS. Supernatant was removed, and IL-8 and TNF-α levels were assessed by ELISA.
Influence of Interferon β on Induction of Endotoxin Tolerance in THP-1 Cells
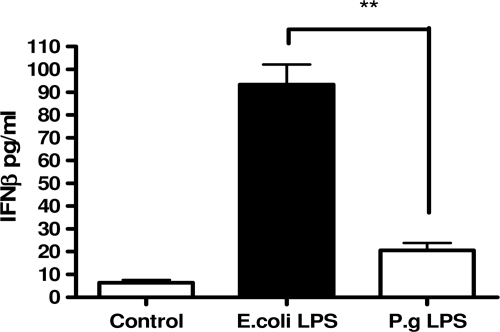
Because P. gingivalis LPS is a TLR2 agonist and activates only the MyD88-dependent pathway, whereas E. coli LPS activates both MyD88-dependent and TRIF pathway, we examined the influence of IFN-β as a product of the TRIF pathway on induction of immune tolerance. First, we observed that the concentration of IFN-β was significantly higher in the THP-1 cells treated with E. coli LPS for 24 h than in those treated with P. gingivalis LPS (Fig. 5). Furthermore, concentrations of IFN-β in supernatants of human gingival fibroblasts, the cells that do not display endotoxin tolerance, treated with either E. coli LPS or P. gingivalis LPS were undetectable (data not shown).
FIGURE 5.
IFN-β production assessed in THP-1 cells supernatants treated with 1 μg/ml E. coli LPS and P. gingivalis LPS for 24 h. Values represent the mean ± S.E. of n = 3.
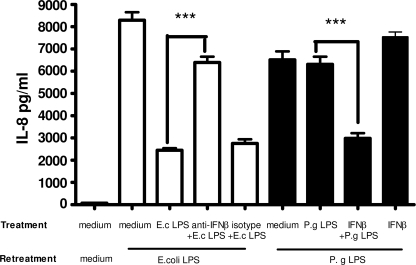
To examine the influence of IFN-β on induction of endotoxin tolerance, recombinant human IFN-β (0.1 ng/ml) was used during P. gingivalis LPS 24-h pretreatment. This particular concentration of IFN-β was chosen as it approximated the concentration of IFN-β detected in THP-1 cells supernatants treated with E. coli LPS for 24 h. On the other hand, the addition of an anti-IFN-β antibody during the E. coli LPS 24-h pretreatment was used to neutralize the IFN-β effect. Subsequently, the cells were re-treated with the same LPS and IκB-α degradation, and IL-8 and TNF-α production were measured.
In THP-1 cells pretreated with 1 μg/ml P. gingivalis LPS in combination with 0.1 ng/ml IFN-β for 24 h and re-treated with the same LPS for 15, 30, and 60 min, IκB-α degradation was delayed in comparison to THP-1 cells pretreated only with P. gingivalis LPS (Fig. 6). ELISA results revealed that the addition of IFN-β during the 24-h P. gingivalis LPS pretreatment caused significantly decreased production of IL-8 in P. gingivalis LPS rechallenged cells. The addition of IFN-β-neutralizing antibody during E. coli LPS pretreatment recovered IL-8 production by THP-1 cells re-treated with the same LPS. Isotype-matched control antibody did not influence IL-8 secretion after repeated challenge with E. coli LPS (Fig. 7). Interestingly, the addition of IFN-β-neutralizing antibody during the 24-h E. coli LPS pretreatment did not alter the endotoxin tolerant state of THP-1 cells with respect to TNF-α production (Fig. 8).
FIGURE 6.
The addition of rIFN-β during P. gingivalis (P.g) LPS pretreatment delays IκB-α degradation after repeated challenge with P. gingivalis LPS. THP-1 cells were pretreated for 24 h with medium alone (a), 1 μg/ml P. gingivalis LPS (b), and a combination of 1 μg/ml P. gingivalis LPS and 0.1 ng/ml IFN-β (c). Cells were washed and re-treated with 1 μg/ml P. gingivalis LPS for the indicated time points. Whole cell lysates were immunoblotted for IκB-α and GAPDH as a loading control. Blots are representative of three separate experiments.
FIGURE 7.
The addition of rIFN-β during 24 h P. gingivalis (P.g) LPS pretreatment significantly decreases production of IL-8 by THP-1 cells after repeated exposure to P. gingivalis LPS. Neutralizing IFN-β antibody included during E. coli (E.c) LPS pretreatment recovered IL-8 production by THP-1 cells re-treated with E. coli LPS. Isotype control antibody did not have influence on establishment of endotoxin tolerance. rIFN-β alone is not able to induce endotoxin tolerance. IL-8 in cell supernatants was assessed by ELISA. Values represent the mean ± S.E. of n = 3 (endotoxin activity of rIFN-β < 0.1 ng/μg).
FIGURE 8.
IFN-β does not have effect on decreased production of TNF-α after repeated challenge with E. coli LPS. THP-1 cells were pretreated with medium alone, 1 μg/ml E. coli (E.c) LPS, 1 μg/ml E. coli LPS + 1 μg/ml anti-IFNβ antibody, and 1 μg/ml E. coli LPS + 1 μg/ml isotype control antibody and re-treated with 1 μg/ml E. coli LPS. TNF-α was measured in the cell supernatants by ELISA. Values represent the mean ± S.E. of n = 3.
Our results show that IL-8 and TNF-α production in endotoxin-tolerant cells is controlled by a different mechanism and at distinct levels. In addition, IFN-β is responsible for induction of endotoxin tolerance with respect to IL-8 production, and the relative absence of IFN-β due to signaling only via the MyD88-dependent pathway may in fact explain the lack of endotoxin tolerance to IL-8 secretion after repeated challenge with P. gingivalis LPS.
Evaluation of E. coli LPS and P. gingivalis LPS Induced Endotoxin Tolerance in Wild-type and IRF3-deficient Mouse Bone Marrow-derived Macrophages
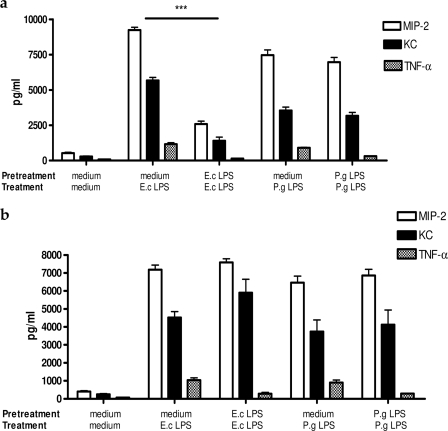
In wild-type mouse bone marrow-derived macrophages, similar to THP-1 cells, TNF-α production was significantly down-regulated in the cells re-treated with either E. coli LPS or P. gingivalis LPS, but production of the chemokines KC and MIP-2 decreased only in E. coli LPS-tolerized cells (Fig. 9a). In contrast to wild type, IRF3−/− macrophages behaved in the same manner when treated with either E. coli or P. gingivalis LPS. Notably, only TNF-α production was down-regulated in tolerized IRF3−/− macrophages, whereas production of KC and MIP-2 remained high in both E. coli and P. gingivalis LPS-re-treated cells (Fig. 9b).
FIGURE 9.
Wild-type mouse bone marrow-derived macrophages reflect the cytokine pattern produced by THP-1 cells, whereas IRF3−/− macrophages down-regulate only TNF-α production after repeated challenge with either E. coli LPS (E.c) or P. gingivalis (P.g) LPS. Wild-type (a) and IRF3 knock-out (b) bone marrow-derived macrophages (5 × 105 cells/ml) were pretreated with medium alone, 100 ng/ml E. coli LPS, or 100 ng/ml P. gingivalis LPS for 24 h and re-treated with the same agonists for 4 h. Mouse MIP-2, KC, and TNF-α were measured in the cells supernatants by ELISA. Values represent the mean ± S.E. of n = 3.
Interferon β Induces Endotoxin Tolerance in Non-tolerant Human Gingival Fibroblasts
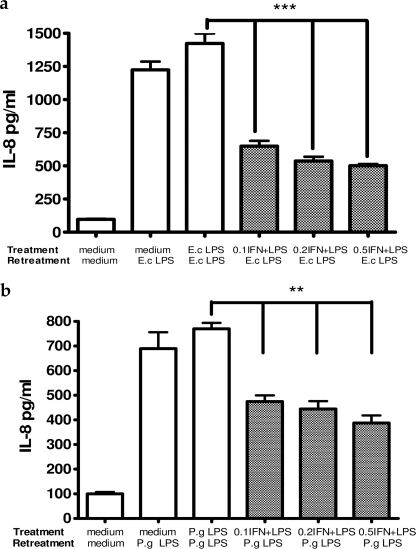
Human gingival fibroblasts, widely known not to display the phenomenon of endotoxin tolerance, were used to further examine the influence of IFN-β on induction of the tolerant state. HGF were pretreated with either 10 ng/ml E. coli LPS or 10 μg/ml P. gingivalis LPS in combination with different concentrations of IFN-β (0.1, 0.2, 0.5 ng/ml) followed by LPS re-treatment for 4 h. IL-8 concentrations detected in HGF supernatants are presented in Fig. 10. IFN-β, included as a co-stimulus during induction of endotoxin tolerance, significantly decreased the production of IL-8 in a dose-dependent manner after repeated challenge with both E. coli and P. gingivalis LPS in otherwise non-tolerant HGFs.
FIGURE 10.
Interferon β included as a co-stimulant during LPS pretreatment induces endotoxin tolerance in human gingival fibrobalsts after repeated challenge with both E. coli LPS (a) and P. gingivalis LPS (b) (concentrations of IFN-β are in ng/ml). HGFs (8 × 104 cells/ml) were pretreated with 10 ng/ml E. coli LPS or 10 μg/ml P. gingivalis LPS with or without the addition of rhIFN-β for 24 h and re-treated with the same LPS for 4 h. IL-8 was measured in the cells supernatants. Values represent the mean ± S.E. of n = 3.
DISCUSSION
The immune system needs to constantly strike a balance between activation and inhibition to avoid detrimental and inappropriate inflammatory responses, and as a result TLR signaling must be tightly regulated to maintain immunological balance. TLR activation is a double-edged sword. It is essential for provoking the innate response and enhancing adaptive immunity against pathogens (30). However, the signal that is transmitted from TLRs must be well controlled, and there is clear evidence that if TLRs are overactivated, infectious and inflammatory disease can result (31). It has been shown that members of the TLR family are involved in the pathogenesis of autoimmune, chronic inflammatory, and infectious diseases such as periodontitis, chronic obstructive pulmonary disease, asthma, atherosclerosis, and systemic lupus erythematosus (32).
Endotoxin tolerance is a mechanism for regulation of the immune response and is defined as transient unresponsiveness to repeated doses of LPS (33). The significance of this refractory state of the hypo-responsiveness has been revealed by studies demonstrating that macrophages from surviving septic shock patients display LPS tolerance (34). Functionally, endotoxin-tolerant monocytes/macrophages exhibit an increased phagocytic ability coupled with a conserved capacity to kill internalized pathogens, albeit with an impaired antigen presentation capacity (35). Considering the in vivo relevance of the above phenotype, poor inflammatory capacity coupled with up-regulation of anti-inflammatory cytokines could contribute to protection against tissue damage, and increased phagocytosis would allow efficient bacterial clearance.
LPS tolerance has traditionally been viewed as a hyporesponsive state of immune cells resulting from receptor desensitization, and the majority of studies have concentrated on decreased production of TNF-α after repeated exposure to canonical E. coli LPS and its interaction with TLR4 (36). However, comparison of the LPS molecules of different organisms revealed that there are structural differences in the chemical composition of naturally occurring lipid A. The number of phosphate groups attached to the glucosamine backbone and the number and length of acyl chains determine whether the LPS will be a TLR4 or TLR2 agonist (37). Based on current evidence, it seems that rather being the exception to the rule, TLR2 signaling LPS may be commonly represented among Gram-negative species (20).
We show here that non-canonical, TLR2-stimulating LPS from P. gingivalis, a key factor in the development of neutrophil-dominated chronic inflammation associated with periodontitis, exhibits impaired endotoxin tolerance compared with E. coli LPS in both the cell line (THP-1) and primary cells (bone marrow-derived macrophages). TLR2 induced immune tolerance by repeated challenge with peptidoglycan, Pam3CSK4, and P. gingivalis LPS has been described before but only with respect to TNF-α production (38, 39). Our current data indicate that the cytokine network produced by THP-1 cells and mouse bone-derived macrophages repeatedly challenged with TLR2-activating LPS (P. gingivalis LPS) differs from those treated with TLR4-activating LPS (E. coli LPS). TNF-α and IL-8 production in THP-1 cells and TNF-α, KC, and MIP-2 in mouse macrophages were significantly reduced after repeated challenge by E. coli LPS, whereas persistent high production of IL-8 as well as KC and MIP-2 accompanied with diminished secretion of TNF-α was observed in P. gingivalis LPS-re-treated cells. In addition, we demonstrated that impaired endotoxin tolerance induced by the LPS-TLR2 complex occurs as a result of the inability of TLR2 agonists to produce significant amounts of IFN-β, which is responsible for inhibition of NF-κB activation.
We also showed that in endotoxin-tolerant cells there are different regulatory mechanisms for the expression of genes that belong to the same functional category (proinflammatory cytokines). Both IL-8 and TNF-α are secreted principally as a response to MyD88-dependent pathway activation and NF-κB nuclear translocation. Nevertheless, IL-8 gene expression in endotoxin-tolerant cells was signaling-specific, whereas transcription of TNF-α gene did not correlate with activation of the NF-κB pathway. The inability of P. gingivalis LPS as a TLR2 agonist to reduce degradation of IκB-α after repeated challenge resulted in a persistent high production of IL-8 in contrast to TNF-α production that was significantly down-regulated. Differences in IκB proteins degradation have also been reported by Martin et al. (40) who observed maintained degradation of IκB-β after repeated challenge by P. gingivalis LPS but not E. coli LPS. High concentrations of IL-8 have been found in gingival crevicular fluid from periodontitis patients, suggesting an important role for this potent chemokine in the disease pathogenesis (41). Furthermore, Muthukuru et al. (42) reported that monocytes from periodontitis patients were more resistant to down-regulation of IL-8 production after repeated challenge with P. gingivalis LPS compared with other cytokines. In a broader view of chemokines, Foster and Medzhitov (43) showed that in mouse bone marrow macrophages, genes for chemokine ligand 9 (Cxcl9) and chemokine ligand 8 (Ccl8) belonged to the “non-tolerizeable” genes group whose promoters are open after repeated challenge with LPS. Transcription of such open promoter requires an LPS signal that induces NF-κB activation and its binding to the DNA promoter regions.
In contrast to IL-8 expression, which is signaling-dependent, it has been shown that production of TNF-α in endotoxin-tolerant cells is controlled at the level of transcription. The silencing of TNF-α production in endotoxin-tolerant THP-1 cells is a result of dimethylation on histone H3 at the TNF-α promoter. This process correlates with diminished binding of the active NF-κB to the promoter and decreased production of TNF-α (28). This could explain differential production of IL-8 and TNF-α by monocytes/macrophages repeatedly challenged with P. gingivalis LPS. We can speculate that despite persistent activation of the NF-κB pathway in P. gingivalis LPS-re-treated monocytes, TNF-α production was reduced due to histone remodeling at its promoter region, whereas IL-8 remained high because of uninterrupted binding of NF-κB to IL-8 non-modified promoter.
Interleukin 8 as well as its mouse counterparts KC and MIP-2 are principal mediators of the inflammatory response that attract leukocytes to the site of infection leading to neutrophil infiltration, which if not controlled may culminate in host tissue damage. Therefore, down-regulation of chemokine production is vital in the prevention of chronic inflammation (45). Results obtained from heterotolerance experiments where THP-1 cells pretreated with E. coli LPS and re-treated with P. gingivalis LPS displayed a decrease in IL-8 production, whereas P. gingivalis LPS pretreatment did not have an effect on IL-8 production after E. coli LPS rechallenge, prompted us to examine the role of IFN-β on chemokine secretion in endotoxin tolerant cells. First, we observed significantly higher production of IFN-β after 24 h of treatment of THP-1 cells with E. coli LPS compared with the same treatment with P. gingivalis LPS. Similar findings were reported by Dobrovolskaia et al. (46) and Toshchakov et al. (47), who found poorly induced IFN-β gene in murine macrophages in response to TLR2 activation. Interestingly, IFN-β concentrations in human gingival fibroblasts supernatants treated for 24 h with either E. coli or P. gingivalis LPS were undetectable, and these cells were even primed to subsequent LPS challenge with respect to IL-8 production. Second, we included recombinant IFN-β during P. gingivalis LPS pretreatment and neutralizing IFN-β antibody during E. coli LPS pretreatment of THP-1 cells. We found that pretreatment with P. gingivalis LPS in combination with rIFN-β delayed degradation of IκB-α and significantly decreased production of IL-8 after repeated challenge with P. gingivalis LPS. Interestingly, the addition of IFN-β neutralizing antibody during the E. coli LPS pretreatment only partially alleviated tolerance to E. coli LPS with respect to IL-8 production, indicating the existence of other regulatory mechanisms. Using a mouse model, Shahangian et al. (48) demonstrated a less effective response with regard to KC and MIP-2 production in wild-type influenza-infected mice secondary challenged with Streptococcus pneumoniae in comparison to mice deficient for type IFN-α/β receptor. IRF3 transcription factor is essential for lipopolysaccharide-induced interferon β gene expression (49). We have shown in IRF3-deficient macrophages that there is no difference in KC and MIP-2 production after repeated challenge with either E. coli or P. gingivalis LPS. Sustained high production of KC and MIP-2 by tolerized IRF3 knock-out macrophages confirms an important role of interferon β in chemokine down-regulation during the state of endotoxin tolerance. In addition, co-stimulation of HGFs with IFN-β and LPS induced significantly lower production of IL-8 after repeated challenge with LPS in these cells, which normally do not display endotoxin tolerance. IFN-β has been shown to prime human gingival fibroblasts to subsequent LPS response but to decrease IL-8 production if incubated at the same time with LPS (50).
An essential function of IFN-β is its antiviral activity, which affects almost all cell types infected with a broad spectrum of viruses. Additionally, and perhaps more important for its involvement in endotoxin tolerance, IFN-β has anti-proliferative and immunomodulatory functions (51). It has been used successfully as one of the therapy options for patients with multiple sclerosis (52). With respect to endotoxin tolerance, it has been shown that in contrast to the MyD88-dependent pathway, which is down-regulated during TLR4-induced endotoxin tolerance, the TRIF pathway and production of IFN-β are up-regulated (53). Furthermore, Biswas and Tergaonkar (8) reported a direct effect on IFN-β on TNF-α production by TLR4-tolerized murine macrophages but not in TLR2-tolerized cells. In our study, pretreatment of human monocytic cells line with IFN-β on its own did not make them tolerant to subsequent E. coli or P. gingivalis LPS challenge, whereas in combination with P. gingivalis LPS it successfully inhibited production of IL-8 after repeated exposure to the same LPS.
TLR2 has been found to be involved in many chronic inflammatory diseases. Elevated expression of proinflammatory cytokines in periodontitis patients has been assigned to TLR2-induced and -amplified response (54). Studies on chronic inflammation in the human gastric mucosa caused by H. pylori infection showed involvement of TLR2 as well (55). The absence of TLR2 protein expression by intestinal epithelial cells has been shown to be important in preventing chronic proinflammatory cytokine secretion in response to commensal bacteria in the gut (56), whereas up-regulation of TLR4 expression in colon protects mice from colitis (44).
Our study indicates that impaired and partial endotoxin tolerance induced in monocytes/macrophages by non-canonical, TLR2-activating LPS, characterized by persistent high secretion of IL-8 due to the lack of immunomodulatory effect of IFN-β, could be responsible for detrimental consequences of chronic TLR2 activation.
Footnotes
- TLR
- Toll-like receptor
- MyD88
- myeloid differentiation factor 88
- HGF
- human gingival fibroblast
- rIFN-β
- recombinant IFN-β
- IRF3
- interferon regulatory factor 3
- TRIF
- TIR domain-containing adaptor-inducing IFN-β
- KC
- keratinocyte-derived cytokine
- MIP-2
- macrophage inflammatory protein-2.
REFERENCES
- 1. Cochran D. L. (2008) J. Periodontol. 79, 1569–1576 [DOI] [PubMed] [Google Scholar]
- 2. Drexler S. K., Foxwell B. M. (2010) Int. J. Biochem. Cell Biol. 42, 506–518 [DOI] [PubMed] [Google Scholar]
- 3. Boyd C. R., Orr S. J., Spence S., Burrows J. F., Elliott J., Carroll H. P., Brennan K., Ní, Gabhann J., Coulter W. A., Jones C., Crocker P. R., Johnston J. A., Jefferies C. A. (2009) J. Immunol. 183, 7703–7709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yeaman M. R., Yount N. Y. (2007) Nat. Rev. Microbiol. 5, 727–740 [DOI] [PubMed] [Google Scholar]
- 5. Beutler B., Hoebe K., Du X., Ulevitch R. J. (2003) J. Leukoc. Biol. 74, 479–485 [DOI] [PubMed] [Google Scholar]
- 6. Pandey S., Agrawal D. K. (2006) Immunol. Cell Biol. 84, 333–341 [DOI] [PubMed] [Google Scholar]
- 7. Kawai T., Akira S. (2007) Semin. Immunol. 19, 24–32 [DOI] [PubMed] [Google Scholar]
- 8. Biswas S. K., Tergaonkar V. (2007) Int. J. Biochem. Cell Biol. 39, 1582–1592 [DOI] [PubMed] [Google Scholar]
- 9. Yang I. V., Alper S., Lackford B., Rutledge H., Warg L. A., Burch L. H., Schwartz D. A. (2010) Am. J. Respir. Cell Mol. Biol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. De Nardo D., Nguyen T., Hamilton J. A., Scholz G. M. (2009) Cell. Signal. 21, 246–252 [DOI] [PubMed] [Google Scholar]
- 11. Xiong Y., Qiu F., Piao W., Song C., Wahl L. M., Medvedev A. E. (2011) J. Biol. Chem. 286, 7905–7916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cavaillon J. M., Adib-Conquy M. (2006) Crit. Care 10, 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ara T., Kurata K., Hirai K., Uchihashi T., Uematsu T., Imamura Y., Furusawa K., Kurihara S., Wang P. L. (2009) J. Periodontal. Res. 44, 21–27 [DOI] [PubMed] [Google Scholar]
- 14. Albrecht V., Hofer T. P., Foxwell B., Frankenberger M., Ziegler-Heitbrock L. (2008) BMC Immunol. 9, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. El Gazzar M., McCall C. E. (2010) J. Biol. Chem. 285, 20940–20951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lepper P. M., Triantafilou M., O'Neill L. A., Novak N., Wagner H., Parker A. E., Triantafilou K. (2011) Mediators Inflamm., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pan H., Ding E., Hu M., Lagoo A. S., Datto M. B., Lagoo-Deenadayalan S. A. (2010) J. Immunol. 184, 5502–5509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brown J., Wang H., Hajishengallis G. N., Martin M. (2011) J. Dent. Res. 90, 417–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Park B. S., Song D. H., Kim H. M., Choi B. S., Lee H., Lee J. O. (2009) Nature 458, 1191–1195 [DOI] [PubMed] [Google Scholar]
- 20. Erridge C., Pridmore A., Eley A., Stewart J., Poxton I. R. (2004) J. Med. Microbiol. 53, 735–740 [DOI] [PubMed] [Google Scholar]
- 21. Muthukuru M., Cutler C. W. (2008) Infect. Immun. 76, 477–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rhee K. J., Wu S., Wu X., Huso D. L., Karim B., Franco A. A., Rabizadeh S., Golub J. E., Mathews L. E., Shin J., Sartor R. B., Golenbock D., Hamad A. R., Gan C. M., Housseau F., Sears C. L. (2009) Infect. Immun. 77, 1708–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dieterle S. (2008) Andrologia 40, 117–119 [DOI] [PubMed] [Google Scholar]
- 24. Zaric S., Bojic B., Jankovic Lj, Dapcevic B., Popovic B., Cakic S., Milasin J. (2009) J. Dent. Res. 88, 946–950 [DOI] [PubMed] [Google Scholar]
- 25. Zaric S., Shelburne C., Darveau R., Quinn D. J., Weldon S., Taggart C. C., Coulter W. A. (2010) Infect. Immun. 78, 4151–4156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kumada H., Haishima Y., Watanabe K., Hasegawa C., Tsuchiya T., Tanamoto K., Umemoto T. (2008) Oral Microbiol. Immunol. 23, 60–69 [DOI] [PubMed] [Google Scholar]
- 27. Takada H., Mihara J., Morisaki I., Hamada S. (1991) Infect. Immun. 59, 295–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. El Gazzar M., Liu T., Yoza B. K., McCall C. E. (2010) J. Biol. Chem. 285, 1259–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Aoshiba K., Yasui S., Hayashi M., Tamaoki J., Nagai A. (1999) J. Immunol. 162, 1692–1700 [PubMed] [Google Scholar]
- 30. Akira S., Hemmi H. (2003) Immunol. Lett. 85, 85–95 [DOI] [PubMed] [Google Scholar]
- 31. Serhan C. N., Brain S. D., Buckley C. D., Gilroy D. W., Haslett C., O'Neill L. A., Perretti M., Rossi A. G., Wallace J. L. (2007) FASEB J. 21, 325–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cook D. N., Pisetsky D. S., Schwartz D. A. (2004) Nat. Immunol. 5, 975–979 [DOI] [PubMed] [Google Scholar]
- 33. Nahid M. A., Pauley K. M., Satoh M., Chan E. K. (2009) J. Biol. Chem. 284, 34590–34599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Adib-Conquy M., Adrie C., Fitting C., Gattolliat O., Beyaert R., Cavaillon J. M. (2006) Crit. Care Med. 34, 2377–2385 [DOI] [PubMed] [Google Scholar]
- 35. Monneret G., Venet F. (2010) Expert Rev. Anti Infect. Ther. 8, 1109–1112 [DOI] [PubMed] [Google Scholar]
- 36. Medvedev A. E., Kopydlowski K. M., Vogel S. N. (2000) J. Immunol. 164, 5564–5574 [DOI] [PubMed] [Google Scholar]
- 37. Erridge C., Bennett-Guerrero E., Poxton I. R. (2002) Microbes Infect. 4, 837–851 [DOI] [PubMed] [Google Scholar]
- 38. Bagchi A., Herrup E. A., Warren H. S., Trigilio J., Shin H. S., Valentine C., Hellman J. (2007) J. Immunol. 178, 1164–1171 [DOI] [PubMed] [Google Scholar]
- 39. Nakayama K., Okugawa S., Yanagimoto S., Kitazawa T., Tsukada K., Kawada M., Kimura S., Hirai K., Takagaki Y., Ota Y. (2004) J. Biol. Chem. 279, 6629–6634 [DOI] [PubMed] [Google Scholar]
- 40. Martin M., Katz J., Vogel S. N., Michalek S. M. (2001) J. Immunol. 167, 5278–5285 [DOI] [PubMed] [Google Scholar]
- 41. Kim Y. J., Viana A. C., Curtis K. M., Orrico S. R., Cirelli J. A., Scarel-Caminaga R. M. (2009) DNA Cell Biol. 28, 185–190 [DOI] [PubMed] [Google Scholar]
- 42. Muthukuru M., Jotwani R., Cutler C. W. (2005) Infect. Immun. 73, 687–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Foster S. L., Medzhitov R. (2009) Clin. Immunol. 130, 7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chaniotou Z., Giannogonas P., Theoharis S., Teli T., Gay J., Savidge T., Koutmani Y., Brugni J., Kokkotou E., Pothoulakis C., Karalis K. P. (2010) Gastroenterology 139, 2083–2092 [DOI] [PubMed] [Google Scholar]
- 45. Shelburne C. E., Coopamah M. D., Sweier D. G., An F. Y., Lopatin D. E. (2007) Cell. Microbiol. 9, 1611–1619 [DOI] [PubMed] [Google Scholar]
- 46. Dobrovolskaia M. A., Medvedev A. E., Thomas K. E., Cuesta N., Toshchakov V., Ren T., Cody M. J., Michalek S. M., Rice N. R., Vogel S. N. (2003) J. Immunol. 170, 508–519 [DOI] [PubMed] [Google Scholar]
- 47. Toshchakov V., Jones B. W., Lentschat A., Silva A., Perera P. Y., Thomas K., Cody M. J., Zhang S., Williams B. R., Major J., Hamilton T. A., Fenton M. J., Vogel S. N. (2003) J. Endotoxin Res. 9, 169–175 [DOI] [PubMed] [Google Scholar]
- 48. Shahangian A., Chow E. K., Tian X., Kang J. R., Ghaffari A., Liu S. Y., Belperio J. A., Cheng G., Deng J. C. (2009) J. Clin. Invest. 119, 1910–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sakaguchi S., Negishi H., Asagiri M., Nakajima C., Mizutani T., Takaoka A., Honda K., Taniguchi T. (2003) Biochem. Biophys. Res. Commun. 306, 860–866 [DOI] [PubMed] [Google Scholar]
- 50. Sakuta T., Tokuda M., Tamura M., Jimi E., Ikebe T., Koba T., Nagaoka S., Takada H. (1998) J. Dent. Res. 77, 1597–1605 [DOI] [PubMed] [Google Scholar]
- 51. Bendtzen K. (2010) J. Interferon Cytokine Res. 30, 759–766 [DOI] [PubMed] [Google Scholar]
- 52. Freedman M. S. (2006) Expert Opin. Pharmacother. 7, S1–S9 [DOI] [PubMed] [Google Scholar]
- 53. Biswas S. K., Lopez-Collazo E. (2009) Trends Immunol. 30, 475–487 [DOI] [PubMed] [Google Scholar]
- 54. Liang S., Hosur K. B., Domon H., Hajishengallis G. (2010) J. Periodontal. Res. 45, 574–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lindgren A., Pavlovic V., Flach C. F., Sjöling A., Lundin S. (2011) Innate Immun. 17, 191–203 [DOI] [PubMed] [Google Scholar]
- 56. Melmed G., Thomas L. S., Lee N., Tesfay S. Y., Lukasek K., Michelsen K. S., Zhou Y., Hu B., Arditi M., Abreu M. T. (2003) J. Immunol. 170, 1406–1415 [DOI] [PubMed] [Google Scholar]