Abstract
The dsRNA genome of mammalian reovirus (MRV), like the dsDNA genomes of herpesviruses and many bacteriophages, is packed inside its icosahedral capsid in liquid-crystalline form, with concentrations near or more than 400 mg/ml. Viscosity in such environments must be high, but the relevance of viscosity for the macromolecular processes occurring there remains poorly characterized. Here, we describe the use of simple viscogens, glycerol and sucrose, to examine their effects on RNA transcription inside MRV core particles. Transcription inside MRV cores was strongly inhibited by these agents and to a greater extent than either predicted by theory or exhibited by a nonencapsidated transcriptase, suggesting that RNA transcription inside MRV cores is unusually sensitive to viscogen effects. The elongation phase of transcription was found to be a primary target of this inhibition. Similar results were obtained with particles of a second dsRNA virus, rhesus rotavirus, from a divergent taxonomic subfamily. Polymeric viscogens such as polyethylene glycol also inhibited RNA transcription inside MRV cores, but in a size-limited manner, suggesting that diffusion through channels in the MRV core is required for their activity. Modeling of the data suggested that the inherent intracapsid viscosity of both reo- and rotavirus is indeed high, two to three times the viscosity of water. The capacity for quantitative comparisons of intracapsid viscosity and effects of viscogens on macromolecular processes in confined spaces should be similarly informative in other systems.
Keywords: Reovirus, RNA Polymerase, RNA Transport, Transcription, Viral Polymerase, Viral Transcription, Virus Structure, Rotavirus, Viscogen, Viscosity
Introduction
The viscosity of a solution affects the rates of reactions within it, as has been shown for a variety of macromolecular processes, including DNA strand separation, DNA translocation through pores, and protein folding (1–4). Under the conditions used for most in vitro biochemical reactions, viscosity is low, not much greater than that of water, and therefore not high enough to be a rate-limiting parameter. On the other hand, intracellular viscosity approaches twice that of water (5, 6), and so for many in vivo processes, the contribution of viscosity to reaction rates may be important. Moreover, there are some biological systems in which viscosity may play an even greater role. Certain such systems are characterized by a high concentration of nucleic acid within a boundaried compartment, such as inside the protein capsid of a virus particle. Inside some bacteriophages, for example, the concentration of dsDNA packed in a liquid-crystalline form approaches 500 mg/ml (7–9). These concentrated solutions of dsDNA should be highly viscous (10), which should in turn impede the movements of DNA during translocation from capsid to cytosol during cell entry and from cytosol to capsid during genome packaging. Similar dsDNA packing and expected consequences of high viscosity are seen in herpesviruses (11).
Other related systems in which viscosity may play an important role are the transcriptionally active particles of dsRNA viruses. Many dsRNA viruses package their genomes to a similarly high concentration as do the dsDNA bacteriophages and herpesviruses. For example, considering the interior cavity diameter of mammalian reovirus (MRV)3 to be 50 nm (12) and its genome molecular weight to be 15 MDa (13), the concentration of dsRNA packed in liquid-crystalline form is calculated to be >380 mg/ml. In fact, upon approximating the fraction of the interior occupied not by dsRNA but by the internally projecting RNA-dependent RNA polymerase and accessory components that constitute the transcriptase of this virus (14), of which there are at least 10 per particle (see below), the concentration of dsRNA more likely exceeds 400 mg/ml. The ability of MRV and other dsRNA viruses to mediate RNA transcription within such concentrated, viscous solutions provides an intriguing system in which to study intracapsid viscosity and its effects. As the process of RNA transcription is associated with both linear and rotational movements of template nucleic acid and nascent RNA product, as well as protein conformational changes and diffusion of NTPs and pyrophosphate, viscosity-based resistance to those movements should be expected.
MRV is the prototype of the genus Orthoreovirus in the family Reoviridae, a diverse family of multisegmented dsRNA viruses to which important pathogens of humans and other vertebrates also belong, including rotavirus. The MRV genome comprises 10 linear segments of dsRNA, ranging in size from 1000 to 4000 bp each. Within infectious virions, these segments are enclosed by two icosahedral layers of proteins: the inner or core capsid and the outer capsid (15). During cell entry, the outer capsid is largely shed, and the remaining core particle (∼52 MDa, including the genome) enters the cytosol (16). There, it proceeds to use the 10 genome segments as templates for reiteratively transcribing the 10 viral mRNAs, each of which is a full-length copy of the respective genomic plus strand. Each core particle is believed to contain 10–12 copies of the 142-kDa viral RNA-dependent RNA polymerase, which are anchored to the inside surface of the inner capsid near the 12 icosahedral 5-fold axes (12, 14, 17). These RNA-dependent RNA polymerase molecules, together with accessory proteins λ1 and μ2, support simultaneous synthesis and release of the 10 mRNAs, whereas both genomic RNA strands are retained in the densely packed core interior (18, 19). The MRV core thus represents a fascinating molecular assemblage from a variety of perspectives, regarding not only its assembly and structure but also its function as a capsid-delimited RNA transcription and transport machine.
In this report, we describe the effects of viscogens on the RNA transcription activity of MRV core particles. We chose glycerol and sucrose for initial studies because they are readily diffusible small molecules that form viscous solutions but have limited chemical effects. Results indicated that each of these agents strongly inhibits transcription inside MRV cores in a concentration-dependent manner with key parameters of the inhibition (critical concentration for onset and concentration for 50% inhibition) occurring at the same viscosities with each agent. The inhibitory effects exceeded those either predicted by theory or exhibited by the nonencapsidated transcriptase of bacteriophage T7, suggesting that transcription inside MRV cores is unusually sensitive to viscogen effects. Transcript elongation was found to be a primary target of this inhibition. Similar results were obtained with transcriptionally active particles of the taxonomically divergent dsRNA virus, rhesus rotavirus (RRV), demonstrating that viscogen effects on RNA transcription hold relevance beyond MRV. Polymeric viscogens, such as PEG, also inhibited transcription inside MRV cores but in a size-limited manner, suggesting that diffusion through channels in the MRV core is required for their activity. Data modeling provided a quantitative estimate of the inherently high intracapsid viscosity of both MRV and RRV particles. The capacity for quantitative comparisons of intracapsid viscosity and effects of viscogens on macromolecular processes in confined spaces should be similarly informative in other systems.
EXPERIMENTAL PROCEDURES
Cells and Viruses
Murine L929 cells were grown in Joklik's modified Eagle's minimal essential medium (Irvine) supplemented with 2% fetal and 2% calf bovine sera (HyClone), 2 mm l-glutamine (Mediatech), 100 units of penicillin, and 100 μg of streptomycin/ml (Irvine). Stocks of MRV strains Type 1 Lang, Type 2 Jones, and Type 3 Dearing were derived from ones obtained from the late B. N. Fields (Harvard Medical School). All experiments were performed with Type 1 Lang unless otherwise indicated. Virions were amplified in L929 cells and purified by CsCl gradient centrifugation (20). Cores were isolated by protease digestion of purified virions followed by CsCl gradient centrifugation (21). Purified virions and cores were stored at 4 °C in virion buffer (150 mm NaCl, 10 mm MgCl2, 10 mm Tris, pH 7.5). Virion and core concentrations were determined by A260 as in previous studies (22). RRV double-layered particles (DLPs) were obtained as a kind gift from S. D. Trask, S. T. Aoki, and S. C. Harrison (Harvard Medical School) after having been generated and purified by standard protocols (23).
Viscogens
Glycerol and sucrose were from Mallinckrodt for most experiments and for all experiments shown in the figures. Glycerol from American Bioanalytical (anhydrous “Super Glycerol”) and sucrose from IBI Scientific were also tested and yielded highly similar results. Viscosities of glycerol or sucrose solutions were calculated according to published equations (24, 25). Linear polyacrylamide of an average molecular weight (MW) of 1500 was purchased from Polysciences, Inc. PEG of average MW 400 or 600 was purchased from Sigma-Aldrich, and PEG of average MW 1000, 2000, 3000, 4000, 6000, or 8000 was purchased from Fluka. Viscosities of polyacrylamide 1500 and of PEG 400 at 45 °C were measured by using a rheometer (TA Instruments, New Castle, Delaware, model AR2000N) with a double concentric cylinder geometry under imposed shear rates.
Transcription Assays
Standard reactions were performed as described previously (26), including the use of 45 °C as a near-optimum temperature for MRV transcription. Briefly, standard transcriptions were carried out by incubating ∼1 × 1010 MRV cores at 45 °C in 10 μl of transcription buffer (100 mm HEPES-KOH (pH 8.1), 10 mm MgCl2, and 0.5 mm EDTA) that also contained 4 mm GTP and 1 mm each of ATP, CTP, and UTP (all from GE Healthcare). Glycerol, sucrose, polyacrylamide, or PEG was added to the transcription buffer at the indicated percentage concentration (w/w) for each sample. In those experiments in which [α-32P]CTP was present, the concentration of nonradiolabeled CTP was lowered to 0.2 mm. Samples were analyzed either by liquid scintillation counting for radiolabel incorporated into acid-insoluble material or by 1% agarose gel electrophoresis. Yields were expressed as a percentage (% transcription) relative to those obtained in the absence of viscogenic agent within each experiment. In limited experiments at 35 °C, we found that glycerol and sucrose inhibited MRV transcription at essentially the same concentrations as at 45 °C. Experiments with RRV DLPs were performed identically to those with MRV cores. Experiments with T7 RNA polymerase (New England Biolabs) were also performed identically, except at 37 °C with a polymerase concentration of 0.5 units/ml and a plasmid template concentration of 5 ng/ml.
To change the reaction medium during transcription, reactions were stopped by mixing samples with an equal volume of ice-cold 100 mm HEPES-KOH (pH 8.1) buffer containing 50 mm EDTA. After microfuge centrifugation at 13,000 rpm for 30 min at 4 °C, supernatant was removed and pellet was washed with 100 μl of 100 mm HEPES-KOH (pH 8.1) buffer. Centrifugation and washing steps were then repeated once or twice. Afterward, cores were resuspended in fresh transcription mix as indicated for each experiment, and transcription was allowed to continue at 45 °C.
Analysis of abortive transcripts was performed as described previously (22) with minor changes. Cores were subjected to a 1-h transcription reaction at 45 °C in the absence or presence of indicated concentration (w/w) of glycerol or sucrose. The reaction was stopped by addition of EDTA to 10 mm followed by 2 min at 100 °C. After cooling to room temperature, EDTA was titrated by addition of MgCl2 to 12.5 mm. Calf intestinal phosphatase (1 unit per 10 μl) was then added, and samples were incubated for 30 min at 37 °C. Products were analyzed by electrophoresis on a 20% sequencing polyacrylamide gel and visualized by phosphorimaging. On a 20% gel, the abortive transcript GC migrated closely below the 10-bp DNA marker.
ATPase Assay
Reactions with MRV cores were set up exactly as for transcription, except that only ATP was present. After 1-h incubation at 45 °C, released inorganic phosphate was measured using a Malachite Green Phosphate Assay Kit (BioAssay Systems).
RESULTS
Glycerol and Sucrose Inhibit Transcription inside MRV Cores
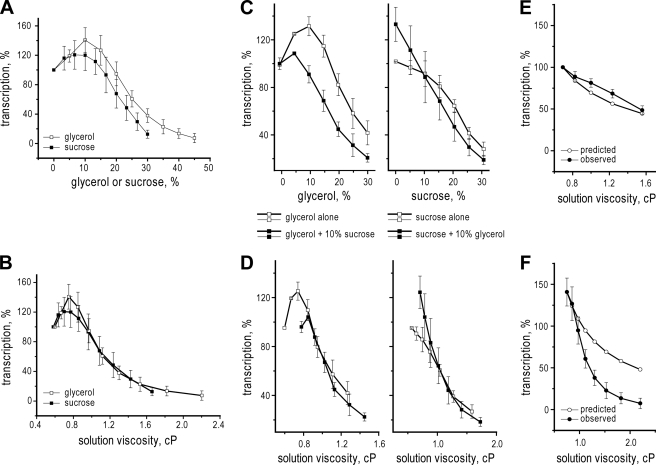
Addition of increasing concentrations of either glycerol or sucrose had large effects on transcription mediated in vitro by MRV cores, and the overall trend of these effects was similar for both agents (Fig. 1A). With each agent, the effects on transcript yields were biphasic: there was no inhibition and usually an activation, until a critical concentration of agent was reached near 10% (w/w), above which transcript yields were decreased progressively. Notably, the inhibition curve with glycerol was shifted to somewhat higher concentrations than that with sucrose (Fig. 1A). Very similar results were obtained with cores of three different MRV strains: Type 1 Lang, Type 2 Jones, and Type 3 Dearing. Although the inhibition phase was highly consistent between experiments, the activation phase was more variable, with extent of activation ranging from 0 to 70%. Because this report is devoted to characterizing the inhibition of MRV transcription by viscogens, the activation phase was not the focus of further experiments described below but is addressed again in discussion.
FIGURE 1.
Effect of simple viscogens on transcription by MRV cores or T7 RNA polymerase. Results are for 1-h reactions with cores of MRV strain Type 1 Lang in the presence of indicated concentrations of glycerol or sucrose. Yields were analyzed by liquid scintillation counting of [α-32P]CTP incorporated into acid-insoluble material and expressed as a percentage (% transcription) relative to those obtained in the absence of viscogenic agent. A, data are plotted relative to the concentration of glycerol (open squares) or sucrose (filled squares). Mean values and standard deviations were calculated from 4–14 and 4–8 independent experiments for glycerol and sucrose, respectively. B, same data as in A, but plotted relative to the viscosity, expressed in cP, that was added to the external solution by each agent. C and D, additive effects of glycerol and sucrose on transcription by MRV cores. Reactions were performed as for A except that both glycerol and sucrose were added to half of the samples as indicated. Left panels, glycerol effects on transcription in the absence (open squares) or presence (filled squares) of 10% sucrose. Right panels, sucrose effects on transcription in the absence (open squares) or presence (filled squares) of 10% glycerol. Mean values and standard deviations were calculated from three independent experiments in each case. In C, data are plotted relative to the concentration of glycerol (left) or sucrose (right), whereas in D, the same data as in C are plotted relative to total external viscosity, expressed in cP. E, observed (filled circles) versus predicted (open circles) results for viscosity dependence of T7 RNA polymerase transcription. Observed results are results are for 1-h reactions at 37 °C in the presence glycerol as viscogenic agent; mean values and S.D. were calculated from three independent experiments. Predicted results are calculated for theoretical 1/η dependence. F, observed (filled circles) versus predicted (open circles) results for viscosity dependence of MRV transcription. Observed results are those from the glycerol curve in B. Only data for external viscosities above the critical concentration (0.8 cP) are shown; data for lower viscosities have been omitted.
Inhibition by Glycerol and Sucrose Correlates with Their Contributed Viscosities
Replotting the results from Fig. 1A as a function of the viscosity contributed to the reaction medium by either glycerol or sucrose revealed that the inhibition curves with these agents are overlapping with respect to solution viscosity (Fig. 1B) not shifted as with respect to concentration (Fig. 1A). The solution viscosity representing the critical concentration for onset of inhibition, ∼0.75 centipoises (cP), was consistent with the two agents and in independent experiments. The solution viscosity providing 50% inhibition of transcription, ∼1.1 cP, was also consistent with the two agents and in independent experiments. These findings suggest that the inhibition of MRV transcription by both glycerol and sucrose is based in their common physical properties as viscogens.
Given these suggestive findings, we considered other explanations for the observed effects. One possibility is that the inhibition could be more chemical than physical, based in specific, direct interaction(s) with one or more component of the MRV core. Although this seems unlikely for either glycerol or sucrose, commercial stocks of these agents might contain impurities that are the true inhibitors. This also seems unlikely because stocks obtained from different vendors gave highly similar results. A related possibility is that the inhibition could be based in hydration effects, such as changes in the content or distribution of water in MRV cores in the presence of either agent. This possibility is given further consideration in discussion, in light of other results presented below.
As a further test of the basis of transcription inhibition by glycerol and sucrose, we addressed whether their effects are additive, as suggested by Hunt et al. (27). Specifically, we compared the effect of glycerol on MRV transcription in the absence or presence of 10% sucrose (Fig. 1, C and D, left), as well as the effect of sucrose on MRV transcription in the absence or presence of 10% glycerol (Fig. 1, C and D, right). In both sets of curves, samples containing 10% (w/w) of the constant agent have the critical concentration for inhibition by the dosed agent shifted to lower values (Fig. 1C), from 10 to 5% glycerol (left) and from 10 to 0% sucrose (right). Moreover, when these effects on MRV transcription are plotted as a function of total viscosity contributed to the reaction medium by the combined agents (Fig. 1D), both curves in each set virtually coincide, suggesting that the inhibitory effects on MRV transcription by glycerol and sucrose are additive specifically with respect to their contributed viscosities. From the results to this point, we therefore tentatively conclude that increased viscosity is the main contributing factor to transcription inhibition by glycerol and sucrose.
Glycerol Also Inhibits Transcription by T7 RNA Polymerase, but at Higher Concentrations More Consistent with Theory
Previous studies of viscogen effects on biochemical processes have shown that reaction rates are approximately halved when solution viscosity is doubled, consistent with theoretical predictions that reactions limited by diffusional events should exhibit a 1/η viscosity dependence (2–4). To validate our studies of viscogen effects on MRV transcription, we therefore attempted to reproduce that predicted result with a nonencapsidated RNA polymerase, that of bacteriophage T7. Indeed, in parallel with predicted behavior (Fig. 1E), transcription by T7 RNA polymerase was progressively inhibited as viscosity was increased by glycerol addition, until 50% inhibition of transcript yields was achieved near 1.5 cP or about twice the initial viscosity in this experiment (Fig. 1E). Notably, this finding contrasts with the MRV transcription results, which deviate much more substantially from predicted behavior (Fig. 1F), not only in having an initial activation phase (see Fig. 1, A–D) but also in undergoing inhibition at much lower solution viscosities than predicted. We conclude from the T7 results that our approach to studies of viscogen effects on transcription has been validated and from the MRV results that a hypersensitivity to viscogens on the part of transcription inside MRV cores has been revealed.
Transcript Elongation inside MRV Cores Is Strongly Inhibited by Glycerol and Sucrose
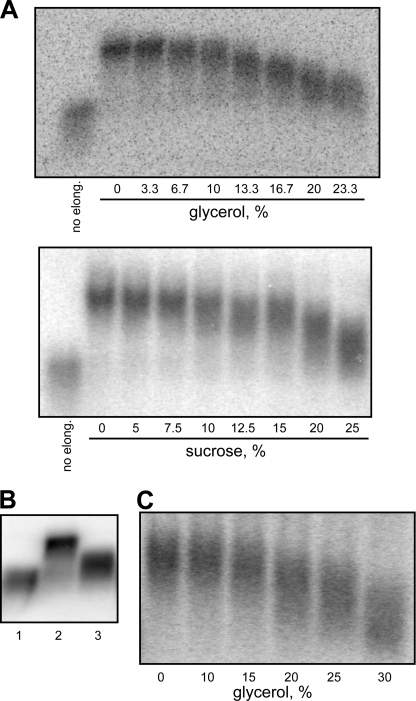
We next turned to identifying which steps in MRV transcription may be inhibited by viscogens. A transcription cycle is divided into four main phases: initiation, promoter escape, elongation, and termination (28). Each phase involves particular types of RNA and protein movements and may thereby be sensitive to increased viscosity. Because elongation comprises the vast bulk of individual events in an MRV transcription cycle (transcripts each 1000–4000 nt in length), it seems likely to be especially vulnerable in this regard. To test this possibility, MRV cores were allowed to transcribe for 10 s in the presence of [α-32P]CTP, but no glycerol, to generate labeled nascent transcripts that had already entered the elongation phase. The reaction was then stopped, and after changing the medium, different aliquots were allowed to continue with elongation for 40 s in the presence of nonradiolabeled CTP, plus increasing concentrations of glycerol, to generate elongated transcripts that might differ in maximum length according to the effects of glycerol. Results are similar to those of the 1-h end point curves shown in Fig. 1A: the first sign of inhibition (reduction in maximum transcript length) is observed near 10% glycerol, making this the critical concentration for inhibition of elongation, and 50% inhibition (maximum length reduced to near halfway between the 0% glycerol and no elongation samples) occurs near 20% glycerol (Fig. 2A, upper panel). Upon repeating the same experiment with sucrose, a similar coincidence of the transcription end point and elongation curves was observed, with a critical concentration near 10% sucrose and a concentration for 50% inhibition near 20% sucrose (Fig. 2A, lower panel). These results indicate that elongation is indeed strongly inhibited by glycerol and sucrose. Moreover, given the near coincidence with results in Fig. 1A, the new results suggest that elongation is the phase of the transcription cycle most sensitive to these viscogens.
FIGURE 2.
Effect of simple viscogens on transcript elongation by MRV cores. Results of representative experiments are shown. Transcripts were analyzed by 1% agarose gel electrophoresis. Transcripts elongated for 1 min are estimated to be ∼600 nt, whereas those elongated for only 10–15 s, as in control samples, are estimated to be only 100–150 nt. A, cores were allowed to transcribe for 10 s (upper) or 15 s (lower) in regular transcription buffer including [α-32P]CTP. The reaction was then stopped, and after changing the medium, samples were allowed to elongate with nonradiolabeled NTPs for 40 s (upper) or 45 s (lower) in the presence of indicated concentrations of glycerol (upper) or sucrose (lower). An aliquot of preinitiated transcripts prior to continued elongation (no elong.) was also analyzed for each experiment. B, cores were allowed to transcribe for 15 s in regular transcription buffer including [α-32P]CTP. The reaction was then stopped, and after changing the reaction medium, samples were allowed to elongate with nonradiolabeled NTPs for 50 s; for this step, UTP concentration was either 1 mm (lane 2) or 0.03 mm (lane 3). An aliquot of preinitiated transcripts prior to continued elongation was also analyzed (lane 1). C, cores were allowed to transcribe for 15 s in regular transcription buffer including [α-32P]CTP. The reaction was then stopped, and after changing the medium, samples were allowed to elongate with nonradiolabeled NTPs including 0.03 mm UTP for 110 s in the presence of indicated concentrations of glycerol.
RNA Movements during Elongation Contribute Little to Viscogen Sensitivity
Having obtained evidence that elongation is greatly affected by viscogens, we considered the possibility that elongation itself may increase viscosity inside the capsid, making it more susceptible to the effects of added viscogens. The idea is that large-scale movements of RNA templates that accompany elongation would be the cause of this “shear thickening,” which should in turn be proportional to the elongation rate. To test this possibility, we analyzed the effect of glycerol on elongation at a lower rate. To reduce the elongation rate, we performed transcription with a lower concentration of UTP. MRV cores were first allowed to transcribe for 15 s in the usual reaction medium including 1 mm UTP and [α-32P]CTP. The reaction was then stopped and, after the reaction medium was changed, divided into three aliquots (Fig. 2B). Aliquot 1 was directly loaded onto a 1% agarose gel and served as a negative control for continued elongation. The other two aliquots were allowed to continue elongation for 45 s in the presence of nonradiolabeled NTPs, including either 1 mm UTP (aliquot 2) or 30 μm UTP (aliquot 3). Because the RNA transcripts from aliquot 3 migrated roughly equally between those from aliquots 1 and 2, we concluded that the elongation rate with 30 μm UTP is roughly half of that with 1 mm UTP. Given that the elongation rate by MRV cores is normally 10–12 nt/s (18, 19, 26), the rate with 30 μm UTP appears to be only ∼5–6 nt/s. If elongation-driven RNA movements contribute substantially to the intracapsid viscosity of MRV cores, one might expect the core interior to be less viscous when the elongation rate is decreased by half, which should be reflected as a shift in the critical concentration of glycerol toward higher values. In the presence of 30 μm UTP, however, the critical concentration of glycerol was again near 10% (Fig. 2C). When repeated with an even lower concentration of UTP, 10 μm, which reduces elongation rate to only 10–20% of that with 1 mm UTP (∼1–3 nt/s), the critical concentration of glycerol was once again near 10%. These results thus fail to support the notion that elongation itself substantially increases intracapsid viscosity.
Transcription Initiation, Promoter Escape, and ATP Phosphohydrolysis Are More Resistant to Glycerol and Sucrose
Two other phases in the transcription cycle that are easy to evaluate are initiation and promoter escape. For examining initiation, the synthesis of abortive transcripts, reflecting failure of the RNA polymerase to mediate promoter escape and enter the elongation phase, is commonly analyzed. In the case of MRV, abortive transcripts represent the first two to four bases (5′-GC(U)(A)) of the conserved 5′-terminal sequence in each MRV plus strand (22, 29, 30). For the current study, we measured synthesis of abortive transcripts by MRV cores by providing only the first two NTPs, GTP and [α-32P]CTP, during a 1-h reaction. The medium and conditions were identical to those for a regular transcription reaction except that ATP and UTP were omitted, making elongation impossible and allowing only the initiation product GC to be produced. In the presence of increasing concentrations of either glycerol (Fig. 3A, upper panel) or sucrose (Fig. 3A, lower panel), production of GC was found to be strongly resistant, tolerating a much higher concentration of either glycerol or sucrose (>20%) than did elongation in the preceding experiments. For examining promoter escape, we subjected MRV cores to a very short (10-s) labeling reaction in the presence of all four NTPs, including [α-32P]CTP and increasing concentrations of glycerol. The reaction was then stopped, the medium was exchanged for one containing only nonradiolabeled NTPs and no glycerol, and the samples were allowed to complete synthesis of the prelabeled transcripts. In this case again, a high concentration of glycerol (>20%) was tolerated before the onset of inhibition (Fig. 3B). Because the steps in the presence of glycerol in this experiment include initiation, promoter escape, and a few bases of elongation, we conclude from these results that both initiation and promoter escape are substantially more resistant to viscogen effects than is elongation.
FIGURE 3.
Effect of simple viscogens on other activities of MRV cores. Results of representative experiments are shown. A, Nonproductive initiation (synthesis of abortive transcripts). Cores were incubated for 1-h in modified transcription buffer containing only GTP and [α-32P]CTP. Indicated concentrations of glycerol (upper) or sucrose (lower) were also present. Samples were analyzed on a 20% sequencing polyacrylamide gel to detect the short transcripts. B, productive initiation including promoter escape and limited elongation. Cores were allowed to transcribe for 10 s in regular transcription buffer including [α-32P]CTP and indicated concentrations of glycerol. The reaction was then stopped, and after changing the medium, samples were allowed to elongate with nonradiolabeled NTPs for 15 min in the absence of glycerol. C, NTPase activity. Cores were incubated for 1-h in modified transcription buffer containing only ATP. The concentration of released inorganic phosphate was then measured and expressed as a percentage relative to that obtained in the absence of viscogenic agent.
Another activity of MRV cores that might contribute in some way to transcription, a relatively well known nucleoside triphosphate phosphohydrolase activity (31, 32), is also easy to examine. In the presence of increasing glycerol or sucrose, a gradual inhibition of ATPase activity was seen, with no activation phase and a concentration for 50% inhibition of >30% for each agent (Fig. 3C). All of these features are strikingly different from those of the other MRV reactions studied above and suggest that the ATPase activity per se contributes little to determining the inhibition of MRV transcription by glycerol and sucrose.
Glycerol Also Inhibits Transcription inside Rotavirus Particles
To determine whether viscogen effects on transcription extend to other dsRNA viruses, we tested RRV, from a different genus (Rotavirus) and subfamily (Sedoreovirinae versus Spinareovirinae for MRV) in the family Reoviridae. Rotavirus DLPs are the transcriptionally active form analogous to MRV cores (23). DLPs of RRV were therefore tested for transcription in the presence of increasing concentrations of glycerol and showed a dose response similar to that of MRV cores, including an activation phase at lower concentrations followed by an inhibition phase at higher ones (Fig. 4A). For RRV, however, inhibition required somewhat higher concentrations of glycerol than seen for MRV: a critical concentration near 15% glycerol (∼0.85 cP of added viscosity) and 50% inhibition near 30% glycerol (∼1.4 cP of added viscosity) for RRV versus 10 and 23% glycerol, respectively, for MRV. These findings suggest that the transcriptional machineries of RRV and MRV differ in their capacities to withstand viscogen effects, with RRV being somewhat more resistant. RRV was again like MRV, however, in being more sensitive to transcription inhibition as a function of solution viscosity than predicted by theory (Fig. 4B). Moreover, for RRV as for MRV, the elongation step of transcription was found to be a major target of this inhibition (Fig. 4C).
FIGURE 4.
Effect of simple viscogens on transcription by RRV DLPs. A and B, results are for 1-h reactions in the presence of indicated concentrations of glycerol. In A, data are plotted relative to the concentration of glycerol; mean values and standard deviations were calculated from three independent experiments. Solution viscosity in cP, calculated as a function of glycerol concentration at the assay temperature (45 °C), is shown as a dashed line. In B, the same data are shown as in A, but plotted relative to viscosity in cP (filled circles) and compared with theoretical predictions calculated for 1/η viscosity dependence of RRV transcription at 45 °C (open circles). C, effect of glycerol on transcript elongation. DLPs were allowed to transcribe for 5 s in regular transcription buffer including [α-32P]CTP. The reaction was then stopped, and after changing the medium, samples were allowed to elongate with nonradiolabeled NTPs for 10 s in the presence of indicated concentrations of glycerol. An aliquot of preinitiated transcripts prior to continued elongation (no elong.) was also analyzed for each experiment.
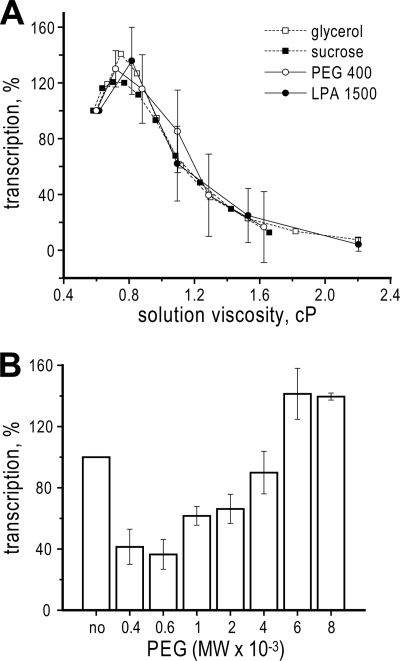
Polymeric Viscogens Inhibit Transcription inside MRV Cores, but in a Size-limited Manner
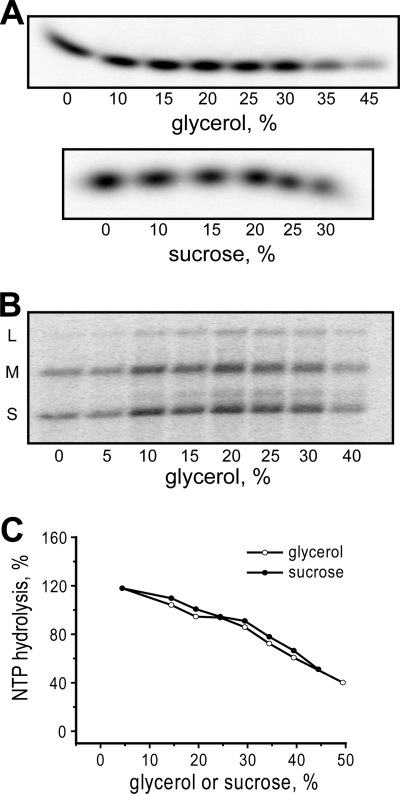
Two polymeric viscogens in common usage are PEG and polyacrylamide. When tested for inhibition of transcription inside MRV cores over a range of viscogen concentrations, the curves for both PEG 400 and polyacrylamide 1500 were found to overlap the curves for glycerol (MW 92) and sucrose (MW 342) with respect to their contributed viscosities (Fig. 5A). These results thus provide further evidence to support the conclusion that inhibition by glycerol and sucrose, as well as by PEG 400 and polyacrylamide 1500, is based in their common physical properties as viscogens. Interestingly, as observed previously with glycerol and sucrose, low concentrations of PEG 400 and polyacrylamide 1500 mildly activated transcription inside MRV cores (Fig. 5A; see “Discussion”).
FIGURE 5.
Effect of polymeric viscogens on transcription by MRV cores. Results are for 1-h reactions in the presence of indicated concentrations of PEG or polyacrylamide. Mean values and S.D. were calculated from four independent samples each. A, concentration curves for PEG 400 and linear polyacrylamide (LPA) 1500, expressed relative to the contributed viscosities of each polymeric viscogen. Accompanying curves for glycerol and sucrose (dashed lines, no error bars) are included for comparison and are the same as those in Fig. 1B. B, effect of PEG size on inhibition activity. Results were obtained with 20% (w/w) PEG preparations of different average MW values as indicated.
PEG has the additional benefit of being commercially available over a range of average MW values. We recognized that there may be a size limitation to the capacity of PEG to diffuse into or through the MRV core for subsequent effects on transcription. Indeed, when PEG preparations of different average MW value were tested, PEG 400 and 600 exhibited similar, maximal effects at inhibiting transcription inside MRV cores; PEG 1000 and 2000 exhibited intermediate levels of inhibition; PEG 4000 exhibited little or no inhibition; and PEG 6000 and 8000 mildly activated transcription (Fig. 5B). The hydrodynamic radii of PEG 1000 and 2000 respectively approximate 11 and 15 Å in aqueous solution (33, 34), and thus the MRV core appears to act as a molecular sieve with maximum channel diameters near 20 to 30 Å.
DISCUSSION
In other recent work, we found that small molecules that increase or decrease RNA duplex stability (e.g. spermidine and dimethyl sulfoxide) have large effects on RNA transcription inside MRV cores, which can be attributed to specific steps in the transcription cycle (26). Those results suggested that future experiments concerning the effects of other small molecules may provide additional insights. The current study follows that suggestion, addressing the effects of different viscogens, whose primary effect might be to increase intracapsid viscosity.
Effect of Simple Viscogens on Particular Steps in MRV Transcription
During RNA transcription, there are many individual events that may be affected by increased viscosity. In addition to conformational changes in the RNA polymerase and other protein factors (28), as well as diffusion of NTPs and pyrophosphate, these events include large-scale linear and rotational (35) movements of template nucleic acid and nascent RNA product (rates of DNA linear translocation have been shown to be affected by viscosity (2)) and movements of template strands away from and toward each other during melting and reannealing (rates of DNA melting have also been shown to be affected by viscosity of the solution (4)). In the case of MRV transcription, there is also template looping (the capped 5′ end of the template plus strand is thought to be continually held by the cap-binding site on the surface of the RNA-dependent RNA polymerase near the template-entry channel so that as rewinding of the duplex proceeds, the rewound regions are thought to form an expanding loop bending away from the template-exit channel) and repositioning of the template for initiation (at the end of each transcription cycle, the 3′ end of template minus strand must be reinserted into the template entry channel) (36). During a single cycle, by far, the bulk of these events accompany the elongation phase, and thus it is not surprising that we find elongation by MRV cores to be most sensitive to glycerol and sucrose among the tested activities. Moreover, protein conformational changes that are known to occur during elongation in other transcriptase complexes (28) are relatively small compared with RNA movements, suggesting the latter as the more relevant target of inhibition by such viscogens.
Activation of MRV Transcription at Lower Concentrations of Viscogens
The characteristic curve of glycerol and sucrose effects on transcription inside MRV cores is biphasic, the first phase involving an activation that peaks ∼0.75 cP of added viscosity (∼10% glycerol or sucrose). Glycerol is the more potent activator, elevating transcription by ∼40% versus 10–20% for sucrose. The mechanism is not clear, but there are indications that it involves promoter escape. First, the rate of elongation is not obviously higher at 10% glycerol or sucrose (see Fig. 3, A and B). Second, the rate of abortive initiation is likewise not obviously higher at 10% glycerol or sucrose (see Fig. 4A). In contrast, productive initiations are clearly higher at 10–30% glycerol, implicating enhanced promoter escape as the source of activation. As we have reported previously, ∼90% of transcriptase complexes in standard preparations of MRV cores are inactive for production of elongated transcripts due to a block in promoter escape, but some of those (called fraction D) can be activated by treatment with dimethyl sulfoxide (26). The activation of transcription inside MRV cores by glycerol and sucrose might similarly result from the activity of new, otherwise silent transcriptase complexes. Notably, lower concentrations of PEG 400 and polyacrylamide 1500 (see Fig. 5A), as well as preparations of larger PEG molecules (Fig. 5B), also activate transcription inside MRV cores, suggesting that the mechanism of activation is common to both simple and polymeric viscogens (see below).
Evidence for Channel-limited Diffusion of Viscogens
One of our starting assumptions for this study was that a viscogen would need to enter the MRV core to be able to inhibit transcription there. During the course of our work, we realized that this assumption should and could be tested by the use of PEG preparations of different average MW values. Our results in this regard (see Fig. 5B) support the initial assumption and indeed suggest that PEG molecules with a hydrodynamic radius larger than 20–30 Å cannot efficiently enter or diffuse through the core. The MRV core structure (12, 15, 17, 37) includes small solvent channels through the 120-subunit, T = 1 capsid at several symmetry-related positions, which may in addition be dynamically expanding and constricting when cores are in solution, e.g. allowing transcription substrates and products to enter and exit. Presumably, these same channels allow access of viscogen molecules to the core interior.
Other Explanations for Viscogen Effects
Our results, in particular those indicating that the inhibitory effects of both simple and polymeric viscogens are correlated with respect to their contributed viscosities, favor the explanation that increased viscosity is the primary factor in their common inhibition of transcription inside MRV cores. Among other possible modes of action by these agents, effects on the hydration of RNA and protein components within the core interior seem perhaps most likely. The addition of viscogens would be expected to change the number and distribution of water molecules in the core interior, with potential inhibitory effects on transcription. On the other hand, it is also possible that these hydration effects are not inhibitory, but rather stimulatory, and might explain the activating effect of viscogens at lower concentrations (see Figs. 1A and 5A). According to this latter suggestion, the hydration-based stimulation of transcription at lower concentrations of viscogens would be overwhelmed by the viscosity-based inhibition of transcription at higher viscogen concentrations. The activating effect of larger PEG molecules (see Fig. 5B) could also be consistent with this explanation, in that by being excluded from the core, these larger molecules would be expected to dehydrate the core interior through an osmotic effect.
Quantitative Estimates of Intracapsid Viscosity
The results in this study led us to attempt a quantitative estimation of the inherent intracapsid viscosity of MRV cores, based on our interpretation that viscosity inside, not outside, the capsid is the relevant determinant of transcript yields in this system. According to theoretical simulations (38), reactions limited by diffusional events should exhibit a 1/η viscosity dependence, and indeed, this dependence has been observed for a number of biochemical processes (1, 3), including approximately by T7 RNA polymerase in this study (see Fig. 1E). For this type of dependence, at the point of 50% transcription inhibition, the overall intracapsid viscosity (viscosity inherent to the MRV core interior plus additional viscosity from added viscogen having diffused inside) should be twice that at the critical concentration for onset of inhibition by the viscogen. Hence, if we assume that intracapsid viscosity increases in direct proportion to solution viscosity (an assumption we readdress below), we would estimate that because MRV transcription is 50% inhibited when ∼1.1 cP of viscosity has been added to the solution (see Fig. 1B), then intracapsid viscosity at the critical concentration of viscogenic agent would likewise be ∼1.1 cP. With glycerol, the critical concentration is ∼10%, which corresponds to ∼0.15 cP of additional viscosity, leaving ∼0.95 cP of intracapsid viscosity inherent to the MRV core at the assay temperature of 45 °C.
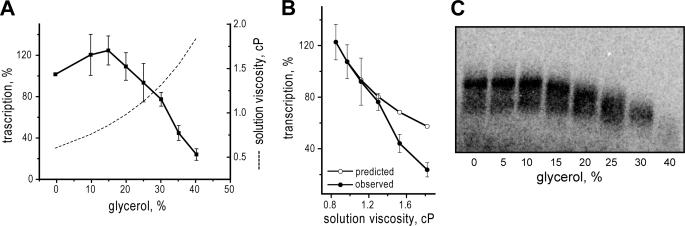
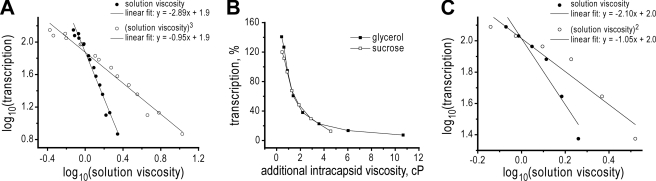
It is not clear, however, that intracapsid viscosity should be assumed to increase in direct proportion to solution viscosity. In an attempt to address this issue, we plotted transcript yields in the presence of glycerol or sucrose, presented as log10 values of the data, versus log10 values of solution viscosity (Fig. 6A). For simplicity, only the data for viscosities at and above the critical concentration are shown. The resulting log-log plot approximates a straight line, supporting the theoretical prediction of a power dependence between transcript yields and solution viscosity; however, the slope of this line is near a value of −3, far from the predicted value of −1 for 1/η viscosity dependence and direct proportionality between solution and intracapsid viscosities. By instead plotting intracapsid viscosity as the cube of solution viscosity in this case, a slope near −1 is restored (Fig. 6A).
FIGURE 6.
Quantitative estimates of intracapsid viscosities. A, log-log presentation of MRV transcription data from Fig. 1A. Data for both glycerol and sucrose were combined for this presentation. Only data for solution viscosities above the critical concentration (0.8 cP) are shown; data for lower viscosities have been omitted. Log10 values of transcript yields are plotted versus log10 values of either solution viscosity (filled circles) or the cube of solution viscosity as described in the text (open circles) (linear curve fits, r2 = 0.97). B, Same data as in Fig. 1B (right) but plotted relative to the cube of solution viscosity, based on the results shown in A. Only data for external viscosities above the critical concentration are shown. C, same type of log-log presentation as in A, but in this case for RRV transcription data from Fig. 4A. Log10 values of transcript yields are plotted versus log10 values of either solution viscosity (filled circles) or the square of solution viscosity as described in the text (open circles) (linear curve fits, r2 = 0.90).
Based on these findings, we should be able to obtain a better estimate of the inherent intracapsid viscosity of MRV cores. Plotting MRV transcript yields relative to the increase in intracapsid viscosity calculated as the cube of solution viscosity (Fig. 6B), we find that 50% inhibition has occurred when intracapsid viscosity has increased by ∼1.4 cP. As mentioned earlier, at the point of 50% inhibition, the additional intracapsid viscosity is expected to equal that at the critical concentration of viscogenic agent, which should therefore also be ∼1.4 cP. It is still not clear how to subtract the contribution of added viscogen at this critical concentration and thereby fully estimate the inherent intracapsid viscosity in the absence of added viscogen, but if we again simply subtract the value of a ∼10% glycerol solution, ∼0.15 cP, we can estimate that the inherent intracapsid viscosity of MRV cores is near 1.25 cP, or ∼2.1 times that of water at the assay temperature of 45 °C.
But why should intracapsid viscosity not increase in direct proportion to solution viscosity, but rather according to a higher power function? We expect this to be true in part because viscosity is known to increase exponentially as a function of the concentration of viscogen (also see Fig. 4A) (24, 25). According to this explanation, as viscogen is added to the solution and then diffuses to equalize concentrations outside and inside the capsid, the already high viscosity inside the capsid causes the total viscosity inside to increase faster than that outside. The viscogen effects inside the capsid would thus be greater than those predicted from the increase in solution viscosity alone. This explanation may be overly simplistic, however, in that we suspect there are other, macromolecular features of the MRV transcription system that further contribute to making it unusually sensitive to viscogens, over and above any disproportionality between solution and intracapsid viscosities.
Significance of RRV Findings
By extending the findings in this study to RRV, we have shown that the viscogen effects on RNA transcription are not unique to MRV, but are instead applicable to at least one other, divergent member of the family Reoviridae. Fig. 6C additionally suggests that a higher-power relationship between solution and intracapsid viscosities may hold true as well for RRV DLPs, though with RRV this relationship is closer to a second-power function, versus a third-power function for MRV cores. By plotting RRV transcript yields relative to the increase in intracapsid viscosity calculated as the square of solution viscosity and correcting for the glycerol contribution as noted above for MRV, we can estimate the inherent intracapsid viscosity of RRV DLPs to be near 1.6 cP, or about 2.7 times that of water at the assay temperature of 45 °C.
In future work, it will be important to extend these types of analyses to even-more divergent dsRNA viruses, in other taxonomic families such as Totiviridae, Partitiviridae, and Cystoviridae, to gain further mechanistic understanding of viscogen effects and what they may tell us about the transcription mechanisms and environments in the different virus interiors. For example, although RNA transcription by both MRV and RRV occurs via a conservative mechanism, RNA transcription by partitiviruses and cystoviruses is semi-conservative (39, 40), and thus it will be especially interesting to compare the latter families of viruses with regard to viscogen effects.
The general features of dsDNA packing in the icosahedral capsids of herpesviruses and some bacteriophages are similar to those of MRV: nucleic acids arranged in liquid-crystalline form, with similar packing densities suggested by center-to-center distances of packaged duplexes (e.g. 26 Å in herpes simplex virus 1 (11), 25 Å in phages λ and T7 (8, 9), and 26 Å in MRV (12, 15, 41)) and nucleic acid concentrations approximating 400–500 mg/ml in many cases (7; also see Introduction). The resulting viscosity in herpesvirus and phage capsids may therefore have substantive effects on translocation of their DNAs both into and from their capsids (42–44). Further investigations on viscogen effects and intracapsid viscosity in such dsDNA viruses may help to clarify this point. Moreover, the findings on viscogen effects in this study would seem to hold relevance for any molecular machine that mediates long-range movements of RNA or DNA in confined settings.
Acknowledgments
We thank Elaine Freimont for technical assistance, other members of the Nibert lab for helpful discussions, and Ian Molineux and the anonymous reviewers for insightful comments about the manuscript. We also thank Scott Aoki, Shane Trask, and Steve Harrison for the kind gift of RRV particles as noted in the text.
This work was supported in part by National Institutes of Health Grant R01 AI47904 (to M. L. N.) and National Science Foundation Grant CBET-0854108 (to T. R. P.).
- MRV
- mammalian reovirus
- RRV
- rhesus rotavirus
- PEG
- polyethylene glycol
- DLP
- double-layered particle
- cP
- centipoise
- MW
- molecular weight
- nt
- nucleotide(s).
REFERENCES
- 1. Davison P. F. (1967) Biopolymers 5, 715–721 [DOI] [PubMed] [Google Scholar]
- 2. Fologea D., Uplinger J., Thomas B., McNabb D. S., Li J. (2005) Nano Lett. 5, 1734–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bhattacharyya R. P., Sosnick T. R. (1999) Biochemistry 38, 2601–2609 [DOI] [PubMed] [Google Scholar]
- 4. Esmann M., Fedosova N. U., Marsh D. (2008) Biophys. J. 94, 2767–2776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cooke R., Kuntz I. D. (1974) Annu. Rev. Biophys. Bioeng. 3, 95–126 [DOI] [PubMed] [Google Scholar]
- 6. Swaminathan R., Hoang C. P., Verkman A. S. (1997) Biophys J. 72, 1900–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Earnshaw W. C., Casjens S. R. (1980) Cell 2, 319–331 [DOI] [PubMed] [Google Scholar]
- 8. Lepault J., Dubochet J., Baschong W., Kellenberger E. (1987) EMBO J. 6, 1507–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cerritelli M. E., Cheng N., Rosenberg A. H., McPherson C. E., Booy F. P., Steven A. C. (1997) Cell 91, 271–280 [DOI] [PubMed] [Google Scholar]
- 10. Kawade Y., Watanabe I. (1956) Biochim. Biophys. Acta 19, 513–523 [DOI] [PubMed] [Google Scholar]
- 11. Bhella D., Rixon F. J., Dargan D. J. (2000) J. Mol. Biol. 295, 155–161 [DOI] [PubMed] [Google Scholar]
- 12. Reinisch K. M., Nibert M. L., Harrison S. C. (2000) Nature 404, 960–967 [DOI] [PubMed] [Google Scholar]
- 13. Farrell J. A., Harvey J. D., Bellamy A. R. (1974) Virology 62, 145–153 [DOI] [PubMed] [Google Scholar]
- 14. Dryden K. A., Farsetta D. L., Wang G., Keegan J. M., Fields B. N., Baker T. S., Nibert M. L. (1998) Virology 245, 33–46 [DOI] [PubMed] [Google Scholar]
- 15. Dryden K. A., Wang G., Yeager M., Nibert M. L., Coombs K. M., Furlong D. B., Fields B. N., Baker T. S. (1993) J. Cell Biol. 122, 1023–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ivanovic T., Agosto M. A., Chandran K., Nibert M. L. (2007) J. Biol. Chem. 282, 12210–12219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang X., Walker S. B., Chipman P. R., Nibert M. L., Baker T. S. (2003) Nat. Struct. Biol. 10, 1011–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Skehel J. J., Joklik W. K. (1969) Virology 39, 822–831 [DOI] [PubMed] [Google Scholar]
- 19. Banerjee A. K., Shatkin A. J. (1970) J. Virol. 6, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Furlong D. B., Nibert M. L., Fields B. N. (1988) J. Virol. 62, 246–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chandran K., Walker S. B., Chen Y., Contreras C. M., Schiff L. A., Baker T. S., Nibert M. L. (1999) J. Virol. 73, 3941–3950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Farsetta D. L., Chandran K., Nibert M. L. (2000) J. Biol. Chem. 275, 39693–39701 [DOI] [PubMed] [Google Scholar]
- 23. Trask S. D., Dormitzer P. R. (2006) J. Virol. 80, 11293–11304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mathlouthi M., Genotelle J. (1995) in Sucrose: Properties and Applications (Mathlouthi M., Reiser P. eds) p. 137, Blackie Academic & Professional, Glasgow, Scotland, UK [Google Scholar]
- 25. Cheng N. S. (2008) Ind. Eng. Chem. Res. 47, 3285–3288 [Google Scholar]
- 26. Demidenko A. A., Nibert M. L. (2009) J. Virol. 83, 5659–5670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hunt A. J., Gittes F., Howard J. (1994) Biophys. J. 67, 766–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Steitz T. A. (2004) Curr. Opin. Struct. Biol. 14, 4–9 [DOI] [PubMed] [Google Scholar]
- 29. Bellamy A. R., Nichols J. L., Joklik W. K. (1972) Nat. New Biol. 238, 49–51 [DOI] [PubMed] [Google Scholar]
- 30. Yamakawa M., Furuichi Y., Nakashima K., LaFiandra A. J., Shatkin A. J. (1981) J. Biol. Chem. 256, 6507–6514 [PubMed] [Google Scholar]
- 31. Kapuler A. M., Mendelsohn N., Klett H., Acs G. (1970) Nature 225, 1209–1213 [DOI] [PubMed] [Google Scholar]
- 32. Noble S., Nibert M. L. (1997) J. Virol. 71, 2182–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bhat R., Timasheff S. N. (1992) Protein Sci. 1, 1133–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee H., Venable R. M., Mackerell A. D., Jr., Pastor R. W. (2008) Biophys. J. 95, 1590–1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Harada Y., Ohara O., Takatsuki A., Itoh H., Shimamoto N., Kinosita K., Jr. (2001) Nature 409, 113–115 [DOI] [PubMed] [Google Scholar]
- 36. Tao Y., Farsetta D. L., Nibert M. L., Harrison S. C. (2002) Cell 111, 733–745 [DOI] [PubMed] [Google Scholar]
- 37. Spencer S. M., Sgro J. Y., Dryden K. A., Baker T. S., Nibert M. L. (1997) J. Struct. Biol. 120, 11–21 [DOI] [PubMed] [Google Scholar]
- 38. Kramers H. A. (1940) Physica 7, 284–304 [Google Scholar]
- 39. Ratti G., Buck K. W. (1978) Nucleic Acids Res. 5, 3843–3854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Van Etten J. L., Burbank D. E., Cuppels D. A., Lane L. C., Vidaver A. K. (1980) J. Virol. 33, 769–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Harvey J. D., Bellamy A. R., Earnshaw W. C., Schutt C. (1981) Virology 112, 240–249 [DOI] [PubMed] [Google Scholar]
- 42. Grayson P., Han L., Winther T., Phillips R. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 14652–14657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fuller D. N., Raymer D. M., Rickgauer J. P., Robertson R. M., Catalano C. E., Anderson D. L., Grimes S., Smith D. E. (2007) J. Mol. Biol. 373, 1113–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Allemand J. F., Maier B. (2009) FEMS Microbiol. Rev. 33, 593–610 [DOI] [PubMed] [Google Scholar]