Abstract
p21Waf1/Cip1 protein levels respond to DNA damage; p21 is induced after ionizing radiation, but degraded after UV. p21 degradation after UV is necessary for optimal DNA repair, presumably because p21 inhibits nucleotide excision repair by blocking proliferating cell nuclear antigen (PCNA). Because p21 also inhibits DNA mismatch repair (MMR), we investigated how p21 levels respond to DNA alkylation by N-methyl-N′-nitro-N-nitrosoguanidine (MNNG), which triggers the MMR system. We show that MNNG caused rapid degradation of p21, and this involved the ubiquitin ligase Cdt2 and the proteasome. p21 degradation further required MSH2 but not MLH1. p21 mutants that cannot bind PCNA or cannot be ubiquitinated were resistant to MNNG. MNNG induced the formation of PCNA complexes with MSH6 and Cdt2. Finally, when p21 degradation was blocked, MNNG treatment resulted in reduced recruitment of MMR proteins to chromatin. This study describes a novel pathway that removes p21 to allow cells to efficiently activate the MMR system.
Keywords: Cell Cycle, DNA Damage, DNA Repair, Protein Degradation, Signal Transduction, Cdt2, MSH2, Mismatch repair, p21
Introduction
p21Waf1/Cip1 is a cell cycle and checkpoint protein that was initially discovered as an inhibitor of cell cycle kinases (1–3). The Vogelstein laboratory (4) established that p21 is a transcriptional target of p53 and that p21 is induced via p53 in response to ionizing radiation, leading to p21 accumulation and a cell cycle block. This cell cycle arrest presumably gives the cell time to repair the DNA double strand breaks caused by ionizing radiation, thus avoiding a mitotic catastrophe that would be caused by an attempt to undergo cell division with unrepaired DNA.
Although p21 accumulates in response to ionizing radiation, we and others have found that UV irradiation results in rapid down-regulation of p21 (5). This is effected by proteolytic degradation of p21 via the ubiquitin/proteasome system. The rationale for this is that different types of irradiation cause characteristic types of DNA damage, which are in turn repaired by a variety of repair systems appropriate to the damage. In response to UV exposure, the resulting base cross-links are repaired by the nucleotide excision repair system, which depends on proliferating cell nuclear antigen (PCNA)2 and DNA polymerase activity. PCNA is a homotrimeric protein that, when bound to chromatin, forms a clamp around DNA that provides docking sites for DNA-manipulating enzymes, including DNA polymerases. The carboxyl-terminal domain of p21 binds to PCNA, blocking its function and thereby inhibiting DNA replication (6). Thus, in UV-irradiated cells, PCNA and DNA polymerase are required for DNA repair, and p21 would inhibit repair by blocking PCNA. It is therefore not surprising that cells have developed a mechanism to degrade p21 in this situation to permit efficient nucleotide excision repair.
p21 has also been found to inhibit another type of DNA repair, namely DNA mismatch repair (MMR) (11, 16, 17). The MMR system maintains genomic integrity by repairing postreplication DNA mismatches and insertion/deletion loops (reviewed in Refs. 7 and 8), and loss of MMR is associated with colorectal cancer (reviewed in Ref. 9). In addition, the MMR system recognizes chemical adducts on DNA such as 6-methylguanine that can be introduced experimentally with alkylating agents such as MNNG (10). A principal component of the MMR system is the MSH2 protein, which forms heterodimers with either MSH6 or MSH3. MSH2/6 (also called MutSα) recognizes base mismatches and alkylation adducts, whereas MSH2/3 (called MutSβ) recognizes insertion/deletion loops. Subsequent steps in MMR involve the recruitment of the MLH1 plus PMS2 heterodimer, excision of the newly synthesized DNA encompassing the mismatch by Exo1, and resynthesis of the excised DNA by polymerase δ or η. PCNA appears to be involved in MMR not only to promote DNA resynthesis but also at a step preceding DNA synthesis (11). In yeast, certain PCNA mutants confer an MMR-deficient phenotype (12). MSH6 and MSH3 contain a PCNA-binding motif called a PIP box that is conserved in many PCNA-binding proteins, including p21 (13–15). In vitro, recombinant p21 as well as a p21 peptide containing a PIP box inhibit MMR, presumably by binding PCNA, as the inhibition is abrogated by adding PCNA (11, 16, 17). p21 also displaces MMR proteins from replicating DNA (18).
Because p21 inhibits MMR, we asked whether cells down-regulate p21 in situations where MMR is needed. We show that in cells treated with MNNG, p21 is rapidly degraded by the ubiquitin/proteasome pathway and that this pathway requires MSH2 and the ubiquitin ligase Cdt2. Failure to degrade p21 resulted in diminished recruitment of MMR proteins to damaged DNA.
EXPERIMENTAL PROCEDURES
Cell Culture
All cell lines were grown in Iscove's modified Dulbecco's medium supplemented with 10% fetal calf serum and penicillin/streptomycin. Transient transfections with plasmids were carried out with Effectene (Qiagen) following the manufacturer's instructions. For protein stability experiments, cells were treated with 20 μg/ml cycloheximide and/or 50 μm N-acetyl-leucyl-leucyl-norleucinal (LLnL) for the indicated times. For puromycin selection, transfected cells were selected by adding 1 μg/ml puromycin 1 day after transfection followed by 2 days of selection. DNA damage was induced either by treatment with 10 μm MNNG plus 10 μm O6-benzylguanine (to inhibit methylguanine methyltransferase (19)) for 1 h unless otherwise noted or by irradiation with UV for 1 min in the tissue culture hood as a control.
Plasmids
p21K6R has been described (5). Both p21wt and p21K6R were subcloned into pCMV-HA-puro, which was created by inserting the puromycin resistance cassette from psiStrike into the MfeI site of pCMV-HA using the oligonucleotides 5′-ATCAATTGTCCAGTCGGGAAACCTGTC-3′ and 5′-ATCAATTGCCTTTGAGTGAGCTGATACC-3′. p21PCNA− was generated by site-directed mutagenesis, starting with p21wt in pCMV-HA-puro and changing Gln-144, Met-147, and Phe-150 to Ala as described in Ref. 20.
RNAi
For most experiments, cells were seeded in 24-well plates at 30 × 103 cells/well and transfected the next day with siRNA (Dharmacon) using Lipofectamine 2000 and Opti-MEM under conditions recommended by Invitrogen. Cells were harvested 2 or 3 days after transfection.
RT-PCR
Total RNA was isolated using RNeasy Plus (Qiagen) to remove genomic DNA and reverse-transcribed using Advantage RT-for-PCR with oligo(dT) primer (Clontech). PCR was performed using HotStarTaq (Qiagen) and gene-specific primers binding different exons, choosing a number of cycles that resulted in a product of moderate quantity.
Cell Cycle
Cells were synchronized by incubating overnight with 100 ng/ml nocodazole. M phase cells were shaken off, washed with PBS, and reseeded in complete medium without nocodazole for the indicated times. DNA content was determined by propidium iodide staining and flow cytometry.
Protein Recruitment to the Triton Non-extractable Fraction
Cells were washed in PBS and extracted with 1 ml (for 24-well plates) of 20 mm Tris-HCl, pH 7.4, 150 mm NaCl, 5 mm EDTA, 10 mm MgCl2, 0.3% Triton X-100, 1 mm DTT, 10 μg/ml each of aprotinin and leupeptin, and 1 mm PMSF for 10 min on ice. The extraction buffer was removed, and the remaining cellular material was lysed in 100 μl of SDS sample buffer.
Antibodies and Western Blotting
For Western blotting, cells in 24-well plates were washed in 1 ml of PBS and lysed in 100 μl of 1× SDS sample buffer containing β-mercaptoethanol to ensure good extraction of nuclear proteins. For Western blots, the following antibodies were used: p21, BD Pharmingen 556431 and Upstate Biotech Millipore 05-655; MSH2, BD Pharmingen 556349; MSH3, BD Transduction Laboratories 611390; MSH6, BD Transduction Laboratories 610918; BRCA1, BD Transduction Laboratories 611842; PCNA, Santa Cruz Biotechnology sc-56; Cdt2, Abcam 72264; Chk2, Santa Cruz Biotechnology sc-2064; HA, Covance 16B12; Myc, S1826; from Clontech kit 631604 TATA box-binding protein (TBP), Abcam ab818; β-actin, Sigma AC-15.
Immunoprecipitations
HeLa cells were seeded at 1.5 × 106 in 100-mm dishes and used at 80% confluency. After MNNG treatment, the cells were washed with PBS and cross-linked with the indicated concentrations of dithiobis(succinimidyl propionate) (DSP) in 5 ml of PBS at 4 °C for 1 h. After cross-linking, the cells were washed with PBS and lysed by scraping in 1 ml of 50 mm Tris, pH 7.4, 150 mm NaCl, 5 mm EDTA, 1% Triton X-100, 10 μg/ml each of aprotinin and leupeptin, and 1 mm PMSF. After a 15-min extraction on ice and repeated vortexing, the cells were sonicated for 15 × 1 s using a Branson Sonifier 150 at a power setting between 1 and 2 to avoid foaming. Cell lysates were clarified by centrifugation at 14,000 × g for 25 min at 4 °C, and the supernatants were subjected to immunoprecipitation with protein G-Sepharose beads and 2 μg of antibodies overnight at 4 °C. The beads were washed four times in cold lysis buffer, and the proteins were eluted with SDS sample buffer containing β-mercaptoethanol, which resulted in cleavage of DSP and allowed migration of proteins at their expected molecular weight.
RESULTS
MNNG Induces Degradation of p21 Protein by the Proteasome
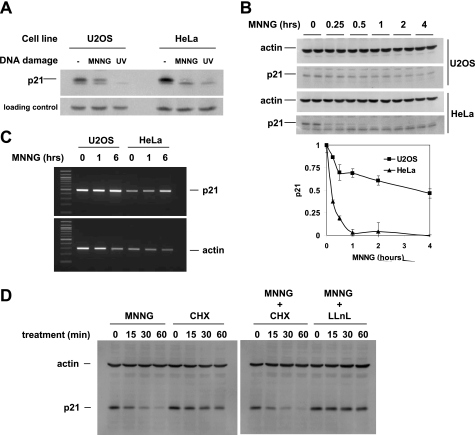
To determine the effect of DNA methylation by MNNG on p21, we treated HeLa and U2OS cells with 10 μm MNNG. After 5 h, p21 was undetectable in HeLa and reduced in U2OS (Fig. 1A). As a control, we irradiated cells with UV, which resulted in complete loss of p21 as shown previously (5). A time course of treatment with MNNG revealed that in HeLa, p21 was lost within 1 h, whereas in U2OS, p21 levels diminished rapidly during the first hour but remained more stable upon longer incubations (Fig. 1B) In a separate experiment in U2OS cells (not shown), there was no further reduction in p21 between 2 and 6 h. To address whether loss of p21 was due to down-regulation of p21 transcription, we performed RT-PCR for p21 on RNA samples taken from selected time points of the same experiment (Fig. 1C). p21 mRNA levels increased moderately in response to MNNG, which ruled out a transcriptional mechanism for p21 down-regulation.
FIGURE 1.
MNNG induces proteasomal degradation of p21. A, endogenous p21 protein in HeLa and U2OS cells was down-regulated after 5 h of continuous treatment with 10 μm MNNG and 5 h after UV irradiation. Total cell extract was immunoblotted with p21 mAb (Upstate Biotech Millipore); different mAbs (BD Pharmingen) verified that the higher Mr band is p21. The nonspecific band was used for loading control; similar results were obtained with β-actin as a loading control. B, time course of MNNG treatment. A Western blot of p21 protein (duplicate samples) in total cell extract is shown. Error bars in the bottom panel indicate S.D. C, p21 mRNA levels were analyzed by RT-PCR. After the indicated duration of treatment with MNNG, total RNA was isolated and reverse-transcribed using oligo(dT) primer, and p21-specific PCR was performed. D, Western blot of p21 protein in total cell lysates from cells treated with the indicated substances for the indicated times. Cycloheximide (CHX) inhibits protein synthesis; a cycloheximide time course measured protein stability. LLnL inhibited protein degradation by the proteasome.
We then determined the effect of MNNG on the stability of p21 at the protein level. HeLa cells were treated for the indicated times with MNNG, cycloheximide (an inhibitor of protein synthesis), or LLnL (an inhibitor of protein degradation by the proteasome) (Fig. 1D). In the presence of cycloheximide, the remaining levels of p21 diminished slowly, demonstrating the known instability of p21. When MNNG was also added, the rate of p21 degradation increased significantly, establishing that MNNG induced proteolytic degradation of p21. The 26 S proteasome is an essential factor in protein turnover and has been shown previously to degrade p21 (21). When we combined the proteasome inhibitor LLnL with MNNG, p21 levels remained stable. Together, these data indicate that MNNG induces proteasomal degradation of p21 protein.
MNNG Induces p21 Degradation Most Efficiently in HeLa Cells Due to the Absence of a Cell Cycle Effect
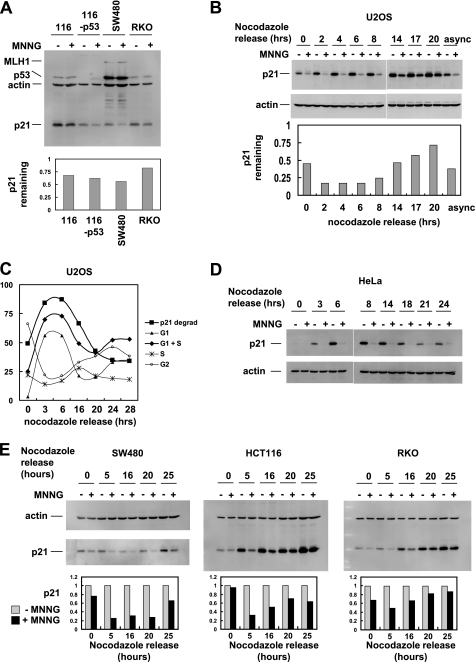
We routinely observed in HeLa cells that p21 was completely degraded after MNNG treatment, whereas in U2OS cells, p21 degradation was incomplete. In an attempt to understand this difference, we examined additional cell lines (Fig. 2A). p21 degradation could be observed in HCT116, HCT116-p53, SW480, and RKO cells but was incomplete in each case. p53 was of interest due to its inactivation by E6 protein in HeLa cells, but HCT116-p53 cells behaved more like parental HCT116 than HeLa cells with respect to MNNG-induced p21 degradation, making it unlikely that the absence of p53 caused more efficient p21 degradation in HeLa.
FIGURE 2.
MNNG leads to p21 degradation in a variety of cell lines. A, the indicated cell lines were treated with 10 μm MNNG or dimethyl sulfoxide (DMSO) control for 1 h followed by Western blot analysis of total cell extracts. Bottom panel, quantitation of p21 normalized to β-actin. B, U2OS cells were synchronized by a nocodazole block, released for the indicated times, and then treated with MNNG for 1 h. p21 Western blot is shown. Bottom panel, quantitation of p21 remaining after MNNG treatment and normalized to β-actin. async, asynchronous cells. C, alignment of p21 degradation (p21 degrad) in U2OS with cell cycle analysis; the extent of p21 degradation correlated best with the percentage of cells in G1 plus S phases. D, in HeLa cells, p21 degradation induced by MNNG occurs throughout the cell cycle. E, cell cycle analysis of MNNG-induced p21 degradation in SW480, HCT116, and RKO cells. Bottom panel, quantitation of p21 normalized to β-actin.
Signaling effects induced by MNNG have been reported to be cell cycle-dependent and may be active only in S phase (22). We therefore tested the response of p21 to MNNG in HeLa and U2OS cells that had been synchronized in M phase by a nocodazole block and then released for various times. In U2OS, MNNG-induced p21 degradation was cell cycle-dependent, with near complete p21 degradation observed from 2 to 6 h after release (Fig. 2B). At later stages, p21 degradation was nearly absent. When we aligned the extent of p21 degradation with cell cycle profiles, p21 degradation correlated best with the proportion of cells in G1 and S phase, suggesting that this pathway is active in G1 and S but not G2 and M phases (Fig. 2C). In contrast, HeLa cells exhibited p21 degradation at every cell cycle stage (Fig. 2D). This difference may explain why non-synchronized populations of HeLa cells degrade p21 more completely than U2OS.
Finally, we performed cell cycle analysis in cell lines that we previously assayed as non-synchronized populations (Fig. 2E). Each cell line showed cell cycle dependence of p21 degradation, and this correlated with the extent of p21 degradation observed in asynchronous cells.
MSH2 Is Required for p21 Degradation
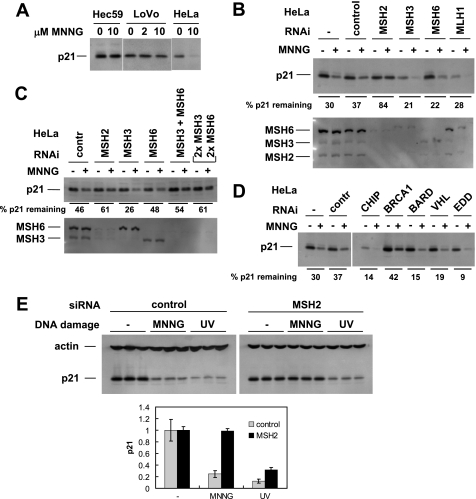
MSH2/6 recognizes O6-methylguanine, the DNA adduct caused by MNNG (10, 23), and is involved in DNA damage signaling in MNNG-treated cells (24). To assess the role of MSH2 in MNNG-mediated p21 degradation, we initially examined cell lines with the MSH2−/− phenotype (Fig. 3A). LoVo and Hec59 cells do not express MSH2 and were deficient in p21 degradation after MNNG treatment. We used RNAi to clarify the involvement of MMR proteins in MNNG-induced p21 degradation. Fig. 3B confirms that MSH2 is required for this pathway. In addition, we tested a series of other mismatch repair proteins and found that MSH3, MSH6, MLH1 (Fig. 3B), as well as PMS1 and PMS2 (not shown) were dispensable for p21 degradation. Surprisingly, when we knocked down both MSH3 and MSH6 simultaneously, p21 degradation was inhibited (Fig. 3C). This suggests that MSH2 can function in a complex with either MSH3 or MSH6 to activate p21 degradation.
FIGURE 3.
MSH2 is required for p21 degradation in MNNG-treated cells. A, cell lines with the absence (Hec59, LoVo) or presence (HeLa) of MSH2 were tested for p21 degradation in response to MNNG treatment. Lack of MSH2 expression correlated with failure to degrade p21. B, in HeLa cells, MMR proteins were individually down-regulated by RNAi followed by MNNG treatment and Western blotting for p21. Only MSH2 knockdown consistently interfered with p21 degradation. C, in HeLa cells, MSH3 and MSH6 were targeted by RNAi individually and in combination. Only cells in which both were knocked down exhibited reduced degradation of p21 in response to MNNG. For the last pair of lanes, twice the amount of both MSH3 and MSH6 siRNAs was used (67 nm final concentration of siRNA on cells). contr, control. D, additional proteins known to be involved in protein degradation were tested in HeLa cells by RNAi for a role in the pathway that leads to p21 degradation in MNNG-treated cells. BRCA1 knockdown had a moderate but reproducible effect on p21 degradation. E, HeLa cells (triplicate samples) were transfected with either control or MSH2 siRNA. Three days later, the cells were treated with either MNNG or UV, and p21 degradation was assayed. Knockdown of MSH2 protected p21 from MNNG but not UV. Bottom panel, quantitation of p21 normalized to β-actin. Error bars in the bottom panel indicate S.D.
p21 is degraded in response to UV irradiation in a ubiquitin/proteasome-dependent manner. Because p21 degradation in MNNG-treated cells similarly involved the proteasome, we tested known ubiquitin ligases for involvement in this pathway. BRCA1 in complex with BARD1 has ubiquitin ligase activity (25). We found that BRCA1 but not BARD1 knockdown interfered with p21 degradation in response to MNNG (Fig. 3D). Although we consistently observed this effect of BRCA1, the lack of BARD1 involvement makes it less likely that BRCA1 acts as a ubiquitin ligase in MNNG signaling. Of the other siRNAs against ubiquitin ligases initially tested, CHIP, VHL, EDD, and Skp2 had no effect. Cul1 siRNA caused an increase in p21 levels in untreated as well as MNNG-treated cells but did not inhibit down-regulation of p21 by MNNG (26, 27).
p21 is degraded after both MNNG-induced and UV-induced DNA damage. Having established that MSH2 is involved in the MNNG pathway, we asked whether MSH2 is also required after UV damage. Fig. 3E shows that MSH2 knockdown stabilized p21 after MNNG treatment but not after UV irradiation. This likely reflects the fact that MSH2 can directly recognize DNA alkylation adducts, but not UV-generated DNA damage, and suggests that MSH2 acts early in the MNNG pathway.
Cdt2 and PCNA Are Involved in p21 Degradation in MNNG-treated Cells
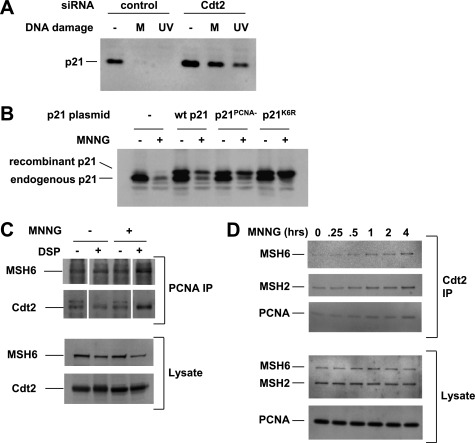
Cdt2, a substrate recognition subunit of the CRL4 ubiquitin ligase complex, has been shown to mediate p21 degradation in UV-irradiated cells (20, 28–30). We used a knockdown approach to demonstrate that Cdt2 was required for p21 degradation in MNNG-treated cells (Fig. 4A). Cdt2-mediated p21 degradation in UV-irradiated cells requires direct binding of p21 to PCNA, which creates a motif (consisting of p21 amino acids 144–155 embedded in PCNA) that is recognized by Cdt2 (31). To address whether the same mechanism applies to p21 degradation after MNNG treatment, we expressed a PIP box mutant of p21 that cannot bind PCNA (p21PCNA−) (20) in HeLa cells and tested its stability. As shown in Fig. 4B, p21PCNA− was not degraded in response to MNNG, thus confirming the requirement for PCNA binding in response to DNA methylation damage.
FIGURE 4.
MNNG-induced p21 degradation requires Cdt2 and PCNA. A, RNAi-mediated knockdown of the ubiquitin ligase adaptor subunit Cdt2 blocked p21 degradation in MNNG-treated (M) and UV-irradiated HeLa cells. B, a p21 mutant unable to bind PCNA (p21PCNA−) was not degraded after MNNG treatment. HeLa cells were transfected with plasmids expressing the WT or mutant p21 proteins as indicated. The recombinant p21 proteins have an N-terminal HA epitope tag resulting in a higher molecular weight than wild-type p21. C, MSH6 and Cdt2 co-immunoprecipitated with PCNA (PCNA IP) after MNNG treatment when protein complexes had been stabilized by chemical cross-linking with DSP. D, reverse immunoprecipitation with Cdt2 antibody (Cdt2 IP) demonstrated the same complex.
Demonstration of protein complexes by co-immunoprecipitation can be difficult in the case of chromatin-associated proteins because they may be non-extractable under mild conditions, or they may dissociate under harsher conditions required to extract them from chromatin. To circumvent these problems, we used a bifunctional cross-linking reagent, DSP, to covalently stabilize protein complexes during harsh extraction conditions involving sonication to shear chromosomal DNA. Fig. 4C shows that MSH6 and Cdt2 co-immunoprecipitated with PCNA in MNNG-treated cells in which protein complexes had been cross-linked with DSP before cell lysis and extraction. This suggests that MNNG-induced DNA alkylation leads to binding of MSH6 to PCNA and of PCNA to Cdt2. The reverse immunoprecipitation with Cdt2 (Fig. 4D) demonstrated binding of MSH6, MSH2, and PCNA to Cdt2 in a time-dependent manner after MNNG treatment and confirmed the formation of a MutS-PCNA-Cdt2 complex in response to MNNG. The similarities between p21 regulation in UV- and MNNG-treated cells therefore include down-regulation by a pathway that involves PCNA, Cdt2, and the proteasome.
Inhibition of p21 Degradation Blocks Translocation of Mismatch Repair Proteins to DNA
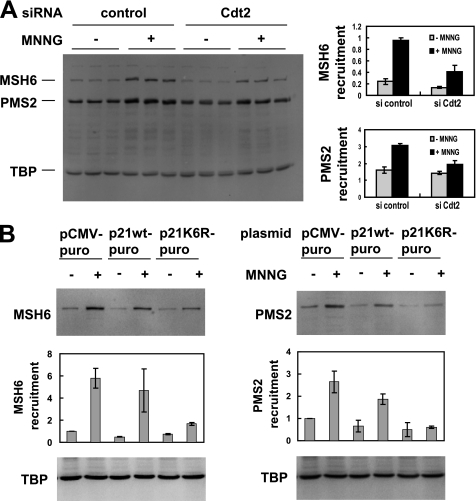
Having established that MNNG leads to the degradation of p21 via a pathway that involves MSH2, BRCA1, PCNA, Cdt2, and the proteasome, we next asked what the consequences for MMR would be if p21 degradation were blocked. MMR proteins translocate to the nucleus in response to MNNG treatment (32, 33) and other forms of DNA damage (34). We knocked down Cdt2 with RNAi and used redistribution of MSH6 and PMS2 to a Triton non-extractable (nuclear) fraction (35, 36) as an indicator of DNA repair in response to MNNG. When Cdt2 was knocked down, we observed that MNNG-induced redistribution of MSH6 and PMS2 was reduced, indicating a need for Cdt2 function and p21 degradation to recruit these proteins to nuclear DNA (Fig. 5A).
FIGURE 5.
Degradation of p21 is required for recruitment of MMR proteins in response to MNNG. A, Cdt2 was knocked down in HeLa cells using RNAi followed by MNNG treatment. Redistribution of MSH6 and PMS2 to DNA was assessed by extracting the cells with a Triton buffer. The remaining, non-extractable (mainly nuclear) proteins were subjected to SDS-PAGE and Western blotting. Cdt2 knockdown resulted in reduced nuclear recruitment of MSH6 and PMS2 in response to MNNG. Triplicate samples (independent wells in a 24-well plate) were quantitated (right panel), and average and S.D. are shown. TATA box-binding protein (TBP) was used as nuclear loading control and normalizer for the quantitation. B, HeLa cells were transiently transfected with either wild-type p21 or non-degradable p21K6R, and transfected cells were selected with puromycin. After MNNG treatment, redistribution of DNA repair proteins was assayed as in A. Expression of non-degradable p21 reduced recruitment of MSH6 (left panel) and PMS2 (right panel) to a Triton non-extractable (nuclear) fraction. Two separate experiments were performed that produced similar results; Western blots are from the first experiment, and quantitations show average and S.D. from both experiments.
Because Cdt2 might potentially regulate other proteins in addition to p21, we also used a more direct approach to assess the necessity for degradation of p21 to allow for DNA repair in response to MNNG. To this effect, we used a mutant of p21 that cannot be degraded via ubiquitination because all six lysine residues (ubiquitin conjugation sites) are mutated to arginine (p21K6R) (5, 37). Fig. 4B demonstrates that this stable p21 mutant is not degraded after MNNG treatment and also confirms the involvement of ubiquitination and the proteasome in p21 degradation. Using this system, we observed a reduction in MSH6 and PMS2 recruitment to a Triton non-extractable fraction when cells were transfected with p21K6R (Fig. 5B). This suggests that p21 degradation allows for enhanced recruitment of DNA repair proteins after MNNG-induced DNA damage.
DISCUSSION
In human cells, p21 is up-regulated in response to ionizing radiation but down-regulated after UV irradiation. The latter response serves to allow for efficient nucleotide excision repair by DNA resynthesis after excision of the damaged strand (5) and may be linked to monoubiquitination of PCNA (38). Monoubiquitinated PCNA is a docking site for DNA polymerase η (39), which has been implicated in MMR (40). MMR, like nucleotide excision repair, involves resynthesis of an excised DNA strand and can therefore be expected to be inhibited by p21. This has indeed been demonstrated using in vitro MMR systems in which p21 inhibited repair reactions by blocking PCNA. The fact that p21 is degraded after UV irradiation to allow for nucleotide excision repair, in combination with the observation that p21 also inhibits MMR, a DNA repair pathway that bears similarities to nucleotide excision repair, prompted us to investigate how p21 levels respond to DNA damage that triggers MMR. MNNG has been used to induce DNA methylation damage that is directly recognized by MMR proteins (10, 23), which results in recruitment of MMR proteins to the nucleus (32, 33).
We show here that MNNG treatment of HeLa cells resulted in the complete loss of p21 protein within 1 h of the addition of MNNG. This loss occurred through proteolytic degradation of p21 via the ubiquitin/proteasome pathway, whereas p21 mRNA levels were unaffected or slightly increased. Using RNAi, we found that the E3 ubiquitin ligase adaptor Cdt2 was required for p21 degradation, similar to the pathway that is initiated by UV treatment. Cdt2 recognizes substrates for ubiquitination that are in complex with PCNA and have a special PIP box with a lysine or arginine four amino acids downstream of the core PIP box (31). We found that the p21 PIP box was required for p21 degradation in MNNG-treated cells, similar to what has been shown in UV-irradiated cells. Furthermore, we found that in response to MNNG, a complex was formed between PCNA and Cdt2, in line with our other data implicating Cdt2 in MNNG-triggered p21 degradation. This complements findings that the CRL4-Cdt2 ubiquitin ligase interacts with PCNA in response to cisplatin, UV, and hydroxyurea (41) and that recruitment of Cdt2 to sites of UV damage was PCNA-dependent (42). On the other hand, siRNA knockdown of Skp2, a subunit of the E3 ubiquitin ligase SCF, which regulates p21 degradation in S phase, resulted in overall elevated levels of p21 but did not abrogate p21 down-regulation in MNNG-treated cells. We therefore conclude that the late stages of p21 degradation are shared between the pathways that are induced by UV and MNNG.
To identify components acting early in the pathway described here, we examined MMR proteins themselves. Both MSH2 and MLH1 have been implicated in the cellular responses to MNNG (24, 43, 44). The best studied effect of MNNG, methylation of O6 in guanine, causes cell cycle arrest or apoptosis, either after the first or after the second S phase following treatment, which may reflect different concentrations of MNNG. This pathway requires MLH1, and cells lacking MLH1 are markedly more resistant to MNNG (44, 45). Comparing HCT116 (MLH1−/−) with HCT116+chr3 (MLH1+), as well as using siRNA to knock down MLH1 in HeLa cells, we found no effect of MLH1 status on p21 degradation within hours of MNNG treatment. This may reflect the possibility that cell cycle arrest and apoptosis are long term effects of MNNG treatment that require DNA replication, whereas p21 degradation described here is a short term effect. Unlike MLH1, MSH2 was involved in degradation of p21 in our system, as shown in cell lines lacking MSH2 (LoVo, Hec59) and using siRNA-mediated MSH2 knockdown in HeLa cells. MSH2, as a heterodimer with MSH6, directly recognizes O6mG:C and O4mT:A base pairs, even before DNA is replicated following MNNG treatment. This makes MSH2 a plausible immediate sensor of alkylation damage. In our hands, individual knockdown of MSH3 or MSH6 did not protect p21 from degradation, whereas their combined knockdown did. This is difficult to reconcile with the lack of binding of recombinant purified MSH2/MSH3 to O6mG/C (23) and may reflect an indirect function of MSH2/MSH3 in vivo or an unidentified posttranslational modification of MSH2/MSH3 in MNNG-treated human cells.
We attempted to identify additional proteins involved in this p21 degradation pathway, and we consistently observed that siRNA-mediated BRCA1 knockdown partially protected p21 from degradation. BRCA1 can bind MSH2 (27), and both are components of the BRCA1-associated genome surveillance complex (26), which makes BRCA1 a candidate for the next component in the pathway that leads from MSH2-mediated recognition of alkylated DNA to CRL4-Cdt2-mediated degradation of p21.
To clarify the importance of p21 degradation for MMR, we blocked p21 degradation by two different approaches: (i) by knocking down Cdt2 and (ii) by overexpressing a non-degradable p21 mutant. In both cases, translocation of MSH6 and PMS2 to a Triton-resistant (chromatin) fraction was reduced. For MSH6, this may be surprising as at least in vitro, MSH2/6 could directly bind O6mG/C base pairs even without PCNA (23), and also in vitro, PCNA did not change the affinity of MSH2/6 for a defined DNA mismatch (46). In vivo, however, RNAi-mediated knockdown of PCNA inhibited association of MSH2, MSH6, MLH1, and PMS2 with chromatin in methylnitrosourea-treated cells (47). Our finding supports the idea of enhanced alkyl adduct binding by a MutS-PCNA complex as compared with MutS alone and emphasizes the importance of p21 as an inhibitor of PCNA function and DNA repair.
Acknowledgment
Full-length human MSH2 plasmid was a kind gift from R. Kolodner, San Diego, CA.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 CA72851 (to C. R. B.). This work was also supported by funds from the Baylor Research Institute.
- PCNA
- proliferating cell nuclear antigen
- MMR
- mismatch repair
- LLnL
- N-acetyl-leucyl-leucyl-norleucinal
- MNNG
- N-methyl-N′-nitro-N-nitrosoguanidine
- DSP
- dithiobis (succinimidyl propionate)
- PIP
- PCNA-interacting protein.
REFERENCES
- 1. Gu Y., Turck C. W., Morgan D. O. (1993) Nature 366, 707–710 [DOI] [PubMed] [Google Scholar]
- 2. Harper J. W., Adami G. R., Wei N., Keyomarsi K., Elledge S. J. (1993) Cell 75, 805–816 [DOI] [PubMed] [Google Scholar]
- 3. Xiong Y., Hannon G. J., Zhang H., Casso D., Kobayashi R., Beach D. (1993) Nature 366, 701–704 [DOI] [PubMed] [Google Scholar]
- 4. el-Deiry W. S., Tokino T., Velculescu V. E., Levy D. B., Parsons R., Trent J. M., Lin D., Mercer W. E., Kinzler K. W., Vogelstein B. (1993) Cell 75, 817–825 [DOI] [PubMed] [Google Scholar]
- 5. Bendjennat M., Boulaire J., Jascur T., Brickner H., Barbier V., Sarasin A., Fotedar A., Fotedar R. (2003) Cell 114, 599–610 [DOI] [PubMed] [Google Scholar]
- 6. Waga S., Hannon G. J., Beach D., Stillman B. (1994) Nature 369, 574–578 [DOI] [PubMed] [Google Scholar]
- 7. Jiricny J. (2006) Nat. Rev. Mol. Cell Biol. 7, 335–346 [DOI] [PubMed] [Google Scholar]
- 8. Kunkel T. A., Erie D. A. (2005) Annu. Rev. Biochem. 74, 681–710 [DOI] [PubMed] [Google Scholar]
- 9. Boland C. R., Goel A. (2010) Gastroenterology 138, 2073–2087.e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Duckett D. R., Drummond J. T., Murchie A. I., Reardon J. T., Sancar A., Lilley D. M., Modrich P. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 6443–6447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Umar A., Buermeyer A. B., Simon J. A., Thomas D. C., Clark A. B., Liskay R. M., Kunkel T. A. (1996) Cell 87, 65–73 [DOI] [PubMed] [Google Scholar]
- 12. Lau P. J., Flores-Rozas H., Kolodner R. D. (2002) Mol. Cell. Biol. 22, 6669–6680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Clark A. B., Valle F., Drotschmann K., Gary R. K., Kunkel T. A. (2000) J. Biol. Chem. 275, 36498–36501 [DOI] [PubMed] [Google Scholar]
- 14. Flores-Rozas H., Clark D., Kolodner R. D. (2000) Nat. Genet. 26, 375–378 [DOI] [PubMed] [Google Scholar]
- 15. Warbrick E., Lane D. P., Glover D. M., Cox L. S. (1995) Curr. Biol. 5, 275–282 [DOI] [PubMed] [Google Scholar]
- 16. Guo S., Presnell S. R., Yuan F., Zhang Y., Gu L., Li G. M. (2004) J. Biol. Chem. 279, 16912–16917 [DOI] [PubMed] [Google Scholar]
- 17. Kleczkowska H. E., Marra G., Lettieri T., Jiricny J. (2001) Genes Dev. 15, 724–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Masih P. J., Kunnev D., Melendy T. (2008) Nucleic Acids Res. 36, 67–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kreklau E. L., Kurpad C., Williams D. A., Erickson L. C. (1999) J. Pharmacol. Exp. Ther. 291, 1269–1275 [PubMed] [Google Scholar]
- 20. Abbas T., Sivaprasad U., Terai K., Amador V., Pagano M., Dutta A. (2008) Genes Dev. 22, 2496–2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Blagosklonny M. V., Wu G. S., Omura S., el-Deiry W. S. (1996) Biochem. Biophys. Res. Commun. 227, 564–569 [DOI] [PubMed] [Google Scholar]
- 22. Yoshioka K., Yoshioka Y., Hsieh P. (2006) Mol. Cell 22, 501–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Berardini M., Mazurek A., Fishel R. (2000) J. Biol. Chem. 275, 27851–27857 [DOI] [PubMed] [Google Scholar]
- 24. Umar A., Koi M., Risinger J. I., Glaab W. E., Tindall K. R., Kolodner R. D., Boland C. R., Barrett J. C., Kunkel T. A. (1997) Cancer Res. 57, 3949–3955 [PubMed] [Google Scholar]
- 25. Hashizume R., Fukuda M., Maeda I., Nishikawa H., Oyake D., Yabuki Y., Ogata H., Ohta T. (2001) J. Biol. Chem. 276, 14537–14540 [DOI] [PubMed] [Google Scholar]
- 26. Wang Y., Cortez D., Yazdi P., Neff N., Elledge S. J., Qin J. (2000) Genes Dev. 14, 927–939 [PMC free article] [PubMed] [Google Scholar]
- 27. Wang Q., Zhang H., Guerrette S., Chen J., Mazurek A., Wilson T., Slupianek A., Skorski T., Fishel R., Greene M. I. (2001) Oncogene 20, 4640–4649 [DOI] [PubMed] [Google Scholar]
- 28. Kim Y., Starostina N. G., Kipreos E. T. (2008) Genes Dev. 22, 2507–2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nishitani H., Shiomi Y., Iida H., Michishita M., Takami T., Tsurimoto T. (2008) J. Biol. Chem. 283, 29045–29052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stuart S. A., Wang J. Y. (2009) J. Biol. Chem. 284, 15061–15070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Havens C. G., Walter J. C. (2009) Mol. Cell 35, 93–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Christmann M., Kaina B. (2000) J. Biol. Chem. 275, 36256–36262 [DOI] [PubMed] [Google Scholar]
- 33. Schroering A. G., Williams K. J. (2008) DNA Repair 7, 951–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hong Z., Jiang J., Hashiguchi K., Hoshi M., Lan L., Yasui A. (2008) J. Cell Sci. 121, 3146–3154 [DOI] [PubMed] [Google Scholar]
- 35. Pagano M., Theodoras A. M., Tam S. W., Draetta G. F. (1994) Genes Dev. 8, 1627–1639 [DOI] [PubMed] [Google Scholar]
- 36. Toschi L., Bravo R. (1988) J. Cell Biol. 107, 1623–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sheaff R. J., Singer J. D., Swanger J., Smitherman M., Roberts J. M., Clurman B. E. (2000) Mol. Cell 5, 403–410 [DOI] [PubMed] [Google Scholar]
- 38. Soria G., Podhajcer O., Prives C., Gottifredi V. (2006) Oncogene 25, 2829–2838 [DOI] [PubMed] [Google Scholar]
- 39. Kannouche P. L., Wing J., Lehmann A. R. (2004) Mol. Cell 14, 491–500 [DOI] [PubMed] [Google Scholar]
- 40. Wilson T. M., Vaisman A., Martomo S. A., Sullivan P., Lan L., Hanaoka F., Yasui A., Woodgate R., Gearhart P. J. (2005) J. Exp. Med. 201, 637–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Terai K., Abbas T., Jazaeri A. A., Dutta A. (2010) Mol. Cell 37, 143–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ishii T., Shiomi Y., Takami T., Murakami Y., Ohnishi N., Nishitani H. (2010) J. Biol. Chem. 285, 41993–42000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. de Wind N., Dekker M., Berns A., Radman M., te Riele H. (1995) Cell 82, 321–330 [DOI] [PubMed] [Google Scholar]
- 44. Koi M., Umar A., Chauhan D. P., Cherian S. P., Carethers J. M., Kunkel T. A., Boland C. R. (1994) Cancer Res. 54, 4308–4312 [PubMed] [Google Scholar]
- 45. Carethers J. M., Hawn M. T., Chauhan D. P., Luce M. C., Marra G., Koi M., Boland C. R. (1996) J. Clin. Invest. 98, 199–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Iyer R. R., Pohlhaus T. J., Chen S., Hura G. L., Dzantiev L., Beese L. S., Modrich P. (2008) J. Biol. Chem. 283, 13310–13319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hidaka M., Takagi Y., Takano T. Y., Sekiguchi M. (2005) Nucleic Acids Res. 33, 5703–5712 [DOI] [PMC free article] [PubMed] [Google Scholar]