Abstract
Insulin-like growth factor-binding protein-3 (IGFBP-3) expression is frequently suppressed in liver cancers and can be reactivated by histone deacetylase (HDAC) inhibition. This study examined the role of IGFBP-3 in mediating the effects of the HDAC inhibitor MS-275 in liver cancer cells and identified IGFBP-3-dependent proteins that regulate proliferation and migration. In HepG2 cells, MS-275 inhibited DNA synthesis, cell cycle activity, and cell viability concomitantly with increased binding of acetylated histone H3 to IGFBP-3 promoter sequences and induction of IGFBP-3 expression. IGFBP-3 down-regulation by siRNA significantly reversed the inhibition of cell viability and DNA synthesis by MS-275, indicating an intermediary role for IGFBP-3. Induction of the cyclin-dependent kinase inhibitor p21 by MS-275 was attenuated by IGFBP-3 down-regulation, providing an explanation for IGFBP-3-dependent effects of MS-275 on cell cycle activity. In contrast, MS-275 stimulated HepG2 cell migration, an effect also inhibited by IGFBP-3 down-regulation. Among genes whose induction by MS-275 was attenuated by IGFBP-3 down-regulation, LYVE1 and THBS2 (thrombospondin-2) were identified as mediators of IGFBP-3-dependent effects of MS-275. Silencing of either protein had no effect on the inhibition of HepG2 viability by MS-275 but reversed its stimulatory effect on cell migration. We conclude that among genes up-regulated by MS-275, IGFBP-3 is a key mediator of effects on hepatoma cell growth and migration, involving IGFBP-3-dependent proteins p21 (proliferation) and LYVE1 and THBS2 (migration). The enhanced cell motility that accompanies reactivation of IGFBP-3 expression in liver cancer by HDAC inhibition suggests the possibility of increased metastatic spread despite inhibited cell proliferation.
Keywords: Cancer Therapy, Cell Death, Cell Migration, Cellular Regulation, Insulin-like Growth Factor (IGF), HDAC Inhibitor, IGFBP-3, LYVE1, Proliferation, Thrombospondin 2
Introduction
Reduced expression of insulin-like growth factor (IGF)-binding protein-3 (IGFBP-3) occurs frequently in human liver cancers. Yumoto et al. (1) detected IGFBP-3 transcripts in only 10% of 57 primary hepatomas, and Luo et al. (2) reported IGFBP-3 mRNA levels almost 80% lower in hepatoma tissues than in normal liver. The decreased IGFBP-3 expression has been associated with epigenetic changes such as DNA methylation and histone deacetylation (3, 4). IGFBP-3 is the most abundant insulin-like growth factor-binding protein in the human circulation, where it forms part of the IGF transport complex that stabilizes IGF-I and IGF-II and limits their access to tissues (5). In addition to its transport function, IGFBP-3 also has multiple biological roles at the cellular level, which depend on its interaction with a wide range of ligands (6, 7). By virtue of its ability to bind IGF-I and IGF-II with high affinity, IGFBP-3 can inhibit signaling through the type I IGF receptor. Other functions, independent of IGF binding, include effects on apoptosis, cell growth, differentiation, and migration, many of which appear dependent on the cell type and context (8–10). For example, IGFBP-3 can induce apoptosis in various cancer cell models, including prostate and breast cancer (11, 12), although it can also have growth-stimulatory effects (13).
Whereas in the rodent liver, IGFBP-3 gene expression is predominantly confined to Kupffer cells (14), in human liver, IGFBP-3 is synthesized by hepatocytes (15) and has been proposed to function as an inhibitor of cell proliferation in hepatocellular carcinoma (HCC)4 (3, 16). MS-275 is a histone deacetylase inhibitor for which a significant anti-cancer role has been demonstrated in vitro and in vivo (17, 18). In this study, we have investigated the involvement of endogenous IGFBP-3 in the anti-tumor effects of MS-275, by down-regulating its expression using siRNA. We describe roles for IGFBP-3 in hepatoma cell proliferation and migration, and we identify IGFBP-3-dependent proteins involved in mediating these effects.
EXPERIMENTAL PROCEDURES
Materials
Cell culture reagents were from Invitrogen. MS-275, trypan blue, propidium iodide, and RNase were purchased from Sigma. Chemically synthesized siRNA against IGFBP-3, THBS2, and LYVE1 and an IGFBP-3 scrambled control were from Qiagen (Doncaster, Victoria, Australia). Recombinant human IGFBP-3 was produced in 911 human retinoblastoma cells (19). The following antibodies were purchased for Western analysis: histone H3, histone H3-acetyl-LYS 9/18, histone H4, and histone H4-acetyl LYS-5/8/12/16 (Upstate Biotechnology, Lake Placid, NY); LYVE1 and THBS2 (R & D Systems Inc., Minneapolis, MN); p21 (Cell Signaling, Beverly, MA); and actin (Sigma).
Cell Culture and siRNA Transfections
Human hepatoblastoma cell lines HepG2 (catalogue no. HB-8065) and HuH6 (catalogue no. RCB-1367) were obtained from American Type Culture Collection and RIKEN Cell Bank, respectively, and maintained in DMEM supplemented with 10% FCS. Both cell lines were authenticated by short tandem repeat profiling (CellBank Australia, Wentworthville, New South Wales, Australia) and found to match repository samples in 100% of tested alleles. Transient transfections with siRNA were achieved by electroporation (Amaxa, Lonza Cologne AG, Cologne, Germany). Cells were grown to 70–90% confluence before transfection, for which 3 × 106 cells were resuspended in 100 μl of Nucleofector Solution V and mixed with 10 μl of siRNA (2.5 μg). The mixed cell suspension was transferred to an Amaxa cuvette inserted into the Nucleofector, and specific programs were activated for different cell lines (HepG2, T-028; HuH6, A-020). After transfection, prewarmed DMEM containing 10% FCS was immediately added to cells, which were then transferred into new plates or flasks.
Quantitative Real Time RT-PCR
Total RNA was isolated using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. RNA was further purified using LiCl (2.5 m final) and incubation at −20 °C for 2 h. Samples were centrifuged at 1500 rpm for 15 min at 4 °C, and the pelleted RNA was resuspended in RNase-free water before storage at −80 °C until analysis. Total RNA (1–3 μg) was reverse-transcribed using Superscript III first-strand synthesis system for RT-PCR (Invitrogen) and random hexamers. Fluorescence-labeled TaqMan probes (Applied Biosystems, Foster City, CA) were used for cDNA amplification, and quantitative RT-PCR (qRT-PCR) was performed and analyzed as described previously (13), using hydroxymethylbilane synthase (HMBS) as the reference gene.
Chromatin Immunoprecipitation Assays
Chromatin was isolated from 1.5 × 107 HepG2 cells using the ChIP-IT assay kit (Active Motif, Carlsbad, CA) according to the manufacturer's protocol and immunoprecipitated with antibodies against acetylated histone H3 or acetylated histone H4 (Upstate Biotechnology, Lake Placid, NY). IGFBP3 primers for PCR were from Sigma. The first IGFBP3 primer pair (−2; +134) was forward 5′-CCAGATGCGAGCACTGCG-3′ and reverse 5′-CATGACGCCTGCAACCG-3′; the second primer pair (+68; +254) was forward 5′-GTGTACTGTCGCCCCATCCC-3′ and reverse 5′-CTCGCAGCGCACCACG-3′. PCR was performed in a DNA Engine Dyad thermal cycler (MJ Research, Incline Village, NV), and products were electrophoresed on 2% agarose gels and visualized after ethidium bromide staining using a FujiFilm Image Reader FLA-3000 (FujiFilm, Tokyo, Japan). Densitometric analysis was performed using MultiGauge version 3.0 analysis software (Fujifilm).
Proliferation and Apoptosis Assays
Cell number and viability were determined by trypan blue exclusion. Cells in 6-well plates were treated with MS-275 for 24 or 48 h, and media and resuspended cells were then pooled and centrifuged. Cell pellets were resuspended in FCS and mixed with an equal volume of trypan blue solution. Viable and nonviable cells were counted separately using a hemocytometer. DNA synthesis was measured by [methyl-3H]thymidine incorporation as described previously (20). Apoptosis was measured on combined adherent and floating cells using the EnzChek caspase 3 assay kit (Molecular Probes, Eugene, OR).
Migration Assays
Cell migration through uncoated 8-μm pore membranes was determined by dispensing HepG2 cells in DMEM containing 10% FCS into the inserts of Transwell plates (Corning Glass), with the same medium in the lower chamber. After 24 h at 37 °C, medium in the inserts was replaced with fresh medium containing 1% FCS and 5 μm cytosine-β-d-arabinofuranoside to inhibit proliferation. Cells that migrated through the membrane over 24 h were stained with Cell Stain Solution (CytoSelect 24-Well cell invasion assay kit, Cell Biolabs Inc, San Diego) for 15 min at 21 °C. Phase contrast micrographs were obtained from three fields per insert (IX70-S1F2 inverted microscope, Olympus Optical Co. Ltd., Japan), and then the stain was extracted into cell extraction solution (CytoSelect 24-Well cell invasion assay kit). Duplicate 100-μl samples were transferred to microtiter plates, and absorbance was measured at 560 nm.
Protein Immunoblotting
Cells were washed twice in PBS and lysed directly in wells by adding sample buffer (62.5 mm Tris-HCl, 2% SDS, 10% glycerol, 50 mm DTT, and 0.01% bromphenol blue). Lysates were sonicated, centrifuged at 12,000 rpm for 10 min at 4 °C, and then heat-denatured for 5 min at 95 °C. Proteins were separated on NuPAGE 4–12% BisTris gels (Invitrogen) and transferred to Hybond C nitrocellulose membranes (GE Healthcare), and membranes were blocked and then incubated with primary antibodies overnight at 4 °C, followed by the appropriate secondary antibody for 1 h at 21 °C. Proteins were visualized using ECL Plus Western blotting detection reagents (GE Healthcare) and an ImageReader LAS-3000 (FujiFilm). Protein expression was normalized to actin, and densitometry was performed using MultiGauge imaging software (FujiFilm).
Statistical Analysis
Experiments were carried out three times, in triplicate unless otherwise stated. Statistical analysis was by StatView (SAS Inc., Cary, NC). Results are expressed as mean values ± S.D., with p < 0.05 considered significant. Post hoc analyses after ANOVA used Fisher's protected least significant difference test.
RESULTS
Effects of IGFBP-3 Induction by MS-275
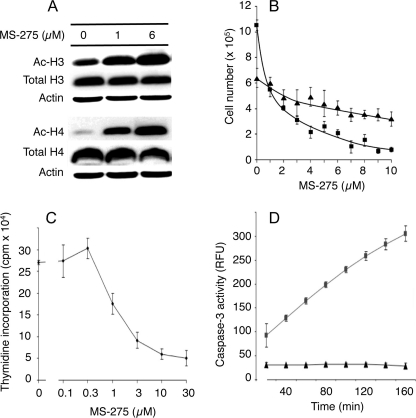
MS-275 is an orally active synthetic benzamide derivative that inhibits HDAC activity in vitro and has been evaluated clinically. It has preferential specificity for class 1 HDACs (21). Fig. 1A shows that exposure of HepG2 cells to 1 or 6 μm MS-275 for 24 h markedly increased the acetylation of histones H3 and H4. A similar result was seen in HuH6 cells (supplemental Fig. 1, A and B). This was associated with a loss of cell viability at 24 h that was more pronounced by 48 h (Fig. 1B). Both inhibition of DNA synthesis (Fig. 1C) and induction of apoptosis (Fig. 1D) appeared to contribute to the loss of cell viability. An alternative HDAC inhibitor, trichostatin A, was also tested for its effect on HepG2 cell growth and was found to inhibit DNA synthesis potently at concentrations of 0.1 μm or higher (supplemental Fig. 2A).
FIGURE 1.
Effects of MS-275 on HepG2 cells. A, HepG2 cells were treated with MS-275 for 24 h, and cell lysates were subjected to Western blot analysis using anti-acetylated histone H3 and H4 antibodies (Ac-H3 and Ac-H4). Total histones H3, H4, and actin were used as loading controls. B, cells were treated with MS-275 for 24 h (triangles) or 48 h (squares). Cell counts are expressed as viable cells per well. C, DNA synthesis was assessed by thymidine incorporation in the presence of the indicated concentrations of MS-275. D, cells were treated with (squares) or without (triangles) 6 μm MS-275 for 24 h, and then floating and adherent cells were collected, pooled, and assayed for caspase-3 activity over 160 min. Mean data are expressed ± S.D. of triplicate determinations, and results are shown from one representative experiment of three.
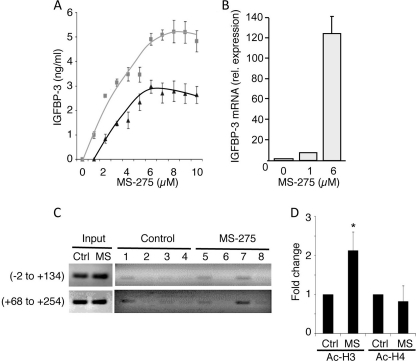
Concomitantly, IGFBP-3 secretion by HepG2 cells, determined by radioimmunoassay (22), was strongly induced by MS-275 (Fig. 2A), and IGFBP-3 mRNA, determined by qRT-PCR, was increased (Fig. 2B). Trichostatin A similarly stimulated IGFBP-3 secretion (supplemental Fig. 2B). In HuH6 cells, secreted IGFBP-3 was undetectably low in conditioned media, but IGFBP-3 mRNA was also inducible by MS-275 (supplemental Fig. 1C). Acetyl histone interaction with the IGFBP3 promoter was therefore determined (Fig. 2C). In control HepG2 cells, immunoprecipitated acetyl histone H3 (Fig. 2C, lane 3) showed only weak association with the IGFBP3 promoter regions from nucleotides −2 to +134 and +68 to +254, whereas immunoprecipitated RNA pol II (lanes 1 and 5), tested as a positive control, showed a positive interaction. Immunoprecipitation with control IgG (Fig. 2C, lane 2) and anti-acetyl histone H4 (lane 4) was negative. Treatment with 6 μm MS-275 significantly increased acetylated histone H3 (Fig. 2C, lane 7) associated with both regions of the IGFBP3 gene (Fig. 2D; p = 0.002 compared with untreated control). However, only weak association was seen when anti-acetyl histone H4 (Fig. 2C, lane 8) or the IgG control (lane 6) was used for immunoprecipitation. This indicates that the increased acetylated histone H3, seen in response to MS-275 treatment, associates with the IGFBP3 promoter, presumably causing direct enhancement of transcription.
FIGURE 2.
MS-275 induces expression of IGFBP-3 in HepG2 cells. A, cells were treated with MS-275 for 24 h (triangles) or 48 h (squares), and IGFBP-3 was measured in conditioned media by radioimmunoassay. Data are means ± S.D. of triplicate determinations from one representative experiment of three. B, cells were treated with MS-275 for 24 h, and IGFBP-3 mRNA was measured by qRT-PCR. IGFBP-3 mRNA was normalized to the reference gene HMBS, and results shown are relative to IGFBP-3 expression in the absence of MS-275. Values are means ± S.D. of triplicate determinations in three separate experiments. C, ChIP analysis of IGFBP3 promoter acetylation in untreated and MS-275-treated HepG2 cells. Immunoprecipitations used RNA pol II antibody (lanes 1 and 5), control rabbit IgG (lanes 2 and 6), or antibodies against acetylated histone H3 (lanes 3 and 7) or acetylated histone H4 (lanes 4 and 8). PCR amplification of IGFBP3 promoter regions −2 to +134 and +68 to +254 is indicated. ChIP assays were performed three times with similar results. D, mean ± S.D. of three independent assays was calculated by densitometric analysis of imaged blots, with changes after MS-275 treatment shown relative to untreated controls (Ctrl). *, p < 0.002 compared with control, determined by ANOVA.
Effects of Down-regulating Endogenous IGFBP-3
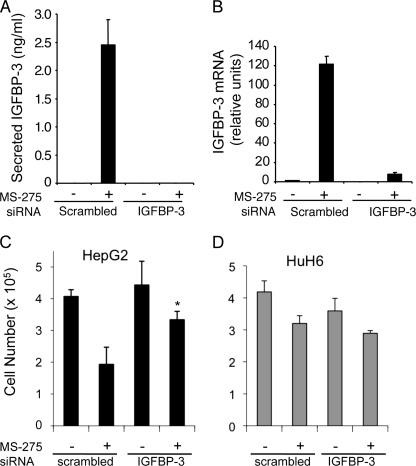
To investigate the role of endogenous IGFBP-3, it was down-regulated in HepG2 and HuH6 cells using siRNA. HepG2 cells transfected with scrambled siRNA and treated with MS-275 secreted ∼2.5 ng/ml IGFBP-3, but IGFBP-3 was undetectable (<0.5 ng/ml) in the absence of MS-275 or when IGFBP-3 was down-regulated (Fig. 3A). Secreted IGFBP-3 was undetectable in HuH6 media (data not shown). After 48 h, IGFBP-3 mRNA from cells transfected with IGFBP-3 siRNA was at least 90% lower than from those cells transfected with scrambled siRNA in HepG2 cells (Fig. 3B), and about 80% lower in HuH6 cells (data not shown). In HuH6 cells without treatment by MS-275, expression of IGFBP-3 mRNA was undetectable.
FIGURE 3.
siRNA-mediated silencing of IGFBP-3. HepG2 cells were transfected with scrambled siRNA or IGFBP-3 siRNA and plated in triplicate for 24 h. Fresh medium containing 6 μm MS-275 was added, and 24 h later conditioned media were collected, and RNA was prepared. A, secreted IGFBP-3 was measured by radioimmunoassay. B, qRT-PCR of IGFBP-3 mRNA was done in triplicate, and the expression relative to untreated, control siRNA was calculated, normalized to the reference gene HMBS. Results are the mean of three independent experiments. C and D, HepG2 and HuH6 cells were treated as described above and then counted using trypan blue to assess viability. Values are means ± S.D. of triplicates from one representative experiment of at least three. *, p < 0.05 versus scrambled siRNA (ANOVA).
Viable HepG2 cells were decreased ∼50% over 24 h by 6 μm MS-275 (Fig. 3C). Down-regulating IGFBP-3 by siRNA significantly reversed the loss of viability caused by MS-275 (p < 0.05), cell numbers no longer being significantly decreased by MS-275 (p = 0.09). IGFBP-3 down-regulation had no effect on viability in the absence of MS-275 (p = 0.5). In contrast, the viability of HuH6 cells was decreased to a smaller extent by MS-275 in the presence of control siRNA (∼24%) and not significantly reversed by knockdown of IGFBP-3 (Fig. 3D).
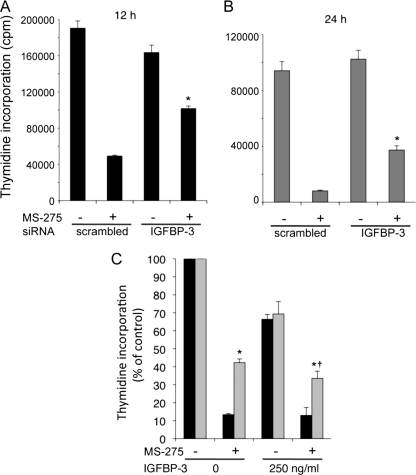
Treatment with 6 μm MS-275 for 12 or 24 h strongly inhibited DNA synthesis (Fig. 4). IGFBP-3 down-regulation had little effect on DNA synthesis in the absence of MS-275, but in its presence, it partially reversed the inhibitory effect of MS-275. After 12 h of MS-275 treatment, DNA synthesis was increased ∼2-fold in cells transfected with IGFBP-3 siRNA compared with the scrambled siRNA-transfected cells (Fig. 4A), and after 24 h of treatment, the increase in DNA synthesis when IGFBP-3 was down-regulated was ∼6-fold (Fig. 4B). Thus, down-regulation of endogenous IGFBP-3 partially protects cells against MS-275-induced inhibition of DNA synthesis. Addition of 250 ng/ml recombinant IGFBP-3 inhibited growth by 30% in the absence of MS-275 and significantly attenuated the protective effect of IGFBP-3 silencing in the presence of MS-275 (p < 0.05). However, the effect was small, and DNA synthesis remained significantly greater than when IGFBP-3 was not silenced (Fig. 4C). This indicates that, although endogenous IGFBP-3 induction accounts for part of the growth-inhibitory effect of MS-275, exogenous IGFBP-3 does not mimic this effect well, suggesting that intracellular IGFBP-3 may be more important than extracellular and/or that there are other actions of MS-275 that are unrelated to IGFBP-3.
FIGURE 4.
Down-regulation of IGFBP-3 partially protects cells against MS-275-induced inhibition of DNA synthesis. HepG2 cells were transfected with scrambled or IGFBP-3 siRNA and incubated ± 6 μm MS-275 for 12 (A) or 24 h (B). C, HepG2 cells transfected with control siRNA (black bars) or IGFBP-3 siRNA (gray bars) were treated ± 6 μm MS-275 and 250 ng/ml IGFBP-3 for 24 h, and thymidine incorporation was determined. Data are means ± S.D. of quadruplicates from one representative experiment of three. *, p < 0.05 versus scrambled siRNA. †, p < 0.05 versus corresponding value in the absence of exogenous IGFBP-3 (ANOVA).
Treatment with 6 μm MS-275 markedly inhibited the progression of HepG2 cells from G0/G1 to S phase (data not shown). Reiterating the DNA synthesis results, when IGFBP-3 was down-regulated by siRNA, MS-275-treated cells had a significantly higher percentage of cells in S phase than when the scrambled siRNA was used (p = 0.03), i.e., MS-275 was less inhibitory to cell cycle progression when IGFBP-3 was down-regulated. This again suggests that IGFBP-3 induction by MS-275 contributed to its effects on cell growth. In HuH6 cells there was no significant decrease in the S phase fraction, or any other cell cycle change, on exposure to MS-275 for 24 or 36 h (data not shown).
Effect of HDAC Inhibition and IGFBP-3 Down-regulation on p21 Expression
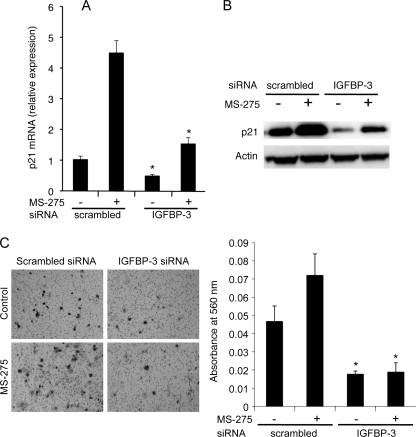
Treatment of cells with MS-275 is associated with an increase in expression of p21(CIP1/WAF1), a cyclin-dependent kinase inhibitor (17, 23–25). Because IGFBP-3 might mediate the antiproliferative effect of MS-275 at least in part by inducing p21, its expression was determined after knockdown of IGFBP-3. In HepG2 cells, p21 mRNA was induced >4-fold by MS-275 over 24 h (Fig. 5A), and IGFBP-3 down-regulation significantly impaired both basal and MS-275-induced p21 expression (both p < 0.001). A similar result was seen if exposure to MS-275 was for 6 or 12 h instead of 24 h (data not shown). In HuH6 cells, p21 mRNA was induced over 60-fold by MS-275 (supplemental Fig. 1D), and similarly IGFBP-3 down-regulation partially impaired the induction of p21 expression (p < 0.001). Western blots confirmed that p21 up-regulation by MS-275 was impaired by IGFBP-3 silencing (Fig. 5B); thus p21 induction by HDAC inhibition is, in part, dependent on IGFBP-3 up-regulation.
FIGURE 5.
MS-275 and IGFBP-3 modulate p21 expression and cell migration. Cells were transfected with scrambled or IGFBP-3 siRNA and incubated for 24 h and then changed to fresh media containing vehicle or 6 μm MS-275 for an additional 24 h. p21 mRNA was analyzed by qRT-PCR (A) and p21 protein by Western blot (B) with actin as loading control. Data are shown as mean fold change ± S.D. (triplicates) relative to the scrambled siRNA-transfected cells without MS-275 treatment and are representative of three experiments. All qRT-PCR values were normalized against HMBS. *, p < 0.001 versus scrambled siRNA. C, migration of cells transfected with control or IGFBP-3 siRNA was analyzed as described under “Experimental Procedures.” Left panel, representative images of one of three fields per insert by phase contrast microscopy. Right panel, absorbance at 560 nm of dye extracted from stained migrated cells. Data are shown as means ± S.D. (triplicates) from one experiment representative of three. *, p < 0.005 versus scrambled siRNA (ANOVA).
Role of Endogenous IGFBP-3 in Cell Migration
In contrast to its negative effect on cell proliferation and viability, exposure to MS-275 for 24 h enhanced cell migration across transwell membranes by 53% (p < 0.01). As shown in Fig. 5C (right panel), IGFBP-3 silencing by siRNA significantly inhibited cell migration in the absence of MS-275 (p < 0.005) and ablated the stimulatory effect of MS-275. These data indicate that endogenous IGFBP-3 enhances HepG2 cell migration and suggest that IGFBP-3 induction alone may account for the stimulatory effect of MS-275 on cell migration.
Genes That Mediate IGFBP-3 Effects
To identify genes that might mediate effects of IGFBP-3 induced by MS-275, gene expression analysis was undertaken using Affymetrix Human Genome U133 Plus 2.0 oligonucleotide arrays. After IGFBP-3 silencing for 24 h, cells were treated with or without MS-275 for 12 or 24 h (see supplemental Methods). MS-275 caused a marked up-regulation of many transcripts (supplemental Fig. 3), with 3267 transcripts up-regulated at least 2-fold after treatment for 24 h, 1439 up-regulated by at least 4-fold, and 367 up-regulated by at least 16-fold. Expression data were analyzed to identify genes up-regulated at least 4-fold by MS-275 in cells treated with scrambled siRNA, for which the up-regulation was attenuated in cells treated with IGFBP-3 siRNA. This would suggest that the MS-275 effect on these genes might be mediated in part through IGFBP-3. The supplemental Fig. 4 illustrates the expression levels of three such genes as follows: IGFBP3 itself, THBS2 encoding thrombospondin-2, and LYVE1 encoding lymphatic vessel endothelial hyaluronan receptor 1. For comparison, expression levels of three other genes, also induced by MS-275 but unaffected by IGFBP-3 silencing, are also shown as follows: HSPA2 (heat shock 70-kDa protein 2), RAB31 (Ras-related protein Rab-31), and FA2H (fatty acid 2-hydroxylase).
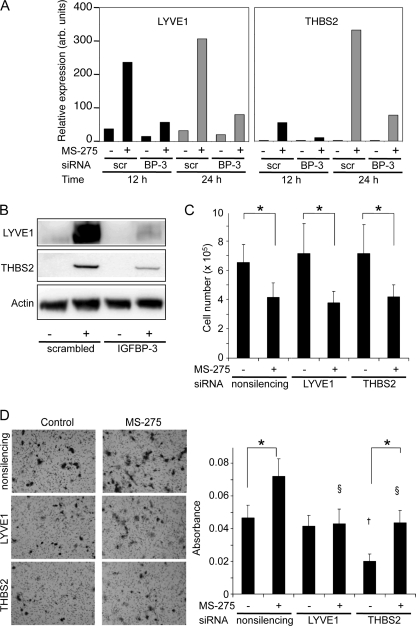
Fig. 6 illustrates the regulation of two proteins selected for further analysis in HepG2 cells, LYVE1 and THBS2. As determined by qRT-PCR, in control cells LYVE1 mRNA expression was increased almost 10-fold after 24 h of exposure to MS-275, but only 3.7-fold when IGFBP-3 was down-regulated, indicating that IGFBP-3 had an intermediary role in the induction of LYVE1 expression (Fig. 6A). Similarly, in control cells, THBS2 mRNA expression was increased 158-fold after 24 h of exposure to MS-275 and reduced to 39-fold when IGFBP-3 was down-regulated. Western blotting confirmed the mediating role for IGFBP-3 in the induction of both proteins by MS-275 in HepG2 cells (Fig. 6B). Under similar conditions in HuH6 cells, neither protein was detectable before or after MS-275 treatment (data not shown).
FIGURE 6.
Effect of LYVE1 and THBS2 silencing on HepG2 cells. 24 h after transfection with control or IGFBP-3 siRNA, cells were changed to fresh medium ± 6 μm MS-275 for an additional 24 h. LYVE1 and THBS2 mRNA were analyzed in triplicate by real time qRT-PCR (A) and protein levels by Western blot (B) with actin as loading control. C, cells transfected with nonsilencing, LYVE1, or THBS2 siRNA were treated with or without 6 μm MS-275 for 24 h, harvested, and counted. Cell counts are expressed as total viable cells per well, mean ± S.D. of triplicate determinations. *, p < 0.05 for MS-275 treatment compared with untreated control. D, migration assays of cells transfected with nonsilencing, LYVE1, or THBS2 siRNA were carried out as described under “Experimental Procedures.” Representative images of one of three fields per insert by phase contrast microscopy (left panel) are shown. Migrated cells were stained, and the absorbance was measured at 560 nm (right). Values are means ± S.D. of quadruplicates from one experiment representative of three. Effect of MS-275 treatment: *, p < 0.05; comparison with control, †, p < 0.001; comparison with control plus MS-275, §, p < 0.001, determined by ANOVA.
To determine whether either LYVE1 or THBS2 was involved in mediating the biological responses to IGFBP-3 induction by MS-275, both proteins were silenced using specific siRNAs. For LYVE1 siRNA, down-regulation of at least 90%, determined after 24 h of MS-275 treatment, was achieved at both the mRNA and protein level, compared with control nonsilencing siRNA, and for THBS2 siRNA, down-regulation of about 70% was achieved at both the mRNA and protein level (data not shown). However, neither LYVE1 nor THBS2 down-regulation affected the ability of MS-275 to inhibit cell viability (Fig. 6C). A similar lack of effect of down-regulating LYVE1 or THBS2 was seen when the end points were thymidine incorporation or cell cycle distribution determined by flow cytometry (data not shown). These data indicate that neither LYVE1 nor THBS2, two proteins whose induction by MS-275 was greatly attenuated by IGFBP-3 down-regulation, was involved in the growth-inhibitory effects of MS-275 that are mediated by IGFBP-3.
In contrast, both LYVE1 and THBS2 appeared to modulate the IGFBP-3-dependent stimulation of HepG2 cell migration by MS-275. MS-275 treatment stimulated HepG2 cell migration by ∼50% (Fig. 6D, left panel). As previously shown in Fig. 5C, this cell migration was substantially inhibited by IGFBP-3 down-regulation. When LYVE1 was down-regulated, basal HepG2 migration was unaffected, but MS-275 no longer stimulated the migration rate (Fig. 6D, right panel). THBS2 down-regulation had a somewhat different effect, causing over 50% inhibition of basal cell migration. MS-275 increased the migration rate, but not above the basal rate seen in nonsilenced cells. These results suggest that the ability of IGFBP-3 to stimulate HepG2 cell migration is at least in part mediated by IGFBP-3 induction of LYVE1, and it also points to a role for the IGFBP-3-dependent protein THBS2 in modulating HepG2 migration.
DISCUSSION
The association between IGFBP-3 expression and cancer cell growth has been difficult to clarify, with conflicting results seen in different studies. There are reports of IGFBP-3 overexpression in squamous cell lung cancer (26), localized prostate cancer (27), pancreatic adenocarcinoma (28), clear cell renal cell carcinoma (29), metastatic melanoma (30), and breast cancers with poor prognosis (31). In contrast, in patients with HCC, IGFBP-3 mRNA is consistently expressed at a lower level in tumor tissue than in healthy liver (3, 16). Serum IGFBP-3 levels are also reduced in patients with HCC (32), although the association between IGFBP-3 levels and disease staging is poor (33). The IGFBP3 gene is epigenetically suppressed in HCC (3), showing transcriptional silencing by methylation of promoter binding sites for p53 (34). HDAC inhibition by trichostatin A has also been shown to cause transcriptional activation of IGFBP3 (4). Together, this pattern of regulation of IGFBP-3 in HCC resembles that of a classic tumor suppressor (3). However, the mechanisms by which IGFBP-3 limits the growth of liver cancer cells are poorly understood.
In the two hepatoma cell lines studied, basal IGFBP-3 protein levels were undetectable although IGFBP-3 mRNA could be detected by qRT-PCR. HDAC inhibition by MS-275 markedly up-regulated IGFBP-3 mRNA and provided an opportunity to determine IGFBP-3 function by suppressing the MS-275-induced levels using siRNA. Both cell lines used in this study express wild type p53, consistent with the majority of HCCs reported in many studies (35, 36). The reactivation of IGFBP-3 expression by HDAC inhibition has been observed in both p53-wild type and mutant cell lines, indicating that this effect is not dependent on p53 (4, 37). Treatment with MS-275 decreased total cell numbers, thymidine incorporation, and G1-S transition and increased apoptosis, accompanied by a marked induction of IGFBP-3 at both the mRNA and protein levels. IGFBP-3 induction appeared to be a direct transcriptional response to HDAC inhibition because increased association of acetylated histone H3 with IGFBP3 promoter sequences was observed. IGFBP-3 down-regulation did not affect cell growth in HepG2 or HuH6 cells in the absence of MS-275 but partially attenuated the growth inhibition by MS-275 in HepG2 cells. The inability of IGFBP-3 down-regulation to completely reverse the MS-275-induced loss of cell viability and DNA synthesis may reflect the incomplete knockdown of IGFBP-3, but it is also likely that factors unrelated to IGFBP-3 contribute to the inhibitory effect of MS-275.
Addition of 250 ng/ml recombinant human IGFBP-3, a relative excess, was unable to reverse the increased DNA synthesis caused by IGFBP-3 silencing, suggesting that the cells were resistant to the exogenous protein, as observed previously in prostate cancer (10) and breast cancer (38) cell cultures. The lack of sensitivity toward exogenous IGFBP-3, relative to endogenous IGFBP-3, might relate to differences in post-translational modification between the two forms or differences in access to the appropriate subcellular compartment. Thus, although exogenous IGFBP-3 can be growth-inhibitory, more potent inhibition is achieved by activating the endogenous protein using agents such as MS-275.
p21, a member of the Cip/Kip family, plays an important role in cell cycle control by regulating cyclin-Cdk kinase activity (39). HDAC inhibitor-mediated activation of p21 has been closely linked to cellular G1-S growth arrest and is a key factor in HDAC inhibitor-mediated prevention of tumor growth (24, 40). Although p21 is induced in a p53-dependent manner in response to DNA damage, it can also be induced in a p53-independent manner (41). For example, increased histone acetylation in response to sodium butyrate in p53-mutant HT-29 colon cancer cells induces p21 (40). Therefore, IGFBP-3 might mediate the anti-proliferative effect of MS-275 in hepatoma cells through stimulation of p21 independently of p53 status. Our observed induction of p21 following inhibition of HDAC by MS-275, seen in both HepG2 and HuH6 cell lines, is consistent with studies in other cell types (23–25). Notably, the enhanced expression of p21 by MS-275 could be blocked by IGFBP-3 siRNA, a novel observation implying an important and previously unrecognized role for the induction of IGFBP-3 in the pathway between MS-275 treatment and cell cycle inhibition by p21.
Like its variable effect on growth parameters in different cell models and contexts, IGFBP-3 has different effects on cell migration in different cell types. Whereas inhibitory effects have been observed in endometrioid ovarian cancer (42), Ewing's sarcoma (43), and non-small cell lung cancer cells (44), IGFBP-3 can induce migration in melanoma (30) and breast cancer (38) cells, as well as umbilical vein (45) and retinal (46) endothelial cells. Furthermore, increased IGFBP-3 expression has been observed in liver and lymph node metastases of pancreatic endocrine neoplasms (47), although it is antimetastatic in lung cancer (44). The factors underlying these contrasting responses are not well understood.
In this study, MS-275 significantly stimulated the migration of HepG2 cells, an effect that was entirely blocked by IGFBP-3 silencing. The pro-migratory effect of MS-275 in this hepatoma cell line, dependent on IGFBP-3, contrasts with its inhibition of cell growth. A possible IGF-independent mechanism by which IGFBP-3 could induce pro-migratory pathways is by up-regulating or activating sphingosine kinase 1, as demonstrated in endothelial (45) and breast epithelial (13) cell lines. Sphingosine kinase 1 activity leads to the generation of sphingosine 1-phosphate, a well known inducer of migration in some cancer cell types (48, 49). Whether this mechanism also leads to stimulation of hepatoma cell migration is unknown. It is notable that in a study in which IGFBP-3 transcripts were detected in only 10% of 57 intrahepatic HCC tumors, lung metastases in four patients showed IGFBP-3 expression (1), consistent with the idea that IGFBP-3 may have a positive effect on cell migration.
A microarray analysis of genes induced by MS-275 and dependent on IGFBP-3 revealed two possible mediators of the stimulatory effect of IGFBP-3 on HepG2 migration. Down-regulation of either LYVE1 or THBS2 substantially ablated the stimulatory effect of MS-275 on migration, possibly through different pathways because LYVE1 down-regulation had no effect on basal migration, whereas THBS2 down-regulation was inhibitory to both basal and MS-275-stimulated migration. Neither protein has previously been described as being regulated either by histone acetylation or by IGFBP-3.
LYVE1 is an integral membrane glycoprotein that is used as a marker of lymphatic vessels (50, 51). Relative to its expression in normal liver, LYVE1 shows progressive down-regulation from polyclonal cirrhotic nodules to monoclonal cirrhotic nodules to HCC (52). Together with IGFBP3, LYVE1 is one of seven down-regulated genes in a 12-marker panel developed to distinguish early HCC from dysplastic nodules (53). LYVE1, which is identical to cell surface retention sequence binding protein-1 (CRSBP-1), has been reported to bind IGFBP-3 and to enhance autocrine growth mediated by IGFBP-3 in lung cancer cells (54). Thus, it is of particular interest that we have identified this protein as being coordinately regulated with IGFBP-3 and suppressed in response to IGFBP-3 silencing. Although we found that LYVE1 down-regulation did not affect HepG2 viability or proliferation in response to MS-275, its apparent role in modulating hepatoma cell migration makes it an important target for further study.
In contrast to LYVE1, THBS2, a large trimeric matricellular protein that was also MS-275-induced and IGFBP-3-dependent, has not been linked functionally to IGFBP-3. However, our demonstration that THBS2 was one of the enabling genes in the IGFBP-3-dependent, MS-275-induced increase in hepatoma cell migration is consistent with a recent computational analysis of metastasis-associated gene expression, showing that THBS2 is among the genes most strongly associated with metastasis across multiple cancer types (55). Surprisingly, in THBS2-null mice, matrix metalloprotease (MMP)-2, a promigratory matrix metalloprotease involved in tumor invasion, is up-regulated, suggesting that MMP-2 is not involved in the observed effect of THBS2 on migration in HepG2 cells, where its down-regulation by siRNA inhibited both basal and MS-275-induced migration. However, it has been proposed that THBS2-MMP-2 complexes may be cleared by low density lipoprotein receptor-related protein (LRP-1) (56), an endocytotic receptor thought to bind and mediate some growth-inhibitory effects of IGFBP-3 (57). It may be speculated that if IGFBP-3, as an LRP-1 ligand, can inhibit the internalization and clearance of THBS2-MMP-2 complexes in HepG2 cells, this may account in part for the promigratory effect of IGFBP-3 in these cells.
In summary, we have shown that expression of IGFBP-3, epigenetically suppressed in liver cancer cells, could be reactivated by the HDAC inhibitor MS-275 through a direct effect on acetylated histone H3 binding to the IGFBP3 promoter. Up-regulation of the cell cycle inhibitor p21 by MS-275 was secondary to the induction of IGFBP-3, because it was significantly attenuated by IGFBP-3 silencing. MS-275 unexpectedly enhanced the migration activity of hepatoma cells, and this effect could also be attributed to IGFBP-3 induction. We identified two MS-275-inducible genes, THBS2 and LYVE1, whose induction was strongly attenuated when IGFBP-3 was down-regulated. Silencing of either of these genes by siRNA was sufficient to prevent the enhancement of cell migration by MS-275. We conclude that reactivation of IGFBP-3 expression in liver cancer by HDAC inhibition, although inhibitory to cell growth, is likely to be accompanied by enhanced cell motility that could be associated with an increased propensity for metastatic spread despite inhibited cell proliferation. The disparate effects of IGFBP-3 reactivation on tumor proliferation and migration may therefore raise questions about the therapeutic benefit of HDAC inhibition in hepatoma.
Supplementary Material

The on-line version of this article (available at http://www.jbc.org) contains supplemental Methods and Figs. S1–S4.
- HCC
- hepatocellular carcinoma
- HDAC
- histone deacetylase
- HMBS
- hydroxymethylbilane synthase
- qRT
- quantitative RT
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- ANOVA
- analysis of variance.
REFERENCES
- 1. Yumoto E., Nakatsukasa H., Hanafusa T., Yumoto Y., Nouso K., Matsumoto E., Onishi T., Takuma Y., Tanaka H., Fujikawa T., Suzuki M., Uemura M., Shiratori Y. (2005) Int. J. Oncol. 27, 1223–1230 [PubMed] [Google Scholar]
- 2. Luo S. M., Tan W. M., Deng W. X., Zhuang S. M., Luo J. W. (2005) World J. Gastroenterol. 11, 4272–4276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hanafusa T., Yumoto Y., Nouso K., Nakatsukasa H., Onishi T., Fujikawa T., Taniyama M., Nakamura S., Uemura M., Takuma Y., Yumoto E., Higashi T., Tsuji T. (2002) Cancer Lett. 176, 149–158 [DOI] [PubMed] [Google Scholar]
- 4. Choi H. S., Lee J. H., Park J. G., Lee Y. I. (2002) Biochem. Biophys. Res. Commun. 296, 1005–1012 [DOI] [PubMed] [Google Scholar]
- 5. Baxter R. C. (1993) Trends Endocrinol. Metab. 4, 91–96 [DOI] [PubMed] [Google Scholar]
- 6. Firth S. M., Baxter R. C. (2002) Endocr. Rev. 23, 824–854 [DOI] [PubMed] [Google Scholar]
- 7. Yamada P. M., Lee K. W. (2009) Am. J. Physiol. Cell. Physiol. 296, C954–C976 [DOI] [PubMed] [Google Scholar]
- 8. Oh S. H., Kim W. Y., Kim J. H., Younes M. N., El-Naggar A. K., Myers J. N., Kies M., Cohen P., Khuri F., Hong W. K., Lee H. Y. (2006) Clin. Cancer Res. 12, 653–661 [DOI] [PubMed] [Google Scholar]
- 9. Kim K. S., Kim M. S., Seu Y. B., Chung H. Y., Kim J. H., Kim J. R. (2007) Aging Cell 6, 535–545 [DOI] [PubMed] [Google Scholar]
- 10. Peng L., Wang J., Malloy P. J., Feldman D. (2008) Int. J. Cancer 122, 558–566 [DOI] [PubMed] [Google Scholar]
- 11. Rajah R., Valentinis B., Cohen P. (1997) J. Biol. Chem. 272, 12181–12188 [DOI] [PubMed] [Google Scholar]
- 12. Butt A. J., Firth S. M., King M. A., Baxter R. C. (2000) J. Biol. Chem. 275, 39174–39181 [DOI] [PubMed] [Google Scholar]
- 13. Martin J. L., Lin M. Z., McGowan E. M., Baxter R. C. (2009) J. Biol. Chem. 284, 25542–25552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chin E., Zhou J., Dai J., Baxter R. C., Bondy C. A. (1994) Endocrinology 134, 2498–2504 [DOI] [PubMed] [Google Scholar]
- 15. Scharf J. G., Schmidt-Sandte W., Pahernik S. A., Koebe H. G., Hartmann H. (1995) J. Hepatol. 23, 424–430 [DOI] [PubMed] [Google Scholar]
- 16. Gong Y., Cui L., Minuk G. Y. (2000) Mol. Cell. Biochem. 207, 101–104 [DOI] [PubMed] [Google Scholar]
- 17. Jaboin J., Wild J., Hamidi H., Khanna C., Kim C. J., Robey R., Bates S. E., Thiele C. J. (2002) Cancer Res. 62, 6108–6115 [PubMed] [Google Scholar]
- 18. Dalgard C. L., Van Quill K. R., O'Brien J. M. (2008) Clin. Cancer Res. 14, 3113–3123 [DOI] [PubMed] [Google Scholar]
- 19. Firth S. M., Ganeshprasad U., Poronnik P., Cook D. I., Baxter R. C. (1999) Protein Expr. Purif. 16, 202–211 [DOI] [PubMed] [Google Scholar]
- 20. Martin J. L., Weenink S. M., Baxter R. C. (2003) J. Biol. Chem. 278, 2969–2976 [DOI] [PubMed] [Google Scholar]
- 21. Riester D., Hildmann C., Schwienhorst A. (2007) Appl. Microbiol. Biotechnol. 75, 499–514 [DOI] [PubMed] [Google Scholar]
- 22. Baxter R. C., Martin J. L. (1986) J. Clin. Invest. 78, 1504–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Saito A., Yamashita T., Mariko Y., Nosaka Y., Tsuchiya K., Ando T., Suzuki T., Tsuruo T., Nakanishi O. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 4592–4597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rosato R. R., Almenara J. A., Grant S. (2003) Cancer Res. 63, 3637–3645 [PubMed] [Google Scholar]
- 25. Baradari V., Höpfner M., Huether A., Schuppan D., Scherübl H. (2007) World J. Gastroenterol. 13, 4458–4466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kettunen E., Anttila S., Seppänen J. K., Karjalainen A., Edgren H., Lindström I., Salovaara R., Nissén A. M., Salo J., Mattson K., Hollmén J., Knuutila S., Wikman H. (2004) Cancer Genet. Cytogenet. 149, 98–106 [DOI] [PubMed] [Google Scholar]
- 27. Massoner P., Haag P., Seifarth C., Jurgeit A., Rogatsch H., Doppler W., Bartsch G., Klocker H. (2008) Prostate 68, 1165–1178 [DOI] [PubMed] [Google Scholar]
- 28. Okami J., Simeone D. M., Logsdon C. D. (2004) Cancer Res. 64, 5338–5346 [DOI] [PubMed] [Google Scholar]
- 29. Chuang S. T., Patton K. T., Schafernak K. T., Papavero V., Lin F., Baxter R. C., Teh B. T., Yang X. J. (2008) J. Urol. 179, 445–449 [DOI] [PubMed] [Google Scholar]
- 30. Xi Y., Nakajima G., Hamil T., Fodstad O., Riker A., Ju J. (2006) Mol. Cancer Ther. 5, 3078–3084 [DOI] [PubMed] [Google Scholar]
- 31. Rocha R. L., Hilsenbeck S. G., Jackson J. G., Lee A. V., Figueroa J. A., Yee D. (1996) J. Natl. Cancer Inst. 88, 601–606 [DOI] [PubMed] [Google Scholar]
- 32. Mattera D., Capuano G., Colao A., Pivonello R., Manguso F., Puzziello A., D'Agostino L. (2003) Clin. Endocrinol. 59, 699–706 [DOI] [PubMed] [Google Scholar]
- 33. Aishima S., Basaki Y., Oda Y., Kuroda Y., Nishihara Y., Taguchi K., Taketomi A., Maehara Y., Hosoi F., Maruyama Y., Fotovati A., Oie S., Ono M., Ueno T., Sata M., Yano H., Kojiro M., Kuwano M., Tsuneyoshi M. (2006) Cancer Sci. 97, 1182–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hanafusa T., Shinji T., Shiraha H., Nouso K., Iwasaki Y., Yumoto E., Ono T., Koide N. (2005) BMC Cancer 5, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang Y. J., Rossner P., Jr., Chen Y., Agrawal M., Wang Q., Wang L., Ahsan H., Yu M. W., Lee P. H., Santella R. M. (2006) Int. J. Cancer 119, 985–991 [DOI] [PubMed] [Google Scholar]
- 36. Yuan R. H., Jeng Y. M., Hu R. H., Lai P. L., Lee P. H., Cheng C. C., Hsu H. C. (2011) J. Gastrointest. Surg. 15, 321–329 [DOI] [PubMed] [Google Scholar]
- 37. Gahr S., Peter G., Wissniowski T. T., Hahn E. G., Herold C., Ocker M. (2008) Oncol. Rep. 20, 1249–1256 [PubMed] [Google Scholar]
- 38. O'Han M. K., Baxter R. C., Schedlich L. J. (2009) Growth Factors 27, 394–408 [DOI] [PubMed] [Google Scholar]
- 39. Harper J. W., Elledge S. J., Keyomarsi K., Dynlacht B., Tsai L. H., Zhang P., Dobrowolski S., Bai C., Connell-Crowley L., Swindell E., et al. (1995) Mol. Biol. Cell 6, 387–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Archer S. Y., Meng S., Shei A., Hodin R. A. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 6791–6796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jung Y. S., Qian Y., Chen X. (2010) Cell. Signal. 22, 1003–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Torng P. L., Lee Y. C., Huang C. Y., Ye J. H., Lin Y. S., Chu Y. W., Huang S. C., Cohen P., Wu C. W., Lin C. T. (2008) Oncogene 27, 2137–2147 [DOI] [PubMed] [Google Scholar]
- 43. Benini S., Zuntini M., Manara M. C., Cohen P., Nicoletti G., Nanni P., Oh Y., Picci P., Scotlandi K. (2006) Int. J. Cancer 119, 1039–1046 [DOI] [PubMed] [Google Scholar]
- 44. Oh S. H., Lee O. H., Schroeder C. P., Oh Y. W., Ke S., Cha H. J., Park R. W., Onn A., Herbst R. S., Li C., Lee H. Y. (2006) Mol. Cancer Ther. 5, 2685–2695 [DOI] [PubMed] [Google Scholar]
- 45. Granata R., Trovato L., Garbarino G., Taliano M., Ponti R., Sala G., Ghidoni R., Ghigo E. (2004) FASEB J. 18, 1456–1458 [DOI] [PubMed] [Google Scholar]
- 46. Chang K. H., Chan-Ling T., McFarland E. L., Afzal A., Pan H., Baxter L. C., Shaw L. C., Caballero S., Sengupta N., Li Calzi S., Sullivan S. M., Grant M. B. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 10595–10600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hansel D. E., Rahman A., House M., Ashfaq R., Berg K., Yeo C. J., Maitra A. (2004) Clin. Cancer Res. 10, 6152–6158 [DOI] [PubMed] [Google Scholar]
- 48. Li M. H., Sanchez T., Yamase H., Hla T., Oo M. L., Pappalardo A., Lynch K. R., Lin C. Y., Ferrer F. (2009) Cancer Lett. 276, 171–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Park K. S., Kim M. K., Lee H. Y., Kim S. D., Lee S. Y., Kim J. M., Ryu S. H., Bae Y. S. (2007) Biochem. Biophys. Res. Commun. 356, 239–244 [DOI] [PubMed] [Google Scholar]
- 50. Cunnick G. H., Jiang W. G., Gomez K. F., Mansel R. E. (2001) Biochem. Biophys. Res. Commun. 288, 1043–1046 [DOI] [PubMed] [Google Scholar]
- 51. McElroy M., Hayashi K., Garmy-Susini B., Kaushal S., Varner J. A., Moossa A. R., Hoffman R. M., Bouvet M. (2009) J. Surg. Res. 151, 68–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Colombat M., Paradis V., Bièche I., Dargère D., Laurendeau I., Belghiti J., Vidaud M., Degott C., Bedossa P. (2003) J. Pathol. 201, 260–267 [DOI] [PubMed] [Google Scholar]
- 53. Llovet J. M., Chen Y., Wurmbach E., Roayaie S., Fiel M. I., Schwartz M., Thung S. N., Khitrov G., Zhang W., Villanueva A., Battiston C., Mazzaferro V., Bruix J., Waxman S., Friedman S. L. (2006) Gastroenterology 131, 1758–1767 [DOI] [PubMed] [Google Scholar]
- 54. Huang S. S., Tang F. M., Huang Y. H., Liu I. H., Hsu S. C., Chen S. T., Huang J. S. (2003) J. Biol. Chem. 278, 43855–43869 [DOI] [PubMed] [Google Scholar]
- 55. Kim H., Watkinson J., Varadan V., Anastassiou D. (2010) BMC Med. Genomics 3, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yang Z., Strickland D. K., Bornstein P. (2001) J. Biol. Chem. 276, 8403–8408 [DOI] [PubMed] [Google Scholar]
- 57. Huang S. S., Ling T. Y., Tseng W. F., Huang Y. H., Tang F. M., Leal S. M., Huang J. S. (2003) FASEB J. 17, 2068–2081 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.