Abstract
The multisubunit membrane protein complex Photosystem II (PSII) catalyzes one of the key reactions in photosynthesis: the light-driven oxidation of water. Here, we focus on the role of the Psb27 assembly factor, which is involved in biogenesis and repair after light-induced damage of the complex. We show that Psb27 is essential for the survival of cyanobacterial cells grown under stress conditions. The combination of cold stress (30 °C) and high light stress (1000 μmol of photons × m−2 × s−1) led to complete inhibition of growth in a Δpsb27 mutant strain of the thermophilic cyanobacterium Thermosynechococcus elongatus, whereas wild-type cells continued to grow. Moreover, Psb27-containing PSII complexes became the predominant PSII species in preparations from wild-type cells grown under cold stress. Two different PSII-Psb27 complexes were isolated and characterized in this study. The first complex represents the known monomeric PSII-Psb27 species, which is involved in the assembly of PSII. Additionally, a novel dimeric PSII-Psb27 complex could be allocated in the repair cycle, i.e. in processes after inactivation of PSII, by 15N pulse-label experiments followed by mass spectrometry analysis. Comparison with the corresponding PSII species from Δpsb27 mutant cells showed that Psb27 prevented the release of manganese from the previously inactivated complex. These results indicate a more complex role of the Psb27 protein within the life cycle of PSII, especially under stress conditions.
Keywords: Bioenergetics, Electron Paramagnetic Resonance (EPR), Electron Transport System (ETS), Mass Spectrometry (MS), Photosynthesis, Cyanobacteria, Photosystem II (PSII), Psb27, Thermosynechococcus elongatus
Introduction
Photosystem II (PSII)2 is the first component of the photosynthetic electron transfer chain located in the thylakoid membranes of cyanobacteria and plant chloroplasts. It catalyzes the light-driven oxidation of water to molecular oxygen (1, 2). The first organisms capable of this reaction were Gram-negative cyanobacteria that evolved 3.5 billion years ago (3).
Active cyanobacterial PSII consists of 20 permanent subunits, which were identified in the x-ray structure of PSII complexes isolated from Thermosynechococcus elongatus BP-1 (henceforth T. elongatus) (4). The central D1 and D2 subunits ligate multiple cofactors that facilitate the water-splitting reaction and mediate the electron transfer to plastoquinone B (QB). The central antenna proteins CP43 and CP47 capture the light energy, which in turn excites the ChlD1 molecule of P680. This causes a charge separation at the primary donor with the electron subsequently transferred to pheophytin (5). After ChlD1+ is reduced by PD1, the electron is transferred from pheophytin to QA and QB, respectively. In turn, PD1+ is rereduced by electrons from the CaMn4 cluster and the water-splitting reaction.
During the early steps of biogenesis that probably takes place in the cytoplasmic membrane (6) a PSII precomplex is formed consisting of the central heterodimer D1 and D2, cytochrome b559, and PsbI (7–9). This precomplex is the smallest known unit capable of charge separation. Before a functional manganese cluster can be assembled, the D1 protein that is expressed as a precursor with a C-terminal extension of several amino acids has to be processed by the CtpA protease that is co-localized in the cytoplasmic membrane (10). After the precomplex is transferred to the thylakoids, the core antenna proteins CP43 and CP47 and several small subunits are assimilated to form an inactive PSII-Psb27 complex, which is in turn activated by incorporation of the CaMn4 cluster and attachment of the extrinsic proteins (11). The details of the assembly model are still controversial, as the mechanism of the PSII transfer from one membrane system to the other is ambiguous.
In particular, under high light conditions, the central D1 subunit is prone to damage and therefore has to be exchanged continuously (12). The damaged PSII complex enters an intricate repair cycle in which PSII is partly disassembled (8, 13). The resulting interstage repair complex is presumably similar to the CP43-less PSII complex RC47, which appears during biogenesis (14). The partial disassembly allows the removal of damaged D1, which is degraded mainly by FtsH protease in cyanobacteria, whereas in plants, several Deg proteases assist this process (15–18). The new pD1 is cotranslationally synthesized into the D1-depleted PSII complex (19–21), and the active holoenzyme is then reassembled similar to the biogenesis process. The complexity of this repair cycle demands a precise and orderly assembly and disassembly of subunits and cofactors, which are managed by the assistance of specific protein factors.
One of these factors, the Psb27 protein, has been shown to play a crucial role within the PSII repair cycle (22). In cyanobacteria, it is an 11-kDa lipoprotein located solely on the luminal side of almost non-oxygen-evolving monomeric PSII (11, 23). It prevents the premature binding of the extrinsic proteins (PsbO, PsbU, and PsbV) to facilitate the formation of the water-oxidizing CaMn4 cluster within the repair cycle of PSII (11, 24). A structural NMR-based analysis of Psb27 from Synechocystis sp. strain PCC 6803 showed that it consists of a right-handed four-helix bundle with a dipolar charge distribution (25, 26), which might be important for binding to PSII at the luminal surface. In Arabidopsis thaliana, two different copies of psb27 are present. One of the two Psb27 homologs is strictly required for D1 processing during PSII assembly (27), whereas the other copy supports the efficient repair of photodamaged PSII (28).
In this study, we generated and characterized a Δpsb27 strain in T. elongatus to investigate the role of Psb27 in PSII biogenesis and repair. We show that Psb27 is essential for the survival of cells grown under environmental stress conditions. Furthermore, analysis of isolated PSII complexes from cold-stressed cells revealed the presence of a novel dimeric PSII-Psb27 species. This complex is involved in processes after the inactivation of PSII and apparently prevents the release of manganese, indicating an extended role of Psb27 within the life cycle of PSII.
EXPERIMENTAL PROCEDURES
Construction of the psb27 Knock-out Mutant
Genomic DNA was isolated from T. elongatus and used as a template to amplify the tll2464 (psb27) gene with additional up- and downstream sequences applying the following oligonucleotide primer pair: TE27KOfor, 5′-GGAATTCGCCGGTGTTGCTA-3′; and TE27KOrev, 5′-GGAATTCAGGCCGTGTCCTT-3′. The product of the PCR was cloned into the pUC18 cloning vector (Fermentas). To inactivate the tll2464 (psb27) gene, a chloramphenicol resistance cartridge was introduced via a BseRI restriction site. Cells of a CP43-His strain of T. elongatus were transformed by electroporation (29, 30) with this vector to produce the Δpsb27 mutant strain. Mutant cells were selected by gradually adding chloramphenicol to the medium up to a concentration of 15 μg/ml. Single colonies were obtained by plating the mutant strain onto selective agar (7 μg/ml chloramphenicol). Segregation was checked via PCR using the TE27KOfor and TE27KOrev primers.
Culture Conditions and Growth Curve
Cells of T. elongatus were grown at 45 °C in BG-11 liquid medium (31) under white fluorescent lamps at a light intensity of 100 μmol of photons × m−2 × s−1 unless noted otherwise. The growth medium was constantly bubbled with air containing 5% CO2. The BG-11 medium used for cultivating the Δpsb27 mutant strain was supplemented with 15 μg/ml chloramphenicol.
For both wild-type and mutant cells, growth was measured in parallel with five cultures (150 ml) in ventilated growth tubes. Precultures were diluted into fresh medium lacking chloramphenicol to OD750 nm = 0.2. Growth was monitored under different light intensities (100 or 1000 μmol of photons × m−2 × s−1) and different temperature conditions (30 or 45 °C) for up to 6 days by taking samples and measuring the turbidity at OD750 nm with a Beckman DU 7400 spectrophotometer.
Oxygen Measurements
Oxygen evolution was measured by dynamic fluorescence quenching with a Fibox 2 system (PreSens). Measurements were carried out in the presence of the electron acceptors 2,6-dichloro-p-benzoquinone (1 mm) and ferricyanide (5 mm potassium ferricyanide). Oxygen evolution of intact cells was measured at 45 and 30 °C, respectively, at 100 μmol of photons × m−2 × s−1. Oxygen evolution rates of cells in at least three different samples in the early exponential growth phase (OD750 nm = 1) were calculated. PSII activities of isolated complexes were measured according to Nowaczyk et al. (11) at a light intensity of ∼12.000 microeinsteins.
77 K Fluorescence Emission Spectra
Cells in the exponential growth phase were harvested, the Chl concentration was set to 5 μg/ml, and samples were frozen in a measuring cuvette using liquid nitrogen. An excitation wavelength of 440 nm was used to induce Chl a fluorescence. Spectra were recorded in 1-nm steps from 630 to 780 nm using an AMINCO-Bowman type II fluorescence spectrometer.
Variable Fluorescence Measurements
The Chl a fluorescence yield from PSII was measured at 45 °C with a modulated fluorometer (DUAL-PAM-100, Walz). Samples containing 2–3 μg/ml Chl were dark-adapted for 10 min at 45 °C in a water bath and then irradiated with low intensity-modulated light (24 μmol of photons × m−2 × s−1) to determine the minimum fluorescence (F0). After illumination of the sample with actinic light (126 μmol of photons × m−2 × s−1), the fluorescence yield (F) was measured. A far-red pulse (720 nm) was introduced in the light-adapted state to obtain the minimum fluorescence (F0′). The maximum fluorescence (Fm′) in the light-adapted state was measured by a single saturating pulse of white actinic light (20,000 μmol of photons × m−2 × s−1, 100 ms). Additionally, the maximum fluorescence (Fm) was obtained by closing all reaction centers with 20 μm 3-(3,4-dichlorophenyl)1–1-dimethylurea, which blocks the acceptor site of PSII. The photochemical quenching parameter (qP) was calculated using the equation of van Kooten and Snel (32).
Isolation of His-tagged PSII
Purification of His-tagged PSII complexes by nickel affinity chromatography (chelating Sepharose fast flow column, GE Healthcare) followed by anion exchange chromatography (UNO Q6 column, Bio-Rad) was performed according to Nowaczyk et al. (11). Cold-stressed PSII complexes were isolated from cells grown at 30 °C for 48 h. The cold stress was introduced in the exponential growth phase at OD750 nm = 1.2.
Oligomerization State of PSII and SDS-PAGE
The monomer/dimer ratio of isolated PSII complexes was determined by Blue native PAGE, which was performed according to Schägger and von Jagow (33) with modifications according to Rexroth et al. (34). Samples of 1 μg of Chl were loaded onto a 3.5–16% acrylamide gradient gel (100 × 70 × 1.5 mm) with a stacking gel (4% acrylamide) on top. Electrophoresis was carried out in a Mini-PROTEAN III electrophoresis chamber (Bio-Rad) at 120 V and 35 mA for 1 h in Coomassie Blue-containing cathode buffer (50 mm Tricine, 15 mm Bistris (pH 7), and 0.02% Serva Blue G) and then for 1 h in Coomassie Blue-free buffer to reduce the gel background. Anode buffer consisted of 50 mm Bistris (pH 7). Denaturing SDS gel electrophoresis according to Schägger and von Jagow (33) was carried out as described by Nowaczyk et al. (11).
15N Pulse-label Experiments
For 15N incorporation analysis, cells were grown on normal BG-11 medium at 45 °C and 80 μmol of photons × m−2 × s−1, and the medium was exchanged with 15N-containing BG-11 medium (NaNO3 replaced with 5 mm 99% 5NH4Cl; Cambridge Isotope Laboratories) during the logarithmic growth phase. Cells were harvested after 24 h, and His-tagged PSII complexes were isolated by immobilized metal ion affinity chromatography and ion exchange chromatography (IEC). Preparation of samples containing tryptic D2 peptides and mass spectrometry analysis were carried out according to Nowaczyk et al. (35). Mass spectrometry data were analyzed using QuPE (36). Data sets were imported into QuPE and searched for proteins using Mascot. The search results for D2 peptides were used in the pulse-chase quantification utilizing peptide elution. The incorporation levels were set to 0.0 fixed, 0.55–0.96 variable, and 0.04 increment. Mass tolerance was set to 0.25 Da, intensity threshold to 1.0, accuracy to 0.1 m/z, and isotopic distribution similarity to 0.8. Elution peaks were searched 60 s before and after the identification spectrum at a maximum allowed peak width of 300. Peaks were detected using exact peak positions with the continuous wavelet transform (CWT)-based method. Statistics were calculated using the t test method of QuPE (five replicates/peptides per sample with a minimum A value of 1.0).
EPR Spectroscopy
Continuous wave EPR spectra were recorded according to Mamedov et al. (23) with a Bruker ELEXSYS E500 spectrometer equipped with a SuperX bridge and SHQE4122 cavity. For the low temperature measurements, an Oxford Instruments E900 cryostat and ITC-4 temperature controller were used. The S2 state multiline EPR signal from the CaMn4 cluster was induced by illumination at 200 K for 6 min (23). The total manganese content in the samples, measured at room temperature, was estimated after extraction with 200 mm HNO3 and 100 mm CaCl2 as described previously (37). Spectral analysis was performed using Bruker Xepr 2.1 software.
RESULTS AND DISCUSSION
Psb27 Is Essential for Photosynthetic Activity under Stress Conditions
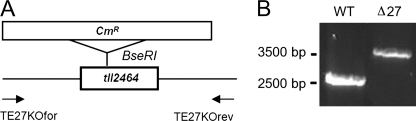
The Δpsb27 strain was generated by disruption of tll2464, which encodes the Psb27 protein in T. elongatus. A chloramphenicol cassette was introduced into the gene sequence via a BseRI restriction site (Fig. 1A), and complete segregation of the mutant allele was confirmed by PCR (Fig. 1B). The 3.5-kilobase pair band originates from the Δpsb27 sequence, and additionally, no wild-type signal (2.4 kilobase pairs) could be detected in the mutant strain.
FIGURE 1.
Generation of the Δpsb27 mutant strain. A, generation of the Δpsb27 mutant (tll2464). The site of insertion of the antibiotic resistance cassette for chloramphenicol (CmR) using the restriction enzyme BseRI and the position of the PCR primers are shown in the diagram. B, segregation analysis by PCR. Genomic DNAs of wild-type (WT) and Δpsb27 mutant cells were isolated and used as templates for amplification with the gene-specific primers TE27KOfor and TE27KOrev.
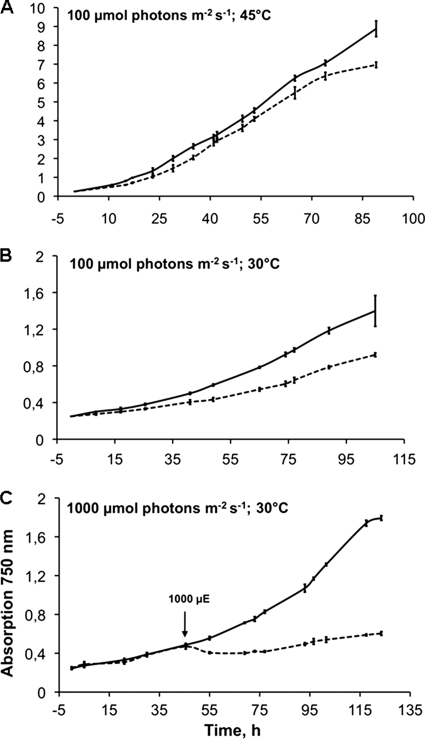
The growth behavior of Δpsb27 and wild-type cells was compared under different growth conditions (Fig. 2). When cells were grown at 45 °C and 100 μmol of photons × m−2 × s−1 (Fig. 2A), the growth rate of the Δpsb27 mutant did not differ significantly from that of wild-type cells, which indicates that the Psb27 protein is not essential for the viability of T. elongatus cells under these conditions. However, upon lowering the temperature to 30 °C, both wild-type and Δpsb27 mutant cells slowed their growth, with the mutant showing a more pronounced effect (Fig. 2B). When cells were incubated at low temperature and high light stress (1000 μmol of photons × m−2 × s−1), the growth of Δpsb27 mutant cells was completely inhibited, whereas wild-type cells were still able to grow (Fig. 2C).
FIGURE 2.
Growth of T. elongatus wild-type (solid lines) and Δpsb27 mutant (dashed lines) cells under different conditions. A, growth at 45 °C and light intensities of 100 μmol of photons × m−2 × s−1. B, growth at 30 °C and light intensities of 100 μmol of photons × m−2 × s−1. C, growth at 30 °C and light intensities of 1000 μmol of photons × m−2 × s−1. High light was introduced after the cultures reached OD750 nm ∼ 0.5. Error bars represent S.D. (n = 3). μE, microeinsteins.
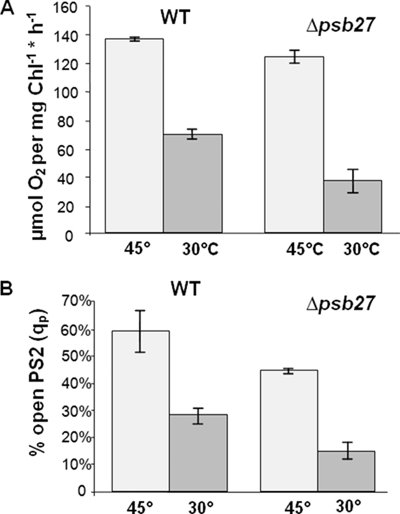
Oxygen evolution measurements were performed to assess the functionality of PSII in whole cells. The measurements were carried out with cells in the early exponential growth phase that were incubated at 45 °C and 100 μmol of photons × m−2 × s−1. Under these conditions, the photosynthetic activity of the wild-type and Δpsb27 mutant cells differed only slightly (120–140 μmol of O2 × mg−1 Chl × h−1) (Fig. 3A). In contrast, the impact of temperature stress (30 °C) was significantly stronger on the Δpsb27 mutant cells than on the wild-type cells (35 and 70 μmol of O2 × mg−1 Chl × h−1, respectively).
FIGURE 3.
Physiological characterization of the Δpsb27 mutant. A, the oxygen-evolving activities of wild-type (WT) and Δpsb27 mutant cells grown at 100 μmol of photons × m−2 × s−1 and 45 and 30 °C are compared. The measurement was carried out with cells in their early logarithmic growth phase (OD750 nm = 0.6). Error bars represent S.D. (n = 3). B, fluorescence yield changes were measured, and qP values for wild-type and Δpsb27 cells were calculated to determine the effect of different growth conditions on photochemical efficiency. Cells were grown at 45 and 30 °C and 100 μmol of photons × m−2 × s−1. Error bars represent S.D. (n = 3).
Chl a fluorescence yield changes in PSII were measured to calculate the photochemical quenching parameter (qP), which indicates the amount of open and closed reaction centers in the cells. On the one hand, qP reflects the closure of reaction centers according to different light intensities, but on the other hand, it also depends on parameters such as temperature and the availability of electron acceptors with direct influence on the electron flow downstream of PSII. To guarantee reproducible results, cultures were kept under identical growth conditions, and only cells harvested in the same growth phase were used in these experiments. Under these conditions, ∼60% of wild-type reaction centers were open compared with 44% in the mutant (Fig. 3B).
Low temperature stress caused an increase in closed reaction centers due to reduced electron transfer downstream of PSII, but the effect was more pronounced in the mutant. Only 28% of wild-type and 12% of mutant reaction centers were open under cold stress conditions. These results reflect, in addition to the varying oxygen evolution rates, a larger amount of inactive centers in the Δpsb27 mutant.
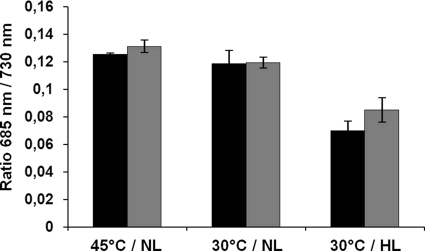
The relative amount of PSII centers compared with PSI was analyzed in a series of 77 K fluorescence emission spectrum measurements (Fig. 4). To examine changes in the PSII/PSI ratio, the spectra were normalized to the fluorescence maximum of PSI (720 nm). Under neither normal nor stress growth conditions could significant variations in the PSII/PSI ratio between wild-type and mutant cells be observed. These results indicate that the total amount of PSII complexes is not dramatically reduced in the mutant compared with wild-type cells even under stress conditions. In conclusion, these results clearly show that Psb27 plays an important role in the viability of the cells under the stress conditions tested.
FIGURE 4.
77 K emission spectra of wild-type (black bars) and Δpsb27 mutant (gray bars) cells after excitation at 440 nm. The relative amount of PSII to PSI is shown by the quotient of 685 nm (maximum value of PSII fluorescence) to 730 nm (maximum value of PSI fluorescence). NL, normal light growth (100 μmol of photons × m−2 × s−1); HL, high light growth (1000 μmol of photons × m−2 × s−1). Error bars represent S.D. (n = 3).
Application of Cold Stress Reveals a Novel Dimeric PSII-Psb27 Complex
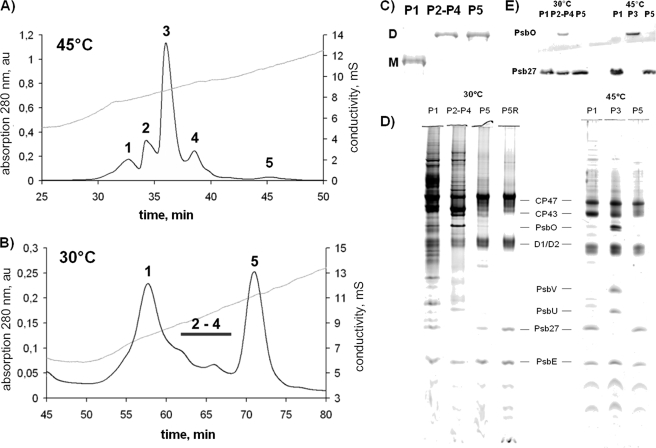
T. elongatus CP43-His cells were incubated at 45 or 30 °C for 48 h, and His-tagged PSII complexes were isolated by two HPLC steps as described previously (11). Five different PSII species could be isolated from cells grown at 45 °C: PSIIM(low) (peak 1), PSIIM(high) (peak 2), PSIID(high) (peak 3), PSIID(low) (peak 4), and a small previously uncharacterized PSII fraction (peak 5) (Fig. 5A). In contrast to these results, the IEC profile of PSII complexes isolated from cells grown at 30 °C showed only two major peaks (peaks 1 and 5 of the standard CP43-His PSII preparation) and several smaller peaks in between (Fig. 5B). Blue native PAGE analysis of the fractions revealed that peak 1 corresponds to monomeric and peak 5 to dimeric PSII complexes, as well as most of the small peaks in between (Fig. 5C).
FIGURE 5.
Purification and characterization of PSII from T. elongatus CP43-His. The elution profiles (280 nm) of IEC purification show five different peaks for cells grown at 45 °C (A) and only two major peaks and several small intermediate peaks for cells grown at 30 °C (B) for 48 h. au, absorbance units; mS, millisiemens. C, the oligomerization state was verified by Blue native PAGE (1 μg of Chl/lane, Coomassie Blue-stained bands). P, peak; D, dimer; M, monomer. D, the subunit composition of the different PSII fractions was analyzed by SDS-PAGE (1 μg of Chl/lane). E, the presence or absence of PsbO and Psb27 was confirmed by Western blot analysis with specific antibodies. P5R, the sample P5 was desalted and purified by IEC a second time.
Oxygen evolution measurements showed activity for neither peak 1 nor peak 5, whereas the pool of the intermediate fractions had a reduced activity of ∼1000 μmol of O2 × mg−1 Chl × h−1. SDS-PAGE and Western blot analysis of these samples revealed the presence of PsbO only in the active pool, whereas the inactive PSII species of peaks 1 and 5 comprised the Psb27 protein instead (Fig. 5, D and E). According to these results, the inactive monomeric PSII complex of peak 1 apparently corresponds to the PSIIM(low) complex, which was previously described (11). Recently, Liu et al. (38) were able to isolate a monomeric PSII complex from the mesophilic cyanobacterium Synechocystis PCC 6803 containing both the PsbO and Psb27 proteins, but according to our results, there is a clear separation between PsbO- and Psb27-containing complexes (Fig. 5E), except for the poorly separated low active intermediate fraction, which was isolated from cells grown at 30 °C.
Most intriguing is the isolated inactive dimeric PSII species (peak 5), hereafter termed PSIID(inact). Analysis of the complex distribution revealed that 45% of all PSII complexes in the cold-stressed membranes belong to the inactive monomer PSIIM(low) and 34% to the inactive dimer PSIID(inact), whereas only 21% of the isolated PSII complexes were active. Table 1 summarizes the characteristics of the different PSII species.
TABLE 1.
Characterization of PSII complexes isolated from cells grown at 45 and 30 °C (48 h)
| IEC CP43-His (45 °C) | P1a | P2 | P3 | P4 | P5 |
|---|---|---|---|---|---|
| Monomer/dimer | Monomer | Monomer | Dimer | Dimer | Dimer |
| Activity (μmol of O2 × mg−1 Chl × h−1)b | 201 ± 88 | 3136 ± 68 | 4853 ± 394 | 2067 ± 144 | 105 ± 46 |
| Relative amount (%) | 15 | 20 | 40 | 20 | 5 |
| Luminal subunits | Psb27 | PsbO, PsbV, PsbU | PsbO, PsbV, PsbU | PsbO, PsbV, PsbU | Psb27 |
| IEC CP43-His (30 °C) | P1 | P2–P4 | P5 | ||
| Monomer/dimer | Monomer | Mainly dimers | Dimer | ||
| Activity (μmol of O2 × mg−1 Chl × h−1)b | 0 | 1083 ± 188 | 0 | ||
| Relative amount (%) | 45 | ∼21 | 34 | ||
| Luminal subunits | Psb27 | PsbO, PsbV, PsbU, Psb27 | Psb27 | ||
a P, peak.
b Mean ± S.E. of three biological replicates.
15N Pulse Labeling Allocates PSIID(inact) to the Disassembly Stage of the PSII Repair Cycle
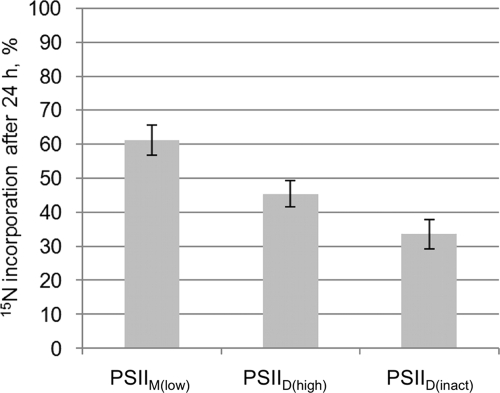
Cells were labeled with 15N in their exponential growth phase, and PSII complexes were isolated after 24 h. Subunits were separated via SDS-PAGE, and peptides of the D2 subunit were analyzed by LC-electrospray ionization-MS/MS. The percentage of D2 peptides enriched in 15N was calculated with QuPE (Fig. 6).
FIGURE 6.
Quantification of 15N incorporation into the D2 subunit of PSII. T. elongatus cells were grown in BG-11 medium, and during the early exponential growth phase, the only nitrogen source was changed from NaNO3 (99.6% 14N) to 15NH4Cl (99% 15N). 24 h after the exchange, the cells were harvested, and PSII was isolated and separated into different complexes. The D2 bands of polyacrylamide gels were digested with trypsin and analyzed in an electrospray ionization-MS/MS mass spectrometer. MS data for different D2 peptides were evaluated with QuPE. The diagram shows the percentage of D2 peptides enriched in 15N, i.e. synthesized after the 14N-to-15N exchange, in different PSII fractions. Higher percentages indicate a younger age of the complex. Five peptides per sample were evaluated. Bars show the arithmetic mean, and error bars indicate S.D., both calculated by QuPE.
Differences in 15N incorporation into the PSII subspecies indicate a different age and therefore a different stage within the PSII life cycle (11). D2 from the Psb27-containing PSIIM(low) complex showed the highest 15N enrichment after 24 h with 61% of all peptides. PSIID(high) contained 45% and PSIID(inact) only 34% 15N-enriched D2 peptides (Fig. 6). This clearly allocates the novel PSIID(inact) complex to PSII disassembly, as it takes a longer time to see the incorporation of 15N into this PSII species.
Characterization of PSII Complexes from Δpsb27 Cells Reveals Functional Differences Compared with Wild-type PSII
The His tag fused to the CP43 subunit allowed the efficient isolation of PSII complexes from the Δpsb27 mutant strain. The preparation yielded five different PSII fractions. Analysis of the oligomerization state, oxygen-evolving activity, and subunit composition revealed a similar PSII subspecies distribution as the wild-type PSII preparation except for the absence of Psb27 (data not shown).
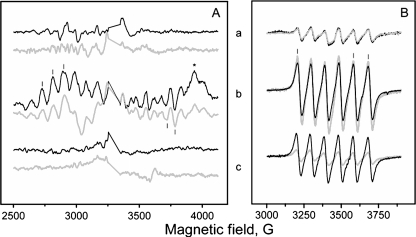
The activity of PSIID(high) complexes isolated from the Δpsb27 mutant strain appeared to be reduced (Table 2). This correlated with the relative amplitude of the induced S2 state multiline EPR signal (Fig. 7A, spectrum b, and Table 2), which indicates an active and structurally intact donor site of PSII. Additionally, the acceptor site seemed to be intact in this PSII fraction as shown by the presence of the QA−QB−Fe2+ interaction signal (Fig. 7A, spectrum b). In PSII complexes lacking O2 evolution, there was no observable S2 state multiline signal.
TABLE 2.
Comparison of isolated PSII complexes from T. elongatus CP43-His and the Δpsb27 strain
| P1a | P3 | P5 | |
|---|---|---|---|
| IEC CP43-His (45 °C) | |||
| Monomer/dimer | Monomer | Dimer | Dimer |
| Activity (μmol of O2 × mg−1 Chl × h−1)b | 201 ± 88 | 4853 ± 394 | 105 ± 46 |
| S2 multiline EPR signal (%)c | None | 100 | None |
| Mn/PSIIb,d | 1.0 ± 0.2 (26%) | 3.8 ± 0.2 (100%) | 3.2 ± 0.4 (85%) |
| IEC Δpsb27:CP43-His (45 °C) | |||
| Monomer/dimer | Monomer | Dimer | Dimer |
| Activity (μmol of O2 × mg−1 Chl × h−1)b | 0 | 2920 ± 298 | 0 |
| S2 multiline EPR signal (%)c | None | 90 | None |
| Mn/PSIIb,d | 0.8 ± 0.3 (20%) | 4.2 ± 0.3 (100%) | 0.8 ± 0.3 (20%) |
a P, peak.
b Mean ± S.E. of three biological replicates.
c The S2 multiline EPR signal was normalized to the YD· concentration. The amount of signal in the PSIID(high) fraction (P3) from the T. elongatus CP43-His strain was taken as 100%.
d The manganese content was calculated based on the assumption that there are 38 Chl molecules/reaction center of PSII according to Nowaczyk et al. (11).
FIGURE 7.
EPR measurements on the different PSII core fractions from wild-type and Δpsb27 mutant cells of T. elongatus. Black spectra correspond to measurements of wild-type PSII, and gray spectra correspond to measurements of PSII purified from the Δpsb27 strain. In detail, the measurements shown represent PSIIM(low) (peak 1; spectra a), PSIID(high) (peak 3; spectra b), and PSIID(inact) (peak 5; spectra c). A, the S2 state multiline signal from the CaMn4 cluster. Spectra shown are light minus dark difference spectra. Bars indicate peaks used for quantification of the signal, and the asterisk indicates the position of the QA−QB−Fe2+ interaction signal. Measuring conditions were as follows: microwave frequency, 9.27 kHz; microwave power, 10 milliwatts; modulation amplitude, 20 G; and temperature, 7 K. B, the free Mn2+ EPR signal originating from manganese ions released after acidification of samples. Bars indicate peaks used for quantification of the signal. Measuring conditions were as follows: microwave frequency, 9.75 kHz; microwave power, 20 milliwatts; modulation amplitude, 20 G; and temperature, 295 K.
Determination of the manganese content of the isolated PSII complexes by EPR revealed striking differences. The manganese content of PSIIM(low) in the mutant was in the same range as the corresponding PSII species in the wild-type cells. In contrast, the manganese content of PSIID(inact) differed significantly (Fig. 7B and Table 2). Only 20% of the maximum manganese content was detected in PSIID(inact) lacking the Psb27 protein, whereas ∼85% was retained in the corresponding PSII species in the wild-type cells. This is remarkable, as both complexes contained no functional manganese cluster according to the missing S2 state multiline signal (Fig. 7A, spectrum c, and Table 2).
Conclusion
The results of this study provide strong evidence that Psb27 is essential for the survival of the cells under specific stress conditions. In particular, low temperature stress leads to a much stronger impact on growth and photosynthetic activity in the Δpsb27 mutant compared with wild-type cells, at least in the thermophilic organism used in this study.
Lowering the growth temperature causes multiple effects on cellular metabolism. In particular, the efficiency of transcription, translation, and photosynthesis is lowered, and also the membrane fluidity is reduced (39). The cell is trying to compensate for these processes by expression of cold-induced proteins that increase the metabolic efficiency. Additionally, the amount of unsaturated fatty acids is increased to enhance the membrane fluidity (40, 41).
In this study, we have shown that reduction of the growth temperature from 45 to 30 °C severely affected the metabolism of the cells. Although the growth rate was reduced, the amount of closed reaction centers increased, and in particular, the ratio of inactive to active PSII complexes was altered. Psb27-containing PSII complexes became the predominant species under low temperature stress, whereas active complexes were reduced to (very) low levels.
Allakhverdiev and Murata (42) suggested that low temperature stress hinders the efficient de novo synthesis of proteins involved in the PSII repair cycle rather than enhancing the rate of photoinhibition itself. This might explain the increased amount of intermediate PSII complexes found in this study. Furthermore, the reduced membrane fluidity might promote the accumulation of intermediate complexes, provided they have to be shuttled between plasma and thylakoid membranes.
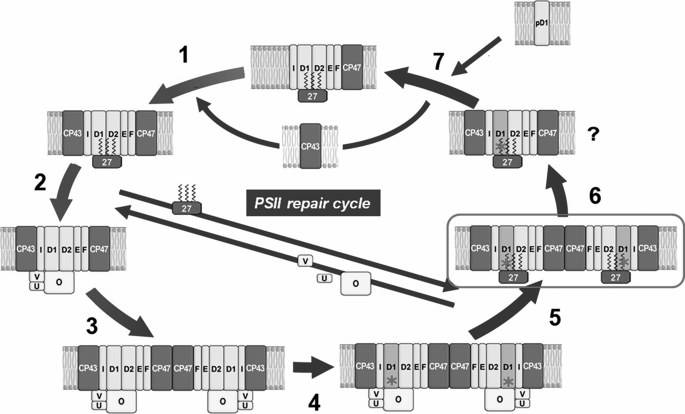
We were also able to isolate and characterize a new Psb27-containing PSII species from cells grown under cold stress. This inactive dimeric PSII-Psb27 complex (PSIID(inact)) was clearly assigned to the repair part of the PSII life cycle by 15N pulse-label experiments. Interestingly, this PSII species has no functional manganese cluster, although it contains remarkable amounts of manganese as shown by EPR spectroscopy. In contrast, the corresponding PSII species isolated from the Δpsb27 mutant contains almost no manganese. These results indicate that, besides being involved in the assembly of the water-oxidizing complex, Psb27 seems to play an additional role after the inactivation of PSII (Fig. 8). This might explain its essential function under stress conditions, but it is not clear why manganese should be retained in the PSIID(inact) complex before the exchange of the D1 protein. One explanation might be that this intermediate PSIID(inact) complex represents a reversible inactive complex, which was postulated recently in the “two-step photoinhibition model” (43–45). This model suggests an inactivation (possibly reversible) of the water-oxidizing complex caused by absorption of UV or blue light, which in turn blocks the electron supply from water to the strong oxidant P680+. As an increased level of P680+ leads to irreversible damage of the reaction center, the D1 protein must be exchanged within the PSII repair cycle. In such a scenario, the PSIID(inact) complex might represent a feasible candidate for the postulated first intermediate complex, but this has to be confirmed in further experiments. In conclusion, our results provide strong evidence that Psb27 is essential for maintaining a sufficient level of functional PSII complexes under environmental stress conditions, in particular under low temperature stress, and that it plays an auxiliary role in the life cycle of PSII after inactivation of the water-oxidizing complex.
FIGURE 8.
Model of the PSII repair cycle in cyanobacteria. After formation of the monomeric PSIIM(low) complex (step 1), this is activated by assembly of the water-oxidizing complex. This process involves release of Psb27 and attachment of the extrinsic proteins PsbO (O), PsbV (V), and PsbU (U) (step 2). Thereafter, the active monomer PSIIM(high) dimerizes (step 3) to the full active PSII complex PSIID(high), which represents the dominant PSII species in the thylakoid membrane. Damage to the D1 subunit in one or both of the monomers (indicated by red asterisks) results in the PSII dimer PSIID(low), with diminished oxygen-evolving activity (step 4). This dimer is transformed into the inactive PSIID(inact) for the exchange of the D1 subunit (step 5). Presumably, this complex monomerizes (step 6) before CP43 is detached and the newly synthesized pD1 protein is inserted (step 7).
Acknowledgments
We thank R. Oworah-Nkruma, C. König, and M. Völkel (Ruhr-Universität Bochum) for excellent technical assistance.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (SFB 480, Project C1; to N. G., M. M. N., and M. R.), the Protein Research Department of the Ruhr-Universität Bochum (to M. M. N. and M. R.), and the European Union (SOLAR-H2) and by the Swedish Research Council (to F. M. and S. S.).
- PSII
- Photosystem II
- QB/QA
- plastoquinone B/A
- Chl
- chlorophyll
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- Bistris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl) propane-1,3-diol
- IEC
- ion exchange chromatography.
REFERENCES
- 1. Goussias C., Boussac A., Rutherford A. W. (2002) Philos. Trans. R. Soc. Lond. B Biol. Sci. 357, 1369–1381; Discussion 1419–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rutherford A. W., Boussac A. (2004) Science 303, 1782–1784 [DOI] [PubMed] [Google Scholar]
- 3. Awramik S. M. (1992) Photosynth. Res. 33, 75–89 [PubMed] [Google Scholar]
- 4. Guskov A., Kern J., Gabdulkhakov A., Broser M., Zouni A., Saenger W. (2009) Nat. Struct. Mol. Biol. 16, 334–342 [DOI] [PubMed] [Google Scholar]
- 5. Holzwarth A. R., Müller M. G., Reus M., Nowaczyk M., Sander J., Rögner M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 6895–6900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zak E., Norling B., Maitra R., Huang F., Andersson B., Pakrasi H. B. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 13443–13448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Komenda J., Reisinger V., Müller B. C., Dobáková M., Granvogl B., Eichacker L. A. (2004) J. Biol. Chem. 279, 48620–48629 [DOI] [PubMed] [Google Scholar]
- 8. Aro E. M., Suorsa M., Rokka A., Allahverdiyeva Y., Paakkarinen V., Saleem A., Battchikova N., Rintamäki E. (2005) J. Exp. Bot. 56, 347–356 [DOI] [PubMed] [Google Scholar]
- 9. Nixon P. J., Michoux F., Yu J., Boehm M., Komenda J. (2010) Ann. Bot. 106, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Anbudurai P. R., Mor T. S., Ohad I., Shestakov S. V., Pakrasi H. B. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 8082–8086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nowaczyk M. M., Hebeler R., Schlodder E., Meyer H. E., Warscheid B., Rögner M. (2006) Plant Cell 18, 3121–3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Aro E. M., Virgin I., Andersson B. (1993) Biochim. Biophys. Acta 1143, 113–134 [DOI] [PubMed] [Google Scholar]
- 13. Mulo P., Sirpiö S., Suorsa M., Aro E. M. (2008) Photosynth. Res. 98, 489–501 [DOI] [PubMed] [Google Scholar]
- 14. Danielsson R., Suorsa M., Paakkarinen V., Albertsson P. A., Styring S., Aro E. M., Mamedov F. (2006) J. Biol. Chem. 281, 14241–14249 [DOI] [PubMed] [Google Scholar]
- 15. Nixon P. J., Barker M., Boehm M., de Vries R., Komenda J. (2005) J. Exp. Bot. 56, 357–363 [DOI] [PubMed] [Google Scholar]
- 16. Komenda J., Barker M., Kuviková S., de Vries R., Mullineaux C. W., Tichy M., Nixon P. J. (2006) J. Biol. Chem. 281, 1145–1151 [DOI] [PubMed] [Google Scholar]
- 17. Komenda J., Tichy M., Prásil O., Knoppová J., Kuviková S., de Vries R., Nixon P. J. (2007) Plant Cell 19, 2839–2854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kapri-Pardes E., Naveh L., Adam Z. (2007) Plant Cell 19, 1039–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tyystjärvi T., Herranen M., Aro E. M. (2001) Mol. Microbiol. 40, 476–484 [DOI] [PubMed] [Google Scholar]
- 20. Zhang L., Paakkarinen V., van Wijk K. J., Aro E. M. (1999) J. Biol. Chem. 274, 16062–16067 [DOI] [PubMed] [Google Scholar]
- 21. Zhang L., Paakkarinen V., van Wijk K. J., Aro E. M. (2000) Plant Cell 12, 1769–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nowaczyk M. M., Sander J., Grasse N., Cormann K. U., Rexroth D., Bernat G., Rogner M. (2010) Eur. J. Cell Biol. 89, 974–982 [DOI] [PubMed] [Google Scholar]
- 23. Mamedov F., Nowaczyk M. M., Thapper A., Rögner M., Styring S. (2007) Biochemistry 46, 5542–5551 [DOI] [PubMed] [Google Scholar]
- 24. Roose J. L., Pakrasi H. B. (2008) J. Biol. Chem. 283, 4044–4050 [DOI] [PubMed] [Google Scholar]
- 25. Cormann K., Bangert J. A., Ikeuchi M., Rögner M., Stoll R., Nowaczyk M. (2009) Biochemistry 48, 8768–8770 [DOI] [PubMed] [Google Scholar]
- 26. Mabbitt P. D., Rautureau G. J., Day C. L., Wilbanks S. M., Eaton-Rye J. J., Hinds M. G. (2009) Biochemistry 48, 8771–8773 [DOI] [PubMed] [Google Scholar]
- 27. Wei L., Guo J., Ouyang M., Sun X., Ma J., Chi W., Lu C., Zhang L. (2010) J. Biol. Chem. 285, 21391–21398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen H., Zhang D., Guo J., Wu H., Jin M., Lu Q., Lu C., Zhang L. (2006) Plant Mol. Biol. 61, 567–575 [DOI] [PubMed] [Google Scholar]
- 29. Iwai M., Katoh H., Katayama M., Ikeuchi M. (2004) Plant Cell Physiol. 45, 171–175 [DOI] [PubMed] [Google Scholar]
- 30. Mühlenhoff U., Chauvat F. (1996) Mol. Gen. Genet. 252, 93–100 [DOI] [PubMed] [Google Scholar]
- 31. Rippka R., Deruelles J., Waterbury J. B., Herdman M., Stanier R. Y. (1979) J. Gen. Microbiol. 111, 1–61 [Google Scholar]
- 32. van Kooten O., Snel J. F. (1990) Photosynth. Res. 25, 147–150 [DOI] [PubMed] [Google Scholar]
- 33. Schägger H., von Jagow G. (1987) Anal. Biochem. 166, 368–379 [DOI] [PubMed] [Google Scholar]
- 34. Rexroth S., Meyer Zu Tittingdorf J. M., Schwassmann H. J., Krause F., Seelert H., Dencher N. A. (2004) Biochim. Biophys. Acta. 1658, 202–211 [DOI] [PubMed] [Google Scholar]
- 35. Nowaczyk M. M., Wulfhorst H., Ryan C. M., Souda P., Zhang H., Cramer W. A., Whitelegge J. P. (2011) Biochemistry 50, 1121–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Albaum S. P., Neuweger H., Fränzel B., Lange S., Mertens D., Trötschel C., Wolters D., Kalinowski J., Nattkemper T. W., Goesmann A. (2009) Bioinformatics 25, 3128–3134 [DOI] [PubMed] [Google Scholar]
- 37. Mamedov F., Gadjieva R., Styring S. (2007) Physiol. Plant. 131, 41–49 [DOI] [PubMed] [Google Scholar]
- 38. Liu H., Roose J. L., Cameron J. C., Pakrasi H. B. (2011) J. Biol. Chem. 286, 24865–24871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Los D. A., Murata N. (1999) J. Mol. Microbiol. Biotechnol. 1, 221–230 [PubMed] [Google Scholar]
- 40. Laczkó-Dobos H., Szalontai B. (2009) Biochemistry 48, 10120–10128 [DOI] [PubMed] [Google Scholar]
- 41. Hongsthong A., Sirijuntarut M., Prommeenate P., Lertladaluck K., Porkaew K., Cheevadhanarak S., Tanticharoen M. (2008) FEMS Microbiol. Lett. 288, 92–101 [DOI] [PubMed] [Google Scholar]
- 42. Allakhverdiev S. I., Murata N. (2004) Biochim. Biophys. Acta 1657, 23–32 [DOI] [PubMed] [Google Scholar]
- 43. Ohnishi N., Allakhverdiev S. I., Takahashi S., Higashi S., Watanabe M., Nishiyama Y., Murata N. (2005) Biochemistry 44, 8494–8499 [DOI] [PubMed] [Google Scholar]
- 44. Tyystjarvi E. (2008) Coord. Chem. Rev. 252, 361–376 [Google Scholar]
- 45. Takahashi S., Murata N. (2008) Trends Plant Sci. 13, 178–182 [DOI] [PubMed] [Google Scholar]