Abstract
Phosphatidic acid (PA) is a critical mediator of mitogenic activation of mammalian target of rapamycin complex 1 (mTORC1) signaling, a master regulator of mammalian cell growth and proliferation. The mechanism by which PA activates mTORC1 signaling has remained unknown. Here, we report that PA selectively stimulates mTORC1 but not mTORC2 kinase activity in cells and in vitro. Furthermore, we show that PA competes with the mTORC1 inhibitor, FK506 binding protein 38 (FKBP38), for mTOR binding at a site encompassing the rapamycin-FKBP12 binding domain. This leads to PA antagonizing FKBP38 inhibition of mTORC1 kinase activity in vitro and rescuing mTORC1 signaling from FKBP38 in cells. Phospholipase D 1, a PA-generating enzyme that is an established upstream regulator of mTORC1, is found to negatively affect mTOR-FKBP38 interaction, confirming the role of endogenous PA in this regulation. Interestingly, removal of FKBP38 alone is insufficient to activate mTORC1 kinase and signaling, which require PA even when the FKBP38 level is drastically reduced by RNAi. In conclusion, we propose a dual mechanism for PA activation of mTORC1: PA displaces FKBP38 from mTOR and allosterically stimulates the catalytic activity of mTORC1.
Keywords: mTOR, mTOR Complex (mTORC), Phosphatidic Acid, Protein Kinases, Signal Transduction, FKBP38
Introduction
The mammalian target of rapamycin (mTOR)3 assembles a signaling network that regulates a myriad of cellular and developmental processes and has emerged as a promising therapeutic target in various diseases, including cancer (1–3). As a protein Ser/Thr kinase, mTOR exists in two biochemically and functionally distinct complexes, mTORC1 and mTORC2, that mediate rapamycin-sensitive and rapamycin-insensitive signaling, respectively. The two complexes are defined by the presence of raptor in mTORC1 and rictor in mTORC2, although they also contain other components (2). The best characterized substrates of mTORC1 kinase are ribosomal S6 kinase 1 (S6K1, phosphorylation at Thr-389) and eukaryotic initiation factor 4E binding protein 1 (4E-BP1, phosphorylation at Thr-37/46), both regulators of protein synthesis and both critically involved in mTOR regulation of cell growth and proliferation (4, 5). mTORC2 phosphorylates Akt at the hydrophobic motif (Ser-473), which is required for Akt activation. In addition, mTORC2 phosphorylates Akt and cPKC at their turn motif, which stabilizes the kinases (6, 7).
Two major types of upstream signals impinge on the mTORC1 pathway in cell growth are mitogens and amino acids. The Rag small G proteins and, separately, the class III phosphatidylinositol 3-kinase hVps34 mediate amino acid signaling to mTORC1 (8). The tuberous sclerosis complex TSC1/TSC2 receives mitogenic signals, among other signals, upstream of mTORC1 (9). The target of the GTPase-activating protein activity of TSC is the small G protein Rheb, which activates mTORC1 signaling (10). Several mechanisms have been proposed for Rheb activation of mTORC1. 1) Direct binding of Rheb to mTOR stimulates the kinase activity of mTORC1 (11). 2) As an effector for Rheb, phospholipase D (PLD) mediates Rheb activation of mTORC1 (12). 3) Rheb displaces FKBP38, an inhibitor of mTORC1 kinase, and consequently activates mTORC1 (13).
Work from our laboratory, and subsequently many others, has established the lipid second messenger phosphatidic acid (PA) as a key mediator of mitogenic activation of mTORC1 (14–16). PLD, an enzyme that converts phosphatidylcholine (PC) to PA (17), is a critical component upstream of the mTORC1 pathway in the regulation of cell growth (15, 16, 18). PA interacts with the FKBP12-rapamycin binding domain (FRB) of mTOR with remarkable specificity, and this interaction is disrupted by FKBP12-rapamycin (14). Recently, a solution structure of the FRB·PA complex (19) has validated the biochemically derived knowledge of FRB-PA interaction.
As a signaling lipid, PA has been found to have many effectors (20). Membrane translocation of the protein upon binding to PA is a major mechanism by which PA regulates its effectors, but PA is also believed to allosterically regulate the enzymatic activities of some of its effectors (20). The exact mechanism or a common mode for the allosteric effect of PA has not emerged, partly because of the lack of any well defined PA-binding module in PA effectors. Although it is well established that PA activates mTORC1 signaling in cells, the mechanism behind this activation has remained a long-standing puzzle. There is no evidence for PA induction of mTOR membrane translocation, and our earlier experimental evidence also argued against the possibility of PA activating mTOR catalytic activity (14, 21). However, recent advances in the understanding of the biochemistry and signaling of mTOR have prompted us to reconsider the role of PA in the context of mTOR kinase activity. Here we report that PA directly activates mTORC1 kinase through a dual mechanism: displacement of the endogenous inhibitor FKBP38 from mTOR and allosteric activation of the kinase.
EXPERIMENTAL PROCEDURES
Reagents
The antibodies used in this study were obtained from the following commercial sources: FLAG M2 (Sigma), HA (16B12) and Myc (9E10.2) (Covance), FKBP38 (R&D Systems), GST and His (Santa Cruz Biotechnology, Inc.), tubulin (Abcam, Inc.), raptor and rictor (Bethyl Laboratory, Inc.), all other antibodies were from Cell Signaling Technology, Inc. C8-PA, 1-palmitoyl-2-oleoyl-PA and 1-palmitoyl-2-oleoyl-PC were from Avanti Lipids. Glutathione beads were from GE Healthcare. Protein G-agarose and His-Akt were from Millipore. All other reagents were from Sigma.
Plasmids
GST-S6K1 (amino acids 332–421) was constructed by inserting the corresponding S6K1 cDNA into pGEX-2T (GE Healthcare). The following plasmids were described previously: Myc-S6K1 (14); FLAG-4EBP1 (23); FLAG-mTOR (24); HA-FKBP38, GST-FKBP38, and GST-mTOR(1967–2191) (13).
Cell Culture
HEK293 cells were grown in DMEM containing 10% FBS at 37 °C with 5% CO2. Transient transfections were performed with PolyFect (Qiagen) or Lipofectamine (Invitrogen) following the manufacturers' recommendations.
Lentivirus-mediated RNAi
All shRNAs were obtained from Sigma-Aldrich in the pLKO.1-puro vector (MISSION® shRNA). Lentivirus packaging and testing were performed as described previously (25). HEK293 cells were infected with the lentiviruses in growth medium containing 6 μg/ml polybrene, followed by selection in 1.5 μg/ml puromycin for 3–4 days. The scramble and PLD1 TRC shRNA clones were described previously (12). Human FKBP38 shRNA was TRCN0000010595.
Lipid Vesicle Formation
PA, PC, and C8-PA vesicles were made by water bath sonication. Lipids in chloroform were dried under nitrogen in a 1.5-ml tube, resuspended in 250 μl of vesicle buffer (150 mm NaCl and 10 mm Tris-Cl (pH8.0)) by vortexing briefly to yield a final lipid concentration of 6 mm. The lipid suspension was then sonicated in a water bath sonicator (Laboratory Supplies, Hicksville, NY, model G112SPIT, 600 volt, 80 kc, and 0.5 A) for 5 min. This procedure is expected to yield small unilamellar vesicles with diameters in the range of 15–50 nm. Lipid vesicles were made freshly before each experiment and were either added directly to cell medium or used in binding assays (see below) at the final concentrations indicated in the figures.
Protein Purification
GST fusion proteins (GST-mTOR(1967–2191), GST-56K1 (332–421), GST-FKBP38, and GST-FKBP12) were expressed in Escherichia coli, purified using glutathione beads, and cleaved of the GST tag as described previously (13, 14).
In Vitro Binding Assays
Purified GST-FKBP38 and mTOR(1967–2191) proteins were mixed at 5 μg each and incubated on ice in 500 μl of binding buffer (40 mm Tris-Cl (pH7.5), 150 mm NaCl, 2 mm EDTA, 2 mm EGTA, 1 mm DTT) for 15 min. Where applicable, PA vesicles (50% PA + 50% PC) or PC vesicles (100% PC) at the final concentrations indicated in the figures were preincubated with mTOR(1967–2191) for 15 min prior to addition of GST-FKBP38. Glutathione beads were used to pull down GST fusion proteins, and the beads were washed with binding buffer followed by boiling in SDS sample buffer and Western blot analysis.
Cell Lysis, Immunoprecipitation, and Western Blot Analysis
Cells were rinsed once with ice-cold PBS and lysed in ice-cold lysis buffer (40 mm HEPES (pH7.2), 120 mm NaCl, 10 mm pyrophosphate, 50 mm NaF, 10 mm β-glycerophosphate, 2 mm EDTA, 1× Sigma protease inhibitor mixture, and 0.3% CHAPS). The supernatant after microcentrifugation at 13,000 rpm for 10 min was collected and subjected to immunoprecipitation at 4 °C with various antibodies in the lysis buffer. The beads were washed three times with lysis buffer and then boiled in SDS sample buffer for 5 min. Proteins were resolved on SDS-PAGE and transferred onto a PVDF membrane (Millipore) followed by incubation with various antibodies according to the manufacturers' recommendations. Detection of horseradish peroxidase-conjugated secondary antibodies was performed with Western LightningTM Chemiluminescence Reagent Plus (PerkinElmer Life Sciences). Quantification of Western band intensities was performed by densitometry of x-ray film images using Image J software.
In Vitro mTOR Kinase Assays
mTORC1 and mTORC2 were immunoprecipitated using anti-raptor and anti-rictor antibodies, respectively, followed by incubation with protein G agarose beads. The kinase assays were performed following procedures described by Ikenoue et al. (26). mTORC1 kinase assays were carried out at 30 °C for 30 min in 25 mm HEPES (pH 7.4), 50 mm KCl, 10 mm MgCl2 and 250 μm ATP, with 100 ng GST-S6K1 as the substrate. mTORC2 kinase assays were carried out at 37 °C for 30 min in 25 mm HEPES (pH 7.4), 100 mm potassium acetate, 1 mm MgCl2, and 500 μm ATP, with 250 ng His-Akt as the substrate. Where applicable, PA or PC vesicles and/or FKBP38 were added to the immunocomplexes 15 min before initiation of the kinase assay by the addition of ATP. Reactions were stopped by the addition of 20 μl of SDS sample buffer and boiling.
RESULTS
PA Stimulates mTORC1 Kinase Activity
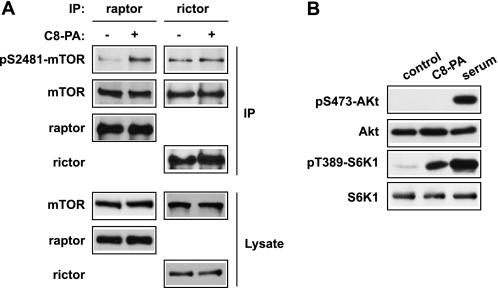
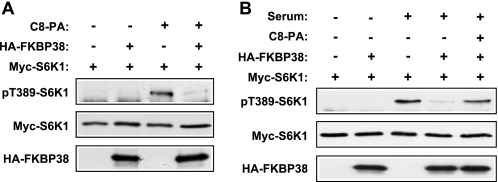
To evaluate a potential effect of PA on the kinase activity of mTOR in cells, we examined the phosphorylation of mTOR on Ser-2481, an autophosphorylation site that has recently been reported to monitor mTORC-specific catalytic activities (27). To avoid potential complications from exogenous PA-derived lysophosphatidic acid (28), which would initiate signaling through the membrane-bound lysophosphatidic acid receptors, we used a short-chain PA (C8-PA) for delivery into cells, which would not be converted into active lysophosphatidic acid (29, 30). mTORC1 and mTORC2 were isolated from HEK293 cells by immunoprecipitation of raptor and rictor, respectively. As shown in Fig. 1A, C8-PA treatment of the cells in the absence of any mitogen induced Ser-2481 phosphorylation of raptor-associated mTOR. Rictor-associated mTOR, on the other hand, displayed a higher basal level of phospho-Ser-2481 that was not affected by PA stimulation. PA activation of mTORC1 signaling was confirmed by S6K1 phosphorylation on Thr-389, whereas phospho-Ser-473-Akt, an indicator of mTORC2 signaling, was not detectable upon PA treatment (Fig. 1B). These results suggest that PA activates mTORC1 but not mTORC2 kinase activity in cells.
FIGURE 1.
PA activates mTORC1 autophosphorylation in cells. A, HEK293 cells were serum-starved overnight and then stimulated with 300 μm C8-PA for 30 min. Vesicle buffer was added as control wherever lipid vesicle was not added. Cells were lysed in lysis buffer, and mTORC1 and mTORC2 were immunoprecipitated (IP) with anti-raptor and anti-rictor antibodies, respectively, and washed with the same buffer, followed by Western blotting. B, cells were serum-starved overnight and then stimulated with 300 μm C8-PA or 20% serum for 30 min followed by cell lysis and Western blotting. Each experiment was performed at least three times, and the representative blots are shown.
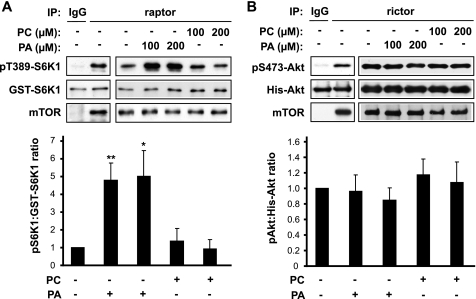
Next, we asked whether PA could directly activate mTORC1 kinase in vitro. We had previously found the FRB domain of mTOR to bind specifically to the PA-containing vesicles in vitro, and not vesicles of other lipid compositions including PC, phosphatidylethanol, phosphatidylserine, and various phosphatidylinositides (14). Kinase assays were performed with immunoprecipitated endogenous mTORC1 and bacterially purified GST-S6K1 as a substrate. Vesicles containing 50% 1-palmitoyl-2-oleoyl-PA and 50% 1-palmitoyl-2-oleoyl-PC were added to the kinase reaction, with 100% PC vesicles as a negative control. As shown in Fig. 2A, PA stimulated the in vitro kinase activity of mTORC1, whereas PC had no effect. Most likely because of a narrow dynamic range of the in vitro assay, the effects of PA vesicles were similar at 100 μm and 200 μm (Fig. 2A) and mild at 20–50 μm (data not shown). The kinase activity of mTORC2, assayed with Akt as a substrate, was unaffected by PA vesicles at the same concentrations (Fig. 2B). The degrees of S6K1 and Akt phosphorylation were measured by densitometry to quantify the kinase activities (Fig. 2, A and B, lower panels). These data demonstrate that PA selectively activates mTORC1 kinase in vitro.
FIGURE 2.
PA stimulates mTORC1 kinase activity in vitro. A, raptor was immunoprecipitated (IP) from HEK293 cells as described in Fig. 1A and subjected to in vitro kinase assays using GST-S6K1 as the substrate. PA or PC vesicles were added at 100 μm and 200 μm prior to kinase assays in the indicated samples. Vesicle buffer was added as control wherever lipid vesicle was not added. The phospho-S6K1 (pS6K1) and GST-S6K1 blots with raptor immunoprecipitates were quantified by densitometry, and the relative ratios of phospho-S6K1 versus GST-S6K1 were calculated with control (no vesicles) designated as 1. B, rictor was immunoprecipitated from HEK293 cells and subjected to in vitro kinase assays using His-Akt as the substrate. The phospho-Akt and His-Akt blots were quantified as described in A. The data shown in the graphs are mean ± S.D. of three independent experiments. Each data point is compared with the control by one-sample t test, and significantly different data points are indicated. *, p < 0.05; **, p < 0.01.
PA Disrupts FKBP38-mTOR Interaction
To probe into the mechanism by which PA activates mTORC1 kinase, we considered the role of FKBP38 as an endogenous inhibitor of mTORC1 (13). Because FKBP38 binds mTOR through a region that overlaps with the PA-binding FRB domain (13, 14), it appeared plausible that PA could compete with FKBP38 for mTOR binding as a mechanism of activating mTORC1. However, although several groups independently demonstrated a role of FKBP38 as a negative regulator of mTORC1 (13, 31, 32), others challenged this conclusion (33, 34). Therefore, we deemed it necessary to reexamine the role of FKBP38 in mTORC1 signaling in the Chen laboratory. We found that overexpression of FKBP38 in HEK293 cells inhibited serum-stimulated phosphorylation of both S6K1 and 4EBP1 (supplemental Fig. S1A), whereas knockdown of endogenous FKBP38 enhanced the phosphorylation of those mTORC1 targets (supplemental Fig. S1B).
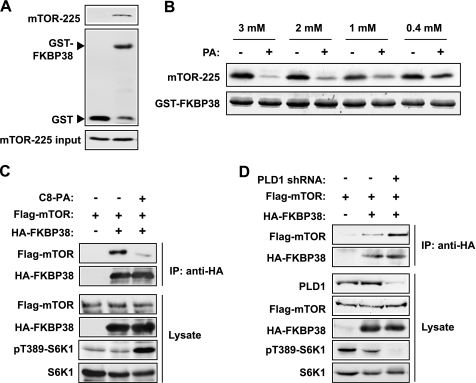
To test the hypothesis that PA competes with FKBP38 for mTOR binding, we first performed in vitro binding assays with bacterially expressed and purified mTOR fragment (amino acids 1967–2191) and GST-FKBP38. The specific interaction between mTOR(1967–2191) and FKBP38 (13) was confirmed by GST pull-down assays (Fig. 3A). Importantly, preincubation with PA vesicles but not PC vesicles disrupted the interaction between GST-FKBP38 and mTOR(1967–2191) in a dose-dependent manner (Fig. 3B). Thus, a competition between PA and FKBP38 for binding to the mTOR fragment is evident in vitro. It is not feasible to mimic physiological concentrations in the in vitro vesicle binding assays, as local concentrations of PA in a cell are not known (but could conceivably be very high).
FIGURE 3.
PA disrupts FKBP38-mTOR interaction. A, GST pull-down assays were performed with purified mTOR(1967–2191) and GST-FKBP38 with GST as a negative control. Western blot analyses are shown. Note that some free GST was present in the GST-FKBP38 protein preparation. B, GST-FKBP38 was preincubated with varying concentrations of PA (+) or PC (−) vesicles prior to addition of purified mTOR-255 and subsequent pull-down assays. C, HEK293 cells were cotransfected with HA-FKBP38 and FLAG-mTOR, followed by serum-starvation and stimulation with 300 μm C8-PA for 30 min. HA-FKBP38 was immunoprecipitated (IP) followed by Western blot analysis. D, cells were infected with lentivirus expressing PLD1 shRNA, puromycin-selected, and then cotransfected with HA-FKBP38 and FLAG-mTOR, followed by immunoprecipitation of HA-FKBP38 and subsequent Western blot analysis. Each experiment was performed at least three times, and the representative blots are shown.
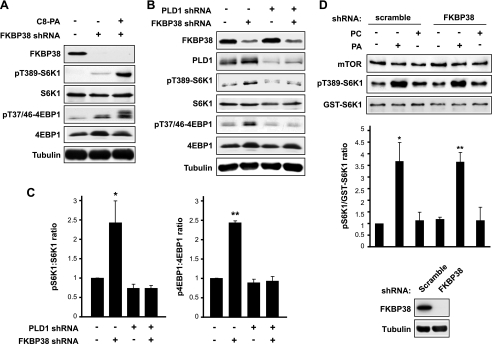
We also confirmed the interaction between FKBP38 and full-length mTOR by coimmunoprecipitation of epitope-tagged FKBP38 and mTOR (Fig. 3C), the latter expressed at a level comparable with endogenous mTOR (data not shown). Moreover, the FKBP38-mTOR interaction was disrupted when cells were exposed to C8-PA (Fig. 3C). PLD1 is responsible for the production of PA upstream of mTORC1 (16, 18). When PLD1 was knocked down, accompanied by diminished S6K1 phosphorylation as expected, an increased amount of mTOR was associated with FKBP38 (Fig. 3D). Collectively, these observations strongly suggest that the FKBP38-mTOR interaction is disrupted by PLD1 signaling and PA.
PA Antagonizes the Inhibitory Effect of FKBP38 on mTORC1 Kinase Activity and Signaling
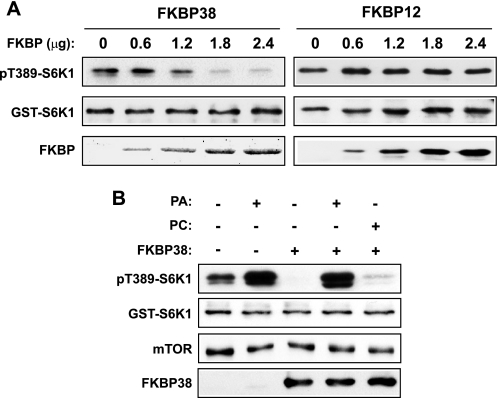
Next, we asked whether the PA/FKBP38 competition for mTOR binding would manifest into an antagonistic relationship on the regulation of mTORC1 kinase activity. Purified FKBP38 inhibited the in vitro kinase activity of mTORC1 in a dose-dependent manner (Fig. 4A), consistent with previous reports (13, 31). FKBP12 added at the same concentrations did not have any effect (Fig. 4A), confirming the specificity of FKBP38 inhibition of mTORC1. Significantly, the presence of PA vesicles but not PC vesicles in the reaction rescued kinase activity from FKBP38 inhibition (Fig. 4B), suggesting that PA directly antagonizes the inhibitory effect of FKBP38 in vitro.
FIGURE 4.
PA antagonizes FKBP38 inhibition of mTORC1 kinase activity in vitro. mTORC1 was immunoprecipitated with anti-raptor antibody from HEK293 cells as described in Fig. 1A and subjected to in vitro kinase assays using GST-S6K1 as a substrate. A, FKBP38 and FKBP12 were added at increasing amounts as indicated prior to kinase assays. B, PA or PC vesicles at 100 μm were added together with FKBP38 prior to kinase assays. Vesicle buffer was added as control wherever lipid vesicle was not added. Each experiment was performed at least three times, and the representative Western blot analyses (or Coomassie blue stain for FKBP38 in A) are shown.
We also examined the relationship between PA and FKBP38 in the context of mTORC1 signaling in cells. As shown in Fig. 5A, C8-PA stimulation of S6K1 phosphorylation in the absence of any mitogen was inhibited by FKBP38 overexpression. On the other hand, inhibition of serum-activation of S6K1 by overexpressed FKBP38 was reversed by exogenous PA (Fig. 5B). We did not observe a reversal of FKBP38 inhibition of mTORC1 signaling with increasing C8-PA concentrations in the absence of any other mitogen (data not shown), possibly because of limited delivery efficiency of exogenous PA. It is also not feasible to estimate or mimic physiological concentrations of PA, as endogenous PA may be highly localized. Nevertheless, our data, taken together, are fully consistent with the model that PA activates mTORC1 by antagonizing FKBP38 both in vitro and in cells.
FIGURE 5.
PA and FKBP38 antagonize each other in the regulation of mTORC1 signaling in cells. A, HEK293 cells were cotransfected with Myc-S6K1 and HA-FKBP38, serum-starved, and then stimulated with 300 μm C8-PA followed by Western blot analysis of cell lysates. B, cells were transfected and starved as in A and stimulated with 10% serum with or without C8-PA followed by Western blot analysis of cell lysates. Vesicle buffer was added as control wherever lipid vesicle was not added. Each experiment was performed at least three times, and the representative blots are shown.
PA Is Also an Allosteric Activator of mTORC1 Kinase
If removing FKBP38 were the sole mechanism for PA activation of mTORC1, one would expect that in the absence of FKBP38 PA would not further stimulate mTORC1. To probe into this issue, we knocked down FKBP38. As shown in Fig. 6A, the level of endogenous FKBP38 was drastically reduced by lentivirus-delivered shRNA, which was accompanied by modestly increased S6K1 and 4EBP1 phosphorylation in serum-starved cells. Interestingly, exogenous C8-PA further stimulated mTORC1 signaling despite the FKBP38 knockdown (Fig. 6A). The dramatic degree of stimulation is unlikely to be explained by any residual FKBP38 protein after knockdown. Rather, these data strongly suggest that displacement of FKBP38 alone is insufficient for PA activation of mTORC1 signaling. Nevertheless, this does not contradict the necessity of FKBP38 displacement by PA for the activation.
FIGURE 6.
PA activates mTORC1 in the absence of FKBP38. A, HEK293 cells were infected with lentivirus expressing FKBP38 shRNA, puromycin-selected, serum-starved, and then stimulated with 300 μm C8-PA followed by Western analysis of lysates. B, cells were infected with lentiviruses expressing shRNA for FKBP38, or PLD1, or both, puromycin-selected, and followed by serum starvation and then Western blot analysis. C, the Western blot analysis results represented by blots in B were quantified by densitometry, and the relative ratios of phospho-S6K1 versus S6K1 and p4EBP1 versus 4EBP1 were calculated with control (no shRNA) designated as 1. D, mTORC1 was immunoprecipitated by anti-raptor as described in Fig. 1A from cells expressing FKBP38 shRNA or a hairpin of scrambled sequence as control and subjected to in vitro kinase assays with or without PA or PC vesicles at 100 μm. Vesicle buffer was added as control wherever lipid vesicle was not added. The Western blot analysis results were quantified by densitometry, and the relative ratios of phospho-S6K1 versus GST-S6K1 were calculated with control (no vesicles) designated as 1. The data shown are mean ± S.D. or representative blots of three independent experiments. Each data point is compared with the control by one-sample t test, and significantly different data points are indicated. *, p < 0.05; **, p < 0.01.
To assess the role of endogenous PA, we knocked down PLD1. If the predominant role of PA were to remove FKBP38, we would expect that in the absence of FKBP38 PLD1 would no longer be essential for mTORC1 signaling. However, we found that PLD1 knockdown abolished the ability of FKBP38 knockdown to induce mTORC1 signaling (Fig. 6B, compare lanes 2 and 4). This is consistent with the observation in Fig. 6A suggesting that PA is required for mTORC1 activation in addition to removing FKBP38. PLD1 knockdown alone did not have an obvious effect on the basal activity of mTORC1 (Fig. 6B), as expected (18). It was noted that FKBP38 knockdown was less efficient, and the effect on mTORC1 signaling less pronounced, when the cells were infected by both FKBP38 and PLD1 shRNA lentiviruses (compare Fig. 6, A and B), likely because selection of cells infected by two types of viruses relied on the same drug (puromycin). Nevertheless, this FKBP38 reduction led to reproducible mTORC1 activation that was eliminated by PLD1 knockdown, as clearly shown by the quantitative measurements of S6K1 and 4E-BP1 phosphorylation (Fig. 6C).
To further validate the observations above, we carried out in vitro kinase assays with mTORC1 isolated from FKBP38 knockdown cells. As shown in Fig. 6D, PA vesicles stimulated the kinase activity of FKBP38-deficient mTORC1, supporting the notion that PA has a positive role in the absence of FKBP38. Of note, the immunoprecipitated mTORC1 activity was indistinguishable between FKBP38 knockdown and control cells both in the presence and absence of added PA vesicles (Fig. 6D), suggesting that even without knockdown the amount of FKBP38 associated with the mTORC1 complex under our experimental conditions was most likely negligible.
Therefore, in addition to displacing FKBP38, PA also activates mTORC1 through another mechanism. The other known inhibitors of mTORC1 are PRAS40 and DEPTOR, both of which would be absent in the mTORC1 immunoprecipitate here as it was subjected to a high salt (500 mm NaCl) wash that removed these two proteins (Refs. 35, 36 and data not shown). The in vitro assay system also made it virtually impossible for PA to recruit an activator. Hence, collectively, the current observations point to the simplest model: that the physical interaction between PA and mTOR exerts an allosteric effect that is required for the kinase activity of mTORC1 after displacement of FKBP38, although the involvement of a third factor (an unknown inhibitor) cannot be formally excluded.
DISCUSSION
Our studies have revealed direct activation of the mTORC1 kinase by phosphatidic acid and identified a dual mechanism by which PA activates mTORC1: displacing FKBP38 and exerting an allosteric effect on the catalytic activity. These findings provide answers to the long-standing question of how PA activates mTOR signaling. The new mechanistic insights may facilitate the exploration of the tremendous therapeutic potential of this signaling network.
It is noteworthy that we had previously failed to observe an effect of PA on mTOR catalytic activity (14, 21). One plausible explanation for the discrepancy may come from the conditions of isolating mTOR for in vitro kinase assays. In previous studies we had used Triton X-100 as the detergent in cell lysis prior to mTOR immunoprecipitation, whereas in this study, CHAPS was used and mTORC1 was isolated by raptor pull-down. As reported by Kim et al. (22), the raptor-mTOR interaction would be disrupted by Triton. The loss of raptor might have prevented PA activation of mTORC1.
We had previously reported a mutation in mTOR (R2109A) that had dampened FRB binding to PA in vitro and mTOR signaling in cells by ∼50% (14). This mTOR mutant, however, did not display differential sensitivity to PA compared with WT mTOR in FKBP38 binding (data not shown). It is possible that limitations in the vesicle binding assay and cellular delivery of C8-PA render insufficient dynamic ranges to discern the partial defect of the R2109A mutant. With the solution structure of the FRB-PA complex (19) as a guide, identification of additional mutations in FRB that drastically disrupt the PA-FRB interaction may be possible. Such mutants would be desirable for future investigations of PA in the regulation of mTORC1.
Although our present data suggest that PA selectively activates mTORC1 and not mTORC2 (Figs. 1 and 2), it has been proposed by Foster and colleagues (37) that PA is required for the assembly of both mTORC1 and mTORC2. These two conclusions need not be mutually exclusive. In this study, we examined PA for its acute effect in stimulating cells and in directly activating mTOR kinase. On the other hand, the effects that Toschi et al. (37) have observed may stem from a basal level of PA in maintaining the integrity of mTOR complexes prior to activation of the kinases. It will be interesting in future investigations to determine whether the activation of mTORC1 and assembly of mTOR complexes share the same mode of PA-mTOR interaction or represent two molecularly distinct mechanisms of PA action.
The recent controversy surrounding the role of FKBP38 in regulating mTORC1 prompted us to reexamine this reported endogenous inhibitor, and our results described here clearly support the model that FKBP38 binds and inhibits mTORC1. We and others (31) observed that recombinant FKBP38 inhibited mTORC1 signaling in cells only when it was highly overexpressed, which may explain the absence of the effect of FKBP38 in similar experiments performed by some groups (33, 34). The requirement of high levels of recombinant FKBP38 to exert an inhibitory effect on mTORC1 does not necessarily mean the FKBP38 mechanism is an inefficient one. One could envision that endogenous FKBP38 might be highly localized for its mTORC1-regulating function and/or that high concentrations of FKBP38 might be necessary to set a threshold to ensure signaling fidelity. In vitro, the inhibitory effect of recombinant FKBP38 on mTORC1 kinase activity was easily detected (Fig. 4A), most likely because of the condition of mTORC1 isolation that led to the dissociation of endogenous FKBP38. The reported ability of Rheb to bind FKBP38 and displace it from mTOR (13) has also been disputed (34, 38). We have not examined the role of Rheb in our studies, as our proposed mode of PA action is independent of Rheb-FKBP38 interaction, although it does not exclude the involvement of Rheb.
Displacement of FKBP38 appears to be a simple and effective way for PA to activate the mTOR kinase, and yet, to the best of our knowledge, this is the first example of PA regulating an effector through removing an inhibitor. mTOR also joins a small roster of PA effectors, the enzymatic activities of which are allosterically regulated by PA binding (20). The only other protein kinase that has been reported to be activated by PA through a possible allosteric effect is Fer, a tyrosine kinase that regulates actin polymerization in cell migration (39). Other than the fact that PA binds at a site N-terminal to the kinase domain, mTOR and Fer do not share any common feature in their PA binding domains. Because PA stimulates mTORC1 activity on an autophosphorylation site but not mTORC2 activity on the same site (Fig. 1), the allosteric effect of PA is unlikely to simply confer catalytic activation or substrate specificity of the kinase domain. Other components in mTORC1, raptor in particular, most likely play an integral role. Future structural studies will be needed to shed light on the exact mode of allosteric regulation by PA of mTOR, or of any other kinase.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health grant AR048914 (to J. C.) CA129821 (to Y. J.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- mTOR
- mammalian target of rapamycin
- S6K1
- S6 kinase 1
- TSC
- tuberous sclerosis complex
- PLD
- phospholipase D
- PA
- phosphatidic acid
- PC
- phosphatidylcholine
- FRB
- FKBP12-rapamycin binding.
REFERENCES
- 1. Martin D. E., Hall M. N. (2005) Curr. Opin. Cell Biol. 17, 158–166 [DOI] [PubMed] [Google Scholar]
- 2. Sarbassov D. D., Ali S. M., Sabatini D. M. (2005) Curr. Opin. Cell Biol. 17, 596–603 [DOI] [PubMed] [Google Scholar]
- 3. Guertin D. A., Sabatini D. M. (2007) Cancer Cell 12, 9–22 [DOI] [PubMed] [Google Scholar]
- 4. Fingar D. C., Blenis J. (2004) Oncogene 23, 3151–3171 [DOI] [PubMed] [Google Scholar]
- 5. Ma X. M., Blenis J. (2009) Nat. Rev. Mol. Cell Biol. 10, 307–318 [DOI] [PubMed] [Google Scholar]
- 6. Ikenoue T., Inoki K., Yang Q., Zhou X., Guan K. L. (2008) EMBO J. 27, 1919–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Facchinetti V., Ouyang W., Wei H., Soto N., Lazorchak A., Gould C., Lowry C., Newton A. C., Mao Y., Miao R. Q., Sessa W. C., Qin J., Zhang P., Su B., Jacinto E. (2008) EMBO J. 27, 1932–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Avruch J., Long X., Ortiz-Vega S., Rapley J., Papageorgiou A., Dai N. (2009) Am. J. Physiol. Endocrinol. Metab. 296, E592–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li Y., Corradetti M. N., Inoki K., Guan K. L. (2004) Trends Biochem. Sci. 29, 32–38 [DOI] [PubMed] [Google Scholar]
- 10. Manning B. D., Cantley L. C. (2003) Trends Biochem. Sci. 28, 573–576 [DOI] [PubMed] [Google Scholar]
- 11. Long X., Lin Y., Ortiz-Vega S., Yonezawa K., Avruch J. (2005) Curr. Biol. 15, 702–713 [DOI] [PubMed] [Google Scholar]
- 12. Sun Y., Fang Y., Yoon M. S., Zhang C., Roccio M., Zwartkruis F. J., Armstrong M., Brown H. A., Chen J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 8286–8291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bai X., Ma D., Liu A., Shen X., Wang Q. J., Liu Y., Jiang Y. (2007) Science 318, 977–980 [DOI] [PubMed] [Google Scholar]
- 14. Fang Y., Vilella-Bach M., Bachmann R., Flanigan A., Chen J. (2001) Science 294, 1942–1945 [DOI] [PubMed] [Google Scholar]
- 15. Foster D. A. (2007) Cancer Res. 67, 1–4 [DOI] [PubMed] [Google Scholar]
- 16. Sun Y., Chen J. (2008) Cell Cycle 7, 3118–3123 [DOI] [PubMed] [Google Scholar]
- 17. Frohman M. A., Sung T. C., Morris A. J. (1999) Biochim. Biophys. Acta 1439, 175–186 [DOI] [PubMed] [Google Scholar]
- 18. Fang Y., Park I. H., Wu A. L., Du G., Huang P., Frohman M. A., Walker S. J., Brown H. A., Chen J. (2003) Curr. Biol. 13, 2037–2044 [DOI] [PubMed] [Google Scholar]
- 19. Veverka V., Crabbe T., Bird I., Lennie G., Muskett F. W., Taylor R. J., Carr M. D. (2008) Oncogene 27, 585–595 [DOI] [PubMed] [Google Scholar]
- 20. Stace C. L., Ktistakis N. T. (2006) Biochim. Biophys. Acta 1761, 913–926 [DOI] [PubMed] [Google Scholar]
- 21. Chen J., Fang Y. (2002) Biochem. Pharmacol. 64, 1071–1077 [DOI] [PubMed] [Google Scholar]
- 22. Kim D. H., Sarbassov D. D., Ali S. M., King J. E., Latek R. R., Erdjument-Bromage H., Tempst P., Sabatini D. M. (2002) Cell 110, 163–175 [DOI] [PubMed] [Google Scholar]
- 23. Kim J. E., Chen J. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 14340–14345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vilella-Bach M., Nuzzi P., Fang Y., Chen J. (1999) J. Biol. Chem. 274, 4266–4272 [DOI] [PubMed] [Google Scholar]
- 25. Yoon M. S., Chen J. (2008) J. Cell Sci. 121, 282–289 [DOI] [PubMed] [Google Scholar]
- 26. Ikenoue T., Hong S., Inoki K. (2009) Methods Enzymol. 452, 165–180 [DOI] [PubMed] [Google Scholar]
- 27. Soliman G. A., Acosta-Jaquez H. A., Dunlop E. A., Ekim B., Maj N. E., Tee A. R., Fingar D. C. (2010) J. Biol. Chem. 285, 7866–7879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Billon-Denis E., Tanfin Z., Robin P. (2008) J. Lipid Res. 49, 295–307 [DOI] [PubMed] [Google Scholar]
- 29. van Corven E. J., van Rijswijk A., Jalink K., van der Bend R. L., van Blitterswijk W. J., Moolenaar W. H. (1992) Biochem. J. 281, 163–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fischer D. J., Nusser N., Virag T., Yokoyama K., Wang D., Baker D. L., Bautista D., Parrill A. L., Tigyi G. (2001) Mol. Pharmacol. 60, 776–784 [PubMed] [Google Scholar]
- 31. Dunlop E. A., Dodd K. M., Seymour L. A., Tee A. R. (2009) Cell. Signal. 21, 1073–1084 [DOI] [PubMed] [Google Scholar]
- 32. Peng L., Liang D., Tong W., Li J., Yuan Z. (2010) J. Biol. Chem. 285, 20870–20881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Maehama T., Tanaka M., Nishina H., Murakami M., Kanaho Y., Hanada K. (2008) J. Biol. Chem. 283, 35053–35059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang X., Fonseca B. D., Tang H., Liu R., Elia A., Clemens M. J., Bommer U. A., Proud C. G. (2008) J. Biol. Chem. 283, 30482–30492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sancak Y., Thoreen C. C., Peterson T. R., Lindquist R. A., Kang S. A., Spooner E., Carr S. A., Sabatini D. M. (2007) Mol. Cell. 25, 903–915 [DOI] [PubMed] [Google Scholar]
- 36. Peterson T. R., Laplante M., Thoreen C. C., Sancak Y., Kang S. A., Kuehl W. M., Gray N. S., Sabatini D. M. (2009) Cell 137, 873–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Toschi A., Lee E., Xu L., Garcia A., Gadir N., Foster D. A. (2009) Mol. Cell Biol. 29, 1411–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Uhlenbrock K., Weiwad M., Wetzker R., Fischer G., Wittinghofer A., Rubio I. (2009) FEBS Lett. 583, 965–970 [DOI] [PubMed] [Google Scholar]
- 39. Itoh T., Hasegawa J., Tsujita K., Kanaho Y., Takenawa T. (2009) Sci. Signal. 2, ra52. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.