Abstract
The rapid emergence and the prevalence of resistance mutations in HIV-1 reverse transcriptase (RT) underscore the need to identify RT inhibitors with novel binding modes and mechanisms of inhibition. Recently, two structurally distinct inhibitors, phosphonoformic acid (foscarnet) and INDOPY-1 were shown to disrupt the translocational equilibrium of RT during polymerization through trapping of the enzyme in the pre- and the post-translocation states, respectively. Here, we show that foscarnet and INDOPY-1 additionally display a shared novel inhibitory preference with respect to substrate primer identity. In RT-catalyzed reactions using RNA-primed substrates, translocation inhibitors were markedly less potent at blocking DNA polymerization than in equivalent DNA-primed assays; i.e. the inverse pattern observed with marketed non-nucleoside inhibitors that bind the allosteric pocket of RT. This potency profile was shown to correspond with reduced binding on RNA·DNA primer/template substrates versus DNA·DNA substrates. Furthermore, using site-specific footprinting with chimeric RNA·DNA primers, we demonstrate that the negative impact of the RNA primer on translocation inhibitor potency is overcome after 18 deoxyribonucleotide incorporations, where RT transitions primarily into polymerization-competent binding mode. In addition to providing a simple means to identify similarly acting translocation inhibitors, these findings suggest a broader role for the primer-influenced binding mode on RT translocation equilibrium and inhibitor sensitivity.
Keywords: Antiviral Agents, DNA Polymerase, Enzyme Catalysis, Enzyme Inhibitors, Enzyme Mechanisms, HIV, Reverse Transcription
Introduction
HIV RT is a multifunctional enzyme that catalyzes the conversion of the single-stranded RNA HIV genome to double-stranded DNA that is then incorporated into the host genome by the HIV integrase. In the process, RT must support RNA-directed and DNA-directed DNA synthesis along with ribonuclease H (RNase H)2 activities to create an integration-competent product. A key step during reverse transcription is the incomplete hydrolysis of the (+)-stranded RNA that leaves behind a purine-rich RNA sequence referred to as the polypurine tract (PPT). The remnant RNA PPT sequence serves as a primer to initiate synthesis of the (+)-strand DNA (second strand) (1–3). The unique structure of the RNA PPT sequence has been shown to be a key determinant of the RNase H cleavage specificity at the PPT/U3 junction (4). A recent report described the mechanism by which RT discriminates between polymerase and RNase H activities thereby enabling more efficient initiation of polymerization from the RNA PPT primer versus other remnant RNA primers (5). Using single-molecule spectroscopy experiments, it was shown that RT binds nucleic acid substrates in two distinct orientations in a manner that is governed by the sugar backbone composition of the four or five nucleotides at each end of the primer. Depending on the binding orientation, RT either initiates polymerization at the 3′-end of the primer (polymerase binding mode on a DNA primer), or alternatively, RNA hydrolysis through the RNase H domain (RNase H binding mode on a RNA primer). Interestingly, whereas RT binds almost exclusively in the RNase H binding orientation on non-PPT RNA primers, RT binds in both orientations when in contact with the RNA PPT primer. As a consequence, RT “flips” or equilibrates between the two binding orientations when the enzyme is in contact with the RNA PPT primer (5).
As reverse transcription is required for viral replication, extensive efforts have been devoted to identifying small molecule inhibitors of RT to treat HIV patients. Indeed nearly half of the anti-HIV drugs target the DNA polymerase activity of RT (reviewed in Ref. 6). The approved inhibitors belong to one of the two classes: nucleoside RT inhibitors (NRTIs) and non-nucleoside RT inhibitors (NNRTIs). NRTIs are structural analogs of natural nucleosides that lack the 3′-OH necessary for continuing polymerization. NRTIs thus act as chain terminators when incorporated into viral DNA by RT (reviewed in Ref. 6). On the other hand, NNRTIs are non-competitive inhibitors (7) that bind to an allosteric site of the RT enzyme known as the NNRTI-binding pocket. The binding of NNRTIs to the NNRTI-binding pocket induces conformational changes that significantly reduce the rate of the polymerization reaction (8, 9).
Despite the availability of potent RT inhibitors for antiretroviral therapy regimens, drug failure arising from the rapid emergence of resistance mutations against both classes of drugs underscores the need to identify novel small molecule inhibitors that act through novel mechanisms. Recently, the inhibitory mechanisms of two structurally distinct RT inhibitors that are neither chain terminators nor NNRTI-binding pocket-directed were described. Both are non-nucleoside inhibitors that block DNA polymerization between two consecutive cycles of nucleotide incorporation by disrupting the translocational equilibrium of RT. Following nucleotide incorporation, RT translocates from the pre-translocational state, to clear the nucleotide binding site (N-site), to the post-translocational state, to bring the 3′-end of the primer to the priming site (P-site) (10, 11). The pyrophosphate analog phosphonoformic acid (PFA or foscarnet) was shown to inhibit RT by trapping the enzyme in the pre-translocational state (12, 13). The observed preference of PFA for the pre-translocational form of the polymerase·DNA complex was recently validated by the first crystal structure of PFA bound to a DNA polymerase, which showed PFA binding and stabilization of the closed enzyme conformation leading to the formation of an untranslocated form of the polymerase·DNA complex (14). In contrast, the more recently discovered scaffold of indolopyridones (INDOPY-1) (15, 16) traps RT in the post-translocational state (15). Owing to its proposed binding mechanism, INDOPY-1 has been referred to as a nucleotide-competing RT inhibit (17).
The extent to which inhibitors with novel mechanisms of inhibition complement or synergize with other classes of inhibitors may depend, in part, on their ability to block a novel and key step or process in reverse transcription. For instance, NNRTIs appear to preferentially inhibit the (+)-strand initiation step of the HIV-1 reverse transcription (18). The structural basis for this ability of NNRTIs to preferentially inhibit RNA-primed DNA synthesis was recently revealed using a single-molecule assay that measured the binding orientation of RT on different substrates (5). Using a substrate that mimicked an RNA PPT primer from which DNA synthesis had initiated, it was shown that NNRTIs decreased polymerization by destabilizing the polymerization-competent orientation of RT in favor of the inverted RNase H-competent orientation. These findings raise questions with respect to putative substrate specificities for other classes of RT inhibitors. Here, we demonstrate that, in contrast to NNRTIs, both PFA and INDOPY-1, which we have operationally defined here as non-nucleoside translocation inhibitors, or NNTIs, specifically inhibit DNA-primed synthesis in vitro without significantly inhibiting (+)-strand initiation polymerization from the RNA PPT primer. Our data suggest that trapping the enzyme in the pre- or post-translocational states is less efficient during RNA-primed polymerization. We show further that the impact of the RNA PPT primer on RT binding dynamics and thus NNRTI and NNTI potency is lost after ∼24 nucleotides have been incorporated. Together, these data provide novel molecular insight into the impact of the primer on conformational dynamics of RT, because it relates to both flipping transitions and translocational equilibrium, and further reconcile observed discrepancies in inhibitor activity measurements across assay platforms.
EXPERIMENTAL PROCEDURES
Reagents
All oligonucleotides were synthesized by Sigma-Aldrich. RT enzyme was purified as described previously (19, 20).
RT Polymerase Assays
A homogeneous scintillation proximity assay (PerkinElmer Life Sciences) was used to characterize the effects of RT inhibitors on PPT-primed (RNA or DNA) polymerization reactions. An assay buffer containing 20 nm primer-template substrate, 50 mm Tris-HCl, pH 7.8, 1 mm dithiothreitol, 6 mm MgCl2, 80 mm KCl, 0.2% polyethylene glycol 8000, 0.1 mm EGTA, 25 μm each of dCTP, dGTP, and dTTP, and 0.5 μm [3H]dATP was pre-incubated with inhibitor (0.5% DMSO) at 37 °C for 5 min. Reactions were initiated by the addition of the HIV-1 RT enzyme to a final concentration of 10 nm and incubated at 37 °C for the indicated times. Reactions were quenched by the addition of scintillation-impregnated streptavidin-coated polyvinyltoluene beads in a quench solution containing 100 mm EDTA. Plates were allowed to rest for 8 h prior to counting radioactivity using a Microbeta 1450 (Trilux) from PerkinElmer Life Sciences. To establish potency of NRTIs, the assay conditions were exactly the same as described above except that a lower concentration (1 μm) of all four dNTPs was used (dCTP, dGTP, dTTP, and [3H]dATP) to minimize competition with NRTIs. Polymerase reaction data were exported from the Microbeta 1450 and imported into a specifically designed data analysis program at Merck Frosst (Kirkland, Quebec, Canada) for further analysis. Inhibition curves and IC50 values (concentration of an inhibitor that is required for 50% inhibition of an enzyme in vitro) were generated using the in-house-developed software (four-parameter logistic fit).
Band Shift Experiment
The formation of ternary complexes was monitored with a Cy5-template (5′-ATTAGATTAGCCCTT(Cy5)CCAGTCCCCCCTTTTCTTTTAAAAAGTGGCGTGGC-3′). The labeled template was pre-annealed to a 3-fold molar excess of the DNA-PPT+2D (5′-ACTTTTTAAAAGAAAAGGGGGGAC-3′) or DNA-PPT+3D (5′-ACTTTTTAAAAGAAAAGGGGGGACT-3′) or an RNA-PPT+2D (5′-rArCrUrUrUrUrArArArArGrArArArArGrGrGrGrGrGAC-3′) or an RNA-PPT+3D (5′-rArCrUrUrUrUrArArArArGrArArArArGrGrGrGrGrGACT-3′) primers. The hybrids were then incubated with a 5-fold excess of wild-type HIV-1 RT enzyme in a buffer containing 50 mm Tris-HCl (pH 7.8) and 60 mm NaCl. Increasing concentrations of PFA or INDOPY-1 (ranging from 0 to 50 μm) or a fixed concentration of the next templated nucleotide (dTTP or dGTP) (50 μm) were added to each sample, and the mixture was incubated for 2 min at room temperature. The complexes were subsequently challenged with 2.5 μg/μl heparin trap, followed by incubation for 15 min at room temperature. The samples were analyzed on 6% non-denaturating polyacrylamide gels.
KOONO Site-specific Footprinting
HIV-1 RT E478Q was generated through site-directed mutagenesis with the Stratagene QuikChangeTM kit using the manufacturer's protocol and purified as previously described (21). A 5′-radiolabeled ([γ-32P]ATP, PerkinElmer Life Sciences) PPT37 template was heat-annealed to a 3-fold molar excess of primer (DNA PPT, RNA PPT, and RNA PPT+12D, +18D, or +24D) in the presence of 50 mm Tris-HCl (pH 7.8) and 50 mm NaCl. It was then added at a concentration of 50 nm to 120 mm sodium cacodylate (pH 7), 1 mm DTT, 10 mm MgCl2, 20 mm NaCl, and 750 nm E478Q RT enzyme. PFA, ddNTPs, and/or dNTPs were added as described in (Fig. 5). The reaction was incubated at 37 °C for 10 min after the addition of ligand (and another 10 min after the addition of dNTPs). Treatment with potassium peroxynitrite (KOONO) was performed essentially as described (22).
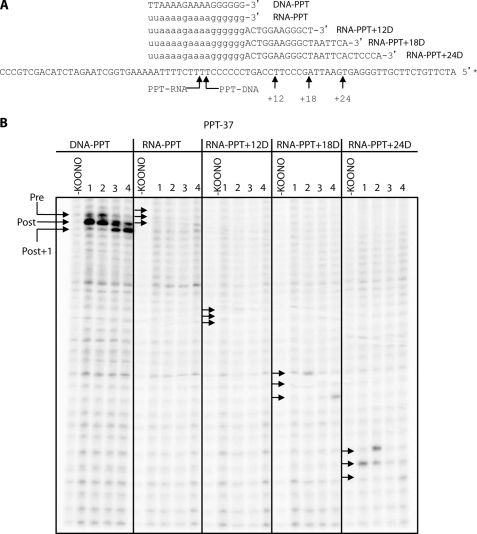
FIGURE 5.
HIV-1 RT binds in a polymerase-competent binding mode only on the RNA PPT primer extended by more than 18 DNA residues. A, nucleic acid sequences used in the footprint. Arrows indicate the position of the post-translocated KOONO-induced cleavages on the PPT37 DNA template. The asterisk indicates the position of the radiolabel. B, site-specific KOONO footprint using the RT mutant E478Q. Control lanes in the absence of KOONO are indicated. Lane 1 contains unliganded RT. Lane 2 contains 100 μm PFA. Lane 3 contains the next templated dideoxynucleotide (20 μm) to act as a chain terminator. Lane 4 is identical to lane 3 except for the presence of the next templated nucleotide after the chain terminator (50 μm); e.g. for DNA PPT, lane 3 contains ddATP (20 μm) and lane 4 contains ddATP (20 μm) and dCTP (50 μm). Arrows indicate the position (or expected position) of the footprint: the top arrow represents a pre-translocated cleavage, the middle arrow represents a post-translocated cleavage, and the bottom arrow represents the post-translocated +1 cleavage after incorporation of a ddNTP.
The following sequences were used as templates. All the templates were PAGE-purified and 5′-biotinylated. The nomenclature consists of numbers after the PPT that refer to the potential maximum number of incorporations after annealing with the PPT primer. PPT3, 5′-AGTCCCCCCTTTTCTTTTAAAAAGTGGCTAAGA-3′; PPT37, 5′-ATCTTGTCTTCGTTGGGAGTGAATTAGCCCTTCCAGTCCCCCCTTTTCTTTTAAAAAGTGGCTAAGATCTACAGCTGCCC-3′; and PPT90, 5′-TTCTGCCAATCAGGGAAGTAGCCTTGTGTGTGGTAGATCCACAGATCAAGGATATCTTGTCTTCGTTGGGAGTGAATTAGCCCTTCCAGTCCCCCCTTTTCTTTTAAAAAGTGGCTAAGA-3′. Each of these templates was annealed to either an RNA PPT primer (5′-rUrUrArArArArGrArArArArGrGrGrGrGrG-3′) or a DNA PPT primer (5′-TTAAAAGAAAAGGGGGG-3′). In the indicated experiments, in addition to the pure RNA or DNA primers, chimeric primers (RNA PPT+12D: 5′-rUrUrArArArArGrArArArArGrGrGrGrGrGACTGGAAGGGCT-3′, RNA PPT+18D: 5′-rUrUrArArArArGrArArArArGrGrGrGrGrGACTGGAAGGGCTAATTCA-3′ or RNA PPT+24D: 5′-rUrUrArArArArGrArArArArGrGrGrGrGrGACTGGAAGGGCTAATTCACTCCCA) were annealed to the PPT90 template and used at a final concentration of 20 nm as described above. All the primers were HPLC-purified.
RESULTS
Mechanistically Distinct RT Inhibitors Display Unique Substrate Preference Profiles
We reported recently that NNRTIs preferentially inhibit RNA PPT-primed DNA synthesis (i.e. (+)-strand initiation) over DNA-primed polymerization reactions (18). Recent single-molecule spectroscopy studies showed that an NNRTI (nevirapine) decreases the frequency with which RT binds an RNA PPT-primed substrate in the DNA polymerization mode (5). Taken together, these studies highlight the interplay between primer, RT·substrate binding mode, and inhibitor binding. In this study, we investigated whether inhibitors of RT translocation (NNTIs) exhibited any preferences in terms of nucleic acid primer/template substrate based on the fact that they do not appear to inhibit reverse transcription in the same manner as NNRTIs (18). To investigate this, we characterized the two known NNTIs, PFA (13) and INDOPY-1 (15), which inhibit ongoing polymerization by trapping RT in the pre-translocation and post-translocation complexes, respectively. Activity and inhibition were characterized using non-PPT-based and PPT-based primers as previously described (18). In this assay system, DNA synthesis from either RNA or DNA primers is quantified by scintillation counting following capture of the radiolabeled product with streptavidin-coated polyvinyltoluene beads. Of note, the DNA-primed polymerization reaction was found to be ∼10 times faster than the RNA-primed reaction (see supplemental Fig. S1). Measurements of activity and inhibition were adjusted accordingly to ensure measurements were taken during the linear phase of the reaction.
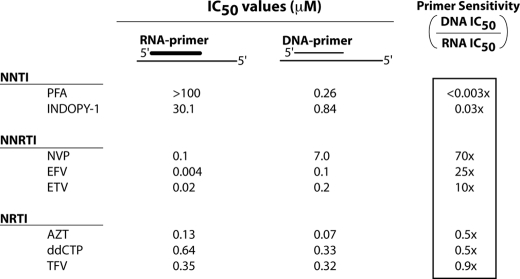
The substrate-preference profile for PFA and INDOPY-1 was tested side-by-side with three NNRTIs from first (Nevirapine), second (Efavirenz), and third generation NNRTIs (Etravirine), two NRTIs (Tenofovir and 3′-azido-3′-deoxythymidine triphosphate), and a dideoxynucleotide (ddCTP). As shown in Fig. 1, each of the three classes of RT inhibitors displayed a unique profile of inhibition on RNA-primed versus DNA-primed reactions. As expected, the potency of NRTIs was unaffected by primer content. NNTIs and NNRTIs, on the other hand, showed significant differences in their ability to inhibit RNA- versus DNA-primed reactions; however, with the opposite pattern. In contrast with NNRTIs, both PFA and INDOPY-1 showed a preference for inhibiting DNA-primed DNA polymerization reactions. The magnitude of the increase in potency for PFA observed (i.e. >100 μm on RNA-primed reactions versus 0.26 μm on DNA-primed reactions) was difficult to quantify as technical limitations prevented an accurate measurement of potency on an RNA primer. Nevertheless, these results highlighted a distinct difference in the mode of inhibition between NNTIs and NNRTIs. Notably, the sensitivity of NNTIs for DNA PPT primer was not related to the DNA PPT sequence itself, because non-PPT DNA primers exhibited the same sensitivity for NNRTIs and NNTIs (data not shown).
FIGURE 1.
NNTIs potently inhibit DNA-primed but not RNA-primed polymerization reaction. Compound potency (IC50 value) was established using an RNA or a DNA PPT primer annealed to the PPT37 template. RNA-primed reactions were performed for 30 min, while the DNA-primed reactions were quenched after 5 min to establish compound potency during the linear phase of the polymerization reaction. IC50 values were calculated by a four-parameter logistic fit.
Differential Stabilization of RT Ternary Complexes Correlates with Ability of NNTIs to Block Polymerization
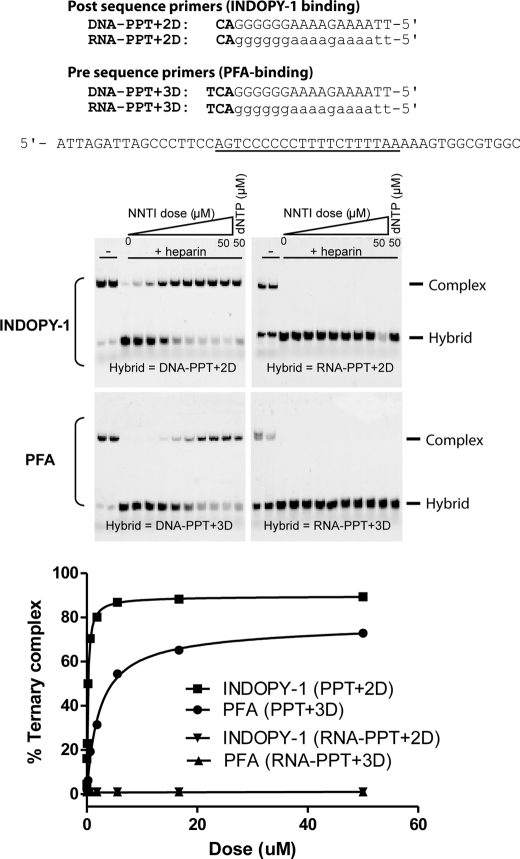
To better understand the observed selectivity of NNTIs for DNA primer over RNA primer, we studied whether a preformed RT-DNA·DNA or RT-RNA·DNA complex exhibited any differences in NNTI-binding sensitivity. Initial attempts using just the PPT primer sequences (DNA or RNA) were unsuccessful (data not shown), as we found that the DNA PPT primer sequence biases RT toward the post-translocational state (23), which precludes its use for measuring binding to PFA, which stabilizes the pre-translocational state. Moreover, although the PPT sequence does stabilize the post-translocational state, it was not suitable for INDOPY-1 measurements, because the PPT primer ends with a purine (dGTP), which is not ideally suited for INDOPY-1 binding (15). For these reasons, we designed tailored chimeric PPT primers that mimic the initiation of the (+)-strand DNA synthesis and favor PFA or INDOPY-1 binding. We thus synthesized a pre-translocated sequence (PPT+3D) to favor PFA binding (13) and a post-translocated sequence (PPT+2D) that ends in a pyrimidine (dCTP) for INDOPY-1 (15).
To investigate the impact of the primer sugar backbone on NNTIs binding to RT, we compared binding of PFA and INDOPY-1 to RT complexed with either a DNA PPT+2D·DNA (INDOPY-1) or a DNA PPT+3D·DNA (PFA) as opposed to an RT-RNA PPT+2D (or +3D)·DNA complex. As shown in Fig. 2, INDOPY-1 and PFA formed stable ternary complexes with the RT-DNA·DNA complex with Kd(app) values of 0.2 and 2.3 μm, respectively, but were unable to form ternary complexes with the RT-RNA·DNA substrate (up to 50 μm). The same trend was observed with the next templated nucleotide (dGTP or dTTP). These data demonstrated that the binding of PFA or INDOPY-1 or binding of the next incoming nucleotide was drastically affected when RT was bound to the RNA·DNA hybrid. This further suggested that RT interaction with the RNA·DNA hybrid did not favor ternary complex formation/stabilization possibly through a reduction of the polymerase-competent binding mode of RT (polymerase active site engaged at the 3′-end of the primer). Consistent with this, it was shown that, when RT interacts with an RNA·DNA hybrid, >60% of the RT population interacts with the hybrid through an RNase H binding mode (5) that reduces dNTP binding (18) and possibly the binding of NNTIs. Taken together, these results further demonstrate selectivity of NNTIs for DNA- over RNA-primed nucleic acid substrate.
FIGURE 2.
INDOPY-1 and PFA are unable to stabilize RT complexed with RNA·DNA substrate. Preformed RT-hybrid (RNA·DNA or DNA·DNA) complexes were incubated with increasing concentrations of INDOPY-1 or PFA or with a fixed concentration (50 μm) of the next complementary nucleotide (dTTP for the PPT+2D or dGTP for the PPT+3D). These complexes were challenged for 15 min at room temperature with heparin to trap dissociated RT molecules before the samples were analyzed on 6% non-denaturating gel.
Primer Impact Is Surmounted When RT Is No Longer in Physical Contact with Primer
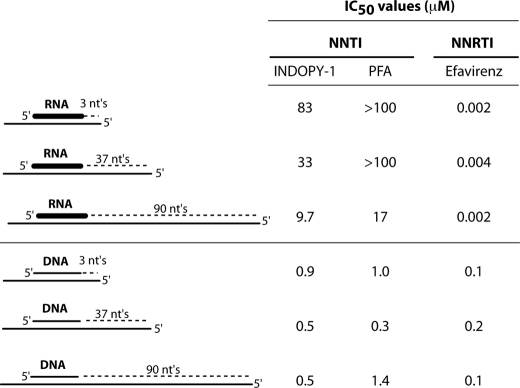
To determine how far-reaching the effects of the RNA primer were on inhibition by NNTIs, we next investigated the impact of template length on the ability of NNTIs to inhibit polymerization. We reasoned that, during the course of an RNA-primed polymerization reaction, at some point, when the replication product was long enough, the impact of the primer would be abrogated. If true, the potency of the NNTIs should increase with increasing template length. To this end, we hybridized an RNA or a DNA PPT primer to a DNA template in which we varied the length to obtain a substrate that allows 3 (PPT3), 37 (PPT37), or 90 deoxyribonucleotide incorporations (PPT90). Before establishing the potency of compounds on these substrates, we performed a time-course experiment to determine the linear phase of each of the polymerization reactions (see supplemental Fig. S1). Based on this time-course study, we selected the polymerization reaction time for each of the nucleic acid substrates such that compound potency was measured within the linear phase of the polymerization reaction.
As shown in Fig. 3, increasing template length had no impact on the ability of NNRTIs to inhibit DNA- and RNA-primed polymerization reactions suggesting that the proposed stochastic inhibitory mechanism of NNRTIs (18) does not apply in the context of PPT-primed DNA synthesis. In contrast, we observed a significant effect of template length on NNTI potency using RNA-primed reactions, as predicted. Despite this increase in potency observed for RNA-primed reactions on a long template, we were initially surprised to see that the absolute potency values for INDOPY-1 and PFA were still at least 12-fold lower on the 90-nt template as compared with a DNA-primed reaction. At first glance, this would seem to suggest that the RNA primer still posed effects on inhibition beyond 90 nucleotides, however, we postulated that, given the slow kinetics of RNA-primed reactions (see above), the reaction likely had not proceeded to completion (i.e. full-length product formation) under the conditions used. To address this in a more direct manner, we employed chimeric primers in which DNA bases were added in defined amounts to the standard RNA PPT primer to simulate early polymerization intermediates.
FIGURE 3.
Template length affects NNTIs potency only on RNA-primed polymerization reaction. Compound potency (IC50 value) was determined for each set of primer/template substrates described under “Experimental Procedures.” Polymerization reaction times for each set of primer/template substrates (RNA-primed = 30 min and DNA-primed = 5 min) were derived from supplemental Fig. S1 to establish IC50 values within the linear phase of the polymerization reaction.
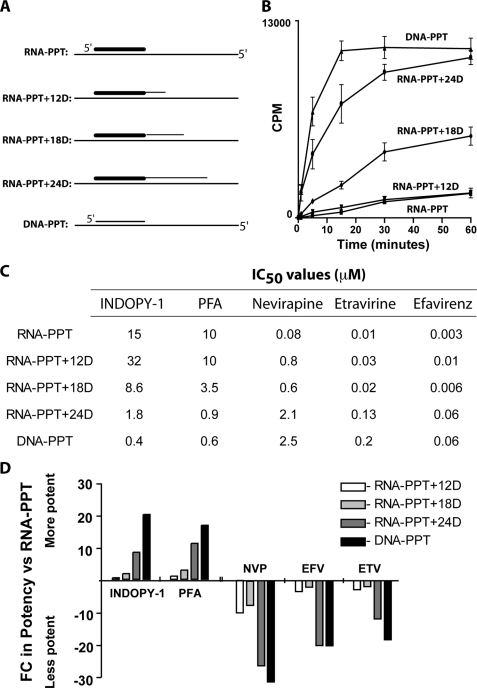
Chimeric substrates composed of RNA PPT primers extended at their 3′-end with 12, 18, and 24 deoxyribonucleotides were synthesized and annealed to the long 90-base DNA template as above (Fig. 4A). Using these chimeric primers (RNA PPT+12D, +18D, and +24D), we could test how far-reaching the impact of the RNA PPT primer was in a more controlled manner. We chose 12 nucleotides to represent a situation where, by definition, RT would be in physical contact with the RNA primer even when engaging the 3′ terminus in polymerization mode. The 24-base extension was selected to represent a case where RT would no longer be in physical contact with the RNA primer when the 3′ terminus was in the DNA polymerization-active site. Finally, an intermediate length (18 nt) was chosen to represent the point at which the two active sites of RT would just be in contact initially as they have been shown to be separated by this precise distance (24–30). We hypothesized that longer chimeric substrates, in which both active sites of RT (polymerase and RNase H) are no longer in contact with the RNA primer during initiation of the polymerization, may shift RT toward a polymerization-competent binding mode similar to that seen for a DNA-primed reaction (5) and, therefore, should lead to RT inhibitor selectivity (NNTIs over NNRTIs).
FIGURE 4.
Extending the RNA PPT primer by more than 18 DNA residues sensitizes the NNTIs while decreasing the potency of the NNRTIs. A, diagram of the chimeric primer/template substrates. B, time course of the different chimeric primer/template substrates. C, compound potency was established for each chimeric primer using the optimal reaction time derived from the time-course experiment showed in B (DNA PPT and RNA PPT+24D = 5 min; RNA PPT, RNA PPT+12D, and RNA PPT+18D = 30 min). D, -fold change in potency of RT inhibitors (IC50 using the chimeric primer/IC50 using the pure RNA PPT primer) upon addition of DNA residues at the 3′-end of the RNA PPT primer.
Initially to see the impact of the chimeric primers on reaction rate, we performed a time-course experiment to determine the optimal polymerization reaction time to use for each substrate. The data show that, although extending the RNA PPT primer with 12 deoxyribonucleotides did not modify the kinetics of the polymerization reaction significantly, the addition of 18 bases gave an intermediate signal between the RNA and the DNA PPT primer controls (Fig. 4B). Interestingly, the addition of 24 deoxyribonucleotides to the 3′-end of the RNA PPT primer almost fully restored the kinetics of the polymerization reaction performed using the DNA PPT primer control. These data showed that the addition of 24 deoxyribonucleotides to the 3′-end of an RNA PPT primer was sufficient to increase the polymerization reaction rate to a similar level to what was observed with a DNA PPT primer. We next wanted to establish potency of NNRTIs and NNTIs on the chimeric substrates and, as shown in Fig. 4, C and D, increasing the DNA content of the RNA PPT primer increased NNTI potency and decreased NNRTI potency. Notably, RNA PPT+24D, i.e. the addition of 24 deoxyribonucleotides, completely restored potency values for both NNRTIs and NNTIs that were seen with DNA-primed reactions. This, combined with the still attenuated potency with RNA-PPT+18D, indicated that the impact of the RNA PPT primer on compound potency was lost somewhere between 18 and 24 deoxyribonucleotides, likely once physical contact between RT and RNA PPT primer was lost.
Primer Impact on RT Translocation Equilibrium
Having refined the impact of the primer on RT·substrate-inhibitor binding and NNTI potency, we next investigated the extent to which the primer impacted on the RT translocation equilibrium status. To this end, we took advantage of the PPT-based RNA·DNA chimeric primers to better draw any correlations between inhibitor potency and primer-induced orientational dynamics of RT. Site-specific footprinting is a powerful technique for measuring the position of HIV-1 RT on a substrate to single-nucleotide resolution. RT·substrate complexes are treated with KOONO, which produces hydroxyl radicals and results in the hyper-reactive cleavage of the DNA substrate via Cys-280 at positions −7/−8, representing the post- and pre-translocational positions of RT, respectively (22, 31). A cleavage at either of these positions indicates that RT is positioned at the 3′ primer terminus (i.e. in a polymerase·competent complex). Thus, in addition of highlighting the pre- or post-state of RT, the site-specific footprint technique also reveals the binding orientation of RT on the nucleic acid substrate. In Fig. 5, we show results where we employed KOONO footprinting to establish the translocational status of RT when interacting with the RNA-PPT chimeric primers. To abolish the RNase H activity of RT that could interfere with the footprint of the RNA·DNA hybrids, we used the RT RNase H mutant E478Q. As shown in Fig. 5B, with the DNA PPT primer, a strong cleavage at position −7 was indicative of a bias toward the post-translocated complex. The pre-translocated state was increased in the presence of 100 μm PFA, which bound to and stabilized pre-translocated complexes (Fig. 5, see DNA PPT/lane 2). Similarly, the post-translocated complex was stabilized upon addition of a chain terminator and the next templated nucleotide (lanes 3 and 4). However, no cleavages were observed either with the RNA PPT primer, or RNA PPT+12D, suggesting that RT had reduced binding affinity for these substrates, or at least did not form a polymerase-competent complex. With the RNA PPT+18D primer, no cleavage was observed with unliganded RT, however a pre- and post-translocated cleavage was observed when the RT·substrate complex was stabilized with PFA and a nucleotide, respectively. These data suggest that RT bound only weakly to this substrate in a polymerase-competent mode but could be stabilized by the creation of a ternary complex. However, with the RNA PPT+24D primer we observed a cleavage with unliganded RT, as well as with the stabilized ternary complexes. RT therefore recognized and bound RNA PPT+24D in a polymerization-competent mode, as was seen with a DNA-PPT substrate. Taken together, these data suggest a link between RT binding orientation, translocation equilibrium, and the ability or inability of NNTI to inhibit polymerization.
DISCUSSION
Recent advances in elucidating RT structures at ever greater resolution have provided tremendous insight into one of nature's most sophisticated enzymes and opened up new avenues for rational design of drugs to treat HIV. Complementary to these structural studies are emerging studies aimed at elucidating the conformational dynamics of RT and its impact on inhibitor binding during the various catalytic steps of reverse transcription. We previously demonstrated that NNRTIs preferentially inhibit the initiation of the (+)-strand DNA synthesis due to their increased ability to inhibit RNA PPT-primed polymerization reaction (18). Abbondanzieri et al. (5) subsequently showed that NNRTIs stabilize the binding of RT in the RNase H binding mode when interacting with an RNA PPT primer. These findings suggested that the flipping transition of RT observed when bound to an RNA PPT primer increases the ability of NNRTIs to stabilize RT in the RNase H binding mode leading to the inhibition of the RT polymerase activity.
In this report, we showed that two inhibitors of RT translocation, PFA and INDOPY-1, were unable to block RNA-primed DNA synthesis (namely analogous to (+)-strand initiation) due to primer-dependent effects on RT structural dynamics. Such orientational dynamics disfavor ternary complex stabilization and establishment of translocational equilibria that are needed for NNTI inhibition. Moreover, we show that the converse is also true: NNTIs preferentially inhibit DNA-primed polymerization and ongoing polymerization reactions, where the translocational equilibria dominate the RT conformational landscape over flipping transitions. Specifically, as revealed by the site-specific footprint profile of RT interacting with the RNA PPT or the RNA PPT+12D chimeric primer, no translocation equilibrium was detected in the absence of inhibitor, and further, PFA was unable to stabilize the pre-translocational state of RT when associated with these substrates. These data suggest that the majority of RT does not bind in a polymerase-competent binding mode when interacting with these RNA·DNA substrates (<18 nt). In addition to this, the bandshift experiment revealed that PFA and INDOPY-1 were not able to form a ternary complex with RT interacting with an RNA·DNA substrate suggesting that NNTIs are not able to bind to this complex or at least are unable to stabilize it. As a result, increased RNase H binding mode or at least improper polymerase binding mode of RT interacting with an RNA·DNA substrate significantly reduces the ability of NNTIs to inhibit (+)-strand initiation synthesis (initiation from the RNA PPT primer).
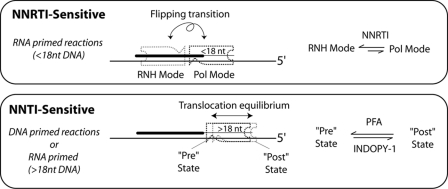
The impact of the flipping transition on the translocational equilibrium of RT may additionally be responsible for the slow reaction rate associated with polymerization reactions initiated from the RNA PPT primer as compared with DNA primer. In fact, when interacting with a DNA·DNA nucleic acid substrate, RT binds almost exclusively in a polymerase-competent mode (5), and so RT can easily translocate from the pre- to the post- state to allow DNA synthesis without interference of the flipping transition (polymerase versus RNase H binding modes). Overall these data demonstrate that, although NNRTIs preferentially inhibit polymerization reaction during the flipping transition of RT by stabilizing RT in the RNase H binding mode (5, 18), NNTIs have an increased ability to inhibit RT during the translocational equilibrium (pre or post) and therefore only when the polymerase active site of RT was engaged at the 3′-end of the primer (Fig. 6).
FIGURE 6.
Model of the sensitivity of RT to NNRTIs and NNTIs dictated by primer-guided binding dynamics. Top, DNA polymerization from an RNA-primed substrate is sensitive to NNRTIs. For RNA-primed substrates, and for substrates in which less than 18 nt of DNA have been incorporated, RT predominantly is in equilibrium between RNase H (RNH) and polymerization (Pol) modes. NNRTIs are able to potently inhibit RT in this regime by favoring the non-polymerization competent RNH mode. In contrast, NNTIs show a reduced ability to inhibit polymerization, which corresponds to a decreased ability to stabilize ternary complexes and an inability to trap RT in a particular polymerization-incompetent translocation state. Bottom, DNA polymerization from a DNA-primed substrate is sensitive to NNTIs. For DNA-primed substrates and for on-going polymerization, RT is primarily in polymerization mode, transitioning between the pre- and post-translocational states facilitating trapping by NNTIs, and disfavoring inhibition by NNRTIs.
In addition to these findings, by using synthetic chimeric RNA·DNA PPT primers, we were able to better characterize the transition between initiation and elongation of the (+)-strand DNA synthesis and thus infer the sensitivity of RT to different classes of non-nucleosides RT inhibitors. In fact, we found previously that the addition of 12 nt did not alter the sensitivity of HIV-1 RT to NNRTIs during RNA PPT-primed polymerization reaction (18), nor did the addition of 9 nt to an RNA primer significantly affect the binding orientation of RT (i.e. RT was mostly in an RNase H binding mode) (5). In our current study, we showed that the addition of more than 18 nt (24 nt) fully restored the ability of the NNTIs to inhibit RT while decreasing the potency of the NNRTIs. In addition to this, the site-specific footprint profile of RT revealed that the addition of more than 18 nucleotides (24 nt) to the RNA PPT primer allowed PFA binding and subsequent trapping of RT in the pre-translocational state (Fig. 5). These data suggested that, beyond 18 nt incorporations, the RNA PPT primer becomes a de facto DNA primer and therefore the vast majority of RT population binds in the polymerase binding mode (Fig. 6). As a consequence, the flipping transition of RT is highly reduced and is replaced by the translocational equilibrium of RT observed during DNA synthesis; NNTIs are then able to inhibit the polymerization reaction by trapping the RT in either the pre- or -post-translocational state. The fact that this was observed only beyond the addition of 18 nucleotides, which corresponds to the distance between the polymerase and RNase H active sites of RT, suggests that both active sites need to be in contact with the DNA part of the primer/template hybrid substrate to favor the polymerase binding orientation. On the other hand, we cannot exclude the possibility that the RNA PPT sequence gets cleaved only after addition of the 18th DNA residue, instead of the 12th as previously demonstrated (32). If this were indeed the case, following the cleavage of the RNA PPT at the RNA·DNA junction by the RNase H activity of RT, the primer would serve as a de facto DNA primer. As observed here and previously, the RNA PPT primer extended by 12 deoxyribonucleotides is still acting like the pure RNA PPT in terms of sensitivity to NNRTIs and NNTIs suggesting that the RNA PPT sequence is still part of the primer/template substrate. In previous work it was shown that RT pauses after the addition of the 12th DNA residue during the initiation of the (+)-strand synthesis from the RNA PPT primer causing the cleavage of the PPT sequence at the RNA PPT-DNA junction (32). One hypothesis that could explain this discrepancy might be that the pausing of RT after the addition of 12 nt, which leads to the cleavage at the RNA PPT-DNA junction, occurs only during ongoing polymerization and not when using a pre-synthesized chimeric primer.
In conclusion, our model suggests that both NNTIs and NNRTIs are able to inhibit the (+)-strand DNA synthesis but do so in a different way and thus at a different step during the course of the polymerization reaction. As proposed earlier, NNRTIs inhibit the initiation from the RNA PPT primer while NNTIs inhibit elongation that can be defined to start after the incorporation of 18 nt or after the removal of the PPT primer by the RNase H activity of RT. These data highlight the ability of the sugar backbone composition of the primer (RNA versus DNA) to dictate the binding orientation of RT (RNase H versus polymerase modes) and, therefore, the sensitivity of the HIV-1 RT enzyme to different classes of non-nucleosides RT inhibitors. Overall, this study further characterizes the mechanism of inhibition of PFA and INDOPY-1 and provides new insights that will facilitate the discovery of novel NNTIs in addition to describing a simple way to quickly discriminate between non-nucleosides RT inhibitors that bind and inhibit RT through novel and distinct mechanisms of action.
Supplementary Material
This work was supported in part by a grant from the Canadian Institutes of Health Research (to M. G.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- RNase H
- ribonuclease H
- PPT
- polypurine tract
- NRTI
- nucleoside RT inhibitor
- NNRTI
- non-nucleoside RT inhibitor
- INDOPY-1
- scaffold of indolopyridones
- nt
- nucleotide(s)
- KOONO
- potassium peroxynitrite
- PFA
- phosphonoformic acid.
REFERENCES
- 1. Smith J. K., Cywinski A., Taylor J. M. (1984) J. Virol. 52, 314–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Omer C. A., Resnick R., Faras A. J. (1984) J. Virol. 50, 465–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Finston W. I., Champoux J. J. (1984) J. Virol. 51, 26–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jones F. D., Hughes S. H. (2007) Virology 360, 341–349 [DOI] [PubMed] [Google Scholar]
- 5. Abbondanzieri E. A., Bokinsky G., Rausch J. W., Zhang J. X., Le Grice S. F., Zhuang X. (2008) Nature 453, 184–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sarafianos S. G., Marchand B., Das K., Himmel D. M., Parniak M. A., Hughes S. H., Arnold E. (2009) J. Mol. Biol. 385, 693–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Spence R. A., Kati W. M., Anderson K. S., Johnson K. A. (1995) Science 267, 988–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Esnouf R., Ren J., Ross C., Jones Y., Stammers D., Stuart D. (1995) Nat. Struct. Biol. 2, 303–308 [DOI] [PubMed] [Google Scholar]
- 9. Rittinger K., Divita G., Goody R. S. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 8046–8049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sarafianos S. G., Clark A. D., Jr., Das K., Tuske S., Birktoft J. J., Ilankumaran P., Ramesha A. R., Sayer J. M., Jerina D. M., Boyer P. L., Hughes S. H., Arnold E. (2002) EMBO J. 21, 6614–6624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yin Y. W., Steitz T. A. (2004) Cell. 116, 393–404 [DOI] [PubMed] [Google Scholar]
- 12. Meyer P. R., Rutvisuttinunt W., Matsuura S. E., So A. G., Scott W. A. (2007) J. Mol. Biol. 369, 41–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Marchand B., Tchesnokov E. P., Götte M. (2007) J. Biol. Chem. 282, 3337–3346 [DOI] [PubMed] [Google Scholar]
- 14. Zahn K. E., Tchesnokov E. P., Götte M., Doublie S. (2011) J. Biol. Chem. 286, 25246–25255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jochmans D., Deval J., Kesteleyn B., Van Marck H., Bettens E., De Baere I., Dehertogh P., Ivens T., Van Ginderen M., Van Schoubroeck B., Ehteshami M., Wigerinck P., Götte M., Hertogs K. (2006) J. Virol. 80, 12283–12292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang Z., Walker M., Xu W., Shim J. H., Girardet J. L., Hamatake R. K., Hong Z. (2006) Antimicrob. Agents Chemother. 50, 2772–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ehteshami M., Scarth B. J., Tchesnokov E. P., Dash C., Le Grice S. F., Hallenberger S., Jochmans D., Götte M. (2008) J. Biol. Chem. 283, 29904–29911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Grobler J. A., Dornadula G., Rice M. R., Simcoe A. L., Hazuda D. J., Miller M. D. (2007) J. Biol. Chem. 282, 8005–8010 [DOI] [PubMed] [Google Scholar]
- 19. Shaw-Reid C. A., Feuston B., Munshi V., Getty K., Krueger J., Hazuda D. J., Parniak M. A., Miller M. D., Lewis D. (2005) Biochemistry 44, 1595–1606 [DOI] [PubMed] [Google Scholar]
- 20. Shaw-Reid C. A., Munshi V., Graham P., Wolfe A., Witmer M., Danzeisen R., Olsen D. B., Carroll S. S., Embrey M., Wai J. S., Miller M. D., Cole J. L., Hazuda D. J. (2003) J. Biol. Chem. 278, 2777–2780 [DOI] [PubMed] [Google Scholar]
- 21. Le Grice S. F., Grüninger-Leitch F. (1990) Eur. J. Biochem. 187, 307–314 [DOI] [PubMed] [Google Scholar]
- 22. Götte M., Maier G., Gross H. J., Heumann H. (1998) J. Biol. Chem. 273, 10139–10146 [DOI] [PubMed] [Google Scholar]
- 23. Scarth B., McCormick S., Götte M. (2011) J. Mol. Biol. 405, 349–360 [DOI] [PubMed] [Google Scholar]
- 24. Furfine E. S., Reardon J. E. (1991) J. Biol. Chem. 266, 406–412 [PubMed] [Google Scholar]
- 25. Wöhrl B. M., Moelling K. (1990) Biochemistry 29, 10141–10147 [DOI] [PubMed] [Google Scholar]
- 26. Gopalakrishnan V., Peliska J. A., Benkovic S. J. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 10763–10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fu T. B., Taylor J. (1992) J. Virol. 66, 4271–4278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. DeStefano J. J., Buiser R. G., Mallaber L. M., Bambara R. A., Fay P. J. (1991) J. Biol. Chem. 266, 24295–24301 [PubMed] [Google Scholar]
- 29. Arnold E., Jacobo-Molina A., Nanni R. G., Williams R. L., Lu X., Ding J., Clark A. D., Jr., Zhang A., Ferris A. L., Clark P. (1992) Nature 357, 85–89 [DOI] [PubMed] [Google Scholar]
- 30. Kohlstaedt L. A., Wang J., Friedman J. M., Rice P. A., Steitz T. A. (1992) Science 256, 1783–1790 [DOI] [PubMed] [Google Scholar]
- 31. Marchand B., Götte M. (2003) J. Biol. Chem. 278, 35362–35372 [DOI] [PubMed] [Google Scholar]
- 32. Götte M., Maier G., Onori A. M., Cellai L., Wainberg M. A., Heumann H. (1999) J. Biol. Chem. 274, 11159–11169 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.