Abstract
Mutations in the ATP13A2 gene are associated with Kufor-Rakeb syndrome (KRS) and are found also in patients with various other types of parkinsonism. ATP13A2 encodes a predicted lysosomal P5-type ATPase that plays important roles in regulating cation homeostasis. Disturbance of cation homeostasis in brains is indicated in Parkinson disease pathogenesis. In this study, we explored the biological function of ATP13A2 as well as the pathogenic mechanism of KRS pathogenic ATP13A2 mutants. The results revealed that wild-type ATP13A2, but not the KRS pathogenic ATP13A2 mutants, protected cells from Mn2+-induced cell death in mammalian cell lines and primary rat neuronal cultures. In addition, wild-type ATP13A2 reduced intracellular manganese concentrations and prevented cytochrome c release from mitochondria compared with the pathogenic mutants. Furthermore, endogenous ATP13A2 was up-regulated upon Mn2+ treatment. Our results suggest that ATP13A2 plays important roles in protecting cells against manganese cytotoxicity via regulating intracellular manganese homeostasis. The study provides a potential mechanism of KRS and parkinsonism pathogenesis.
Keywords: Lysosomes, Manganese, Neurodegeneration, Neurological Diseases, Parkinson Disease, ATP13A2, Kufor-Rakeb Syndrome, Parkinsonism
Introduction
Mutations in ATP13A2 were initially identified in patients with Kufor-Rakeb syndrome (KRS),2 an atypical form of inherited parkinsonism. KRS is characterized by juvenile-onset autosomal recessive nigro-striatal-pallidal-pyramidal neurodegeneration with clinical features of Parkinson disease (PD) plus spasticity, supranuclear upgaze paresis, and dementia (1). Homozygous and heterozygous mutations in ATP13A2 are also found in patients with various parkinsonism, including juvenile parkinsonism, young-onset PD, early-onset PD, and familial PD (2–9). ATP13A2 encodes a predicted lysosomal P5-type cation-transporting ATPase with multiple transmembrane domains. It is highly expressed in the brain, especially in the substantia nigra, the region with characteristic dopaminergic neuronal loss in PD. Ypk9, a yeast ortholog of ATP13A2, was shown to function as a manganese transporter to protect cells from excess Mn2+ exposure, whereas loss of Ypk9 increases the sensitivity of yeast to Mn2+ toxicity (10, 11). Previous studies have suggested that cation disturbance is involved in pathogenesis of PD neurodegeneration. Increased levels of cations, including iron and aluminum, are found in the substantial nigra of the PD patient brain (12, 13). Chronic occupational exposure to copper and/or manganese is associated with higher incidence of PD in a case-control study (14). Furthermore, excess levels of Mn2+ accumulation in brains associated with occupational exposure, psychostimulant drug abuse, and liver disease result in an atypical form of parkinsonism in human (15).
PD and parkinsonism are believed to be consequences of interactions between both genetic and environmental components (16, 17). In this study, we aimed to explore the connection between the KRS-associated ATP13A2 genetic defect and manganese-associated toxicity. Our results show that wild-type ATP13A2 (ATP13A2WT), but not KRS-associated pathogenic ATP13A2 mutants, protected cells from Mn2+-induced toxicity in mammalian cell lines and primary rat neuronal cultures. Cells overexpressing ATP13A2WT exhibited lower level of intracellular manganese than cells transfected with the control or KRS pathogenic ATP13A2 mutants. Furthermore, manganese treatment up-regulated expression of endogenous ATP13A2. Our results suggest that ATP13A2 plays important roles in protecting cells against manganese toxicity via regulating intracellular manganese homeostasis.
EXPERIMENTAL PROCEDURES
Cell Culture, Transfection, and Stable Cell Line Generation
HEK293 and N2a cells were obtained from American Type Culture Collection and maintained as recommended by the provider. Stable cell lines expressing ATP13A2 variants were generated as described (18). Expression levels of ATP13A2 variants were determined by immunoblotting. Primary neuronal cultures were prepared from embryonic day 17 Sprague-Dawley rat hippocampi. Briefly, hippocampi were dissected in Hanks' balanced salt solution and digested with 0.5 mg/ml trypsin for 12 min at 37 °C, followed by trituration through serial Pasteur pipettes with gradually decreased tip diameters. Trypsinized cells were plated at 100,000 cells/cm2 on glass coverslips precoated with polylysine (50 μg/ml). Cells were cultured with Neurobasal medium supplemented with vitamin B27 supplement (2%), GlutaMAX (500 μm), and penicillin/streptomycin (50 μg/ml) at 37 °C and 5% CO2. At 6 days in vitro, the primary hippocampal neurons were transfected with calcium phosphate as described previously (19).
Immunofluorescence
Cells were cultured on glass coverslips for 24–48 h, followed by fixation with 4% paraformaldehyde for 10 min and permeabilization with 0.1% Triton X-100 in PBS for 10 min at room temperature (25 °C). After blocking with 3% BSA for 30 min, cells were blocked with primary antibodies and detected with Alexa-conjugated secondary antibodies. The antibodies used included mouse anti-V5 monoclonal antibody (Invitrogen), rabbit anti-V5 polyclonal antibody (Sigma), mouse anti-LAMP1 monoclonal antibody (Santa Cruz Biotechnology, Inc.), rabbit anti-calnexin polyclonal antibody (Sigma), and Alexa 488- or Alexa 633-labeled donkey anti-mouse or anti-rabbit IgG (Invitrogen). Lysosome and Golgi were stained with LysoTracker Red (Invitrogen) or wheat germ agglutinin for 45 min, respectively. Cell nuclei were stained with DAPI (Invitrogen). Images were taken under a Leica confocal microscope with appropriate excitation and emission filter pairs.
Immunoblotting
Cells were lysed with 2× SDS sample buffer (63 mm Tris-HCl, 10% glycerol, and 2% SDS). The supernatant was collected. The protein concentration was determined using a Bio-Rad protein assay kit. Proteins (20 μg) were separated on an SDS-polyacrylamide gel and transferred to a PVDF membranes (Millipore). The membranes were incubated for 1 h in blocking solution (5% nonfat dry milk in 0.1% Triton X-100/PBS buffer), followed by incubation with the appropriate primary antibodies in blocking solution. After washing in 0.1% Triton X-100/PBS buffer, the membrane was incubated with the appropriate secondary antibodies for 1 h and visualized via an enhanced chemiluminescence kit (GE Healthcare) according to the manufacturer's instruction. The antibodies used to detect ATP13A2 and cytochrome c were from Sigma and Abcam, respectively.
Manganese Concentration Measurement
Intracellular Mn2+ concentrations were measured by graphite furnace atomic absorption spectroscopy (PE Analyst 800, PerkinElmer Life Sciences). Briefly, cells were digested in 900 μl of 0.2% ultrapure nitric acid for 1 h and vortexed for 10 s (3000 rpm). Samples (20 μl) were mixed with 10 μl of 0.2% HNO3 and 5 μl of 2% NH4H2PO4 for ashing at 1100 °C then atomized at 2100 °C for 5 s. The absorption at 279.5 nm was recorded.
Cell Death Detection
HEK293 and N2a cells stably expressing ATP13A2WT or KRS mutants were treated with 2 or 1 mm MnCl2 for 12 h, respectively. Adherent cells were collected by trypsin digestion, whereas floating cells were harvested from the medium. Resuspended single cells were stained with propidium iodide (1 μg/ml) without fixation for detection of dead cells. The total numbers of cells and those positive for propidium iodide staining were counted in randomly selected fields, with at least 1000 cells counted per coverslip. Hippocampal neurons at 6 days in vitro were transfected with pEGFP-N1+ pcDNA3.1, pEGFP-N1 + pcDNA3.1-ATP13A2WT-V5, or pEGFP-N1 + pcDNA3.1-ATP13A2DUP22-V5 using calcium phosphate. 24 h after transfection, cells were treated with 0 and 800 nm MnCl2 for another 24 h, followed by fixation with 4% paraformaldehyde and staining with DAPI (2 μg/ml). The morphological changes in nuclear chromatin in apoptotic neurons were counted under a fluorescence microscope at an excitation wavelength of 340/380 nm.
Assays for Cytochrome c Release
Cells were treated with 1 and 2 mm MnCl2 for 12 h, followed by collection, washing twice with PBS, and incubation in lysis buffer (68 mm sucrose, 200 mm mannitol, 50 mm KCl, 1 mm EDTA, 1 mm EGTA, 1 mm dithiothreitol, and protease inhibitor mixture) on ice for 30 min. Cells were further lysed with 40 passages through a 25-gauge 5/8 needle and centrifuged at 1500 × g for 10 min. Cytosolic extracts were recovered by centrifugation at 13,000 × g for 20 min. For each condition, 10 μg of mitochondrial and cytosolic proteins were separated on a 15% SDS-polyacrylamide gel and immunoblotted with an anti-cytochrome c antibody.
Real-time PCR
Total RNA was isolated using TRIzol reagent (Invitrogen) as suggested by the manufacturer. The cDNA was synthesized from total RNA using a TaqMan cDNA synthesis kit (Applied Biosystems). Real-time PCR was performed using SYBR Green Master Mix with rhodamin X (ROX) (Takara) and a 7900HT fast real-time PCR system (Applied Biosystems). Primers used for quantitative PCR included the following: ATP13A2, TGCCTCTGAACAGGACAGTG (forward) and ACGAAGTTGAGGGTGACCAG (reverse); and β-actin, ACTCTTCCAGCCTTCCTTCC (forward) and GTACTTGCGCTCAGGAGGAG (reverse). Each cycle was at 95 °C for 5 s and at 60 °C for 30 s for 40 cycles.
Statistical Analysis
Results are presented as means ± S.D. Statistical significance of differences was evaluated with one-way analysis of variance, followed by Tukey's test and Dunnett's test. A probability of p < 0.05 was taken as statistically significant.
RESULTS
ATP13A2WT and KRS Pathogenic ATP13A2 Are Differentially Distributed in Cells
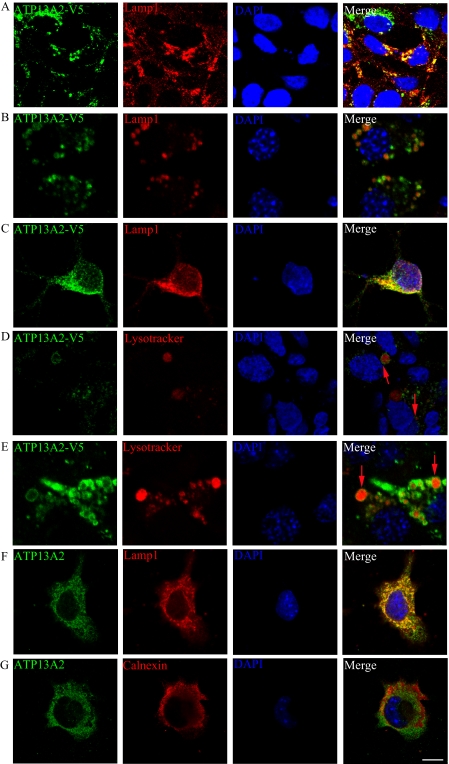
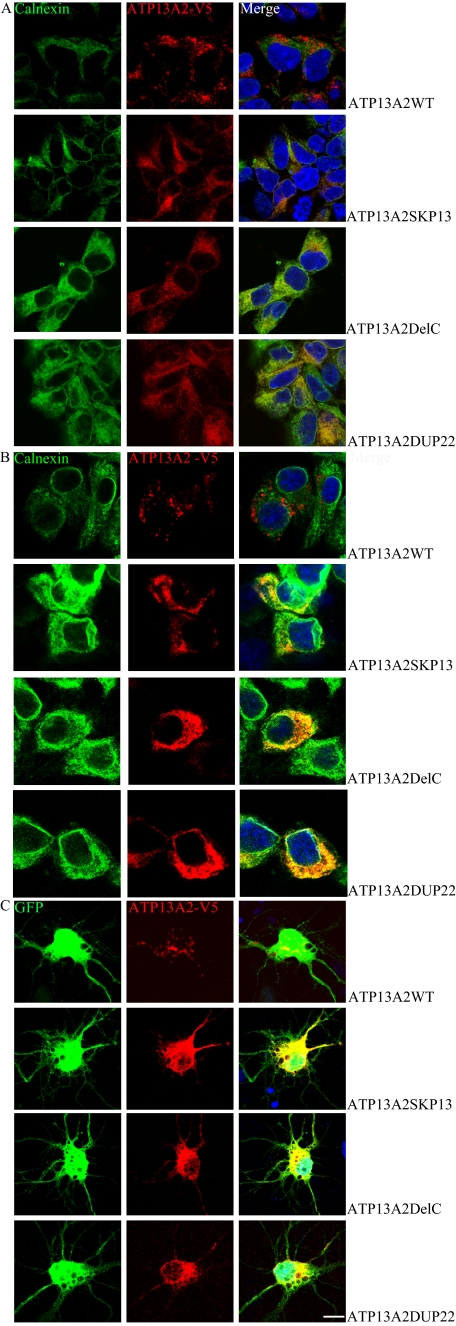
To investigate the pathophysiological function of ATP13A2, we stably expressed ATP13A2WT and KRS pathogenic ATP13A2 mutants in HEK293 cells and mouse neuroblastoma N2a cells. For each ATP13A2 variant, several stable clones were isolated and maintained in G418-containing medium. Expression of ATP13A2 variants was verified by immunoblotting (supplemental Fig. 1). Consistent with a previous report (1), the pathogenic ATP13A2 mutants showed lower steady-state levels of expression compared with their wild-type counterpart, due mainly to the shorter half-life times of these proteins (supplemental Fig. 2). Immunofluorescence revealed predominant co-localization of exogenous ATP13A2WT and lysosomal protein LAMP1 in HEK293 and N2a cells and primary cultured rat neurons (Fig. 1, A–C). ATP13A2WT was detected largely on the membrane-like structures of lysosomes and LysoTracker Red-positive organelles in both HEK293 (Fig. 1D) and N2a (Fig. 1E) cells. Little ATP13A2WT was found in other membrane structures, such as the plasma membrane, Golgi, and mitochondria (data not shown). Likewise, endogenous ATP13A2 was co-localized with LAMP1 (Fig. 1F), but not the endoplasmic reticulum (ER) marker calnexin (Fig. 1G) in SH-SY5Y cells. These results suggest that ATP13A2WT is localized predominantly on the lysosomal membrane. In contrast to the lysosomal localization of ATP13A2WT, the KRS pathogenic ATP13A2 mutant proteins were detected predominantly in the ER with co-localization with the ER protein calnexin (Fig. 2, A and B). Similar ER localization was observed in rat primary neurons expressing the KRS pathogenic ATP13A2 mutants (Fig. 2C). Thus, pathogenic ATP13A2 mutants are abnormally distributed in neurons.
FIGURE 1.
Subcellular localization of ATP13A2WT. HEK293 (A) and N2a (B) cells stably expressing V5-tagged ATP13A2WT and primary rat hippocampal neurons transfected with V5-tagged ATP13A2WT (C) were immunostained with anti-LAMP1 (red) and anti-V5 (green) antibodies. ATP13A2 surrounded LysoTracker, a lysosome-specific dye (red), in HEK293 (D) and N2a (E) stable cell lines (red arrows). Endogenous ATP13A2 in SH-SY5Y cells was detected using an anti-ATP13A2 antibody. Endogenous ATP13A2 (green) co-localized with the lysosome marker protein LAMP1 (red; F), but not with the ER marker calnexin (red; G). Nuclei were stained with DAPI (blue). Scale bar = 10 μm.
FIGURE 2.
Subcellular localization of KRS ATP13A2 mutants. Representative confocal images of HEK293 (A) and N2a (B) cells and rat hippocampal neurons (C) expressing the KRS pathogenic ATP13A2 mutants are shown. ATP13A2 mutant proteins were labeled with an anti-V5 tag antibody (red). The ER was stained with an anti-calnexin antibody (green). Nuclei were stained with DAPI (blue). Scale bar = 10 μm.
ATP13A2WT, but Not the KRS Pathogenic ATP13A2 Mutants, Protects Cells against MnCl2-induced Cytotoxicity
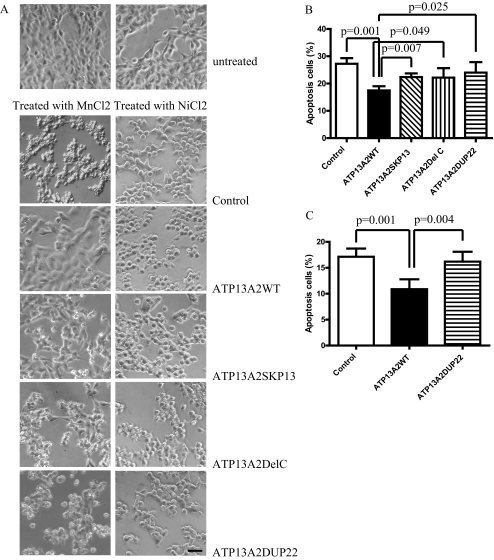
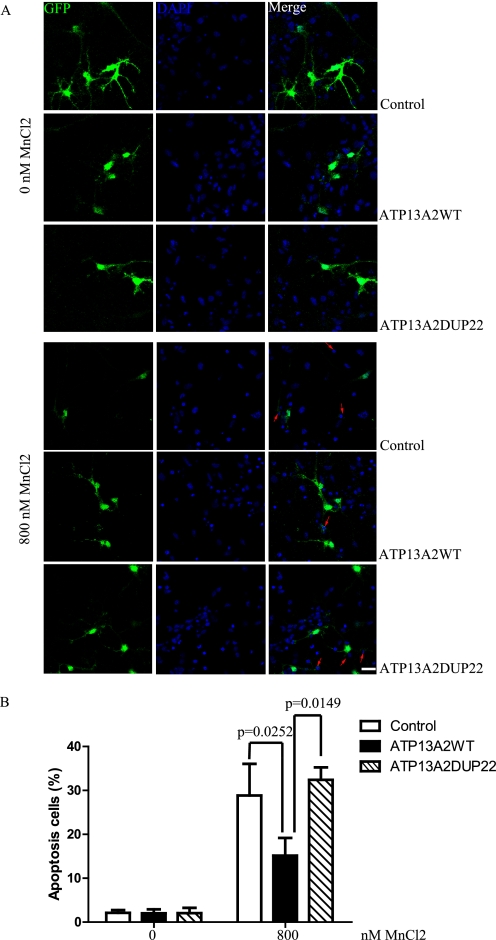
Inactivation of Ypk9, a yeast ortholog of ATP13A2, induces hypersensitivity of yeast cells to Mn2+ toxicity (10, 11). Overexpression of wild-type Ypk9 protects cells against Mn2+ toxicity. However, human ATP13A2 fails to rescue the Mn2+-sensitive phenotype in yeast cells with Ypk9 inactivation. To examine whether human ATP13A2 functions in preventing Mn2+ cytotoxicity in mammalian cells, we treated cells stably expressing ATP13A2WT and pathogenic ATP13A2 mutants with either 2 mm (HEK293 cells) or 1 mm (N2a cells) MnCl2. After treatment, morphological changes and eventual death with apoptotic characteristics, including membrane blebbing, nuclear fragmentation, and shrinkage, were observed in the KRS pathogenic mutant expressers (Fig. 3A, left panel). In contrast, cells expressing ATP13A2WT showed resistance to MnCl2-induced cytotoxicity and remained viable with normal morphology. Cells expressing the KRS mutants were not sensitive to treatment with NiCl2 and FeCl2, demonstrating specificity (Fig. 3A and supplemental Fig. 3). Quantitative results revealed that ATP13A2WT protected HEK293 cells from Mn2+ cytotoxicity with a reduced apoptosis rate of 17.48 ± 1.545% compared with the control vector (27.26 ± 2.056%, p = 0.001), ATP13A2SKP13 (22.37 ± 1.362%, p = 0.007), ATP13A2DelC (22.18 ± 3.458%, p = 0.049), and ATP13A2DUP22 (24.03 ± 3.813%, p = 0.025) (Fig. 3B). Similar results were obtained in N2a cells treated with 1 mm MnCl2 for 12 h. The apoptosis rates of cells expressing the control vector, ATP13A2WT, and ATP13A2DUP22 were 17.13 ± 1.581, 10.87 ± 1.908, and 16.19 ± 1.902%, respectively, with significant differences between ATP13A2WT and the control vector (p = 0.001) and between ATP13A2WT and the ATP13A2DUP22 mutant (p = 0.004) (Fig. 3C). Likewise, ATP13A2WT, but not the pathogenic ATP13A2DUP22 mutant, suppressed MnCl2-induced death of rat neuronal cultures (Fig. 4). The apoptosis rate of cells overexpressing ATP13A2WT (15.16 ± 4.023%) was significantly lower than that of cells overexpressing the control vector (28.86 ± 7.182%, p = 0.0252) and the pathogenic ATP13A2DUP22 mutant (32.40 ± 2.836%, p = 0.0149) (Fig. 4B). Thus, overexpression of ATP13A2WT, but not the KRS pathogenic mutants, suppresses Mn2+ cytotoxicity specifically in various cell lines and primary neuronal cultures.
FIGURE 3.
ATP13A2 protects cells from manganese-induced apoptosis. HEK293 cells stably expressing ATP13A2WT and KRS pathogenic ATP13A2 mutants were treated with either 2 mm MnCl2 or 1 mm NiCl2 for 12 h. Representative images of cells after treatment are shown (A). Scale bar = 20 μm. Note that dying cells have morphologies characterized as shrinkage and membrane blebbing. The results from quantification of cell death in three independent experiments with HEK293 (B) and N2a (C) cells are shown. Results are presented as means ± S.D.
FIGURE 4.
ATP13A2 protects neurons from manganese-induced apoptosis. A, rat hippocampal neurons were cotransfected with GFP and ATP13A2WT or the ATP13A2DUP22 mutant and treated with 0 and 800 nm MnCl2 for 24 h. Representative images of DAPI staining are shown. Apoptotic cells with nuclear condensation and fragmentation in GFP-positive cells are indicated (red arrows). Scale bar = 10 μm. B, the results from quantification of three independent experiments are show. Results are presented as means ± S.D.
ATP13A2 Regulates Intracellular Mn2+ Concentration
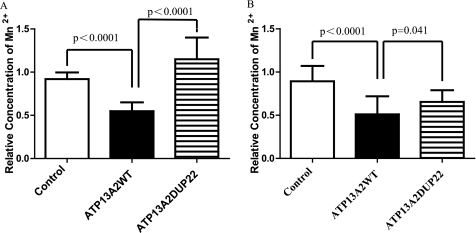
To elucidate the mechanism of ATP13A2WT-mediated suppression of Mn2+ cytotoxicity, we determined the intracellular Mn2+ concentration using atomic absorption spectrophotometry. The intracellular Mn2+ concentrations of HEK293 cells expressing ATP13A2 variants were measured after preloading 2 mm MnCl2 for 12 h. The results revealed that the intracellular Mn2+ concentration was significantly lower in cells expressing ATP13A2WT (2.812 ± 0.529 μg/107 cells) than in cells transfected with the control vector (4.714 ± 0.398 μg/107 cells, p < 0.0001) or in cells expressing the pathogenic ATP13A2DUP22 mutant (5.899 ± 1.289 μg/107 cells, p < 0.0001) (Fig. 5A). A similar observation was made with N2a cells. With 1 mm MnCl2 treatment, the intracellular Mn2+ concentrations were 0.592 ± 0.243 μg/107 cells in ATP13A2WT-expressing cells, 1.037 ± 0.206 μg/107 cells in control vector-transfected cells (p < 0.0001 compared with ATP13A2WT cells), and 0.759 ± 0.157 μg/107 cells in ATP13A2DUP22 mutant-expressing cells (p = 0.041 compared with ATP13A2WT cells) (Fig. 5B). Therefore, ATP13A2WT, but not the pathogenic ATP13A2DUP22 mutant, regulates intracellular Mn2+ homeostasis potentially by controlling the intracellular Mn2+ concentration.
FIGURE 5.
ATP13A2WT, but not the KRS pathogenic ATP13A2DUP22 mutant, regulates intracellular Mn2+ concentration. The intracellular Mn2+ concentrations of HEK293 (A) and N2a (B) cells were quantified, followed by 2 and 1 mm MnCl2 treatment for 12 h, respectively. The intracellular Mn2+ concentrations of manganese-treated cells are plotted relative to those of the corresponding control cells transfected with vector. Results are presented as means ± S.D.
ATP13A2 Protects Cytochrome c Release from Mitochondria Induced by Mn2+
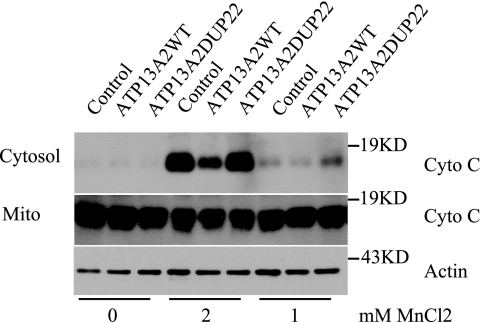
Mn2+-induced cell death involves cytochrome c release (20, 21). To determine whether ATP13A2 protects cytochrome c release from mitochondria, cytochrome c in the cytoplasm stimulated by Mn2+ was analyzed. HEK293 cells stably expressing ATP13A2WT or the ATP13A2DUP22 mutant were treated with 1 and 2 mm MnCl2 for 12 h, followed by detection of cytochrome c in the cytoplasmic and mitochondrial fractions. Cytoplasmic cytochrome c level was markedly lower in ATP13A2WT cells than in ATP13A2DUP22 and control cells (Fig. 6). Thus, overexpression of ATP13A2WT, but not the pathogenic ATP13A2DUP22 mutant, suppresses Mn2+-induced cytochrome c release.
FIGURE 6.
ATP13A2 protects manganese-induced cytochrome c release. HEK293 cells stably transfected with vector alone (Control), ATP13A2WT, or the ATP13A2DUP22 mutant were treated with 1 and 2 mm MnCl2 for 12 h. Cytochrome c (Cyto C) in the cytosol and mitochondrial (Mito) fractions was detected by immunoblotting. Actin in the cytosol was detected as a loading control. Note that cytosolic cytochrome c level in cells expressing ATP13A2WT was markedly lower than in cells transfected with the control vector and in cells expressing the pathogenic ATP13A2DUP22 mutant.
MnCl2 Potentiates Expression of ATP13A2 in Cells
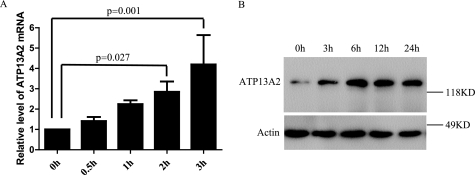
We further investigated whether Mn2+ regulates expression of ATP13A2. Cells were incubated with 2 mm MnCl2 for different time intervals, and the expression level of endogenous ATP13A2 was determined by real-time quantitative PCR. As shown in Fig. 7, Mn2+ treatment induced time-dependent expression of ATP13A2. With three independent experiments, the relative levels of ATP13A2 mRNA were increased by 1.411 ± 0.2032-, 2.250 ± 0.1827-, and 2.855 ± 0.5008-fold (p = 0.027) and 4.199 ± 1.446-fold (p = 0.001) with Mn2+ treatment for 30 min, 1 h, 2 h, and 3 h, respectively (Fig. 7A). Thus, expression of ATP13A2 is significantly up-regulated by Mn2+ treatment for 2 h or longer. Consistent with the RNA expression results, the dose-dependent up-regulation of endogenous ATP13A2 protein induced by Mn2+ was also detected using an anti-ATP13A2 antibody (Fig. 7B). The results suggest that expression of endogenous ATP13A2 is regulated by Mn2+.
FIGURE 7.
Regulation of endogenous ATP13A2 expression by Mn2+. HEK293 cells were incubated in 2 mm MnCl2 for 0–3 h. Expression of ATP13A2 mRNA was detected by real-time PCR. Mean ± S.D. of three independent experiments are shown (A). Endogenous ATP13A2 protein was also detected by immunoblotting (B). Note that MnCl2 treatment induced time- and dosage-dependent increased expression of endogenous ATP13A2. Results are presented as mean ± S.D.
DISCUSSION
We have shown in this study that KRS-associated ATP13A2 suppresses Mn2+-induced cell death and cytochrome c in two different cell lines and in primary rat neuronal cultures. Pathogenic ATP13A2 mutants exhibited reduced cytotoxicity protection activity compared with their wild-type counterpart. Overexpression of ATP13A2 reduced the intracellular Mn2+ concentration. Moreover, Mn2+ treatment induced expression of endogenous ATP13A2. These findings suggest that ATP13A2 plays important roles in protecting cells against manganese cytotoxicity via regulating the intracellular Mn2+ homeostasis that likely contributes to the KRS pathogenesis.
Manganese is a biologically important metal that functions as a cofactor for many enzymes, including carboxylases and phosphatases in the cytosol, sugar transferases in the Golgi, and superoxide dismutase in mitochondria (22–26). Excessive accumulation of Mn2+ in the cell interferes with calcium metabolism and increases replication errors of mitochondrial DNA (27). In human, Mn2+ overexposure results in “manganism,” characterized by symptoms resembling those of PD. KRS is a type of early- or juvenile-onset parkinsonism (28–30). Thus, our finding that KRS-associated ATP13A2 promotes neuronal survival and regulates homeostasis of intracellular Mn2+ suggests a potential mechanism of ATP13A2 involvement in the pathogenesis of parkinsonism.
The lysosomal compartment is essential for a series of cellular functions, including turnover of most long-lived proteins and metal ion storage and release. A number of P-type ATPase ion channels have been suggested to transport Mn2+ in yeast, Caenorhabditis elegans, and other lower organisms (31–35). Interestingly, ATP13A2 is a P5-type ATPase localized in lysosomal membranes and is expressed ubiquitously in various tissues with particularly high levels in brain (1, 36). It is possible that ATP13A2 regulates Mn2+ in neuron by mediating Mn2+ transportation. Consistent with this postulation, overexpression of ATP13A2 resulted in a reduced intracellular Mn2+ concentration. Moreover, neurodegeneration with brain iron accumulation and other neuronal system diseases are caused by defects in lysosomal membrane proteins (37, 38). Even though no specific substrate of ATP13A2 has been identified, brain MRIs indicate that patients with ATP13A2 mutations show generalized atrophy of the putaminal and caudate with bilateral iron accumulation (2, 9). Previous studies indicated that Mn2+ and iron share common transport systems (39–45). Our finding suggests that increased iron uptake in the brains of patients harboring ATP13A2 mutants is likely associated with increased cellular Mn2+ and increased cell susceptibility to Mn2+ cytotoxicity. Epidemiological studies have shown a correlation between PD and working in the iron industry (14) or dietary iron intake (46). Nevertheless, a defect in ATP13A2 does not lead to accumulation of iron in yeast (10, 11). In this study, overexpression of ATP13A2 did not protect HEK293 cells from iron-induced cell death (supplemental Fig. 3). The functional relationship between iron and Mn2+ remains to be elucidated.
Manganese-induced cell death involves DNA damage (47), oxidative stress (48), disruption of Ca2+ and iron homeostasis (49, 50), and mitochondrial dysfunction (20, 21). Consistent with these mechanisms, we found that expression of ATP13A2 suppressed Mn2+-induced cytotoxicity and inhibited cytochrome c release. Furthermore, manganese exposure induces α-synuclein expression in cell lines (51, 52) and triggers structural transformation, aggregation, and fibrillation of α-synuclein in vitro (53). A previous study suggested that overexpression of ATP13A2 suppresses the cytotoxicity of α-synuclein in yeast, C. elegans, and rat dopaminergic neurons (10). Although no change in α-synuclein was found in Mn2+-treated cells (supplemental Figs. 4–6), our results indicated that Mn2+ treatment markedly up-regulated expression of endogenous ATP13A2. The increased expression of ATP13A2 may provide cells with a protective function against increased Mn2+ in cells and maintain the mitochondrial function. This finding of Mn2+-involved cell survival offers a potential pathogenic mechanism of ATP13A2 mutation-associated KRS. Further studies on the roles of lysosomes in maintaining Mn2+ homeostasis are needed to address the molecular function of ATP13A2 in neurons.
Supplementary Material
Acknowledgment
We thank Christian Kubisch (University of Cologne, Cologne, Germany) for kindly providing ATP13A2WT and KRS mutant plasmids.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 NS057289 and P01 ES016738 (to Z. Z.). This work was also supported by Chinese National 973 Projects Grant 2011CB510000 (to Z. Z.) and Natural Science Foundation of China Grants 30730052 (to Z. Z.) and 30971469 and 30800586 (to D. W.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–6.
- KRS
- Kufor-Rakeb syndrome
- PD
- Parkinson disease
- ATP13A2WT
- wild-type ATP13A2
- ER
- endoplasmic reticulum.
REFERENCES
- 1. Ramirez A., Heimbach A., Gründemann J., Stiller B., Hampshire D., Cid L. P., Goebel I., Mubaidin A. F., Wriekat A. L., Roeper J., Al-Din A., Hillmer A. M., Karsak M., Liss B., Woods C. G., Behrens M. I., Kubisch C. (2006) Nat. Genet. 38, 1184–1191 [DOI] [PubMed] [Google Scholar]
- 2. Behrens M. I., Bruggemann N., Chana P., Venegas P., Kagi M., Parrao T., Orellana P., Garrido C., Rojas C. V., Hauke J., Hahnen E., Gonzalez R., Seleme N., Fernandez V., Schmidt A., Binkofski F., Kompf D., Kubisch C., Hagenah J., Klein C., Ramirez A. (2010) Movement Disord. 25, 1929–1937 [DOI] [PubMed] [Google Scholar]
- 3. Di Fonzo A., Chien H. F., Socal M., Giraudo S., Tassorelli C., Iliceto G., Fabbrini G., Marconi R., Fincati E., Abbruzzese G., Marini P., Squitieri F., Horstink M. W., Montagna P., Libera A. D., Stocchi F., Goldwurm S., Ferreira J. J., Meco G., Martignoni E., Lopiano L., Jardim L. B., Oostra B. A., Barbosa E. R., Bonifati V. (2007) Neurology 68, 1557–1562 [DOI] [PubMed] [Google Scholar]
- 4. Djarmati A., Hagenah J., Reetz K., Winkler S., Behrens M. I., Pawlack H., Lohmann K., Ramirez A., Tadić V., Brüggemann N., Berg D., Siebner H. R., Lang A. E., Pramstaller P. P., Binkofski F., Kostić V. S., Volkmann J., Gasser T., Klein C. (2009) Movement Disord. 24, 2104–2111 [DOI] [PubMed] [Google Scholar]
- 5. Lees A. J., Singleton A. B. (2007) Neurology 68, 1553–1554 [DOI] [PubMed] [Google Scholar]
- 6. Lin C. H., Tan E. K., Chen M. L., Tan L. C., Lim H. Q., Chen G. S., Wu R. M. (2008) Neurology 71, 1727–1732 [DOI] [PubMed] [Google Scholar]
- 7. Mao X. Y., Burgunder J. M., Zhang Z. J., Chang X. L., Peng R., Burgunder J. M., Yang Y., Wang Y. C., Li T., Zhang Z. J. (2010) Parkinsonism Relat. Disord. 16, 235–236 [DOI] [PubMed] [Google Scholar]
- 8. Ning Y. P., Kanai K., Tomiyama H., Li Y., Funayama M., Yoshino H., Sato S., Asahina M., Kuwabara S., Takeda A., Hattori T., Mizuno Y., Hattori N. (2008) Neurology 70, 1491–1493 [DOI] [PubMed] [Google Scholar]
- 9. Schneider S. A., Paisan-Ruiz C., Quinn N. P., Lees A. J., Houlden H., Hardy J., Bhatia K. P. (2010) Movement Disord. 25, 979–984 [DOI] [PubMed] [Google Scholar]
- 10. Gitler A. D., Chesi A., Geddie M. L., Strathearn K. E., Hamamichi S., Hill K. J., Caldwell K. A., Caldwell G. A., Cooper A. A., Rochet J. C., Lindquist S. (2009) Nat. Genet. 41, 308–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schmidt K., Wolfe D. M., Stiller B., Pearce D. A. (2009) Biochem. Biophys. Res. Commun. 383, 198–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dexter D. T., Wells F. R., Agid F., Agid Y., Lees A. J., Jenner P., Marsden C. D. (1987) Lancet 2, 1219–1220 [DOI] [PubMed] [Google Scholar]
- 13. Hirsch E. C., Brandel J. P., Galle P., Javoy-Agid F., Agid Y. (1991) J. Neurochem. 56, 446–451 [DOI] [PubMed] [Google Scholar]
- 14. Gorell J. M., Johnson C. C., Rybicki B. A., Peterson E. L., Kortsha G. X., Brown G. G., Richardson R. J. (1997) Neurology 48, 650–658 [DOI] [PubMed] [Google Scholar]
- 15. Guilarte T. R. (2010) Environ. Health Perspect. 118, 1071–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Di Monte D. A. (2003) Lancet Neurol. 2, 531–538 [DOI] [PubMed] [Google Scholar]
- 17. Lees A. J., Hardy J., Revesz T. (2009) Lancet 373, 2055–2066 [DOI] [PubMed] [Google Scholar]
- 18. Tang B., Xiong H., Sun P., Zhang Y., Wang D., Hu Z., Zhu Z., Ma H., Pan Q., Xia J. H., Xia K., Zhang Z. (2006) Hum. Mol. Genet. 15, 1816–1825 [DOI] [PubMed] [Google Scholar]
- 19. Zhang Z., Hartmann H., Do V. M., Abramowski D., Sturchler-Pierrat C., Staufenbiel M., Sommer B., van de Wetering M., Clevers H., Saftig P., De Strooper B., He X., Yankner B. A. (1998) Nature 395, 698–702 [DOI] [PubMed] [Google Scholar]
- 20. Tamm C., Sabri F., Ceccatelli S. (2008) Toxicol. Sci. 101, 310–320 [DOI] [PubMed] [Google Scholar]
- 21. Prabhakaran K., Chapman G. D., Gunasekar P. G. (2009) Neurotoxicology 30, 414–422 [DOI] [PubMed] [Google Scholar]
- 22. Jabalquinto A. M., Laivenieks M., Zeikus J. G., Cardemil E. (1999) J. Protein Chem. 18, 659–664 [DOI] [PubMed] [Google Scholar]
- 23. Kinnett D. G., Wilcox F. H. (1982) Int. J. Biochem. 14, 977–981 [DOI] [PubMed] [Google Scholar]
- 24. Wiggins C. A., Munro S. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 7945–7950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang P., Anglade P., Hirsch E. C., Javoy-Agid F., Agid Y. (1994) Neuroscience 61, 317–330 [DOI] [PubMed] [Google Scholar]
- 26. Naranuntarat A., Jensen L. T., Pazicni S., Penner-Hahn J. E., Culotta V. C. (2009) J. Biol. Chem. 284, 22633–22640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Malecki E. A. (2001) Brain Res. Bull. 55, 225–228 [DOI] [PubMed] [Google Scholar]
- 28. Najim al-Din A. S., Wriekat A., Mubaidin A., Dasouki M., Hiari M. (1994) Acta Neurol. Scand. 89, 347–352 [DOI] [PubMed] [Google Scholar]
- 29. Williams D. R., Hadeed A., al-Din A. S., Wreikat A. L., Lees A. J. (2005) Movement Disord. 20, 1264–1271 [DOI] [PubMed] [Google Scholar]
- 30. Hampshire D. J., Roberts E., Crow Y., Bond J., Mubaidin A., Wriekat A. L., Al-Din A., Woods C. G. (2001) J. Med. Genet. 38, 680–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Furune T., Hashimoto K., Ishiguro J. (2008) Genes Genet. Syst. 83, 373–381 [DOI] [PubMed] [Google Scholar]
- 32. Cho J. H., Ko K. M., Singaravelu G., Ahnn J. (2005) FEBS Lett. 579, 778–782 [DOI] [PubMed] [Google Scholar]
- 33. Bates S., MacCallum D. M., Bertram G., Munro C. A., Hughes H. B., Buurman E. T., Brown A. J., Odds F. C., Gow N. A. (2005) J. Biol. Chem. 280, 23408–23415 [DOI] [PubMed] [Google Scholar]
- 34. Johnson N. A., Liu F., Weeks P. D., Hentzen A. E., Kruse H. P., Parker J. J., Laursen M., Nissen P., Costa C. J., Gatto C. (2009) Arch. Biochem. Biophys. 481, 157–168 [DOI] [PubMed] [Google Scholar]
- 35. Wu Z., Liang F., Hong B., Young J. C., Sussman M. R., Harper J. F., Sze H. (2002) Plant Physiol. 130, 128–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schultheis P. J., Hagen T. T., O'Toole K. K., Tachibana A., Burke C. R., McGill D. L., Okunade G. W., Shull G. E. (2004) Biochem. Biophys. Res. Commun. 323, 731–738 [DOI] [PubMed] [Google Scholar]
- 37. Dong X. P., Cheng X., Mills E., Delling M., Wang F., Kurz T., Xu H. (2008) Nature 455, 992–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ruivo R., Anne C., Sagné C., Gasnier B. (2009) Biochim. Biophys. Acta 1793, 636–649 [DOI] [PubMed] [Google Scholar]
- 39. Gunshin H., Mackenzie B., Berger U. V., Gunshin Y., Romero M. F., Boron W. F., Nussberger S., Gollan J. L., Hediger M. A. (1997) Nature 388, 482–488 [DOI] [PubMed] [Google Scholar]
- 40. Fleming M. D., Trenor C. C., 3rd, Su M. A., Foernzler D., Beier D. R., Dietrich W. F., Andrews N. C. (1997) Nat. Genet. 16, 383–386 [DOI] [PubMed] [Google Scholar]
- 41. Tabuchi M., Yoshimori T., Yamaguchi K., Yoshida T., Kishi F. (2000) J. Biol. Chem. 275, 22220–22228 [DOI] [PubMed] [Google Scholar]
- 42. Fleming M. D., Romano M. A., Su M. A., Garrick L. M., Garrick M. D., Andrews N. C. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 1148–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yokel R. A. (2006) J. Alzheimers Dis. 10, 223–253 [DOI] [PubMed] [Google Scholar]
- 44. Yokel R. A. (2002) Environ. Health Perspect. 110, Suppl. 5, 699–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Malecki E. A., Devenyi A. G., Beard J. L., Connor J. R. (1999) J. Neurosci. Res. 56, 113–122 [DOI] [PubMed] [Google Scholar]
- 46. Logroscino G., Marder K., Graziano J., Freyer G., Slavkovich V., Lojacono N., Cote L., Mayeux R. (1998) Movement Disord. 13, 13–16 [PubMed] [Google Scholar]
- 47. Oikawa S., Hirosawa I., Tada-Oikawa S., Furukawa A., Nishiura K., Kawanishi S. (2006) Free Radic. Biol. Med. 41, 748–756 [DOI] [PubMed] [Google Scholar]
- 48. Chen C. J., Liao S. L. (2002) Exp. Neurol. 175, 216–225 [DOI] [PubMed] [Google Scholar]
- 49. Kwik-Uribe C. L., Reaney S., Zhu Z., Smith D. (2003) Brain Res. 973, 1–15 [DOI] [PubMed] [Google Scholar]
- 50. Gavin C. E., Gunter K. K., Gunter T. E. (1990) Biochem. J. 266, 329–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cai T., Yao T., Zheng G., Chen Y., Du K., Cao Y., Shen X., Chen J., Luo W. Brain Res. 1359, 201–207 [DOI] [PubMed] [Google Scholar]
- 52. Li Y., Sun L., Cai T., Zhang Y., Lv S., Wang Y., Ye L. (2010) Brain Res. Bull. 81, 428–433 [DOI] [PubMed] [Google Scholar]
- 53. Uversky V. N., Li J., Fink A. L. (2001) J. Biol. Chem. 276, 44284–44296 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.