Abstract
Although essential for T cell function, the identity of the T cell receptor (TCR) “inside-out” pathway for the activation of lymphocyte function-associated antigen 1 (LFA-1) is unclear. SKAP1 (SKAP-55) is the upstream regulator needed for TCR-induced RapL-Rap1 complex formation and LFA-1 activation. In this paper, we show that SKAP1 is needed for RapL binding to membranes in a manner dependent on the PH domain of SKAP1 and the PI3K pathway. A SKAP1 PH domain-inactivating mutation (i.e. R131M) markedly impaired RapL translocation to membranes for Rap1 and LFA-1 binding and the up-regulation of LFA-1-intercellular adhesion molecule 1 (ICAM-1) binding. Further, N-terminal myr-tagged SKAP1 for membrane binding facilitated constitutive RapL membrane and Rap1 binding and effectively substituted for PI3K and TCR ligation in the activation of LFA-1 in T cells.
Keywords: Cell Adhesion, Immunology, Integrin, Lymphocyte, PI3K, Protein Translocation, LFA-1, SKAP1, T Cells, Inside-out Signaling
Introduction
Integrins regulate T cell migration to sites of inflammation, movement in lymph nodes, and conjugate formation between T cells and antigen-presenting cells (1–3). They comprise as many as 20 αβ heterodimers that are activated by changes in conformation and clustering (2, 3). T cell function is primarily regulated by the β2 family members, such as lymphocyte function-associated antigen (LFA-1,2 αL (CD11a)/β2 (CD18)) and α4 integrins such as α4β1 (Very Late Antigen-4 (VLA-4)). Binding to intercellular adhesion molecule 1 (ICAM-1) is mediated by the LFA-1 α and β subunits that interact to create a headpiece in the β subunit, termed the I-like domain (4). LFA-1 binds to ICAM-1 and ICAM-2 on major histocompatibility complex bearing antigen-presenting cells (5).
While in low-affinity conformation on resting T-cells, TCR and chemokine receptor ligation generates “inside-out” signals that activate integrins (3, 6, 7). Generic upstream regulators of the pathway include the CD4- and CD8-p56lck complexes (8–10), IL-2-inducible T cell kinase (11), the guanine nucleotide exchange factor Vav-1 (12), PI3K (13), and GTP binding proteins such as Rac/Rho (14).
Despite this, a major challenge has been to identify the direct downstream mediators of the pathway. Prime candidates include the GTP binding protein Rap1 (15, 16) and its binding partner RapL (regulator of cell adhesion and polarization enriched in lymphoid tissues) and RIAM (Rap1-interacting adaptor molecule) (17, 18). RapL (MAXP1, NORE1B, RASSF3) is an immune cell-specific isoform of the RASSF5 (Ras association domain family 5) family. It has an unique N terminus followed by a Ras-associating domain (RA) and C-terminal coiled-coil (Sav/RassF/Hpo domain (SARAH)) domain (17). The RA domain binds GTP-bound active Rap1, whereas the SARAH domain binds the coiled-coil domain of the serine kinase macrophage-stimulating 1 (MST-1) (17, 18). RapL binds directly to the LFA-1 α chain cytoplasmic tail (17, 18).
At the same time, immune cell-specific adaptor proteins SLP-76 (76-kDa src homology 2 domain-containing leukocyte phosphoprotein) (19), ADAP (adhesion and degranulation-promoting adaptor protein) (HUGO official designation: Fyb) (20–25) and SKAP1 (HUGO official designation: src kinase-associated phosphoprotein 1, also SKAP-55, src kinase-associated phosphoprotein 55) are needed for T cell adhesion (26–29). SLP-76 binds Vav1 as mediated by ZAP-70 phosphorylation (30, 31), whereas ADAP is comprised of a unique NH2 terminus, an internal SH3 domain, two nuclear localization sequences, and a C-terminal SH3-like domain (20, 32). Src kinase p59fynT phosphorylates two YDDV motifs on ADAP that are needed for SLP-76 SH2 domain binding (33, 34). Mutation of the YDDV sites reduces LFA-1-ICAM1 adhesion, T cell-antigen-presenting cell conjugate formation and formation of the peripheral supramolecular activation complex (26, 35). At the same time, ADAP forms a complex with SKAP1, an adaptor with a unique NH2-terminal region followed by a PH domain and a carboxy-terminal SH3-like domain (36–38). The complex is mediated by SKAP1 SH3 domain binding to a proline residue in the ADAP and, to a lesser extent, the ADAP-SH3-like domain binds to unique sequences in SKAP1 (37–39). One key function of ADAP is to protect SKAP1 from proteolytic degradation (40). Retroviral transduction and shRNA knockdown first implicated SKAP1 in adhesion (21, 22, 27, 41), whereas Skap1−/− mice possess T cells with defective β and β2 integrin function at levels similar to that observed in Adap−/− (Fyb−/−)-deficient T cells (42). Unlike with Adap−/− T cells, which show the concurrent loss of SKAP1 (23, 24), Skap1−/− T cells retain ADAP expression (42). This phenotype pointed to SKAP1 as the effecter of LFA-1 activation in the ADAP-SKAP1 module.
Despite the initial evidence for separate pathways, a recent study by us demonstrated that SKAP1 and Rap1-RapL are interconnected in the context of TCR-induced inside-out signaling (43). SKAP1 was found to act as the upstream regulator of RapL-Rap1 complex formation on the basis of the observation that RapL-Rap1 complexes fail to form in the absence of SKAP1 (43). SKAP1 also binds directly to the coiled-coiled SARAH domain of RapL and can compete for MST1 binding to the same region (18, 43). Further, a RapL SARAH domain mutation that disrupts SKAP1 binding without affecting MST-1 impaired T-cell binding to ICAM1 on dendritic cells and reversed the ability of RapL to reduce T cell motility (i.e. slowing) in lymph nodes (34). Further, the SKAP1 pathway appears preferentially coupled to TCR inside-out signaling because Skap1−/− T-cells show only a mild loss of migration to chemokines such as CXCL12 (44).
Despite these advances, the manner by which SKAP1 regulates Rap1-RapL complex formation and its connection to the PI3K pathway has been unclear. In this paper, we show that SKAP1 is needed for RapL binding to membranes in a manner dependent on the PH domain of SKAP1 and the PI3K pathway.
EXPERIMENTAL PROCEDURES
Cells and Antibodies
Primary T cells and Jurkat cells were cultured in RPMI 1640 medium with 10% (v/v) fetal calf serum and 1% (w/v) penicillin/streptomycin. Murine hybridoma T8.1-expressing TCR specific for Ttox (830–843) was a gift of Professor O. Acuto, Oxford University. Transfection was performed by electroporation (Bio-Rad). Anti-SKAP1 (BD Transduction Laboratories), anti-V5 (Invitrogen), anti-Rap1 and anti-p-glycogen synthase kinase 3 (GSK3) (Cell Signaling Technology, Inc.), anti-RapL (GenWay Biotech, Inc.), anti-FLAG and anti-β-actin (Sigma), anti-GFP (Santa Cruz Biotechnology, Inc.), anti-human CD3 (American Type Culture Collection), anti-mouse CD3 (2C11, hamster anti-mouse CD3), and anti-CD18 (anti-LFA-1) (Epitomics, Inc.). Wortmannin and LY294002 (Cell Signaling Technology, Inc.) and anti-murine ICAM1-FC was purchased from R&D Systems (MN).
Generation of Plasmids and Mutagenesis
Full-length human SKAP1 cDNA were cloned into the pSRa expression vector and in-frame with the NH2 terminus of the GFP gene (Promega Corp.) and in the pcDNA 3-FLAG vector (Invitrogen). Human RapL was cloned into the pcDNA3.1-V5 expression vector (Invitrogen). The SKAP1-R131M mutant and the myr-tagged version were generated by site-directed mutagenesis (Stratagene).
Immunoprecipitation Blotting
Precipitation was conducted by incubation of the lysate with the antibody for 1 h at 4 °C, followed by incubation with 30 μl of protein G-Sepharose beads (10% w/v) for 1 h at 4 °C. Immunoprecipitates were washed three times with ice-cold lysis buffer and subjected to SDS-PAGE. For blotting, precipitates were separated by SDS-PAGE and transferred onto nitrocellulose filters (Schleicher and Schuell). Bound antibody was revealed with horseradish peroxidase-conjugated rabbit anti-mouse antibody using enhanced chemiluminescence (ECL, Amersham Biosciences). For purification of membrane fractions, Jurkat or primary T cells were sheared in hypotonic buffer and the nuclei removed by low-speed centrifugation (1500 rpm, 10 min), and the supernatant was recentrifuged at high speed (25,000 rpm) for 1 h. The cytosolic fraction comprised the supernatant, whereas membranes remained in the pellet.
Integrin Adhesion Assay
For ICAM-1 binding, flat-bottomed 96-well plates were coated with 4 μg/ml murine ICAM-1 human Fc in PBS overnight at 4 °C, washed with RPMI medium, and blocked with 2.5% BSA in PBS for 1 h at 37 °C. Transfected T8.1 hybridoma cells were stimulated by incubation with 5 μg/ml anti-CD3 (mAb 2C11) followed by cross-linking with 2.5 μg/ml of goat anti-hamster IgG for 30 min at 37 °C. Stimulated cells (1–2 × 105 cells/well) were added to the murine ICAM-1-Fc-coated plates. Plates were incubated for 30 min at 37 °C. Nonadherent cells were removed by washing. The number of adherent cells were counted.
RESULTS
SKAP1 Binding and RapL Translocation to Membranes Is PH Domain-dependent
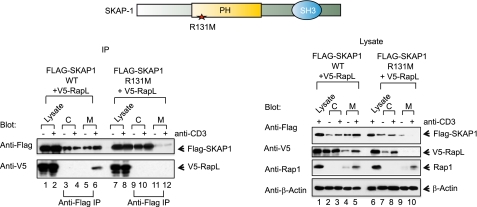
To test for the role of the SKAP1 PH domain in the formation of the SKAP1-RapL-Rap1 complex, Flag-tagged SKAP1 WT and a mutant with a PH domain inactivating mutation at 131 (i.e. R131M) were generated and expressed in Jurkat cells with V5-tagged RapL (Fig. 1). Cells were left untreated or ligated with anti-CD3 for 5 min. Anti-FLAG SKAP1 readily coprecipitated SKAP1 from membranes of resting and anti-CD3-ligated cells (Fig. 1, left panel, lanes 5 and 6). The presence of SKAP1 increased with anti-CD3 ligation (Fig. 1, left panel, lane 6 versus lane 5). By contrast, markedly reduced amounts of SKAP1 R131M were precipitated from resting and anti-CD3-ligated cells (Fig. 1, left panel, lanes 11 and 12). A slight increase in R131M was occasionally observed in response to anti-CD3 (Fig. 1, left panel, lane 12 versus lane 11). However, this occurred at levels far lower than observed with SKAP1 WT (i.e. < 10%). Similarly, anti-SKAP1 coprecipitated RapL from membranes of anti-CD3-ligated cells (Fig. 1, left panel, lane 6) but not resting cells (lane 5) or cells transfected with R131M (lanes 11 and 12). As a control, the blotting of solubilized cytosolic and membrane fractions showed that anti-CD3 induced an increase in the presence of SKAP1 and RapL in the membrane fraction of SKAP1 WT (Fig. 1, right panel, lane 5 and 6) but not in R131M-transfected cells (lanes 9 and 10). Further, expression of R131M prevented the appearance of RapL in the membrane fraction (Fig. 1, right panel, lanes 9 and 10 versus lanes 4 and 5). In this manner, R131M acted as a dominant negative in overriding the positive effect of endogenous SKAP1 on the translocation of RapL to the membranes.
FIGURE 1.
SKAP1 PH domain mediates anti-CD3 induced SKAP1 and RapL localization to membranes. FLAG-tagged SKAP1 WT or the mutant form of SKAP1 at R131M were coexpressed in Jurkat cells with V5-tagged RapL. Cells were either left untreated or ligated to anti-CD3 for 5 min prior to precipitations (IP) with anti-FLAG followed by blotting with anti-FLAG (upper panel) or anti-V5 (lower panel). Left panel, shown are FLAG-SKAP1-WT and V5-RapL (lanes 1–6) and FLAG-SKAP1-R131M and V5-RapL (lanes 7–12). Right panel, lysates transfected with FLAG-SKAP1 WT or FLAG-SKAP1 R131M and V5-RapL were blotted with anti-FLAG, anti-V5, anti-Rap1, and anti-β-actin.
Expression of R131M had much less effect on the localization of Rap1 to the membrane fraction (Fig. 1, right panel). Anti-CD3 increased levels of Rap1 in the membrane fraction in SKAP1 WT and R131M-expressing cells (Fig. 1, right panel, lanes 4 and 5 and lanes 9 and 10). The increase in R131M cells was occasionally observed to be slightly less than in WT SKAP1-expressing cells. As a control, actin did not change in the fractions from transfected cells (Fig. 1, right panel). These observations showed that SKAP1 membrane localization in response to anti-CD3 is PH domain-dependent and, secondly, that RapL membrane localization is dependent on SKAP1 membrane binding and its PH domain.
SKAP1 Binding and RapL Translocation to Membranes Is PI3K-dependent
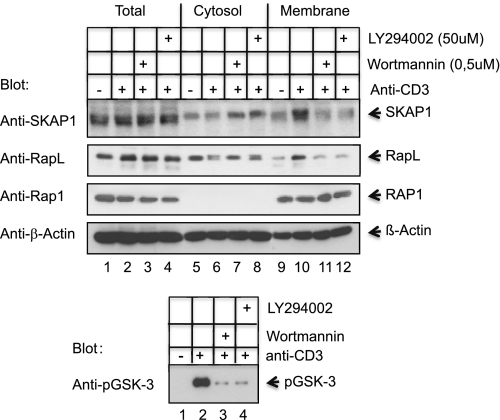
PH domains bind to D-3 lipids generated by PI3K (45). To confirm that PI3K activity was needed for SKAP1 translocation, the effect of wortmannin and LY294002, inhibitors of PI3K, was assessed (Fig. 2). Mouse splenocytes were incubated with the PI3K inhibitors for 30 min prior to ligation with anti-CD3 followed by isolation of cytosolic and membrane fractions and immunoblotting using the designated antibodies. The anti-CD3-induced translocation of WT SKAP1 to the membrane fraction was blocked by each inhibitor (Fig. 2, upper panel, lanes 11 and 12 versus lane 10). There was a concurrent increase in its presence in the cytosolic fraction (Fig. 2, upper panel, lanes 7 and 8 versus lane 6). Significantly, again, this effect on SKAP1 was mirrored by impaired RapL translocation to the membranes (Fig. 2, upper panel, lanes 11 and 12 versus lane 10). Further, consistent with previous results, the presence of Rap1 was unaffected by inhibition of PI3K (Fig. 2, upper panel, lanes 9–12). Inhibition of PI3K was confirmed by inhibition of protein kinase B (PKB)/AKT phosphorylation of GSK3 (Fig. 2, lower panel, lanes 3 and 4 versus lane 2). These results confirmed the importance of PI3K activity in the translocation of SKAP1 and RapL to the membranes of cells.
FIGURE 2.
SKAP1 PH domain membrane association and regulation of RapL is PI3K-dependent. T cell splenocytes were ligated with anti-CD3 for 5 min in the presence or absence of wortmannin or LY294002 (i.e. 30-min preincubation), followed by separation into cytosolic and membrane fractions. Immunoblotting was conducted using the designated antibodies. Upper panel, lysates were blotted with anti-SKAP1, anti-RapL, anti-Rap1, and anti-β-actin. Lower panel, control for LY294002 and wortmannin inhibition of PI3K. Splenocytes were ligated as described above, followed by immunoblotting with anti-pGSK3.
SKAP1 PH Domain Is Needed for LFA-1 Binding and TCR-induced ICAM-1 Adhesion
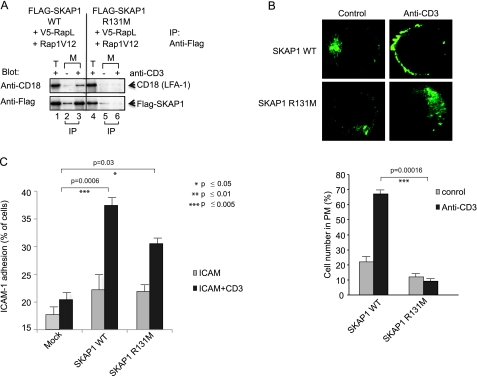
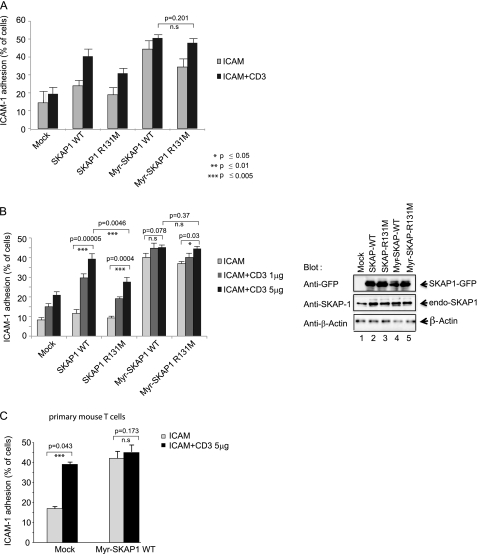
We next asked whether the inability of the R131M mutant to translocate to the membranes affected binding to CD18 (Fig. 3A). We showed previously that SKAP1 forms a trimeric complex with RapL and Rap1 that binds to LFA-1 (43). Jurkat cells were transfected with FLAG-SKAP1, V5-RapL, and Rap1V12 and cross-linked with anti-CD3 followed by membrane isolation and immunoprecipitation with anti-FLAG. Blotting was then conducted using anti-CD18 (Fig. 3A, upper panel) or anti-FLAG (lower panel). Although anti-FLAG-SKAP1 precipitated CD18 in a CD3 ligation-dependent manner (Fig. 3, lane 3 versus lane 2), it failed to precipitate CD18 from cells transfected with the R131M mutant (upper panel, lanes 6 and 5). Further, the absence of R131M translocation to the cell surface for binding to LFA-1 was seen by fluorescence microscopy (Fig. 3B, left panels). Although anti-CD3 induced the localization of GFP-SKAP1 WT to the periphery of cells, it failed to induce the translocation of the GFP-R131M mutant, which remained in the cytosol. The lower histogram shows the relative difference in the cell number with cell surface staining.
FIGURE 3.
SKAP1 PH domain mutant R131M shows reduced CD18 binding and LFA-1 adhesion. A, Jurkat cells were transfected with FLAG-tagged SKAP1 WT or FLAG-tagged SKAP1 R131M plus V5-RapL and Rap1V12, followed by precipitation with anti-FLAG and blotting with anti-CD18 (upper panel) or anti-FLAG (lower panel). B, Jurkat cells transfected with GFP-SKAP1 or GFP-SKAP1 R131M were imaged for membrane localization (upper panels). The lower panels shows the percent cell number with SKAP1 in the plasma membrane. C, T8.1 cells transfected with SKAP1 WT or SKAP1 R131M were assessed for binding to ICAM-1 on plates as described under “Experimental Procedures.” Error bars represent S.D. For paired T-tests, experimental groups were compared with their respective groups. Significant differences (p ≤ 0.05) are indicated with asterisks.
To test for TCR-induced inside out signaling, the adhesion of T cells to ICAM-1 on plates was tested (Fig. 3C). For this, T8.1 cells were transfected and plated on ICAM-1-coated plates followed by the addition of anti-CD3, as described previously (26, 27, 42, 43, 44). Although suboptimal anti-CD3 induced a marginal increase of the binding of cells to ICAM-1 on plates in mock-transfected T8.1 T cells (i.e. from 17 to 20% of cells), transfected WT SKAP1 induced a major increase of binding involving 37% of cells (p = 0.0006). By contrast, the R131M mutant, although providing some increase in adhesion, was impaired relative to WT SKAP1 (38 versus 30%). These observations confirm that the PH domain of SKAP1 mediates localization and promotion of RapL to membranes and is needed for optimal augmentation of TCR-induced LFA-1 adhesion to ICAM-1.
Myr-SKAP1 Substitutes for TCR in the Induction of Constitutively Enhanced T cell Adhesion
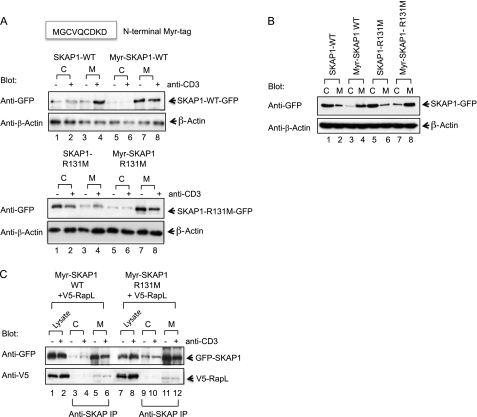
The next question was whether the involvement of the PH domain was strictly due to its role in membrane localization and whether constitutive membrane localization could overcome the need for the PH domain. To assess this, a MGCVQCDKD-myristoylation tag was fused to the N terminus of WT SKAP1, and the R131M mutant was followed by expression in T8.1 T cells. Myr-SKAP1 was found constitutively associated with the membrane fraction of resting and anti-CD3-ligated cells (Fig. 4A, upper panel, lanes 7 and 8). The level of binding in resting cells was comparable for the WT SKAP1 induced by anti-CD3 (Fig. 4A, upper panel, lanes 7 and 8 versus lane 4), and the level of myr-SKAP1 did not increase further with anti-CD3 ligation (lane 8 versus lane 7). Similarly, myr-SKAP1 R131M constitutively associated with membranes (Fig. 4A, lower panel, lanes 7 and 8). The level was much greater than the small increase in anti-CD3-induced SKAP1 R131M (Fig. 4A, lower panel, lane 4 versus lane 3) (i.e. a reduction of 60%).
FIGURE 4.
Myr-SKAP1 binds to the membrane and RapL in an anti-CD3-independent manner. A, T8.1 T cells were transfected with SKAP1 WT or myr-SKAP1 (upper panel) or SKAP1 mutant R131M or myr-SKAP1 mutant R131M (lower panel) and assessed for association with the cytosol or membranes. Lysates were blotted with anti-GFP and anti-β-actin (lanes 1–8). B, 293 T cells were transfected with SKAP1 WT or myr-SKAP1 and assessed for association with the cytosol or membranes. Lysates were blotted with anti-GFP and anti-β-actin (lanes 1–8). C, T8.1 T cells were transfected with the myr-SKAP1 WT or myr-SKAP1 mutant R131M plus V5-RapL. Anti-SKAP1 was used to precipitate GFP-SKAP1 (upper panel) or to coprecipitate V5-RapL (lower panel). Blotting was conducted with anti-GFP (upper panel) and anti-V5 (lower panel).
Similar results were obtained in 293T cells, where both myr-SKAP1 WT and the myr-SKAP1 R131M mutant constitutively associated with membranes (Fig. 4B, lanes 4 and 8). SKAP1 WT and SKAP1 R131M were found mostly in the cytosolic fraction (Fig. 4B, lanes 1 and 5). In the case of T cells, this constitutive binding of myr-SKAP1 WT and myr-SKAP1 R131M supported coprecipitation of cotransfected RapL from the membrane fraction of resting and anti-CD3-ligated cells (Fig. 4C, lower panel, lanes 5 and 6 and lanes 11 and 12). These data indicate that myr-tagged SKAP1 can bypass the need for the PH domain in RapL and membrane binding.
To test for the effect of myr-SKAP1 on adhesion, transfected T8.1 T cells were assessed for binding to ICAM-1 on plates (Fig. 5A). Myr-SKAP1 WT showed constitutively elevated ICAM-1 adhesion, effectively bypassing the need for anti-CD3 (i.e. 45–50%). Myr-SKAP1 R131M showed similarly high levels of constitutive binding (i.e. 35%). A small additional increase was observed with anti-CD3 (i.e. 47%). A titration of 1 versus 5 μg/ml anti-CD3 showed that myr-SKAP1 or myr-SKAP1-R131M induced the same level of adhesion in the absence of anti-CD3 that was normally induced with anti-CD3 (Fig. 5B). Immunoblotting showed the level of expression of the transfected and endogenous SKAP1 proteins (Fig. 5B, right panel).
FIGURE 5.
Myr-SKAP1 induced LFA-1 adhesion in an anti-CD3-independent manner. A, T8.1 cells were transfected with either mock, SKAP1 WT, SKAP1 R131M, myr-SKAP1-WT, or myr-SKAP1-R131M and assessed for binding to ICAM-1 on plates as described under “Experimental Procedures.” B, as above, but with anti-CD3 titration (left panel). Lysates of transfected T8.1 cells were blotted with anti-GFP, anti-SKAP1, and anti-β-actin (right panel). C, primary mouse T cells were transfected with either mock or myr-SKAP1 WT and assessed for binding to ICAM-1 on plates. Error bars represent S.D. For paired t tests, experimental groups were compared with their respective groups. Significant differences (p ≤ 0.05) are indicated with asterisks.
Identical results were obtained with primary murine T-cells (Fig. 5C). Preactivated murine spleen T cells were rested for 12 h prior to transfection with myr-SKAP1 wild type. Anti-CD3 induced an increase in binding to ICAM-1 on plates (i.e. 15 to 38%). Myr-SKAP1 transfected T cells showed binding to ICAM-1 at the same level as with anti-CD3 ligation of non-transfected cells (i.e. 41 versus 38%). Anti-CD3 was not able to increase adhesion more than supported by transfection of cells with myr-SKAP1 (i.e. 43 versus 41%, p = 0.173). These data show that myr-SKAP1 can substitute for anti-CD3 in the activation of T cell adhesion.
SKAP1 Bypasses the Need for PI3K Regulation of T Cell Adhesion
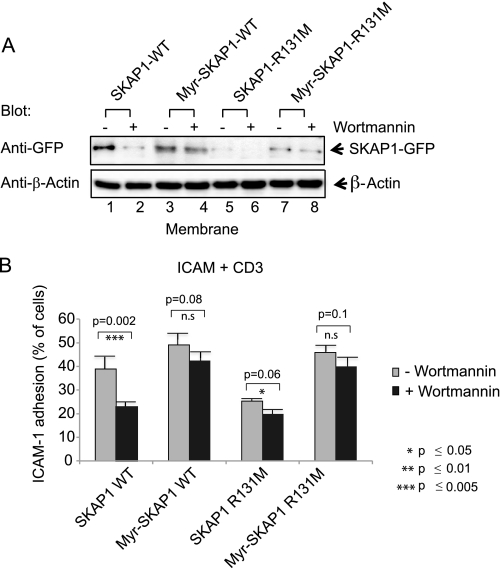
Previous studies have implicated PI3K in the TCR induction of LFA-1 adhesion (13, 46). Given the PH domain binding to D-3 lipids and the ability of myr-SKAP1 to replace the TCR signal for adhesion, we next examined whether the SKAP1-RapL connection could account for PI3K regulation of T cell adhesion (Fig. 6). T8.1 T cells were incubated with the PI3K inhibitor wortmannin for 30 min prior to anti-CD3 ligation and isolation of membranes from transfected cells. This was followed by blotting with anti-GFP for detection of SKAP1. Although the movement of WT SKAP1 to membranes was inhibited by wortmannin (Fig. 6A, lane 2 versus lane 1), the presence of myr-SKAP1 WT (lanes 3 and 4) and myr-SKAP1-R131M (lanes 7 and 8) was unaffected (upper panel). SKAP1-R131M failed to associate with membranes in the presence or absence of wortmannin (Fig. 6A, lanes 5 and 6). These observations confirmed that PI3K activity was not needed for myr-SKAP1 binding with the membrane fraction.
FIGURE 6.
Myr-SKAP1 overrides the need for PI3K in the regulation of LFA-1 adhesion. T8.1 cells were transfected with SKAP1-WT, myr-SKAP1 WT, SKAP1-R131M, or myr-SKAP1-R131M; ligated with anti-CD3 in the presence or absence of wortmannin; followed by an assessment of membrane binding by blotting (A) or binding to ICAM-1 on plates as described under “Experimental Procedures” (B). Lysates were blotted with anti-GFP and anti-β-actin (A, lanes 1–8). Error bars represent S.D. For paired t tests, experimental groups were compared with their respective groups. Significant differences (p ≤ 0.05) are indicated with asterisks.
In terms of ICAM-1 binding, wortmannin inhibited the binding to ICAM-1 induced by the combination of anti-CD3 with SKAP1-WT expression (Fig. 6B). The level of binding was similar to that observed with SKAP1-R131M expression with or without wortmannin (i.e. ∼20%). By contrast, the expression of myr-SKAP1 WT or myr-SKAP1-R131M induced high levels of T cell binding to ICAM-1 at 40–50% of the population. This level of binding was observed in the presence or absence of wortmannin. Further, the level was similar or slightly higher than that observed with anti-CD3 plus SKAP1 WT in the absence of the drug. These findings show that SKAP1 induces a major increase in LFA-1 function when constitutively bound to membranes and effectively overrides the need for PI3K in T cells.
DISCUSSION
We previously showed that the immune cell adaptor SKAP1 is an essential upstream regulator of RapL-Rap1 complex formation induced by ligation of the TCR complex (43). Despite this, the detailed mechanism by which SKAP1 regulates complex formation has been unclear. In this study, we have shown that the SKAP1 PH domain is needed for the translocation of itself and RapL to membranes of T cells in response to anti-CD3 ligation. This, in turn, was needed for optimal binding to Rap1 and activation of LFA-1. Myr-tagged SKAP1 and R131M resulted in constitutive membrane binding of SKAP1 and RapL and, remarkably, substituted for TCR ligation and PI3K in the induction of LFA-1 adhesion in T8.1 and primary T cells. These observations underscore the importance of SKAP1 in TCR and PI3K up-regulation of LFA-1 adhesion.
We showed previously that RapL-Rap1 complex formation and its binding to LFA-1 failed to occur in Skap1−/− T cells and, secondly, that TCR regulation of adhesion depended on SKAP1 N-terminal binding to the SARAH domain of RapL (43). The presence of a PH domain in SKAP1 suggested the possibility that it might respond to PI3K-generated PIP3 for membrane localization and, in the process, indirectly control the membrane localization of RapL for an interaction with Rap1 and LFA-1. The PH domain of related SKAP2 has been shown previously to bind D-3 lipids (47). We have found that SKAP1 and RapL translocate to the membrane upon TCR ligation (43) (Fig. 1). The SKAP1-R131M PH domain mutation abrogated this anti-CD3-induced translocation of SKAP1, as detected biochemically from isolated membranes (Fig. 1) and by impaired plasma membrane localization seen by immunofluorescence staining of cells (Fig. 3). Further, SKAP1-R131M also impaired the localization of RapL to the membrane fraction. Translocation was reduced by more than 80% in the presence of R131M. The remaining translocation is likely due to the presence of endogenous wild-type SKAP1 in cells.
Conversely, myr-SKAP-1 and myr-SKAP1-R131M constitutively associated with membrane binding and, concurrently, restored RapL membrane binding, confirming the essential role of SKAP1 membrane binding for the membrane localization of RapL. These observations support a model where SKAP1 acts as a shuttle for RapL translocation to membranes, such as in the case of RIAM and talin (48) or, alternatively, acts as a membrane-linked anchor for RapL binding. This latter interpretation is supported by our observation that it was difficult to detect SKAP1-RapL complexes in the cytosol prior to their appearance in the membrane. On the other hand, it is possible that these complexes are transported too rapidly to the membrane fraction for detection using biochemical approaches.
By contrast, SKAP1-R131M had far less effect on the anti-CD3-induced increase in Rap1. TCR ligation is known to activate membrane-associated Rap1 to an active GTP-bound form (15, 16). A substantial increase in membrane Rap1 was noted in the presence of SKAP1-WT and SKAP1-R131M. This is consistent with our previous finding that Rap1 is found in the membranes of cells in Skap1−/− T-cells (43). Occasionally, the level of Rap1 was reduced by some 20–30%, consistent with some effect of SKAP1 on Rap1 membrane localization (28). At the same time, anti-CD3-induced activation of Rap1 is needed for SKAP1-RapL binding to the protein, leading to the formation of the SKAP1-RapL-Rap1 complex (43). In this context, Rap1 can also bind to other proteins, such as RIAM. Distinct pools of Rap1 are likely to exist that regulate distinct events. It remains to be determined whether Rap1 is limited in expression so that its involvement in the SKAP1-RapL might compete for RIAM-talin binding and vice versa, or whether it is expressed at sufficiently high levels to accommodate the dual coordinate regulation of both sets of complexes.
The importance of SKAP1 for LFA-1 activation was further underscored by the effect of myr-SKAP1 on adhesion (Figs. 4–6). Constitutive membrane binding of myr-SKAP1 was able to substitute for TCR ligation in the up-regulation of LFA-1 adhesion (i.e. via inside-out signaling). This was achieved with only 2- to 3-fold over-expression relative to endogenous levels of SKAP1 and was observed in T8.1 and primary T cells. Expression of either myr-SKAP1-WT or myr-SKAP1-R131M induced a level of adhesion in the absence of anti-CD3 ligation that was achieved by a combination of anti-CD3 plus SKAP1-WT or anti-CD3 alone. Myr-SKAP1 was also able to associate with RapL in the membrane in an anti-CD3-independent fashion (Fig. 4C). These results show that the constitutive binding of SKAP1 to the membranes of cells can suffice to replace the anti-CD3-induced inside-out pathway for LFA-1 adhesion.
Further, our findings also demonstrate for the first time the connection between SKAP1-RapL-Rap1 and the PI3K pathway. Although inhibitors of PI3K effectively impaired TCR adhesion, expression of myr-SKAP1 simply overrode this blockade and supported normal levels of adhesion. This was a surprise, given the ability of PI3K to regulate the translocation and activity of numerous proteins with PH domains in cells. For example, one would have expected that other PH domains, such as RIAM, would be needed. This was under conditions where wortmannin inhibited more than 98% of GSK-3 phosphorylation by AKT/PKB, an accepted surrogate for PI3K activity (Fig. 2). Although it would be unrealistic to claim that SKAP1 is the sole mediator of PI3K dependence in the regulation of adhesion, these observations confirm the potent role of SKAP1 in promoting PI3K-dependent adhesion. It also begins to suggest that there may exist distinct pathways (e.g. SKAP-1-RapL-Rap1 and RIAM-talin) that can activate LFA-1 adhesion.
Further work is needed to establish the degree to which the SKAP1-RapL pathway accounts for the TCR inside-out pathway in T cells. In the context of low-intermediate strength TCR signals, our previous work has shown that the pathway accounts for 50–70% of adhesion (42). Similar levels of impaired inside-out-driven LFA-1 adhesion have also been observed in Fyb−/− (Adap−/−) T cells to peptides of differing affinities (48, 49). However, the dependence on SKAP1 and ADAP varies with the strength of the TCR signal. Higher-strength anti-CD3 signaling can override the need for SKAP1 in the up-regulation of adhesion, indicating that an alternate pathway exists (3, 42, 48). ADAP binding to SLP-76 upstream also regulates outside-in signaling for costimulation (25). Further work will be needed to establish the identity of other possible inside-out pathways coupled to the TCR complex.
Footnotes
- LFA-1
- lymphocyte function-associated antigen
- ICAM-1
- intercellular adhesion molecule 1
- RapL
- regulator of cell adhesion and polarization enriched in lymphoid tissues
- RIAM
- Rap1-interacting adaptor molecule
- ADAP
- adhesion- and degranulation-promoting adaptor protein
- SLP-76
- 76-kDa src homology 2 domain-containing leukocyte phosphoprotein
- SKAP1
- src kinase-associated phosphoprotein 1
- PH
- Pleckstrin homology domain
- TCR
- T cell receptor.
REFERENCES
- 1. Dustin M. L., Bivona T. G., Philips M. R. (2004) Nat. Immunol. 5, 363–372 [DOI] [PubMed] [Google Scholar]
- 2. Carman C. V., Springer T. A. (2003) Curr. Opin. Cell Biol. 15, 547–556 [DOI] [PubMed] [Google Scholar]
- 3. Wang H., Rudd C. E. (2008) Trends Cell Biol. 18, 486–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Takagi J., Petre B. M., Walz T., Springer T. A. (2002) Cell 110, 599–611 [DOI] [PubMed] [Google Scholar]
- 5. Kupfer A. (2006) Immunity 25, 11–13 [DOI] [PubMed] [Google Scholar]
- 6. Kinashi T. (2005) Nat. Rev. Immunol. 5, 546–559 [DOI] [PubMed] [Google Scholar]
- 7. Mazerolles F., Hauss P., Barbat C., Figdor C. G., Fischer A. (1991) Eur. J. Immunol. 21, 887–894 [DOI] [PubMed] [Google Scholar]
- 8. Rudd C. E. (1990) Immunol. Today 11, 400–406 [DOI] [PubMed] [Google Scholar]
- 9. Rudd C. E., Trevillyan J. M., Dasgupta J. D., Wong L. L., Schlossman S. F. (1988) Proc. Natl. Acad. Sci. U.S.A. 85, 5190–5194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Barber E. K., Dasgupta J. D., Schlossman S. F., Trevillyan J. M., Rudd C. E. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 3277–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Labno C. M., Lewis C. M., You D., Leung D. W., Takesono A., Kamberos N., Seth A., Finkelstein L. D., Rosen M. K., Schwartzberg P. L., Burkhardt J. K. (2003) Curr. Biol. 13, 1619–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Collins T. L., Deckert M., Altman A. (1997) Immunol. Today 18, 221–225 [DOI] [PubMed] [Google Scholar]
- 13. Shimizu Y., Mobley J. L., Finkelstein L. D., Chan A. S. (1995) J. Cell Biol. 131, 1867–1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mor A., Campi G., Du G., Zheng Y., Foster D. A., Dustin M. L., Philips M. R. (2007) Nat. Cell Biol. 9, 713–719 [DOI] [PubMed] [Google Scholar]
- 15. Bivona T. G., Wiener H. H., Ahearn I. M., Silletti J., Chiu V. K., Philips M. R. (2004) J. Cell Biol. 164, 461–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Katagiri K., Hattori M., Minato N., Irie S., Takatsu K., Kinashi T. (2000) Mol. Cell. Biol. 20, 1956–1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Katagiri K., Maeda A., Shimonaka M., Kinashi T. (2003) Nat. Immunol. 4, 741–748 [DOI] [PubMed] [Google Scholar]
- 18. Katagiri K., Imamura M., Kinashi T. (2006) Nat. Immunol. 7, 919–928 [DOI] [PubMed] [Google Scholar]
- 19. Kuhné M. R., Lin J., Yablonski D., Mollenauer M. N., Ehrlich L. I., Huppa J., Davis M. M., Weiss A. (2003) J. Immunol. 171, 860–866 [DOI] [PubMed] [Google Scholar]
- 20. da Silva A. J., Li Z., de Vera C., Canto E., Findell P., Rudd C. E. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 7493–7498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Geng L., Pfister S., Kraeft S. K., Rudd C. E. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 11527–11532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Geng L., Raab M., Rudd C. E. (1999) J. Immunol. 163, 5753–5757 [PubMed] [Google Scholar]
- 23. Griffiths E. K., Krawczyk C., Kong Y. Y., Raab M., Hyduk S. J., Bouchard D., Chan V. S., Kozieradzki I., Oliveira-Dos-Santos A. J., Wakeham A., Ohashi P. S., Cybulsky M. I., Rudd C. E., Penninger J. M. (2001) Science 293, 2260–2263 [DOI] [PubMed] [Google Scholar]
- 24. Peterson E. J., Woods M. L., Dmowski S. A., Derimanov G., Jordan M. S., Wu J. N., Myung P. S., Liu Q. H., Pribila J. T., Freedman B. D., Shimizu Y., Koretzky G. A. (2001) Science 293, 2263–2265 [DOI] [PubMed] [Google Scholar]
- 25. Wang H., Wei B., Bismuth G., Rudd C. E. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 12436–12441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang H., Moon E. Y., Azouz A., Wu X., Smith A., Schneider H., Hogg N., Rudd C. E. (2003) Nat. Immunol. 4, 366–374 [DOI] [PubMed] [Google Scholar]
- 27. Jo E. K., Wang H., Rudd C. E. (2005) J. Exp. Med. 201, 1733–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kliche S., Breitling D., Togni M., Pusch R., Heuer K., Wang X., Freund C., Kasirer-Friede A., Menasche G., Koretzky G. A., Schraven B. (2006) Mol. Cell. Biol. 26, 7130–7144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rudd C. E. (1999) Cell 96, 5–8 [DOI] [PubMed] [Google Scholar]
- 30. Bubeck Wardenburg J., Fu C., Jackman J. K., Flotow H., Wilkinson S. E., Williams D. H., Johnson R., Kong G., Chan A. C., Findell P. R. (1996) J. Biol. Chem. 271, 19641–19644 [DOI] [PubMed] [Google Scholar]
- 31. Raab M., da Silva A. J., Findell P. R., Rudd C. E. (1997) Immunity 6, 155–164 [DOI] [PubMed] [Google Scholar]
- 32. Musci M. A., Hendricks-Taylor L. R., Motto D. G., Paskind M., Kamens J., Turck C. W., Koretzky G. A. (1997) J. Biol. Chem. 272, 11674–11677 [DOI] [PubMed] [Google Scholar]
- 33. Raab M., Kang H., da Silva A., Zhu X., Rudd C. E. (1999) J. Biol. Chem. 274, 21170–21179 [DOI] [PubMed] [Google Scholar]
- 34. Veale M., Raab M., Li Z., da Silva A. J., Kraeft S. K., Weremowicz S., Morton C. C., Rudd C. E. (1999) J. Biol. Chem. 274, 28427–28435 [DOI] [PubMed] [Google Scholar]
- 35. Wang H., McCann F. E., Gordan J. D., Wu X., Raab M., Malik T. H., Davis D. M., Rudd C. E. (2004) J. Exp. Med. 200, 1063–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Marie-Cardine A., Bruyns E., Eckerskorn C., Kirchgessner H., Meuer S. C., Schraven B. (1997) J. Biol. Chem. 272, 16077–16080 [DOI] [PubMed] [Google Scholar]
- 37. Liu J., Kang H., Raab M., da Silva A. J., Kraeft S. K., Rudd C. E. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 8779–8784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kang H., Freund C., Duke-Cohan J. S., Musacchio A., Wagner G., Rudd C. E. (2000) EMBO J. 19, 2889–2899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Marie-Cardine A., Hendricks-Taylor L. R., Boerth N. J., Zhao H., Schraven B., Koretzky G. A. (1998) J. Biol. Chem. 273, 25789–25795 [DOI] [PubMed] [Google Scholar]
- 40. Huang Y., Norton D. D., Precht P., Martindale J. L., Burkhardt J. K., Wange R. L. (2005) J. Biol. Chem. 280, 23576–23583 [DOI] [PubMed] [Google Scholar]
- 41. Duke-Cohan J. S., Kang H., Liu H., Rudd C. E. (2006) J. Biol. Chem. 281, 13743–13750 [DOI] [PubMed] [Google Scholar]
- 42. Wang H., Liu H., Lu Y., Lovatt M., Wei B., Rudd C. E. (2007) Mol. Cell. Biol. 27, 6863–6875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Raab M., Wang H., Lu Y., Smith X., Wu Z., Strebhardt K., Ladbury J. E., Rudd C. E. (2010) Immunity 32, 541–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang H., Lu Y., Rudd C. E. (2010) Immunol. Lett. 128, 148–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lee J. Y., Engelman J. A., Cantley L. C. (2007) Science 317, 206–207 [DOI] [PubMed] [Google Scholar]
- 46. Zell T., Hunt S. W., 3rd, Mobley J. L., Finkelstein L. D., Shimizu Y. (1996) J. Immunol. 156, 883–886 [PubMed] [Google Scholar]
- 47. Swanson K. D., Tang Y., Ceccarelli D. F., Poy F., Sliwa J. P., Neel B. G., Eck M. J. (2008) Cell 32, 564–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lee H. S., Lim C. J., Puzon-McLaughlin W., Shattil S. J., Ginsberg M. H. (2009) J. Biol. Chem. 284, 5119–5127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wu J. N., Gheith S., Bezman N. A., Liu Q. H., Fostel L. V., Swanson A. M., Freedman B. D., Koretzky G. A., Peterson E. J. (2006) J. Immunol. 176, 6681–6689 [DOI] [PubMed] [Google Scholar]