Abstract
HSP100 proteins are molecular chaperones involved in protein quality control. They assist in protein (un)folding, prevent aggregation, and are thought to participate in precursor translocation across membranes. Caseinolytic proteins ClpC and ClpD from plant chloroplasts belong to the HSP100 family. Their role has hitherto been investigated by means of physiological studies and reverse genetics. In the present work, we employed an in vitro approach to delve into the structural and functional characteristics of ClpC2 and ClpD from Arabidopsis thaliana (AtClpC2 and AtClpD). They were expressed in Escherichia coli and purified to near-homogeneity. The proteins were detected mainly as dimers in solution, and, upon addition of ATP, the formation of hexamers was observed. Both proteins exhibited basal ATPase activity (Km, 1.42 mm, Vmax, 0.62 nmol/(min × μg) for AtClpC2 and Km ∼19.80 mm, Vmax ∼0.19 nmol/(min × μg) for AtClpD). They were able to reactivate the activity of heat-denatured luciferase (∼40% for AtClpC2 and ∼20% for AtClpD). The Clp proteins tightly bound a fusion protein containing a model transit peptide. This interaction was detected by binding assays, where the chaperones were selectively trapped by the transit peptide-containing fusion, immobilized on glutathione-agarose beads. Association of HSP100 proteins to import complexes with a bound transit peptide-containing fusion was also observed in intact chloroplasts. The presented data are useful to understand protein quality control and protein import into chloroplasts in plants.
Keywords: ATPases, Chloroplast, Heat Shock Protein, Protein Purification, Protein Translocation, Chaperones, Hsp100, Protein Import, Transit Peptide
Introduction
Molecular chaperones are a diverse group of proteins involved in a plethora of biological activities. They usually target hydrophobic stretches present in unfolded polypeptides, which arise as a consequence of protein synthesis in the ribosome (1), protein unfolding by stress insults (2), or import across membranes (3–5). By masking these aggregation-prone regions, molecular chaperones prevent the wasteful formation of inactive protein aggregates (6). Many chaperones can act as independent units and/or interact with proteases to degrade irreversibly damaged proteins (7, 8). Other specialized types of chaperones can disassemble unfolded polypeptides present in inclusion bodies or quaternary protein structures (9). Overall, they are key players in regulating the protein quality control system.
The Clp/HSP100 family of molecular chaperones is found in all kingdoms of life. There are two classes of HSP100 proteins according to the presence of two (Class I) or one (Class II) nucleotide-binding domains in their structure (10). Class I is further subdivided (ClpA-E and L) based on the length and amino acid composition of the spacer region between the nucleotide binding domains. ClpA from Escherichia coli (EcClpA)3 was the first member to be discovered and was shown to regulate the activity of the ClpP protease (11). ClpB from bacteria and eukaryotes can disassemble protein aggregates and plays a pivotal role in thermotolerance (12). ClpE and ClpL are found in some Gram-positive bacteria. ClpC is found in Gram-positive bacteria and photobionts, while ClpD is restricted to plants (10).
In A. thaliana, members of the ClpC (AtClpC1 and AtClpC2), ClpD (AtClpD) and ClpB (AtClpB3) families are located in chloroplasts, along with five different ClpP proteases (AtClpP1,3–6), four ClpP-like proteins with no protease activity (ClpR1–4) and three adaptor/modulator proteins (AtClpT1/2 and AtClpS) (13). The current knowledge about their function came primarily from physiological studies of wild-type plants and insertional mutants in the corresponding genes. AtClpC1/2 and AtClpD are constitutively expressed and are highly conserved among different plant species (14). While insertional mutants in the Atclpc2 gene have no obvious phenotype, ΔAtclpc1 plants showed retarded growth and moderate chlorosis throughout all developmental stages (15–17). Double mutants are not viable (18). This evidence indicates that AtClpC1 and 2 are housekeeping chaperones, which may regulate the activity of the proteolytic ClpP core. Much less is known about AtClpD, apart from its expression profile (19, 20).
Interestingly, precursor import into chloroplasts was also impaired in the ΔAtclpc1 mutant (16, 17), although a third report showed otherwise (15). Additionally, in Pisum sativum, a fraction of ClpC is localized in the stromal side of the chloroplast inner membrane and interacts with the import machinery when a precursor is being translocated (21). This interaction stimulates its ATPase activity (22), which may provide the necessary energy for translocation. Based on this data, current models of protein import into chloroplasts place HSP100 proteins as components of the molecular motor driving protein import. Yet, researchers recently alerted that evidence showing HSP100 proteins having direct contacts with precursors was still lacking (23, 24).
Despite ongoing efforts to characterize the purified members of the Clp complex from lower organisms, none of its counterparts from plants has been studied at the molecular level. In the present work, we were able to clone, express in E. coli, and purify AtClpC2 and AtClpD, which allowed us to analyze their structural and functional properties. As established by gel filtration chromatography, both proteins were recovered mainly as dimers, and, upon addition of ATP; the formation of hexamers was detected. They presented ATP-hydrolyzing activity without the need of any adaptor protein. We also established their chaperone activity by luciferase renaturation assays. Finally, AtClpC2 and AtClpD interacted with the transit peptide (TP) of ferredoxin-NADP+ reductase.
EXPERIMENTAL PROCEDURES
Plasmid Construction
AtClpC1, AtClpC2, and AtClpD cDNAs were obtained from the RIKEN DNA Bank (pda08715, 05508, and 00833, respectively). The cDNAs were used as templates for amplification using Pfu DNA polymerase. In all cases, the region coding for the mature protein (i.e. without the TP) was amplified. The primers contained restriction sites that allowed directional cloning into the multiple cloning site of plasmid pET28a. These were as follows: for AtClpC1, 5′-GGT GAA AGC AAT GCA TAT GAG ATT TAC GGA G-3′ and 5′-CCC CCA TGT ATC TGA ATT CAT AAC CTT CTC TT-3′ (restriction sites for NdeI and EcoRI, respectively, are underlined); for AtClpC2, 5′-CCA AAG CTA GCT TCG AGC GTT TTA CCG AGA AAG-3′; and 5′-AAT CAT AAG CGG CCG CAT AGT CAA TTC AAT AAC AC-3′ (restriction sites for NheI and NcoI, respectively, are underlined); for AtClpD, 5′-TTT CAG CGG CTA GCG AGC GGT TCA CCG-3′ and 5′-TAA GTA TAA GAC GCG GCC GCA TAA TCA AAG AAT CT-3′(restriction sites for NheI and NotI, respectively, are underlined). The pET28a plasmid allows the expression of the proteins as fusions to an N-terminal hexahistidine tag followed by a thrombin cleavage site. After digestion, AtClpC1 would retain four amino acids from the plasmid, while AtClpC2 and AtClpD would both keep six. All constructs were checked by DNA sequencing.
Expression and Purification of AtClpC2 and AtClpD
Each resulting plasmid was transformed into the E. coli BL21(DE3)pLysS strain. An overnight culture was used to inoculate 1 liter of Luria-Bertani medium (+ 50 μg/ml kanamycin, 25 μg/ml chloramphenicol, and 10 mm MgCl2) distributed in five 1-liter flasks. The cultures were grown at 37 °C until an A600 of 0.6–0.7. The temperature was lowered to 16 °C and after 1 h; IPTG was added to a final concentration of 0.5 mm. After 16 h of induction, cells were recovered by centrifugation and resuspended in cold lysis buffer (50 mm Tris-HCl, pH 8.0, 400 mm NaCl, 10 mm MgCl2, 10 mm imidazole, 1 mm PMSF, 1 mm benzamidine, 10% v/v glycerol) at a 30:1 ratio (ml culture:ml lysis buffer). After this point, every subsequent step was performed at 4 °C. Cells were lysed by three passages through a French Press and the soluble fraction was recovered by centrifugation (30,000 × g, 1 h). The supernatant was supplemented with 500 μl of Ni2+-NTA-agarose resin and incubated in batch for 30 min. The mixture was transferred to a column and washed with lysis buffer (40 column volumes). Lysis buffer + 200 mm imidazole was used to elute the recombinant proteins from the column in 500-μl fractions. These fractions were desalted according to the method of gel filtration/centrifugation of Penefsky (25) in Sephadex G-75 columns equilibrated with P buffer (50 mm Tris-HCl, pH 8.0, 100 mm NaCl, 10 mm MgCl2, 10% v/v glycerol). To remove the polyhistidine tag, 1 mg of recombinant protein was incubated in the presence of 5 units of thrombin at 12 °C for 16 h. The samples were then loaded onto a Ni2+-NTA-agarose column to remove the tags and undigested protein. The protein solution was further purified by gel filtration/centrifugation as described. The chaperones were detected by 10% SDS-polyacrylamide gels stained with Coomassie Brilliant Blue. Even though this procedure was often sufficient to achieve a level of 95% purity, in cases where this criterion was not met, the samples were subjected to gel filtration chromatography as described below. Protein concentration was determined using the Bradford method (26) and BSA as standard protein.
Gel Filtration Chromatography
Purified samples were applied onto a Superdex-200 10/30 GL column attached to an ÄKTA Prime chromatography system. The runs were performed at a flow rate of 0.5 ml/min using degassed P buffer without glycerol. When the effect of ATP on oligomerization was analyzed, 5 mm ATP was added to the running buffer. The molecular weight range covered by each collected fraction (0.5 ml) was inferred from the separation of molecular weight standards (MWGF1000 kit for molecular weights 29,000–700,000, Sigma-Aldrich). Each fraction was precipitated overnight with 10% v/v TCA and 0.002% w/v sodium deoxycholate. The precipitates were resuspended in 1× loading buffer, separated in 10% SDS-polyacryamide gels, and immunoblotted to nitrocellulose membranes. Bands were detected with commercial antibodies raised against ClpC from Synechococcus elongatus (SyClpC, Agrisera AB), which cross-react with AtClpC proteins. In the case of AtClpD, antibodies were raised in rabbits against the purified protein. Antibodies were purified from sera according to Plaxton (27). Both antibodies do not cross-react with E. coli proteins.
ATPase Assays
The amount of inorganic phosphate resulting from ATP hydrolysis was determined using the Malachite green method (28). The measurement of ATPase activity at different pH values was done according to Schirmer et al. (29). For magnesium ion dependence studies, proteins were desalted by gel filtration/centrifugation into P buffer minus MgCl2, added to the reaction mixture (50 mm Tris-HCl, pH 7.5, 100 mm NaCl, 5 mm ATP), and assayed at 37 °C for ATPase activity, which was then compared with that of the proteins in the presence of 5 mm MgCl2. In all cases, proteins were at a concentration of 0.01 mg/ml and reactions were carried out for 30 min (within the linear range of the assay) in a 50-μl reaction mixture.
Reactivation of Heat-denatured Luciferase
Firefly luciferase (200 nm) was heat-denatured for 15 min at 43 °C. Next, it was diluted 1:1 with a reaction mixture containing either chaperone (0.6 μm) and 5 mm ATP. The mixture was incubated at 30 °C. Luciferase activity was determined at various time points by adding 1 μl of the reaction mixture into 99 μl of luciferase assay reagent (Promega). Light emission was measured over a period of 2 s in a microplate luminometer.
Purification of TP-glutathione-S-transferase (GST)
The transit peptide of pea ferredoxin-NADP+ reductase was amplified from plasmid pGF202-TP (30) using primers containing NcoI and SacI sites. The product was digested and cloned in-frame with GST in plasmid pETGEXCT (31). BL21(DE3) cells expressing TP-GST were grown for 16 h at 16 °C after induction with 0.5 mm IPTG. Next, cells were harvested by centrifugation, resuspended in cold B buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1 mm PMSF, 1 mm benzamidine, 1 mm DTT, 5 mm EDTA, 10% v/v glycerol) and sonicated six times (total time: 2 min) in an ice-water bath. The soluble fraction obtained by centrifugation was incubated in the presence of glutathione-agarose resin (35 μl resin/ml soluble extract) at 4 °C for 30 min. Afterward, the beads were washed three times with 10 volumes of B buffer, three times with 3 volumes of B buffer containing 5 mm ATP and 0.15 mg/ml E. coli-denatured proteins (as described in Rial et al., 32), and finally, three times with 10 volumes of P buffer. The fusion protein was eluted using P buffer + 10 mm glutathione and used immediately. GST without TP was purified as described elsewhere (33).
Interaction of Chaperones with TP-GST
Equimolar quantities of either purified chaperone and TP-GST or GST (1 μm) were incubated for 30 min at room temperature in the presence of 5 mm ATP. The mixture was then loaded into the Superdex-200 column, under the same conditions described previously. The running P buffer (minus glycerol) contained 5 mm ATP. Fractions were collected every 0.5 ml, precipitated, and analyzed by immunoblotting using antibodies against AtClpC and GST.
Binding Assays
The TP-GST fusion protein was bound to glutathione-agarose beads from cell lysates as described above. Then, 500 μl of loaded matrix were added to 500 μl of AtClpC2 or AtClpD containing 5 mm ATP and incubated for 30 min at 25 °C. After mild centrifugation, the supernantant was discarded, and the beads were resuspended in 500 μl of P buffer. A 100-μl aliquot was taken from the mixture and centrifuged to collect the resin. The beads were boiled in 50 μl of 2× loading buffer and analyzed by SDS-polyacryamide gels and immunoblotting. This washing procedure was repeated four more times using the remaining loaded resin. Resin without recombinant proteins or loaded with GST served as controls.
Isolation of Intact Chloroplasts
Chloroplasts were isolated exactly as described in Kubis et al. (34) from 14-day-old A. thaliana plants (ecotype Col-0) grown in long day conditions. Chloroplasts were suspended in HMS buffer (50 mm HEPES-NaOH, pH 8.0, 3 mm MgSO4, 300 mm sorbitol) at a concentration of 1 mg chlorophyll/ml.
Precursor Binding to Chloroplasts and Pull-down Assays
50 μl of the chloroplast suspension were mixed with either purified TP-GST or GST (final concentration 0.1 mg/ml) in the presence of 100 μm ATP. Chloroplasts with no added exogenous protein served as controls. Mixtures were incubated in the dark for 10 min at room temperature. Chloroplasts were reisolated by a passage through a 40% v/v Percoll cushion and lysed hypotonically by resuspension in 200 μl of lysis buffer (25 mm HEPES-KOH, pH 8.0, 4 mm MgCl2) for 5 min on ice in the dark. The supernatant and membrane fractions were separated by ultracentrifugation (5 min, 100,000 × g). The supernatant was removed, and membranes were washed twice with lysis buffer and suspended in 1 ml of IPES-DDM (25 mm HEPES-KOH, pH 7.5, 50 mm NaCl, 2 mm EDTA, 2 mm EGTA, 1 mm PMSF, 1% w/v dodecylmaltoside). Insoluble material was removed by an ultracentrifugation step as described above. The supernatant consisting of detergent-solubilized membranes was incubated with 50 μl of glutathione-agarose resin for 30 min at 4 °C. Then, the resin was washed twice with 1 ml of IPES-DDM, twice with 1 ml of IPES without DDM and boiled in 50 μl of 2× loading buffer.
RESULTS
Expression of the Recombinant Proteins
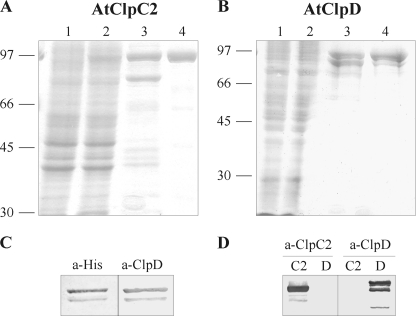
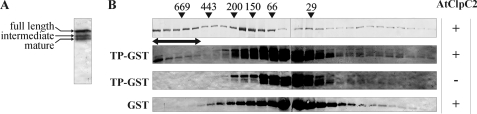
For the expression and purification of three chloroplastic Clp/HSP100 chaperones from A. thaliana, we used two common strategies: expression in E. coli from a T7 promoter-based expression vector and recovery of the recombinant proteins by immobilized-metal affinity chromatography. As we previously reported, AtClpC2 and AtClpD were mainly insoluble (35), but the soluble fraction could be recovered by affinity chromatography (Fig. 1, A and B). Despite our efforts, AtClpC1 could not be expressed in any condition tested. For that reason, we continued our studies using AtClpC2 and AtClpD. Their molecular masses were as expected (93 kDa for AtClpC2 and 95 kDa for AtClpD). Interestingly, when purified AtClpD was detected in denaturing gels and immunoblots using specific antibodies, two bands were consistently spotted: one corresponding to the full-length protein and the other corresponding to a product of lower molecular mass (∼81 kDa). These two forms appeared as soon as IPTG was added to induce protein expression (results not shown) and could not be separated. These two bands were detected by antibodies raised against the hexahistidine tag (Santa Cruz Biotechnology) or AtClpD, which indicates that AtClpD suffered proteolysis at its C terminus (Fig. 1C). All subsequent analyses of AtClpD were performed with this mixture. Anti-AtClpD antibodies do not recognize AtClpC2 and vice versa (Fig. 1D).
FIGURE 1.
Expression and purity of AtClpC2 and AtClpD. Expression and purification of the recombinant proteins were monitored by gel electrophoresis. The content of each lane is identical for AtClpC2 (A) or AtClpD (B). Soluble extracts from uninduced and induced cultures were loaded in lanes 1 and 2, respectively. Lane 3 shows the eluted proteins after affinity chromatography and lane 4, the purified proteins after the whole purification procedure. Molecular weight standards are depicted on the left. C, purified AtClpD was transferred to two nitrocellulose sheets and detected by antibodies raised against the polyhistidine tag (a-His) or against AtClpD (a-AtClpD). D, purified proteins were blotted after electrophoresis and detected with anti-ClpC (left) or anti-AtClpD (right) antibodies. C2 and D indicate lane content (AtClpC2 and AtClpD, respectively).
Oligomerization of AtClpC2 and AtClpD
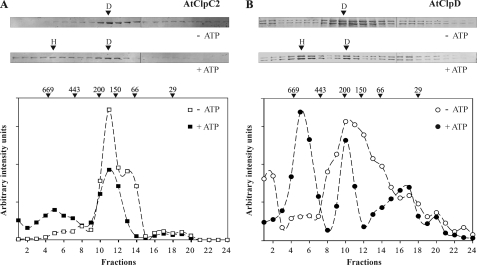
All HSP100 proteins studied to date are able to oligomerize into higher order complexes, which were observed using mainly electron microscopy and gel filtration chromatography. By the use of the latter technique, the oligomeric status of the proteins was analyzed. In the absence of ATP, both proteins were detected mainly as dimers (Fig. 2 and supplemental Fig. S1). The peak containing AtClpC2 was centered on 180 kDa, while AtClpD migrated with a molecular mass of about 220 kDa. However, it should be noted that reactive bands covered the majority of the column separating range. Upon addition of 5 mm ATP to the running buffer, the intensity of the bands corresponding to fractions eluting in a size range of 700–500 kDa was higher, indicating hexamerization. In addition, band intensity in the zone corresponding to the presence of dimers and monomers diminished, suggesting that a portion of this population assembled into higher order complexes.
FIGURE 2.
Oligomerization status of AtClpC2 and AtClpD. The purified proteins were subjected to gel filtration chromatography in the absence or presence of ATP. Fraction collection began after 8 ml (void volume, fraction 1). All fractions were collected every 0.5 ml, precipitated and electrophoresed. AtClpC2 (panel A) and AtClpD (panel B) were detected by immunoblots. The intensity of the bands was quantified using the software GelPro and plotted below the blots. For AtClpD, the intensity value in the graph is the sum of the intensity of both bands, because their migration pattern was identical. Arrows above the band intensity plots indicate the migration of molecular weight standards. Bands that correspond to peaks in the band intensity plots are indicated by arrows (H, hexamers; D, dimers).
ATPase Activity of the Recombinant Proteins
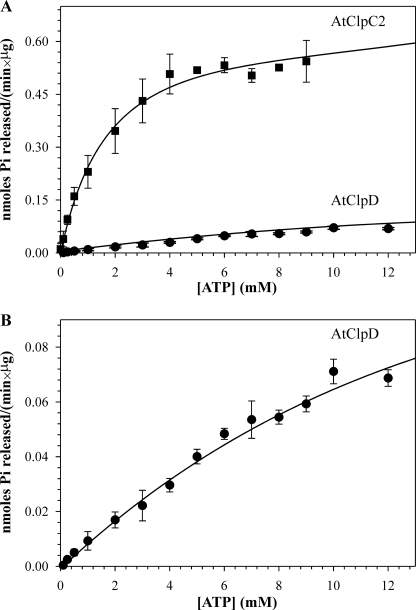
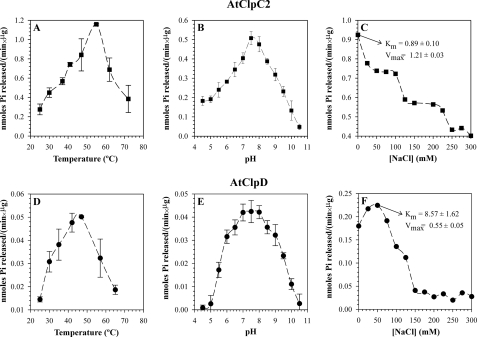
The ATP-hydrolyzing activity of the proteins was measured by the colorimetric detection of inorganic phosphate complexed with moblidate ions and Malachite green. Both proteins presented Michaelis-Menten kinetics with different parameters (Fig. 3). The Km and Vmax of AtClpC2 were 1.42 mm and 0.62 nmol/(min × μg protein), respectively, and those of AtClpD were ∼19.80 mm and ∼0.19 nmol/(min × μg protein) at 37 °C. These parameters were the same for different batches of protein; even those further purified by gel filtration chromatography (see “Experimental Procedures”). It is worth noting that AtClpD activity did not reach saturation. Amounts of ATP larger than 15 mm produced unacceptable high absorbance values of the blanks, so ATP concentration could not be increased further. For that reason, the kinetic parameters of AtClpD should be taken as rough estimates. We also analyzed the optimal pH and temperature values where ATPase activity was maximal (Fig. 4). Both proteins had detectable ATPase activity at temperatures up to 75 °C, being the temperature optimum for AtClpC2 (55 °C) 10 °C higher than that of AtClpD (45 °C) and presented pH optima centered at 7.5. Additionally, the ATPase activity of the proteins was ionic strength-dependent. As the salt content of the buffers increased, ATPase activity decreased (Fig. 4, C and F). We also calculated the kinetic parameters under these low-salt conditions. While the Km dropped for both proteins, the Vmax was higher. Other parameters, such as the pH and temperature optima, remained unaffected (results not shown).
FIGURE 3.
Kinetic analysis of ATP hydrolysis by AtClpC2 and AtClpD. Release of inorganic phosphate was monitored spectrophotometrically by the Malachite green method. A, enzymatic activity of each protein is represented as a function of ATP concentration. B, same kinetic analysis of ATP hydrolysis by AtClpD shown in panel A is depicted with a different scale on the y-axis. Data points represent the mean of triplicate experiments, and bars show the standard error. The curve was fitted to the Michaelis-Menten equation (strong lines) using Sigma Plot.
FIGURE 4.
ATPase activity parameters of AtClpC2 and AtClpD. ATPase activity for each protein was monitored under different environmental conditions, such as temperature (A and D), pH (B and E), and ionic strength (C and F). The kinetic parameters Km and Vmax shown in panels C and F were measured at the NaCl concentration where ATPase activity was maximal. Data points represent the mean of triplicate experiments, and bars show the standard error.
In strong contrast with other proteins of the HSP100 family, the ATPase activity of both proteins was only marginally higher in the presence of the model substrate casein. Additionally, the enzymes did not display ADPase activity (results not shown). Removal of the proteins with protein A-Sepharose beads loaded with the corresponding specific antibodies resulted in a complete loss of ATPase activity, measured at 37 °C and pH 7.5 in the presence of 5 mm ATP (supplemental Fig. S2). This confirmed that the samples were free of other contaminating ATPases. Under these same conditions, the dependence of magnesium ions on ATPase activity was studied. Chaperones were desalted by gel filtration/centrifugation into P buffer minus Mg2+ and the ATPase activity of these samples was measured. ATPase activity could not be detected, showing that ATP hydrolysis requires the divalent cation (data not shown).
Renaturation of Heat-denatured Luciferase
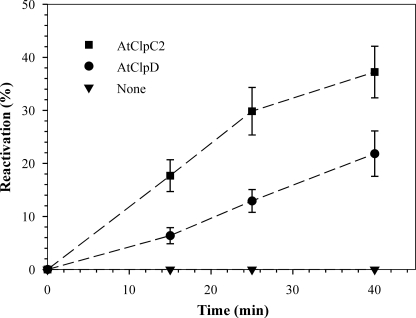
To establish the ability of the recombinant proteins to assist in protein folding, we tested the chaperone activity of AtClpC2 and AtClpD using heat-denatured luciferase as a model substrate. The reactivation of luciferase activity with time was measured in the presence of the chaperones. Both proteins were able to recover luciferase activity, albeit at different extents: AtClpD had a reduced activity (∼20% reactivation after 40 min) when compared with AtClpC2 (∼40% reactivation after 40 min, Fig. 5).
FIGURE 5.
Reactivation of luciferase activity. Heat-denatured luciferase was added to reaction mixtures containing AtClpC2 (■), AtClpD (●) or without additional proteins (▾). The mixtures were incubated at 30 °C, and aliquots were taken for luciferase activity measurements at the indicated time points. The results are expressed as a percent of native luciferase activity (100%) incubated at 30 °C for the same time as the experimental point. Data points represent the mean of triplicate experiments, and bars show the standard error.
Interaction of the Recombinant Proteins with TP-GST
Molecular chaperones can interact with unstructured hydrophobic patches such as those present in transit peptides. For that reason, they are proposed to play a crucial role in protein import across membranes. To analyze if a direct physical interaction exists between the AtClpC2 and AtClpD chaperones and a model transit peptide, we used a fusion protein consisting of the TP of pea ferredoxin-NADP+ reductase and GST. The fusion was consistently purified as three different forms. A major one corresponds to the full-length TP-fusion protein, another to a truncated form of intermediate molecular weight and the last one corresponding to mature GST (Fig. 6A).
FIGURE 6.
Interaction between AtClpC2 and TP-GST. A, typical immunoblot of a TP-GST preparation detected by antibodies raised against GST. B, mixtures of AtClpC2 with TP-GST or GST were subjected to gel filtration chromatography. Aliquots were taken every 0.5 ml, precipitated and analyzed by SDS-polyacrylamide gels and immunoblotting. All blots were cut in two, and the top halves were revealed with anti-ClpC antibodies (top blot), while anti-GST antibodies (remaining blots) were used for the bottom halves. The first two blots are from the same membrane, but the elution profile of AtClpC2 for the control GST + AtClpC2 (fourth blot) is not included for the sake of simplicity, as it was identical to the one shown in the top blot. The third blot shows the migration pattern of TP-GST alone. The double-headed arrow indicates the elution range where TP-GST was detected as a high molecular weight complex. Arrows above the top blot indicate the migration of molecular weight standards. Representative results from three different experiments are shown.
To analyze the possible interaction between the fusion protein and the chaperones, TP-GST and AtClpC2 were incubated in the presence of 5 mm ATP for 30 min at 25 °C. Then, the mixture was subjected to gel filtration chromatography. The elution profile of TP-GST in the presence of the chaperones was analyzed by precipitating the collected fractions, followed by immunoblotting and then, by comparing the resulting migration pattern to that of TP-GST alone (Fig. 6B). For the TP-GST/AtClpC2 mixture, we observed that a population of TP-GST (full-length and intermediate) migrated as a 700–500 kDa protein complex, in the same range where AtClpC2 hexamers were found. Interestingly, the amount of TP-GST intermediate form migrating in this range was much higher than the amount of full-length fusion protein. This suggests that this form is a better substrate than the full-length TP-GST fusion. To confirm that this retardation was caused by the formation of a complex between hexameric AtClpC2 and TP-GST, the elution profile of the fusion was obtained in the absence of AtClpC2. In this case, no bands were detected in the 700–500 kDa zone. Moreover, to show that AtClpC2 interacted with the fusion specifically by its TP extension, the migration pattern of GST was analyzed in the presence of the chaperone. Again, no bands were detected in the high molecular weight range, showing that the GST moiety of the fusion does not interact with AtClpC2 hexamers. This experiment was also carried out with a TP-GST/AtClpD mixture, but TP-GST could not be detected in the zone of AtClpD multimers (data not shown).
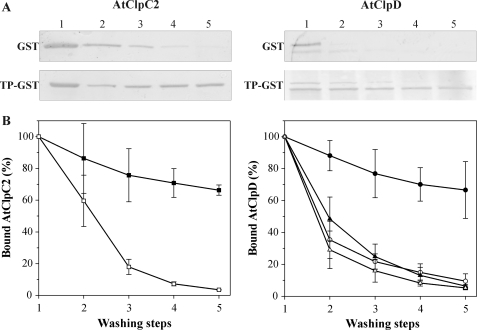
To confirm the association of the chaperones with TP-GST, binding assays were performed. TP-GST or GST was loaded to glutathione-agarose beads. Then, the loaded resins were supplemented with either chaperone + 5 mm ATP, incubated for 30 min at 25 °C and subjected to successive washing steps (i.e. pelleting and resuspending of the beads). After each step, a portion of the matrix was removed, boiled in 2× loading buffer and used for gel electrophoresis and immunoblot analysis. The amount of chaperone bound to the loaded resins after each step was measured by band densitometry of the blots. For AtClpC2, less than 5% remained associated to the GST-containing resin after the extraction protocol, while a higher amount (more than 65%) was found in the TP-GST-containing resin after the washing steps (Fig. 7A). When this experiment was replicated with AtClpD, a surprising result was observed. Similar to AtClpC2, less than 5% of AtClpD in its two variants remained associated to the GST-containing resin after the extraction protocol (Fig. 7B, open and closed triangles). However, when AtClpD was incubated with the TP-GST-containing resin, only the lower band corresponding to the C terminus-processed form remained associated (more than 65%, closed circles). The full-length form was almost completely washed away (open circles). Taken together, these results indicate that both chaperones remained attached to the TP extension with greater affinity when compared with GST alone.
FIGURE 7.
Chaperone binding assays with immobilized TP-GST and GST. A, TP-GST or GST were bound to a glutathione-agarose resin and then, AtClpC2 or AtClpD were added. After a 30-min incubation, the beads were washed five times, and aliquots of the resins were taken after each washing step. The beads were boiled in loading buffer to unbind the proteins, which were then analyzed by SDS-polyacrylamide gels followed by immunoblotting using antibodies against AtClpC (left) or AtClpD (right). Representative blots are shown. B, digitized bands from six identical experiments were quantified using the software GelPro. The graphs show the percentage of residual chaperone in the GST-containing resin (open symbols) or in the TP-GST-containing resin (closed symbols) after each washing step. For AtClpD (left), the results for the upper band are represented by triangles, while circles were used for the lower band.
Association of HSP100 Proteins with Import Complexes and Precursors
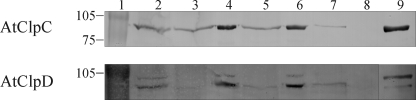
Having established that the recombinant proteins interact with TP-GST but not with GST, we sought to determine if this interaction could also be detected in isolated chloroplasts. Intact chloroplasts were obtained from 14-day-old A. thaliana plants and incubated with TP-GST (or GST as a control) in the presence of 100 μm ATP. This experimental condition is known to promote precursor docking to the import machinery but does not allow its import (36). If HSP100 proteins stably interact with translocation complexes containing TP-GST engaged in translocation, its recovery with glutathione-agarose resin would also extract the chaperones. To test this hypothesis, envelope membranes were isolated after hypotonic lysis of intact chloroplasts followed by ultracentrifugation. Membranes were solubilized with a mild detergent and the TP-GST bound to the import complexes was pulled down. The presence of AtClpC proteins or AtClpD was assessed by SDS-PAGE followed by immunoblotting. It is important to note that antibodies against ClpC proteins cannot discriminate between AtClpC1 and AtClpC2. For this reason, these chaperones are referred collectively in these experiments as AtClpC proteins. The results are shown in Fig. 8. Lane content is identical for both blots, the only difference being the primary antibody used for detection and the control of recombinant protein (lane 9). The presence of the chaperones in intact chloroplasts was analyzed (lane 2); bands of expected molecular weight were observed. In addition, aliquots of membrane and supernatant fractions from lysed chloroplasts without any exogenous protein added were analyzed (lanes 3 and 4, respectively). We observed that AtClpD was mainly detected in the supernatant fraction, while a fraction of AtClpC proteins could also be detected in the membrane fraction in the absence of precursor. When TP-GST was added before lysis, the ratio of HSP100 proteins in the membrane and supernatant fraction seemed not to be altered (lanes 5 and 6). Capture of TP-GST with glutathione-agarose resin co-extracted AtClpC proteins and AtClpD (lane 7), indicating that the chaperones formed a stable association with the import machinery. Interestingly, the processed form of AtClpD was recovered in a larger amount than the unprocessed full-length form, as judged by the intensity of both bands. The TP moiety of the fusion was responsible for HSP100 interaction, as addition of GST to the chloroplasts resulted in no association of HSP100 proteins after the experimental procedure (lane 8).
FIGURE 8.
Association of HSP100 proteins with import complexes containing TP-GST. TP-GST was incubated with intact chloroplasts in the presence of 100 μm ATP for 10 min at room temperature. After lysis, chloroplastic membranes were separated from soluble material by ultracentrifugation and solubilized in buffer containing DDM. Glutathione-agarose beads were added to capture TP-GST and associated proteins from the mixture. Samples corresponding to 50 μl of chloroplasts were analyzed by SDS-PAGE followed by immunoblotting using antibodies against AtClpC proteins (upper blot) or AtClpD (lower blot). Lane 1: molecular weight standards, lane 2: isolated chloroplasts, lanes 3 and 4: membrane and supernatant fractions from lysed chloroplasts, lanes 5 and 6: membrane and supernatant fractions from lysed chloroplasts with bound TP-GST, lane 7: import complexes containing bound TP-GST captured with glutathione-agarose resin, lane 8: as lane 7 but using GST instead of TP-GST, lane 9: purified recombinant protein. Representative blots from three different experiments are shown.
DISCUSSION
In this report, we show for the first time that HSP100 proteins from plant chloroplasts function as molecular chaperones. We chose E. coli as a host for the recombinant expression of AtClpC1, AtClpC2, and AtClpD. This host was successfully used for the expression of SyClpC (37), ClpC1 from Mycobacterium tuberculosis (MtClpC1) (38) and many others from bacterial origin. AtClpC1 could not be detected under any condition tested. This result was surprising, as AtClpC2 was readily obtained. Both proteins share 92% identity and 96% similarity at the amino acid level, so it is puzzling why AtClpC1 should not be expressed. AtClpD was consistently expressed and purified as two distinct forms, one corresponding to the full-length protein and other with lower molecular weight. A previous work studying the expression profile of AtClpD using leaf material, also showed the presence of two reactive bands in the immunoblots (14). The cleavage occurred at the C terminus, as the processed protein retained the hexahistidine tag. It is known that the C terminus of EcClpA is unstructured and prone to degradation (39). It could be possible that AtClpD also shares this feature.
HSP100 proteins self-assemble into ring-shaped hexamers in the presence of ATP, being ClpV from Salmonella typhimurium a notable exception to the ATP requirement (40). In absence of the nucleotide, a monomer:dimer equilibrium is observed for EcClpA (41). Both AtClpC2 and AtClpD eluted in the range of 200 kDa when subjected to gel filtration chromatography. When ATP was included in the running buffer, a fraction eluted in the 700–500 kDa zone, which indicates the presence of hexamers. In contrast, the oligomerization process for EcClpA, HSP104 from Saccharomyces cerevisiae or ClpV from S. typhimurium is an all-or-none phenomenon: upon addition of ATP, the proteins completely assemble into hexamers, which elute as a single 600 kDa-centered peak (40, 42). Our data and that of others reflect a large variety in the oligomerization behavior of these proteins.
The ATPase activity of the HSP100 proteins is also diverse between different subtypes. While the majority displays basal ATPase activity, ClpC from Bacillus subtilis is not able to hydrolyze ATP in absence of the adaptor MecA (43). Both AtClpC2 and AtClpD presented intrinsic ATPase activity; i.e. without the need of any adaptor protein. The kinetic parameters for AtClpC2 were comparable to those of EcClpA (44), ClpB from E. coli (45) and the related SyClpC (37) (supplemental Table S1). The Km for AtClpC2 is 1.42 mm, in the range of the physiological concentration of ATP in illuminated chloroplasts (46). In contrast, the calculated Km for AtClpD is the highest reported to date (∼19.80 mm) and well outside the in vivo concentration of ATP. This value suggests that AtClpD would not hydrolyze ATP under physiological conditions. One explanation is that, in vivo, AtClpD may need a special environment or an unidentified adaptor to show significant activity. Either way, the different kinetic parameters of AtClpC2 and AtClpD implies that these proteins may be subjected to different regulatory mechanisms and may have distinct roles in protein quality control. Other parameters, such as pH and temperature optima, and activity dependence on magnesium ions were similar to those reported. In addition, the ATPase activity was influenced by ionic strength, as was also reported for HSP104 from S. cerevisiae (29).
We next tested the ability of the recombinant proteins to act as chaperones using heat-aggregated luciferase as a model substrate. Both proteins were able to reactivate luciferase activity to different extents (∼40% for AtClpC2 and ∼20% for AtClpD after 40 min). Again, members of the HSP100 family show different performance with regard to this activity. It was thought that HSP100 proteins did not possess disaggregating capabilities by themselves, as that was the case for EcClpA (47), ClpB from E. coli (48) and ClpC from B. subtilis (49). Conversely, other recently characterized HSP100 proteins can bind to aggregated substrates and promote their folding, such as SyClpC and MtClpC1. It is interesting to note that while SyClpC could recover up to 4% of luciferase activity in 2 h (37), AtClpC2 most resembles MtClpC1 in that it could reactivate luciferase activity in more than 30% in less than 1 h, while AtClpD showed less disaggregase activity.
This foldase activity would be of extreme importance during protein import, as the hydrophobic stretches that penetrate into organelles are targeted by chaperones. It has been long confirmed that HSP70 is the motor driving protein import into mitochondria (50) and the endoplasmic reticulum (51). On the other hand, in chloroplasts, several results indicated that chaperones other than HSP70 could also participate in the process. First, Nielsen et al. (21) found that pea HSP93 was tightly associated with chloroplast translocons during active import. Recently, it was shown that pea HSP93 interacts with Tic40, a stromal co-chaperone that facilitates protein translocation across the inner membrane (22). In addition, A. thaliana plants lacking AtClpC1 present import defects (16–18). Finally, a mutant transit peptide lacking all its HSP70 recognition sites was efficiently translocated (52). These results led to the notion that HSP100 proteins could assist in protein import into chloroplast, yet there was little in vitro evidence to support this assumption. To fill the void in this area, we studied the possible interaction between AtClpC2 and AtClpD with the transit peptide of pea ferredoxin-NADP+ reductase (fused to GST). This strategy was previously used in our laboratory to study the association between DnaK from E. coli and pea HSP70 with the precursor of ferredoxin-NADP+ reductase (30). We noted that the purification of these TP-containing fusions results in a combination of full-length TP-fusion protein, mature protein and an intermediate fusion in which the TP is truncated from the N terminus. In gel filtration studies, this mixture elutes in the 70–50 kDa range (being the molecular weight of the GST dimer 52 kDa). When the mixture was incubated in the presence of AtClpC2 and then injected into the column, a fraction of TP-GST and a larger quantity of the intermediate form was found in the fractions were AtClpC2 hexamers eluted. This indicates that both fusions were “trapped” by the oligomers. Interestingly, the intermediate form was preferentially bound to AtClpC2 hexamers, suggesting that this form is a better substrate.
In another set of experiments, TP-GST was loaded to a gluthatione-agarose resin and incubated in the presence of either chaperone. Then, the beads were successively washed and the amount of each chaperone still bound to the resin was analyzed. For AtClpC2, more than 65% was retained after five washing steps. For AtClpD, only the C terminus-processed variant remained bound to the TP-GST-containing resin (more than 65%), which suggests that this would be the active form and not the full-length protein, at least under these conditions. For EcClpA, processing of the flexible C terminus protects the protein against autodegradation, but this modification is not necessary for its activity (39). In fact, there is no evidence of any HSP100 protein whose chaperone activity is regulated by C-terminal processing, which could represent a novel mechanism to adjust the quantity of active chaperones. Finally, when GST was loaded to the matrix instead, both chaperones were completely washed after no more than three steps. Taken as a whole, these results clearly show that both chaperones can stably bind to the TP-GST fusion.
We finally tested if the observed interaction could also be replicated with the natural chaperones. Intact chloroplasts were isolated from young A. thaliana plants by isopycnic centrifugation in Percoll gradients. Membrane and supernatant fractions were separated after ultracentrifugation of lysed chloroplasts when no exogenous protein was added and analyzed by immunoblotting with antibodies raised against the chaperones. This allowed us to study the distribution of the chaperones in these fractions in normal conditions. A portion of AtClpC proteins was detected in the membrane fraction while the majority was found in the supernatant (composed mainly of stroma), while AtClpD was found almost exclusively in the stromal fraction. When TP-GST was added to the chloroplasts before lysis in the presence of ATP 100 μm, the distribution of the HSP100 proteins in the two fractions did not appear to be altered. Under these limiting ATP concentrations, the TP is inserted across both envelope membranes and can interact with stromal components (21). When TP-GST docked to the import machinery was recovered from detergent-solubilized membranes by capture with glutathione-agarose resin, AtClpC proteins and AtClpD were detected. The association could be a result of direct interaction of HSP100 proteins with TP-GST, or interaction with a component in the translocation apparatus. Nevertheless, the results point to HSP100 chaperones as being key pieces of import complexes. Interestingly, the processed form of AtClpD was preferentially recovered over the full-length form, further indicating that the former may be the physiologically relevant variant.
These data add more evidence in support of the model in which HSP100 proteins could act as motors pulling precursor chains into the chloroplast. Moreover, this would explain why ΔAtclpc1 plants are viable and their chloroplasts are able to import precursors, albeit at a lower rate. AtClpC2 (or AtClpD) could compensate for the AtClpC1 deficiency, being the sick phenotype a result of the lower amount of AtClpC2 with respect to AtClpC1. The latter contributes to 65% of the ClpC wild-type levels (15). This hypothesis is supported by the work of Kovacheva et al., in which overexpression of AtClpC2 in ΔAtclpc1-deficient plants resulted in complementation of the mutation (18). Despite these findings, we cannot rule out that HSP70 also takes part in the translocation process. In fact, recent work showed that HSP70 interacts with chloroplasts translocons in the same manner as pea HSP93 (23, 53). Plants lacking AtClpC1 and one of the stromal HSP70s (Hsc70-1) show more severe import defects than the single mutants. In addition, anti-Hsc70 antibodies stoichiometrically precipitated AtClpC1 from solubilized envelope membranes (23). Hence, the current models of protein translocation into chloroplasts place both HSP70 and HSP100 proteins as molecular motors driving protein import. This is not surprising, as chaperone redundancy has been well documented (54).
AtClpC2 and AtClpD are the first molecular chaperones of the HSP100 group from plants to be characterized. They display chaperone activity in vitro and the ability to associate with a transit peptide. However, more experiments are necessary to establish their exact role in precursor translocation.
Supplementary Material
This study was supported by grants from CONICET and Agencia de Promoción Científica y Tecnológica (ANPCyT, Argentina).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and Table S1.
- EcClpA
- E. coli ClpA protein
- AtClpC1
- A. thaliana ClpC1 protein
- AtClpC2
- A. thaliana ClpC2 protein
- AtClpD
- A. thaliana ClpD protein
- cDNA
- complementary DNA
- DDM
- dodecylmaltoside
- GST
- glutathione S-transferase
- IPTG
- isopropyl β-d-1-thiogalactopyranoside
- MtClpC1
- M. tuberculosis ClpC protein
- NTA
- nitrilotriacetic acid
- pda
- accession number for a RIKEN clone
- PMSF
- phenylmethylsulfonyl fluoride
- SyClpC
- Synechococcus elongatus ClpC protein
- TP
- transit peptide.
REFERENCES
- 1. Hartl F. U., Hayer-Hartl M. (2002) Science 295, 1852–1858 [DOI] [PubMed] [Google Scholar]
- 2. Feder M. E., Hofmann G. E. (1999) Annu. Rev. Physiol. 61, 243–282 [DOI] [PubMed] [Google Scholar]
- 3. Jackson-Constan D., Akita M., Keegstra K. (2001) Biochim. Biophys. Acta 1541, 102–113 [DOI] [PubMed] [Google Scholar]
- 4. Neupert W., Brunner M. (2002) Nat. Rev. Mol. Cell Biol. 3, 555–565 [DOI] [PubMed] [Google Scholar]
- 5. Brodsky J. L., Werner E. D., Dubas M. E., Goeckeler J. L., Kruse K. B., McCracken A. A. (1999) J. Biol. Chem. 274, 3453–3460 [DOI] [PubMed] [Google Scholar]
- 6. Martin J., Hartl F. U. (1997) Curr. Opin. Struct. Biol. 7, 41–52 [DOI] [PubMed] [Google Scholar]
- 7. Gottesman S., Wickner S., Maurizi M. R. (1997) Genes Dev. 11, 815–823 [DOI] [PubMed] [Google Scholar]
- 8. Hayes S. A., Dice J. F. (1996) J. Cell Biol. 132, 255–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Doyle S. M., Wickner S. (2009) Trends Biochem. Sci. 34, 40–48 [DOI] [PubMed] [Google Scholar]
- 10. Schirmer E. C., Glover J. R., Singer M. A., Lindquist S. (1996) Trends Biochem. Sci. 21, 289–296 [PubMed] [Google Scholar]
- 11. Gottesman S., Clark W. P., Maurizi M. R. (1990) J. Biol. Chem. 265, 7886–7893 [PubMed] [Google Scholar]
- 12. Sanchez Y., Taulien J., Borkovich K. A., Lindquist S. (1992) EMBO J. 11, 2357–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Adam Z., Rudella A., van Wijk K. J. (2006) Curr. Opin. Plant Biol. 9, 234–240 [DOI] [PubMed] [Google Scholar]
- 14. Zheng B., Halperin T., Hruskova-Heidingsfeldova O., Adam Z., Clarke A. K. (2002) Physiol. Plant 114, 92–101 [DOI] [PubMed] [Google Scholar]
- 15. Sjögren L. L., MacDonald T. M., Sutinen S., Clarke A. K. (2004) Plant Physiol. 136, 4114–4126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kovacheva S., Bédard J., Patel R., Dudley P., Twell D., Ríos G., Koncz C., Jarvis P. (2005) Plant J. 41, 412–428 [DOI] [PubMed] [Google Scholar]
- 17. Constan D., Froehlich J. E., Rangarajan S., Keegstra K. (2004) Plant Physiol. 136, 3605–3615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kovacheva S., Bédard J., Wardle A., Patel R., Jarvis P. (2007) Plant J. 50, 364–379 [DOI] [PubMed] [Google Scholar]
- 19. Nakashima K., Kiyosue T., Yamaguchi-Shinozaki K., Shinozaki K. (1997) Plant J. 12, 851–861 [DOI] [PubMed] [Google Scholar]
- 20. Weaver L. M., Froehlich J. E., Amasino R. M. (1999) Plant Physiol. 119, 1209–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nielsen E., Akita M., Davila-Aponte J., Keegstra K. (1997) EMBO J. 16, 935–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chou M. L., Chu C. C., Chen L. J., Akita M., Li H. M. (2006) J. Cell Biol. 175, 893–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Su P. H., Li H. M. (2010) Plant Cell 22, 1516–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li H. M., Chiu C. C. (2010) Annu. Rev. Plant Biol. 61, 157–180 [DOI] [PubMed] [Google Scholar]
- 25. Penefsky H. S. (1977) J. Biol. Chem. 252, 2891–2899 [PubMed] [Google Scholar]
- 26. Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 27. Plaxton W. C. (1989) Eur. J. Biochem. 181, 443–451 [DOI] [PubMed] [Google Scholar]
- 28. Lanzetta P. A., Alvarez L. J., Reinach P. S., Candia O. A. (1979) Anal. Biochem. 100, 95–97 [DOI] [PubMed] [Google Scholar]
- 29. Schirmer E. C., Queitsch C., Kowal A. S., Parsell D. A., Lindquist S. (1998) J. Biol. Chem. 273, 15546–15552 [DOI] [PubMed] [Google Scholar]
- 30. Rial D. V., Arakaki A. K., Ceccarelli E. A. (2000) Eur. J. Biochem. 267, 6239–6248 [DOI] [PubMed] [Google Scholar]
- 31. Sharrocks A. D. (1994) Gene 138, 105–108 [DOI] [PubMed] [Google Scholar]
- 32. Rial D. V., Ceccarelli E. A. (2002) Protein Expr. Purif. 25, 503–507 [DOI] [PubMed] [Google Scholar]
- 33. Harper S., Speicher D. W. (2008) Curr. Protoc. Protein Sci., John Wiley & Sons, Inc, Hoboken, NJ [Google Scholar]
- 34. Kubis S. E., Lilley K. S., Jarvis P. (2008) Methods Mol. Biol. 425, 171–186 [DOI] [PubMed] [Google Scholar]
- 35. Rosano G. L., Ceccarelli E. A. (2009) Microb. Cell Fact. 8, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Olsen L. J., Theg S. M., Selman B. R., Keegstra K. (1989) J. Biol. Chem. 264, 6724–6729 [PubMed] [Google Scholar]
- 37. Andersson F. I., Blakytny R., Kirstein J., Turgay K., Bukau B., Mogk A., Clarke A. K. (2006) J. Biol. Chem. 281, 5468–5475 [DOI] [PubMed] [Google Scholar]
- 38. Kar N. P., Sikriwal D., Rath P., Choudhary R. K., Batra J. K. (2008) FEBS J. 275, 6149–6158 [DOI] [PubMed] [Google Scholar]
- 39. Maglica Z., Striebel F., Weber-Ban E. (2008) J. Mol. Biol. 384, 503–511 [DOI] [PubMed] [Google Scholar]
- 40. Schlieker C., Zentgraf H., Dersch P., Mogk A. (2005) Biol. Chem. 386, 1115–1127 [DOI] [PubMed] [Google Scholar]
- 41. Maurizi M. R., Singh S. K., Thompson M. W., Kessel M., Ginsburg A. (1998) Biochemistry 37, 7778–7786 [DOI] [PubMed] [Google Scholar]
- 42. Kress W., Mutschler H., Weber-Ban E. (2007) Biochemistry 46, 6183–6193 [DOI] [PubMed] [Google Scholar]
- 43. Turgay K., Hamoen L. W., Venema G., Dubnau D. (1997) Genes Dev. 11, 119–128 [DOI] [PubMed] [Google Scholar]
- 44. Maurizi M. R., Thompson M. W., Singh S. K., Kim S. H. (1994) Methods Enzymol. 244, 314–331 [DOI] [PubMed] [Google Scholar]
- 45. Woo K. M., Kim K. I., Goldberg A. L., Ha D. B., Chung C. H. (1992) J. Biol. Chem. 267, 20429–20434 [PubMed] [Google Scholar]
- 46. Heineke D., Riens B., Grosse H., Hoferichter P., Peter U., Flügge U. I., Heldt H. W. (1991) Plant Physiol 95, 1131–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wickner S., Gottesman S., Skowyra D., Hoskins J., McKenney K., Maurizi M. R. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 12218–12222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Goloubinoff P., Mogk A., Zvi A. P., Tomoyasu T., Bukau B. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 13732–13737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schlothauer T., Mogk A., Dougan D. A., Bukau B., Turgay K. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 2306–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gambill B. D., Voos W., Kang P. J., Miao B., Langer T., Craig E. A., Pfanner N. (1993) J. Cell Biol. 123, 109–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Matlack K. E., Misselwitz B., Plath K., Rapoport T. A. (1999) Cell 97, 553–564 [DOI] [PubMed] [Google Scholar]
- 52. Rial D. V., Ottado J., Ceccarelli E. A. (2003) J. Biol. Chem. 278, 46473–46481 [DOI] [PubMed] [Google Scholar]
- 53. Shi L. X., Theg S. M. (2010) Plant Cell 22, 205–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Frydman J. (2001) Annu. Rev. Biochem. 70, 603–647 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.