Abstract
12-O-tetradecanoylphorbol-13-acetate (TPA) has been shown to induce transcriptional activation of human manganese superoxide dismutase (MnSOD) mRNA in human lung carcinoma cells, A549, mediated by a protein kinase C (PKC)-dependent activation of cAMP-responsive element-binding protein (CREB)-1/ATF-1-like factors. In this study, we showed that MnSOD protein expression was elevated in response to TPA or TNF-α, but not to hydrogen peroxide treatment. TPA-induced generation of reactive oxygen species (ROS) was blocked by pretreatment of the PKC inhibitor BIM and NADPH oxidase inhibitor DPI. Small interfering RNA (siRNA) experiments indicated that knocking down the NADPH oxidase components e.g. Rac1, p22phox, p67phox, and NOXO1 in A549 cells impaired TPA-induced MnSOD expression. To identify the PKC isozyme involved, we used a sod2 gene response reporter plasmid, pSODLUC-3340-I2E-C, capable of sensing the effect of TNF-α and TPA, to monitor the effects of PKC isozyme-specific inhibitors and siRNA-induced knockdown of specific PKC isozyme. Our data indicate that TPA-induced MnSOD expression was independent of p53 and most likely mediated by PKC-α-, and -ϵ-dependent signaling pathways. Furthermore, siRNA-induced knock-down of CREB and Forkhead box class O (FOXO) 3a led to a reduction in TPA-induced MnSOD gene expression. Together, our results revealed that TPA up-regulates, in part, two PKC-dependent transcriptional pathways to induce MnSOD expression. One pathway involves PKC-α catalyzed phosphorylation of CREB and the other involves a PKC-mediated the PP2A catalyzed dephosphorylation of Akt at Ser473 which in turn leads to FOXO3a Ser253 dephosphorylation and its activation.
Keywords: Akt, Akt PKB, CREB, Foxo, Protein Kinase C (PKC), Superoxide Dismutase (SOD), TPA
Introduction
Manganese superoxide dismutase (MnSOD)4 is a nuclear-encoded antioxidant enzyme that is imported into the mitochondrial matrix (1), to catalyze the dismutation of superoxide radical anions (O2˙̄) into hydrogen peroxide (H2O2) and oxygen (2). MnSOD is considered as the primary defensive enzyme against oxidative stress within mitochondria (3). Targeted disruption of murine sod2 causes dilated cardiomyopathy and neonatal lethality (4). In addition, a low level of MnSOD has been implicated in causing various human tumors (5), whereas its overexpression suppresses tumorigenicity (6, 7).
A number of studies have demonstrated the induction of MnSOD in various cell lines and tissues following oxidative stress induced by treatments with TNF-α (8–12), interleukin-1 (8, 10–12), lipopolysaccharide (10, 12), interferon-γ (11), 12-O-tetradecanoylphorbol-13-acetate (TPA) (12, 13), or irradiation (14, 15). The induction of MnSOD is transactivated via two parts of the sod2 gene in various species following oxidative stress. One part is the 5′-flanking promoter region regulated by Sp-1 (16–18), and with early growth response factor (Egr-1) after treatment with TPA (19), or by AP-2 (16, 17, 20–22). The other part is the enhancer within the second intron regulated by the CCAAT/enhancer-binding protein (C/EBP) and NF-κB in response to TNF-α and interleukin-1 (IL-1) (23, 24) or TPA (25, 26). We have previously identified the manganese superoxide dismutase TPA-responsive element (MSTRE), in the 5′-flanking region, located between −1292 and −1202, which contains a cAMP-responsive element (CRE)-like sequence, and demonstrated that CREB/ATF-1 bound to MSTRE and TPA treatment induced CREB phosphorylation (27).
It has been proposed that the tumor-promoting phorbol ester, TPA, (28, 29) induces MnSOD expression in a protein kinase C (PKC)-dependent manner (27, 30–32). Our previous study using A549 cells reveals that PKC may be involved in CREB phosphorylation (27), and another study has shown that transcription factor FOXO3a (also known as FKHRL1) can elevate the expression of MnSOD in response to oxidative stress (33). In addition, FOXO transcription factors are known to trigger a variety of cellular processes by up-regulating a series of target genes, including sod2, in response to different cellular stresses (34). However, the mechanism by which PKC regulates MnSOD induction remains unclear.
In this study we investigated the roles of PKC in TPA-induced MnSOD in A549 cells. To this end we identified PKC-α as the PKC isozyme that catalyzes the phosphorylation of CREB. However, TPA treatment also causes a reduction in phosphorylated Akt (at Ser473) and FOXO3a (at Ser253), and the PKC inhibitor restored the phosphorylation level suppressed by TPA. In addition, we showed that knock-down of four components of NADPH oxidase diminished TPA-mediated MnSOD induction, suggesting that NADPH oxidase is involved in the early stage of MnSOD gene induction. This observation suggests to us that superoxide radical anions could be the upstream signal for TPA induction. Together, our data showed that PKC is involved in regulating the activation of transcription factors induced by TPA, via the phosphorylation of CREB on one hand and dephosphorylation of FOXO3a on the other hand.
EXPERIMENTAL PROCEDURES
Materials
DMSO, TPA, TNF-α, diphenyleneiodonium chloride (DPI), N-acetylcysteine (NAC), and 2′,7′-dichlorofluorescein diacetate (DCFH-DA) were purchased from Sigma. From Calbiochem (La Jolla, CA), we purchased PKC inhibitors BIM, Gö6983, Gö6976, and Rottlerin; and protein phosphatase 1 and 2A (PP1 and PP2A) inhibitor okadaic acid. The PH domain leucine-rich repeat protein phosphatase (PHLPP) inhibitor, NCS 45586 (#13) was a generous gift from Dr. Alexandria C. Newton, at the University of California San Diego, La Jolla, CA (35).
Cell Line
The human lung adenocarcinoma cell line A549 (ATCC CCL-185) was purchased from American Type Culture Collection. Cells were cultured in RPMI 1640 (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS) and antibiotics (Invitrogen) at 37 °C under 5% CO2.
Subcellular Fractionations
Subcellular fractions were prepared from A549 cells according to Enoksson et al. (36) with minor modification (37). In brief, fresh or frozen cells were lysed in MSH buffer (210 mm mannitol, 70 mm sucrose, 5 mm Hepes, pH 7.5) in the presence of 1 mm EDTA, incubated for 30 min on ice and homogenized with a tight-fitting glass-Teflon motorized homogenizer (500 rpm, 30 strokes). Homogenates were centrifuged at 600 × g for 8 min at 4 °C. The pellet was solubilized in lysis buffer (10 mm HEPES, pH 7.9, 10 mm KCl, 0.1 mm EDTA, 0.1 mm EGTA, 1 mm DTT, 0.5 mm PMSF, 2 μg/ml leupeptin, and 2 μg/ml aprotinin). After 30 min incubation on ice, 0.3% Nonidet P-40 was added, vortexed for 1 min, and centrifuged at 12,000 × g for 1 min. The pellet was lysed with nuclear extraction buffer (25 mm HEPES, pH 7.9, 0.5 mm EDTA, 0.5 mm EGTA, 0.4 m NaCl, 1 mm DTT, 1 mm PMSF, 2 μg/ml leupeptin, 2 μg/ml aprotinin). After centrifugation at 12,000 × g for 5 min at 4 °C, the nuclear fraction (supernatant) was collected. The supernatant from the first centrifugation was centrifuged at 5,500 × g for 15 min to obtain the mitochondrial fraction. Membrane and cytosolic fractions were prepared as described previously (30).
Western Blotting and Immunoprecipitation
Samples were electrophoresed in 10–20% Tris-Glycine gels (Invitrogen) and transferred onto polyvinylidene difluoride (PVDF) membranes (Invitrogen). After incubating with Li-Cor Blocking Buffer (Li-Cor Biosciences, Lincoln, NE) for 30 min, blots were incubated with specific primary antibodies against PKC-α (polyclonal, C-20), PKC-βI (polyclonal, C-16), PKC-βII (polyclonal, C-18), PKC-δ (polyclonal, C-20), PKC-ϵ (polyclonal, C-15), PKC-μ (polyclonal, C-20), p53 (monoclonal, DO-1), or α-tubulin (monoclonal, TU-02) from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA), or CREB (rabbit monoclonal, 48H2), phospho-CREB at Ser133 (rabbit monoclonal, 87G3), FOXO3a, phospho-FOXO3a (Ser253), Akt (polyclonal,), and phospho-Akt (Ser473, polyclonal) from Cell Signaling Technology (Danvers, MA), Rac1 (monoclonal, 05–389), and p67phox (polyclonal, 07–502) from Upstate (Lake Placid, NY), NOXO1 (goat polyclonal, IMG-3062) from IMGENEX (San Diego, CA), MnSOD (polyclonal, RDI-RTSODMabR) from Fitzgerald (Concord, MA), mtHSP70 (monoclonal, MA3–028) from Affinity Bioreagents (Golden, Co), β-actin (monoclonal (AC-74)) from Sigma or Lamin B (NA12) from Calbiochem in Li-Cor Blocking Buffer containing 0.1% Tween 20 for overnight at 4 °C. Blots were then incubated with either goat anti-rabbit IRDye800CW secondary antibodies (611-731-127, Rockland Immunochemicals, Gilbertsville, PA) or goat anti-mouse Alexa Fluor 680 secondary antibodies (A21058, Molecular Probes, Eugene, OR) and visualized using Odyssey Infrared Imaging System (Li-Cor Biosciences).
For immunoprecipitation, cells were lysed in lysis buffer (Cell Signaling Technology) containing Complete Mini, protease inhibitor mixture tablets (Roche Diagnostics, Indianapolis, IN) and phosphatase inhibitor cocktails (Set II and VI, Calbiochem) and were incubated with either normal rabbit IgG (sc-2027, Santa Cruz Biotechnology), or rabbit monoclonal phospho-Akt at Ser473 (193H12) antibody (Cell Signaling Technology) for 1 h and followed by an additional 30 min of incubation with protein A-agarose (Millipore, Lake Placid, NY). Immunoprecipitation and 2% of total cell lysates separated by SDS-PAGE and transferred onto PVDF were then incubated with phospho-Akt (Ser473) (193H12) antibody or PP2A C subunit antibody (2038, Cell Signaling Technology).
Assessment of Intracellular ROS
Production of ROS was assessed by oxidation-sensitive fluorescent probe DCFH-DA as described previously (38). Hydrogen peroxide (1 mm) was used as a positive control. After washing twice with PBS to remove the extracellular dye, cells were analyzed with a flow cytometry (FACSCalibur, BD Biosciences, San Jose, CA). DCFH-DA fluorescence was excited at 488 nm, and the emission was monitored at 530 nm.
Dual Priming Oligonucleotide (DPO) PCR System
mRNAs from A549 cells were anayzed using dual priming oligonucleotide (DPO) PCR system (39). DPO primers for NADPH oxidase component (supplemental Table S2) and for PKC family (supplemental Table S3), and housekeeping gene primers (SM1001) were purchased from Seegene Institute of Life Science (Seoul, Korea). They included dual oxidase (DUOX) 1 and 2; p47phox and its homologue Nox organizer 1 (NOXO1), p67phox and its homologue and Nox activator 1 (NOXA1); p40phox, Rac1, Rac2, and p22phox; classical PKC (PKC-α, -βI, -βII, and -γ), novel PKC (PKC-δ, -θ, -ϵ, and -η), atypical PKC (PKC-ζ and -λ), and PKD (PKC-μ).
Small-interfering RNA (siRNA)
Sixty hours after transfection with siRNAs (250 nm) using Amaxa Nucleofector System (Amaxa Biosystems, Gaithersburg, MD), cells were treated with TPA for the indicated times. In this study, both siGENOME SMART pool and ON-TARGETplus SMART pool (Dharmacon/Thermo Fisher, Chicago, IL) were used; Rac1 (M-003560-02), p22phox (L-011020-00), p67phox (L-004529-00), DUOX1 (L-008126-00), and NOXO1 (M-016237-00) for knockdown of NADPH oxidase components; PKC-α (L-003523-00), PKC-βI (L-003758-00), and PKC-δ (L-003524-00) for knockdown of PKC family; and CREB (L-003619-00), FOXO3a (L-003007-00) and p53 (M-003329-01, and M-003329-02 from Dharmacon; sc-29435 from Santa Cruz Biotechnology). A non-targeting pool was used as control RNA (ON-TARGETplus siCONTROL, Dharmacon).
Luciferase Assay
Small enhancer region in intron 2 of sod2 gene (I2E), which is responsive to tumor necrosis factor-α (TNF-α) and interleukin-1β (IL-1β), was found in mouse (23) and in human (24). Combined with 3340 base pairs of 5′-flanking region of the human sod2 gene promoter (27), three plasmids were constructed, in which I2E were located in N-terminal end (pSODLUC-3340-I2E-N) and C-terminal end (pSODLUC-3340-I2E-C) of the promoter, and between the promoter and the luciferase gene (Luc+) (pSODLUC-3340-I2E-ORF) (supplemental Fig. S1a). A549 cells were transfected with pSODLUC plasmids using Amaxa Nucleofector System. Twenty-four hours after transfection, cells were treated with TPA and incubated for another 24 h. The firefly and Renilla luciferase activities were measured using the Dual Luciferase assay system (Promega, Madison, WI).
Statistical Analysis
An unpaired two-tailed distribution Student's t test (Microsoft Excel) and a one-way ANOVA were used to analyze data, and results were expressed as means ± S.E. A value of p < 0.05 was considered significant.
RESULTS
Induction of MnSOD in Mitochondria by TPA-mediated Production of Superoxide Radical Anion
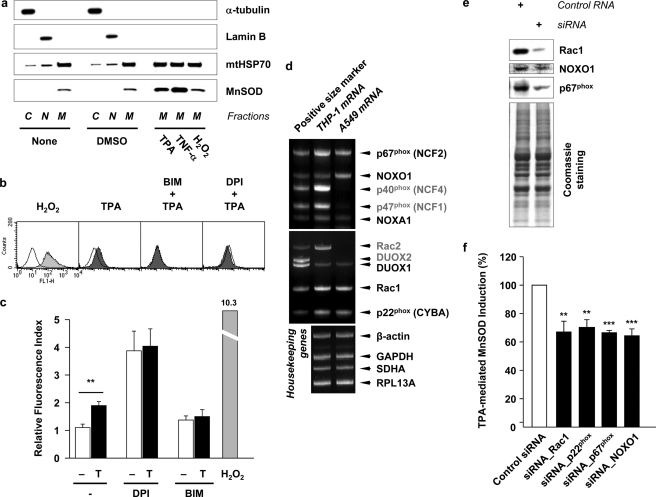
Previously we showed that the induction of MnSOD mRNA by TPA treatment was mediated by the manganese superoxide dismutase TPA-responsive element (MSTRE) in the 5′-flanking region of the human MnSOD gene promoter (27). Fig. 1a shows that MnSOD protein in mitochondria was significantly elevated by treatment of A549 cells with either TPA or TNF-α, but not with the externally added 1 mm hydrogen peroxide. It should be pointed out that when A549 cells were incubated in the presence of 5 mm H2O2 for 24 h at 37 °C, the cells were found dead and detached. However, when the concentration of H2O2 was reduced to 0, 0.5, and 1 mm under similar experimental conditions the cells exhibited similar confluency. The effect of TPA is consistent with the observed increase in MnSOD mRNA level, while the effect of TNF-α is in agreement with that reported by Wong et al. (9) using a human embryonic kidney (HEK) 293 cell line. Together these data suggest that the reactive oxygen species (ROS) produced by TPA or TNF-α treatments to induce MnSOD in mitochondria, is not H2O2.
FIGURE 1.
Superoxide radical anion generated by NADPH oxidase in response to TPA induces MnSOD in mitochondria. a, A549 cells were fractionated into cytosolic (C, α-tubulin as a marker), nuclear (N, LaminB as a marker), and mitochondrial (M, mtHSP70 as a marker) fractions after treatment of TPA (32.4 nm, (20 ng/ml)), TNF-α (10 ng/ml) or hydrogen peroxide (H2O2) (1 mm) for 24 h at 37 °C. Each fraction sample was separated using 10–20% Tris-glycine gel and analyzed by immunoblotting. b, after incubation with either PKC inhibitor BIM (1 μm) or NADPH oxidase inhibitor DPI (100 nm) for 1 h, cells were treated with TPA for 30 min. The cells were further incubated in the presence of 40 μm 2′,7′-dichlorofluorescein diacetate (DCFH-DA) for 30 min at 37 °C. Stained cells were washed with PBS and analyzed by flow cytometry (FACS). H2O2 (1 mm) was treated as a positive control. c, DCF fluorescence index was presented as a ratio to the cells not exposed to both inhibitors and TPA (T). Values are means ± S.E. (n ≥ 4, **, p < 0.01). d, components of NADPH oxidase in A549 cells are examined by dual priming oligonucleotide (DPO) PCR system (missing components in A549 cells are presented as gray letters). For the data in e and f, small interfering RNA (siRNA) for Rac1, p22phox, p67phox, and NOXO1 (250 nm each) were used to knockdown each components of NADPH oxidase, which exist in A549 cells. Sixty hours after transfection of siRNAs, cells were treated with TPA for additional 24 h at 37 °C. e, Western blots for Rac1, NOXO1 and p67phox after knockdown by each siRNAs. Coomassie-stained gel shows that equal amounts of sample were loaded. f, inhibitory effect of siRNAs on the TPA-mediated MnSOD induction was presented as a percentage to the MnSOD induction in cells transfected with control siRNA and treated with TPA. Values are means ± S.E. (n ≥ 3, **, p < 0.01 and ***, p < 0.001 versus control siRNA by ANOVA; n.s., not significant).
The results in Fig. 1c show the intracellular concentration of ROS was elevated by 1.9-fold after A549 cells were treated with 32.4 nm (20 ng/ml) of TPA for 1 h. However this observed increase of intracellular ROS induced by TPA, monitored by DCF fluorescence, was suppressed either by PKC inhibitor BIM (1 μm) or by NADPH oxidase inhibitor DPI (100 nm) (Fig. 1, b and c). It should be pointed out that DPI by itself causes a significant increase in ROS, a phenomenon due to DPI-elicited inhibition of cell redox metabolism and augmented oxidative stress (40). These results suggest that ROS produced in response to TPA is a superoxide radical anion (O2˙̄) generated by NADPH oxidase or flavoproteins in a PKC-dependent manner.
It is generally believed that TPA-induced ROS is mediated by PKC activation which in turn activates NADPH oxidase. To identify which components of NADPH oxidase are crucial in TPA-mediated MnSOD induction in A549 cells, a dual priming oligonucleotide (DPO) PCR system was used. As shown in Fig. 1d, we observed the mRNAs for p67phox, NOXO1 (p47phox homologue) NOXA1 (p67phox homologue), DUOX1, Rac1, and p22phox in A549 cells. However, mRNA for p40phox, p47phox (NCF1), Rac2, or DUOX2 was not detected. To show the effect of knocking down the observed components of NADPH oxidase, such as Rac1, p22phox, p67phox, and NOXO1, on TPA-induced MnSOD protein expression in A549 cells siRNA methods were used. Fig. 1e shows the efficiency of siRNA in reducing the protein level of Rac1, NOXO1 and p67phox. The results in Fig. 1f show that knocking down Rac1, p22phox, p67phox, or NOXO1, each contributes to a reduction of 30% in the amplitude of TPA-induced MnSOD expression, relative to that obtained with control siRNA-transfected cells. These results suggest that not only cytosolic regulatory factors, such as p67phox, NOXO1 (p47phox), and Rac1, but membrane-bound p22phox is also required to induce MnSOD expression.
PKC-dependent TPA-mediated MnSOD Induction
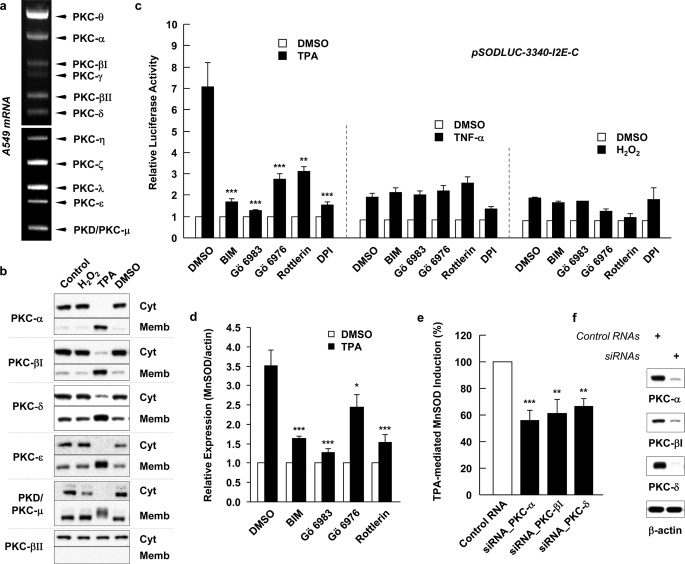
To investigate which PKC isozyme is responsible for the TPA induced MnSOD in A549 cells, the PKC isozymes in these cells were identified using the DPO PCR method (Fig. 2a). These results reveal that PKC-α, -βI, -βII, -γ, -δ, -θ, -ϵ, -η, -ζ, -λ, and -μ, are expressed in A549 cells. Fig. 2b shows the effect of TPA and H2O2 on the activation and membrane translocation of PKC isozyme in A549 cells. Using six different PKC polyclonal antibodies, we detected that PKC-α, -βI, -δ, -ϵ, and -μ (PKD) but not PKC-βII were translocated to the membrane fractions in TPA-treated cells. Interestingly none of these PKC isoforms was affected by H2O2 treatment. The TPA-mediated translocations of PKC-α, -βI, -δ, and -ϵ to the membrane fraction were somewhat similar to those in the previous study (30), except in our hands the membrane translocation of PKC-α is much more robust and the membrane translocated PKC-μ appeared to be covalently modified since the band is more diffused and migrated at a slower rate.
FIGURE 2.
TPA-induced MnSOD expression was blocked by PKC inhibitors and siRNA knockdown on specific isozyme. a, in A549 cell, PKC isoforms were identified using DPO PCR system. b, translocations of PKCs to subcellular membrane were analyzed by immunoblotting after treatment with 32.4 nm TPA or 1 mm H2O2 for 30 min. Cyt, cytosolic fraction; Memb, membrane fraction. c, A549 cells were preincubated with BIM (1 μm, inhibits PKC-α, -βI, -βII, -γ, -δ, and -ϵ), Gö 6983 (1 μm, inhibits PKC-α, -βI, -βII, -γ, -δ, and -ζ), Gö 6976 (1 μm, inhibits PKC-α, -βI, and -μ), or Rottlerin (5 μm, inhibits PKC-δ and -θ) and treated with 32.4 nm TPA, 10 ng/ml of TNF-α or 1 mm H2O2 for 24 h. The effect of PKC inhibitors on the TPA-, TNF-α-, or H2O2-induced reporter gene activity was presented as relative luciferase activities to its activities in cells transfected with pSODLUC-3340-I2E-C and treated with the same volume of DMSO (vehicle) used in other samples. Values shown are means ± S.E. (n ≥ 4, **, p < 0.01 and ***, p < 0.001 versus TPA-only treated group by ANOVA). d, after treatment as described previously, whole cell lysates were subjected to SDS-PAGE and blotted with anti-MnSOD antibody. The effect of PKC inhibitors on the TPA-mediated MnSOD induction was presented as a ratio to the MnSOD induction in cells pretreated with DMSO and then treated with TPA. Values are means ± S.E. (n ≥ 3, *, p < 0.05 and ***, p < 0.001 versus TPA-only treated group by ANOVA). e, inhibitory effect of siRNAs for PKC-α, -βI, and -δ (250 nm each) on the TPA-mediated MnSOD induction was presented as a percentage to the MnSOD induction in cells transfected with control siRNA and treated with TPA. (n ≥ 6, **, p < 0.01 and ***, p < 0.001 versus control siRNA by ANOVA). f, reduction of PKC-α, -βI, and -δ protein expression by siRNA.
To assess the effect of a PKC inhibitor on the TPA-induced transcription of sod2, a luciferase reporter plasmid was used. Because MnSOD was induced by TPA and TNF-α (Fig. 1a), and the pSODLUC-3340 plasmid did not respond to TNF-α treatment (27), new plasmids, which include the intron 2 enhancer (I2E) fragment, known as a responsive element to TNF-α in murine (23) and human (24), were constructed. The three plasmids shown in supplemental Fig. S1a were constructed as described under “Experimental Procedures.” The new plasmids allow one to monitor the possible synergic effect of I2E on TPA-mediated luciferase activity. supplemental Fig. S1b shows that A549 cells transfected with pSODLUC-3340 plasmid did not exhibit any increase in luciferase activity due to TNF-α treatment. However, cells transfected with pSODLUC-3340-I2E-C, -N, and -ORF plasmids induced sod2 reporter gene expression by 2.4-, 3.6-, and 3.5-fold, respectively, due to TNF-α treatment, consistent with previous reports (23, 27). Moreover, the insertion of I2E also enhanced the TPA-mediated luciferase activity by 6.1-fold (pSODLUC-3340-I2E-C), 8.5-fold (pSODLUC-3340-I2E-N) and 5.3-fold (pSODLUC-3340-I2E-ORF) (supplemental Fig. S1b). Because I2E of pSODLUC-3340-I2E-C is located at a similar distance from the promoter as the original site of I2E and only pSODLUC-3340-I2E-C construct showed the inhibitory effect of Rottlerin (data not shown), it was selected for further promoter studies. To identify the effective PKCs, among those activated and translocated to membrane fraction by TPA, on MnSOD induction, four different PKC inhibitors, BIM (1 μm), Rottlerin (5 μm), Gö 6983 (1 μm), and Gö 6976 (1 μm), were used. Their Ki or IC50 for a given PKC isozyme are summarized in supplemental Table S1. Fig. 2c shows that treatment with 20 ng/ml of TPA causes a 7-fold increase in transcription activation of sod2 and this induction was blocked by all four PKC inhibitors used and by DPI. Among the PKC inhibitors, general PKC inhibitors, BIM and Gö 6983 are the most potent relative to Gö 6976, which inhibits PKC-α and -βI, and to Rottlerin. However, sod2 gene expression in response to TNF-α or H2O2 treatment was not significantly inhibited by any PKC inhibitors or DPI.
Fig. 2d shows that TPA-induced MnSOD protein generation was also inhibited by the PKC inhibitors. BIM and Gö 6983 yielded a 53.5 and 63.8% inhibition, respectively; while Gö 6976 and Rottlerin yielded a 30.1 and 56.3% inhibition, respectively. The effect of an individual PKC isozyme on the TPA-induced MnSOD generation was also investigated using the siRNA knockdown method. In this case, the siRNAs for PKC-α, PKC-βI and PKC-δ were transfected to reduce their expression. The effectiveness of the knockdown is shown in Fig. 2f. The inhibitory effects of knocking down PKC-α, -βI, and -δ are shown in Fig. 2e. The results indicated that knocking down PKC-α, -βI, and -δ individually leads to 44.2, 38.9, and 33.5% inhibition respectively of TPA-induced MnSOD formation.
TPA-induced PKC-α-catalyzed CREB Phosphorylation
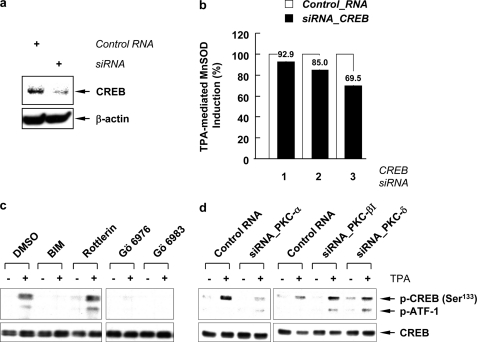
Previously we reported that transcription activation of MnSOD in A549 cells induced by TPA is mediated via the binding of MSTRE to a CREB-1/ATF-1-like transcription factor, which is activated by PKC-catalyzed phosphorylation of CREB at Ser133 (27). The participation of CREB in the TPA-mediated MnSOD induction was further determined using siRNA technique. When siRNA for CREB was transfected into A549 cells, it successfully reduced the CREB protein (Fig. 3a). The reduction of the CREB protein led to a 30.5% suppression of the TPA-mediated MnSOD induction at 60 h (Fig. 3b). To link the upstream PKCs with the downstream transcription factors, we examined the effect of PKC inhibitors or siRNA on the phosphorylation of CREB. When cells were pretreated with BIM, Gö 6983, or Gö 6976, the TPA-stimulated phosphorylations of CREB were completely eliminated by inhibitors of classical PKCs, but not by Rottlerin, an inhibitor of PKC-δ and -θ (Fig. 3c). In addition, knockdown of PKC-α by siRNA also abolished CREB phosphorylation (Fig. 3d), while the reduction of PKC-βI and PKC-δ had no effect on the TPA-mediated phosphorylation of CREB. These data suggest that TPA-stimulated phosphorylation of CREB is mediated by PKC-α, and other PKCs induce expression of MnSOD by a path that does not involve CREB.
FIGURE 3.
Phosphorylation of CREB catalyzed by PKC in TPA mediated MnSOD induction. a, Western blots for CREB after knockdown by siRNA. b, inhibitory effect of CREB siRNA on the TPA-mediated MnSOD induction was presented as a percentage to the MnSOD induction in cells transfected with control siRNA using the Amaxa Nucleofector System and treated with TPA. Lane 1, transfected with 100 nm siRNA for 48 h; lanes 2 and 3, transfected with 250 nm siRNA for 48 h in lane 2 and 60 h in lane 3. c, cells were pretreated with BIM (1 μm), Gö 6983 (1 μm), Gö 6976 (1 μm), or Rottlerin (5 μm) for 1 h and treated with TPA for 30 min. d, siRNA for PKC-α, -βI, and -δ (100 nm each) were used to knockdown each gene product. Two days after transfection of siRNAs, cells were treated with TPA for 30 min. Total cell lysates were subjected to SDS-PAGE and blotted with phospho-CREB (Ser133) and CREB antibodies.
PKC-dependent Dephosphorylation of Akt and FOXO3a Induced by TPA Treatment
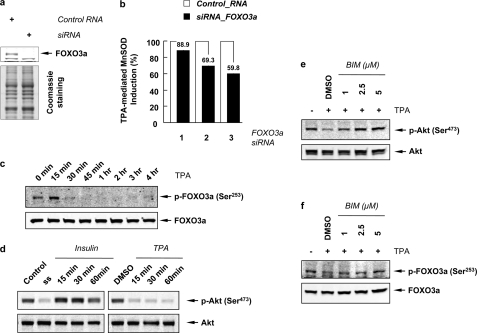
It has been shown that MnSOD protein expression is regulated by a PI3K-Akt-Forkhead signaling pathway (33). Compared with other transcription factors, the molecular mechanism by which Forkhead transcription factors regulates MnSOD gene expression after TPA treatment is not well characterized. In order to examine whether the Forkhead transcription factor FOXO3a (FKHRL1) is involved in the TPA-mediated MnSOD induction, siRNA for FOXO3a was transfected to block its expression. Fig. 4a shows that siRNA for FOXO3a successfully abolished its target protein. As a result of the FOXO3a knockdown, TPA-mediated MnSOD induction was inhibited by up to 40.2% (Fig. 4b). Fig. 4c shows that in the nuclear fraction, TPA treatment led to the dephosphorylation of Ser253 in FOXO3a within 30 min.
FIGURE 4.
TPA induces the dephosphorylation of Akt and FOXO3a in a PKC-dependent manner. a, Western blots for FOXO3a after knockdown by siRNA. Coomassie-stained gel shows that equal amounts of sample were loaded. b, inhibitory effect of FOXO3a siRNA on the TPA-mediated MnSOD induction is presented as percent of MnSOD generated relative to that observed with control siRNA. Lane 1, transfected with 100 nm siRNA for 48 h; lanes 2 and 3, transfected with 250 nm siRNA for 48 h in lane 2 and 60 h in lane 3. c, nuclear fractions of A549 cells treated with TPA for the indicated time (up to 4 h), were separated in 8% Tris-glycine gel, and blotted with phospho-FOXO3a (Ser253) and FOXO3a antibodies. d, phosphorylation of Akt was examined by Western blotting using phospho-antibody against Ser473 residue of Akt after treated serum-starved (ss) cells with insulin (200 nm) (left panel) or TPA (right panel). e and f, A549 cells were preincubated with BIM (1, 2.5, and 5 μm) for 1 h and treated with TPA for an additional 1 h. Total cell lysates were blotted with phospho-Akt (Ser473) and Akt antibodies (e), and nuclear fractions were blotted with phospho-FOXO3a (Ser253) and FOXO3a antibodies (f). Note that these blots represent one of three independent experiments.
Because Akt is a known upstream kinase for FOXO3a (41), we next investigated the phosphorylation of Akt after TPA treatment. Fig. 4d shows that Akt phosphorylation at Ser473 decreased due to serum starvation and elevated rapidly due to insulin treatment. In contrast, Akt phosphorylation decreased dramatically within 15 min induced by TPA treatment and sustained for up to 24 h (data not shown). These observations are in accord with the reports showing that TPA attenuated Akt phosphorylation in A549 cells (42) and PKC-δ and PKC-ϵ negatively regulated the phosphorylation of Akt in mouse keratinocytes (43). To investigate the effect of PKC on the the dephosphorylation of Akt and FOXO3a, cells were preincubated with BIM and then treated with TPA. As shown in Fig. 4, e and f, the TPA-stimulated dephosphorylation of Akt at Ser473 and of FOXO3a at Ser253 were restored in a BIM concentration-dependent manner. These results suggest that the dephosphorylation of Akt and FOXO3a is mediated by PKC.
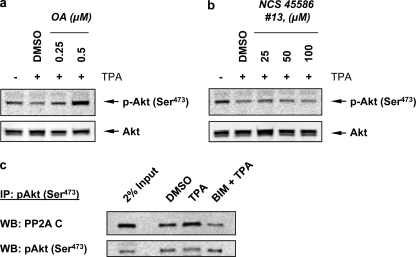
The phosphorylated Akt at the hydrophobic motif Ser473 is known to be dephosphorylated by okadaic acid-sensitive phosphatases such as PP2A (44–47) and by PHLPP (48). To differentiate which of these family of phosphatases is involved in regulating the activity of Akt mediated by TPA treatment, A549 cells were treated with either PP1 and PP2A inhibitor okadaic acid (46) or PHLPP inhibitor NCS 45586 (35). Fig. 5a shows that the dephosphorylation of Akt was inhibited by preincubation with okadiac acid in a dose-dependent manner. However NCS 45586 failed to inhibit Akt dephosphorylation (Fig. 5b). To investigate the mechanism by which PKC mediated the dephosphorylation of Akt catalyzed by PP2A, we observed that TPA induced immuno-coprecipitation of pAkt complex with the catalytic subunit of PP2A and BIM inhibited the complex formation (Fig. 5c). This observation is in accord with the finding of Li et al. using mouse keratinocytes (43). Together, these results suggest that PKC induces the complex formation between pAkt and the catalytic subunit of PP2A to facilitate the dephosphorylate Akt, which in turn activates FOXO3a by reducing the population of the phosphorylated FOXO3a at Ser253.
FIGURE 5.
TPA induces PKC-mediated Akt dephosphorylation catalyzed by PP2A and not by PHLPP. Cells were pretreated with either PP2A inhibitor okadaic acid (OA) (a), or PHLPP inhibitor NCS 45586 (#13) (b) for 1 h, and then treated with TPA for another 1 h. Total cell lysates were blotted with phospho-Akt (Ser473) and Akt antibodies. c, cells treated with DMSO (vehicle), TPA only, or BIM plus TPA were lysed, and immunoprecipitation (IP) was performed using phospho-Akt (Ser473) antibodies. The immunoprecipitants and total cell lysates were analyzed by Western blotting (WB) for phospho-Akt (Ser473) and PP2A C subunit. Two percentages of total cell lysates were saved before IP and were also immunoblotted as a control (2% Input). Note that these blots represent one of three independent experiments.
TPA-induced MnSOD Expression in A549 Cells Is Independent of p53
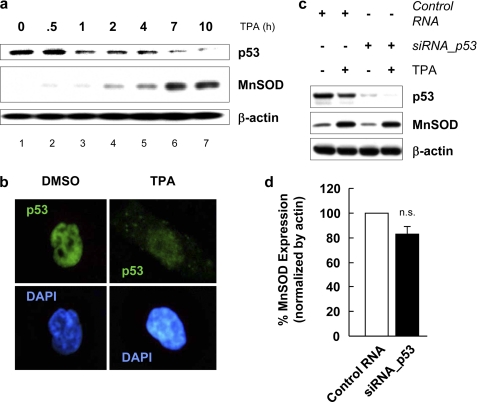
p53 has been shown to suppress MnSOD mRNA and protein levels by forming a complex with the Sp1 site at the 5′-flanking promoter region of the sod2 gene and led to the reduction of transcription activity under both the constitutive and TPA-stimulated conditions in human hepatoma cell line HepG2 (18). With A549 cells, TPA induced a time-dependent reduction of p53 protein, while the MnSOD protein was elevated (Fig. 6a). A histochemical analysis revealed that the observed reduction of p53 was due to its translocation from the nucleus to the cytosol to be degraded (Fig. 6b). This TPA-mediated translocation and degradation of p53 was blocked by pretreatment of the cells with a potent and membrane permeable proteasome inhibitor, MG132, and a nuclear export inhibitor, leptomycin B (LMB) (data not shown). This apparent correlation, which could lead one to conclude that p53 also suppresses MnSOD expression in A549 cells, was uncoupled with experiments using siRNA to reduce the p53 protein expression. The results shown in Fig. 6c reveal p53 siRNA effectively lowering the p53 protein. However it exhibits little differential effect on the level of MnSOD protein expression in the absence or presence of p53. The quantitative effect of p53 siRNA knockdown is shown in Fig. 6d. Together these results indicate that while the variation of p53 and MnSOD induced by TPA suggests that p53 suppresses MnSOD expression in A549 cells, the siRNA data suggest otherwise, that p53 does not suppress MnSOD expression.
FIGURE 6.
p53 is not a suppressor of MnSOD gene expression in human lung adenocarcinoma cell line A549. a, whole lysates of A549 cells treated with TPA for the indicated time period (up to 10 h), were separated in 10% Tris-glycine gel, and blotted with anti-p53 and anti-MnSOD antibodies. b, after TPA treatment, A549 cells on microscope slides in 6-well plates were fixed with 4% paraformaldehyde for 15 min at room temperature, permeabilized with 0.1% Triton X-100 for 5 min, and incubated with 2% BSA for 1 h to block nonspecific staining. The cells were then immunostained with anti-p53 antibody in 2% BSA for 1 h at room temperature and further incubated with fluorescence-conjugated secondary antibodies (Jackson ImmunoResearch, West Grove, PA) for 1 h at room temperature. To visualize nuclei, the cells were stained with 4′,6′-diamidino-2-phenylindole (DAPI, Sigma) for 5 min. Finally, the cells were mounted onto slides using FluoroGuard antifade reagent (Bio-Rad). Immunofluorescence was examined using fluorescence microscope Axiovert (Carl Zeiss, Oberkochen, Germany). c, effect of p53 siRNA on the TPA-mediated MnSOD induction was monitored by Western blotting. d, quantitative effect of p53 siRNA on the TPA-induced MnSOD production is shown as percent MnSOD formed normalized by actin, with the value of 100% set for cells transfected with control siRNA followed with TPA treatment. Values shown are means ± S.E. (n = 3, n.s., not significant).
DISCUSSION
Phorbol esters, the pharmacological analogs of DAG, act as tumor promoters through activation of PKC. Among them TPA is the most widely used in models to study carcinogenesis (28). In this study, we investigated the mechanism by which TPA induces expression of MnSOD, a primary anti-oxidant enzyme, expression in A549 cells. p53, a tumor suppressor, has been reported to function as a pro-apoptotic factor under cellular response to stress conditions, and it has been shown that p53 and sod2 genes are reciprocally regulated in HepG2 and MCF-7 cells (18, 49). However, despite the fact that our data also revealed an apparent reciprocal effect on the expression of p53 and MnSOD induced by TPA treatment, the results obtained from the siRNA study are inconsistent with the notion that p53 suppresses MnSOD expression (Fig. 6, c and d). The reduction of p53 protein observed was caused by the TPA-mediated translocation of p53 from the nucleus to the cytosol for degradation.
We have previously shown that TPA induced MnSOD expression was blocked by inhibitors of flavoproteins and NADPH oxidase suggesting that superoxide radical anion generation is likely an upstream signal (27). Here we show that A549 cell contains a number of components needed for constituting the reactive NADPH oxidase (Fig. 1d). In accord with the notion that the super oxide radical anion is an upstream signal, the results of siRNA knockdown of Rac1, p67phox, and NOXO1 (p47phox) significantly reduced the amplitude of MnSOD expression, while hydrogen peroxide failed to support the TPA-induced MnSOD generation.
It has been demonstrated that transcriptional activation of sod2 mediated by TPA in A549 cells involves the binding of CREB-1/ATF-1-like transcription factor to a CRE-related sequence, MSTRE, in the promoter region of the gene (27). However TPA treatment did not lead to any change in the abundance or binding activity of CREB-1/ATF-1 complexes. This observation is in accord with the known behavior of CREB as a signal-dependent activator, whose trans-activation potential is specifically affected by phosphorylation instead of its nuclear targeting or its DNA binding activity (50). The transcription factor, CREB, is known to be regulated by a number of protein kinases, including PKA and PKC (51, 52). Because we found that the specific inhibitor for PKC, BIM, rather than the PKA inhibitor, H89, blocked the TPA-mediated induction of MnSOD mRNA (27), we used different PKC inhibitors and siRNA techniques to knockdown specific PKC isozymes in an attempt to identify the PKC isozyme which catalyzes the phosphorylation of CREB in response to TPA treatment. The results show that knocking down PKC-α leads to a 44.2% reduction in TPA-induced MnSOD expression (Fig. 2e). Under these conditions, the phosporylation of CREB at Ser133 is nearly eliminated (Fig. 3, c and d). Together these results indicate that PKC-α is the PKC isozyme that catalyzed the phosphorylation of CREB and lead to its transcriptional activation in response to TPA treatment (Fig. 7). In addition, the data in Fig. 2e show that knocking down PKC-δ and PKC-βI also leads to a reduction of MnSOD expression. However the PKC-δ inhibitor Rottlerin and the siRNA knockdown of PKC-βI and of PKC-δ exhibit little effect on CREB Ser133 phosphorylation. Nevertheless, PKC-δ has been shown to up-regulate the NF-kB-mediating MnSOD induction pathway (26, 30, 31).
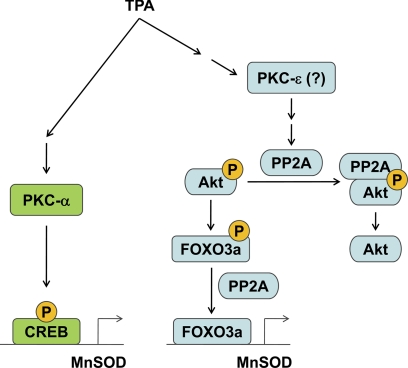
FIGURE 7.
A simplified pathway to depict the TPA-induced MnSOD expression in A549 cells. Here TPA was shown to activate PKC-α, which in turn phosphorylates the CREB transcription factor, and PKC-ϵ, which induces the formation of PP2A-pAkt complex to facilitate the dephosphorylation of pAkt and subsequently lead to the accumulation the dephosphorylated and active FOXO3a.
The mammalian FOXO family of Forkhead transcription factors contains four family members, namely: the FOXO1/FKHR, FOXO3/FKHRL1, FOXO4/AFX, and FOX6. They play an important role in longevity and growth/tumor suppression by up-regulating target genes involved in stress response (e.g. MnSOD), metabolism, cell cycle arrest (p21Waf1/Cip1 and p27Kip1), DNA repair (GADD45 and DDB1), apoptosis, and autophagy (Bim and FasL) (34, 53). In our study, we found that A549 cells were arrested at G1 phase after 24 h of TPA treatment (supplemental Fig S2a), with an increase in the levels of p27Kip1, GADD45, FasL, as well as MnSOD. Among them, MnSOD was the only target gene whose induction by TPA was blocked by BIM (supplemental Fig. S2b). These observations indicate that TPA induces FOXO transcription factor activation in A549 cells to trigger up-regulation of target genes. They include p27, fasl, gadd45, and sod2, and the induction of sod2 is inhibited by PKC inhibitor, BIM. The increase of p27Kip1 may, in part, responsible for the observed cell cycle arrest at G1 phase (54). Furthermore, siRNA knockdown of CREB and FOXO3a led to a 30.5, and a 40.2% inhibition, respectively, (Figs. 3b and 4b) in TPA-mediated MnSOD induction. Together, these results indicate that PKC is also involved in up-regulating FOXO3a transcription activity.
The activity of FOXO3a is regulated by reversible covalent modification, including reversible phosphorylation at three conserved residues, Thr32, Ser253, and Ser315. The phosphorylation is catalyzed by Akt, GSK, and AMPK, while the dephosphorylation is catalyzed by PP2A (34, 47). Because Ser253 overlaps with the nuclear localization signaling (NLS) sequence, phosphorylation on this residue will likely interfere with nuclear import by disrupting the positively charged NLS, and impair its ability to react with its target genes. Akt, a known kinase that phosphorylates FOXO3a at Ser253, is activated by the phosphorylation of its Ser473. The dephosphorylation of Ser473 is catalyzed by PP2A (44, 45, 47) and by PHLPP (48). Experiments using overexpression of Akt and PHLPP in 293T cells or overexpression of PHLPP in H157 or MDA-MB-231 cells revealed that PHLPP specifically dephosphorylated pSer473 relative to pThr308 in Akt (48). However, the specificity for the dephosporylation of pSer473 and pThr308 by PP2A appears to be regulated by the nature of its B regulatory subunit (45, 55). PP2A-B55 and PP2A-B56 holoenzymes have been shown to preferentially dephosphorylates pThr308 and pSer473, respectively. Under our experimental conditions, pAkt (Ser473) was dephosphorylated by okadaic acid-sensitive PP2A and not by PHLPP (Fig. 5, a and b). Therefore for PKC to activate FOXO3a, it must somehow activate PP2A activity. To this end, we found that TPA induces a complex formation between the phosphorylated Akt and the catalytic subunit of PP2A, and the stability of this complex is mediated by the activity of PKC (Fig. 5c). This observation is in acord with reports showing that activation of PKC-δ and PKC-ϵ provides a negative regulation for Akt phosphorylation and its kinase activity in mouse keratinocytes and TPA sitimulated the association PP2A with Akt (43).
Investigating which of the PKC isozymes, PKC-δ or PKC-ϵ or both, is responsible for inducing the PP2A-mediated dephosphorylation of pAkt (Ser473) and pFOXO3a (Ser253), we found that BIM, a PKC inhibitor for PKC-α, -βI, -βII, -γ, and -ϵ, inhibited the TPA-induced dephosphorylation of pAkt (Ser473) and pFOXO3a (Ser253) in a concentration dependent manner (Fig. 4, e and f). However, when A549 cells were pre-treated with either 1 μm Gö 6976, which inhibits PKC-α and -βI, or with 5 μm Rottlerin, an inhibitor for PKC-δ and -θ, no inhibition of TPA-induced dephosphorylation of pAkt (Ser473) and pFOXO3a (Ser253) was detected (data not shown). Because TPA does not activate PKC-βII (Fig. 2b), an isozyme inhibited by BIM, that leaves PKC-ϵ and PKC-γ as the only PKC isozymes inhibited by BIM but not by Gö 6976 and Rottlerin. Between these two PKC isozymes, PKC-ϵ has been shown, together with PKC-δ, as the PKC isozymes negatively regulated the phosphorylation of Akt and its kinase activity in mouse keratinocytes (43). However, PKC-δ can be ruled out because Rottlerin failed to inhibit TPA-induced Akt dephosphorylation. Therefore PKC-ϵ is likely the PKC isozyme responsible for mediating the dephosphorylation of Akt and FOXO3a (Fig. 7). This simplified model depicted in Fig. 7 shows that activation of PKC-ϵ would lead to complex formation between PP2A and pAkt to facilitate the dephosphorylation of pAkt and pFOXO3a and lead to transcriptional activation of FOXO3a.
In summary, the elevation of MnSOD expression was mediated by NADPH oxidase pathway in A549 cells in response to TPA or TNF-α treatment, while treatment with external hydrogen peroxide fails induce a similar effect. In addition, while the effects of TPA on MnSOD induction and on p53 suppression appear to suggest that p53 suppresses MnSOD expression, our results from siRNA knockdown of p53 indicate otherwise, namely that p53 does not suppress MnSOD expression. Furthermore, the TPA-induced MnSOD generation was abolished by a general PKC inhibitor, BIM. Using various PKC isozyme inhibitors and siRNA knockdown methods, we reveal that TPA-induced MnSOD expression is, in part, regulated by a PKC-α-catalyzed phosphorylation of CREB transcription factor, and by a PKC mediated dephosphorylation of Akt at Ser473 catalyzed by PP2A, which in turn lead to FOXO3a dephosphorylation at Ser253. Thus, PKC, activated by a TPA-mediated pathway, is involved in activating at least two transcription factors, one via the phosphorylation of CREB and the other via a PKC-mediated dephosphorylation of Akt and FOXO3a by PP2A.
Supplementary Material
Acknowledgment
We thank Dr. Alexandria C. Newton of the University of California San Diego, La Jolla, CA for the generous gift of the PHLPP-specific inhibitor, NCS 45586 (#13).
This work was supported, in whole or in part, by the Intramural Research Program of NHLBI, National Institutes of Health.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and Tables S1–S3.
- MnSOD
- manganese superoxide dismutase
- PKC
- protein kinase C
- CREB
- cAMP-responsive element-binding protein
- FOXO
- Forkhead box class O
- TPA
- 12-O-tetradecanoylphorbol-13-acetate
- ROS
- reactive oxygen species
- siRNA
- small interfering RNA
- PHLPP
- PH domain leucine-rich repeat protein phosphatase.
REFERENCES
- 1. Weisiger R. A., Fridovich I. (1973) J. Biol. Chem. 248, 4793–4796 [PubMed] [Google Scholar]
- 2. Fridovich I. (1995) Annu. Rev. Biochem. 64, 97–112 [DOI] [PubMed] [Google Scholar]
- 3. Fridovich I. (1978) Science 201, 875–880 [DOI] [PubMed] [Google Scholar]
- 4. Li Y., Huang T. T., Carlson E. J., Melov S., Ursell P. C., Olson J. L., Noble L. J., Yoshimura M. P., Berger C., Chan P. H., Wallace D. C., Epstein C. J. (1995) Nat. Genet. 11, 376–381 [DOI] [PubMed] [Google Scholar]
- 5. Oberley L. W., Buettner G. R. (1979) Cancer Res. 39, 1141–1149 [PubMed] [Google Scholar]
- 6. St Clair D. K., Wan X. S., Oberley T. D., Muse K. E., St Clair W. H. (1992) Mol. Carcinog. 6, 238–242 [DOI] [PubMed] [Google Scholar]
- 7. Church S. L., Grant J. W., Ridnour L. A., Oberley L. W., Swanson P. E., Meltzer P. S., Trent J. M. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 3113–3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wong G. H., Goeddel D. V. (1988) Science 242, 941–944 [DOI] [PubMed] [Google Scholar]
- 9. Wong G. H., Elwell J. H., Oberley L. W., Goeddel D. V. (1989) Cell 58, 923–931 [DOI] [PubMed] [Google Scholar]
- 10. Visner G. A., Dougall W. C., Wilson J. M., Burr I. A., Nick H. S. (1990) J. Biol. Chem. 265, 2856–2864 [PubMed] [Google Scholar]
- 11. Harris C. A., Derbin K. S., Hunte-McDonough B., Krauss M. R., Chen K. T., Smith D. M., Epstein L. B. (1991) J. Immunol. 147, 149–154 [PubMed] [Google Scholar]
- 12. Fujii J., Taniguchi N. (1991) J. Biol. Chem. 266, 23142–23146 [PubMed] [Google Scholar]
- 13. Whitsett J. A., Clark J. C., Wispé J. R., Pryhuber G. S. (1992) Am. J. Physiol. 262, L688–693 [DOI] [PubMed] [Google Scholar]
- 14. Oberley L. W., St Clair D. K., Autor A. P., Oberley T. D. (1987) Arch. Biochem. Biophys. 254, 69–80 [DOI] [PubMed] [Google Scholar]
- 15. Akashi M., Hachiya M., Paquette R. L., Osawa Y., Shimizu S., Suzuki G. (1995) J. Biol. Chem. 270, 15864–15869 [DOI] [PubMed] [Google Scholar]
- 16. Yeh C. C., Wan X. S., St Clair D. K. (1998) DNA Cell Biol. 17, 921–930 [DOI] [PubMed] [Google Scholar]
- 17. Xu Y., Porntadavity S., St Clair D. K. (2002) Biochem. J. 362, 401–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dhar S. K., Xu Y., Chen Y., St Clair D. K. (2006) J. Biol. Chem. 281, 21698–21709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Porntadavity S., Xu Y., Kiningham K., Rangnekar V. M., Prachayasittikul V., St Clair D. K. (2001) DNA Cell Biol. 20, 473–481 [DOI] [PubMed] [Google Scholar]
- 20. Xu Y., Krishnan A., Wan X. S., Majima H., Yeh C. C., Ludewig G., Kasarskis E. J., St Clair D. K. (1999) Oncogene 18, 93–102 [DOI] [PubMed] [Google Scholar]
- 21. Zhu C. H., Huang Y., Oberley L. W., Domann F. E. (2001) J. Biol. Chem. 276, 14407–14413 [DOI] [PubMed] [Google Scholar]
- 22. Xu Y., Fang F., Dhar S. K., Bosch A., St Clair W. H., Kasarskis E. J., St Clair D. K. (2008) Mol. Cancer Res. 6, 1881–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jones P. L., Ping D., Boss J. M. (1997) Mol. Cell Biol. 17, 6970–6981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xu Y., Kiningham K. K., Devalaraja M. N., Yeh C. C., Majima H., Kasarskis E. J., St Clair D. K. (1999) DNA Cell Biol. 18, 709–722 [DOI] [PubMed] [Google Scholar]
- 25. Kiningham K. K., Xu Y., Daosukho C., Popova B., St Clair D. K. (2001) Biochem. J. 353, 147–156 [PMC free article] [PubMed] [Google Scholar]
- 26. Dhar S. K., Lynn B. C., Daosukho C., St Clair D. K. (2004) J. Biol. Chem. 279, 28209–28219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kim H. P., Roe J. H., Chock P. B., Yim M. B. (1999) J. Biol. Chem. 274, 37455–37460 [DOI] [PubMed] [Google Scholar]
- 28. Nishizuka Y. (1984) Nature 308, 693–698 [DOI] [PubMed] [Google Scholar]
- 29. Cerutti P. A. (1985) Science 227, 375–381 [DOI] [PubMed] [Google Scholar]
- 30. Das K. C., Guo X. L., White C. W. (1998) J. Biol. Chem. 273, 34639–34645 [DOI] [PubMed] [Google Scholar]
- 31. Kiningham K. K., Daosukho C., St Clair D. K. (2004) Biochem. J. 384, 543–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Storz P., Döppler H., Toker A. (2005) Mol. Cell Biol. 25, 8520–8530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kops G. J., Dansen T. B., Polderman P. E., Saarloos I., Wirtz K. W., Coffer P. J., Huang T. T., Bos J. L., Medema R. H., Burgering B. M. (2002) Nature 419, 316–321 [DOI] [PubMed] [Google Scholar]
- 34. Calnan D. R., Brunet A. (2008) Oncogene 27, 2276–2288 [DOI] [PubMed] [Google Scholar]
- 35. Sierecki E., Sinko W., McCammon J. A., Newton A. C. (2010) J. Med. Chem. 53, 6899–6911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Enoksson M., Robertson J. D., Gogvadze V., Bu P., Kropotov A., Zhivotovsky B., Orrenius S. (2004) J. Biol. Chem. 279, 49575–49578 [DOI] [PubMed] [Google Scholar]
- 37. Chung Y. W., Jeong D. W., Won J. Y., Choi E. J., Choi Y. H., Kim I. Y. (2002) Biochem. Biophys. Res. Commun. 293, 1248–1253 [DOI] [PubMed] [Google Scholar]
- 38. Jeong D. W., Yoo M. H., Kim T. S., Kim J. H., Kim I. Y. (2002) J. Biol. Chem. 277, 17871–17876 [DOI] [PubMed] [Google Scholar]
- 39. Chun J. Y., Kim K. J., Hwang I. T., Kim Y. J., Lee D. H., Lee I. K., Kim J. K. (2007) Nucleic Acids Res. 35, e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Riganti C., Gazzano E., Polimeni M., Costamagna C., Bosia A., Ghigo D. (2004) J. Biol. Chem. 279, 47726–47731 [DOI] [PubMed] [Google Scholar]
- 41. Brunet A., Bonni A., Zigmond M. J., Lin M. Z., Juo P., Hu L. S., Anderson M. J., Arden K. C., Blenis J., Greenberg M. E. (1999) Cell 96, 857–868 [DOI] [PubMed] [Google Scholar]
- 42. Wen H. C., Huang W. C., Ali A., Woodgett J. R., Lin W. W. (2003) Cell Signal 15, 37–45 [DOI] [PubMed] [Google Scholar]
- 43. Li L., Sampat K., Hu N., Zakari J., Yuspa S. H. (2006) J. Biol. Chem. 281, 3237–3243 [DOI] [PubMed] [Google Scholar]
- 44. Millward T. A., Zolnierowicz S., Hemmings B. A. (1999) Trends Biochem. Sci. 24, 186–191 [DOI] [PubMed] [Google Scholar]
- 45. Rocher G., Letourneux C., Lenormand P., Porteu F. (2007) J. Biol. Chem. 282, 5468–5477 [DOI] [PubMed] [Google Scholar]
- 46. Ishihara H., Martin B. L., Brautigan D. L., Karaki H., Ozaki H., Kato Y., Fusetani N., Watabe S., Hashimoto K., Uemura D., et al. (1989) Biochem. Biophys. Res. Commun. 159, 871–877 [DOI] [PubMed] [Google Scholar]
- 47. Singh A., Ye M., Bucur O., Zhu S., Tanya Santos M., Rabinovitz I., Wei W., Gao D., Hahn W. C., Khosravi-Far R. (2010) Mol. Biol. Cell 21, 1140–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gao T., Furnari F., Newton A. C. (2005) Mol. Cell 18, 13–24 [DOI] [PubMed] [Google Scholar]
- 49. Drane P., Bravard A., Bouvard V., May E. (2001) Oncogene 20, 430–439 [DOI] [PubMed] [Google Scholar]
- 50. Brindle P., Linke S., Montminy M. (1993) Nature 364, 821–824 [DOI] [PubMed] [Google Scholar]
- 51. Mayr B., Montminy M. (2001) Nat. Rev. Mol. Cell Biol. 2, 599–609 [DOI] [PubMed] [Google Scholar]
- 52. Muthusamy N., Leiden J. M. (1998) J. Biol. Chem. 273, 22841–22847 [DOI] [PubMed] [Google Scholar]
- 53. Ramaswamy S., Nakamura N., Sansal I., Bergeron L., Sellers W. R. (2002) Cancer Cell 2, 81–91 [DOI] [PubMed] [Google Scholar]
- 54. Medema R. H., Kops G. J., Bos J. L., Burgering B. M. (2000) Nature 404, 782–787 [DOI] [PubMed] [Google Scholar]
- 55. Kuo Y. C., Huang K. Y., Yang C. H., Yang Y. S., Lee W. Y., Chiang C. W. (2008) J. Biol. Chem. 283, 1882–1892 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.